Abstract
ST3Gal1 is a key sialyltransferase which adds α2,3‐linked sialic acid to substrates and generates core 1 O‐glycan structure. Upregulation of ST3Gal1 has been associated with worse prognosis of breast cancer patients. However, the protein substrates of ST3Gal1 implicated in tumor progression remain elusive. In our study, we demonstrated that ST3GAL1‐silencing significantly reduced tumor growth along with a notable decrease in vascularity of MCF7 xenograft tumors. We identified vasorin (VASN) which was shown to bind TGF‐β1, as a potential candidate that links ST3Gal1 to angiogenesis. LC‐MS/MS analysis of VASN secreted from MCF7, revealed that more than 80% of its O‐glycans are sialyl‐3T and disialyl‐T. ST3GAL1‐silencing or desialylation of VASN by neuraminidase enhanced its binding to TGF‐β1 by 2‐ to 3‐fold and thereby dampening TGF‐β1 signaling and angiogenesis, as indicated by impaired tube formation of HUVECs, suppressed angiogenesis gene expression and reduced activation of Smad2 and Smad3 in HUVEC cells. Examination of 114 fresh primary breast cancer and their adjacent normal tissues showed that the expression levels of ST3Gal1 and TGFB1 were high in tumor part and the expression of two genes was positively correlated. Kaplan Meier survival analysis showed a significantly shorter relapse‐free survival for those with lower expression VASN, notably, the combination of low VASN with high ST3GAL1 yielded even higher risk of recurrence (p = 0.025, HR = 2.967, 95% CI = 1.14–7.67). Since TGF‐β1 is known to transcriptionally activate ST3Gal1, our findings illustrated a feedback regulatory loop in which TGF‐β1 upregulates ST3Gal1 to circumvent the negative impact of VASN.
Keywords: ST3Gal1, vasorin, TGF‐β1, breast cancer, angiogenesis, sialylation
Short abstract
What's new?
The addition of sialic acid to glycoproteins is dysregulated in many cancers, and enhanced expression of one key enzyme, the sialyltransferase ST3Gal1, is associated with poor prognosis. Here, the authors identified the membrane protein vasorin as a new ST3Gal1 substrate and connect it with TGF‐β1‐induced signaling and angiogenesis in breast cancer. As silencing of ST3Gal1 dampened TGF‐β1 signaling and suppressed angiogenesis, development of ST3Gal1 inhibitors might be clinically useful to improve the prognosis of breast cancer patients.
Abbreviations
- CI
Confidence Interval
- CM
conditioned media
- ER
Estrogen Receptor
- HR
Hazard Ratio
- PNA
peanut lectin agglutinin
- PR
Progesterone Receptor
- RFS
Relapse free survival
- sc
scramble siRNA control
- si
ST3Gal1 siRNA
- ST3Gal1
ST3 beta‐galactoside alpha‐2,3‐sialyltransferase 1
- sVASN
souble VASN
- VASN
Vasorin
- WB
western blot
Introduction
In breast cancer, the upregulation of ST3 beta‐galactoside alpha‐2,3‐sialyltransferase 1 (ST3Gal1) terminates core 1 structure by adding α2,3‐linked sialic acid to Gal1‐3GalNac‐1‐O‐Ser/Thr,1, 2 while in normal breast tissue, C2GnT1 is the key enzyme adding GlcNAc to GalNAc on core 1 (GalB1–3GalNAc‐Ser/Thr) to generate extended core 2 structures. Accordingly, ST3Gal1 and increased amount of truncated O‐glycans were detected in glioblastomas,3 hepatocellular carcinoma,4 colorectal cancer,5 bladder cancer6 and conferred poor prognosis in patients.7 Particularly, elevation of ST3Gal1 was detected in breast cancer and correlated with increased tumor grade.8 However, the protein substrates of ST3Gal1 involved in tumor progression remain elusive.
In our study, we identified vasorin (VASN) as a ST3Gal1 substrate protein. VASN is a type I membrane protein of 673 amino acids containing tandem arrays of leucine‐rich repeat motif, an epidermal growth factor‐like motif, and a fibronectin type III‐like motif at the extracellular domain. The soluble or secreted VASN released by ADAM179 was reported to affect vascular pathophysiology by trapping TGF‐β1 at the extracellular level without significantly impacted TGF‐β receptor complexes.10 However, the role of VASN in tumor angiogenesis has never been explored. Up to date, only a few studies on VASN have been associated with cancer. Li et al., reported an elevated circulating level of VASN was detected in patients with hepatocellular carcinoma.11 Huang et al., found that VASN‐containing exosomes promoted the migration of recipient human umbilical vein endothelial cells (HUVECs).12 Recently, a report reveal that VASN is critical to maintaining glioma stem‐like cells in hypoxia.13 However, the role of VASN in cancer remains enigmatic.
In our study, we identified VASN as the ST3Gal1 target protein. Sialyl‐3 T and disialyl‐T are the majority of the O‐glycan of VASN. Overexpression of VASN in ST3Gal1‐silenced cells greatly inhibited TGF‐β1‐induced Smad2/3 signaling in HUVEC, implicating that ST3Gal1 mediated sialylation is critical for VASN bind to TGF‐β1. Finally, the gene expression of ST3Gal 1 (encoded by ST3GAL1), VASN (VASN) and TGF‐β1 (TGFB1) in breast cancer samples and their clinical correlations were evaluated.
Material and Method
Clinical specimens
Fresh primary breast cancer tumor and adjacent normal tissue specimens were collected from 114 patients during surgical resections performed at the Tri‐Service General Hospital (Taipei, Taiwan). Informed consent was obtained from all subjects before their tissue were deposited. The sample were fully encoded and used under a protocol approved by the Institutional Review Board of Human Subjects Research Ethics Committee of the Tri‐Service General Hospital and Chang Gung Memorial Hospital (Taoyuan, Taiwan). The clinicopathologic information is described by Fan et al.14
Experimental procedures
Female NSG mice (8–12 weeks old) were purchased from the Jackson Laboratory (Maine, USA). All animals were housed under specific pathogen‐free conditions in the Animal Center of Chang Gung University and studies were approved by the Institutional Animal Care and Use Committee of the University.
Cells, antibodies and recombinant proteins
Human breast cancer cell lines MCF7 was purchased from American Type Culture Collection (Rockville, Maryland USA) and maintained in DMEM supplemented with 10% FBS. The after antibodies were used in the study: Biotinylated antihuman Vasorin antibody (BAF2140, R&D Systems, Minneapolis, MN), SLITL2 (#7468–1, Abcam, CA), antimouse CD31 (BD Pharmingen, San Jose, CA, 550274), Streptavidin Peroxidase Labeled (474–3,000, KPL, Gaithersburg, MD USA), Smad2/3 (#5678, Cell Signaling, Danvers, MA USA), and pSmad2 (#8828 Cell Signaling), pSmad3 (#1880‐S‐Ex, Epitomics, Burlingame, CA). Recombinant Human Transforming Growth Factor ‐β1/TGF‐β1 (rhTGFβ‐1; CA59) and Human Vasorin (C394) were purchased from Novoprotein (Summit, NJ).
Small interference RNA (siRNA) or short hairpin RNA (shRNA) plasmid transfection
Control pLAS.Void (pVoid), and ST3Gal1 shRNA (TRCN0000231843) plasmids were purchased from the RNAi core (Academia Sinica, Taipei, Taiwan). The transfection was performed as described.14 MCF7 cells stably expressing ST3Gal1 (AGAGACTTGAGTGGCGATTAC) or control pVoid shRNAs were maintained in media with puromycin (2 μg/mL). Control scramble siRNA (sc) and siRNA (si) oligonucleotides (UCACUCUGAUCUUUGCAGGAACCGG and CCGGUUCCUGCAAAGAUCAGAGUGA.) directed against ST3Gal1 were purchased from Invitrogen (Carlsbad, CA). Cells were transfected with siRNA oligonucleotides using Lipofectamine RNAiMAX (Invitrogen) and incubated at 37 °C for 48 h.
In‐gel reductive β‐elimination
sVASN from MCF7 medium was immunopurified using anti‐VASN antibody and separated by SDS‐PAGE. VASN bands were excised from the PAGE and cut into (1 mm × 1 mm) pieces, followed by PNGase F (New England BioLabs, Hertfordshire,UK) digestion overnight at 37 °C.16 The released N‐glycan was removed by washing several times with pure water and 50% acetonitrile solution. VASN in the gel was then subjected to β‐elimination to remove O‐glycans as previously described.17 In short, the gel was soaked in 1 M sodium borohydride in 100 mM sodium hydroxide solution overnight at 45 °C, and the reaction was quenched by adding drops of glacial acetic acid. The solution containing the O‐glycans was aliquoted and desalted using Dowex 50Wx8 (H+ form) ion exchange resin. Excess borate was removed in a nitrogen blow dryer by co‐evaporation with 10% (v/v) acetic acid in methanol. Prior to permethylation and MS analysis, all extracts were dried in a SpeedVac and kept at −30 °C.
Glycan permethylation, nanoflow LC–MS/MS analysis, and the MS data mining
Permethylation of O‐glycans was conducted using the NaOH/DMSO slurry method and iodomethane.16 The permethylated derivatives were extracted using a chloroform/water method and purified by ZipTip‐C18 tips. The permethylated glycans nanoflow LC–MS/MS experiments were performed using a reverse phase C18 capillary column (75 μm × 25 cm packed with ReproSil‐Pur basic C18, 1.9 μm) and an Orbitrap Fusion mass spectrometer (Thermo Scientific) equipped with a nanoelectrospray ion source (New Objective) and an UltiMate 3,000 RSLCnano system pump (Thermo Scientific Dionex). The nanoflow LC for the separation of O‐glycans was conducted at a flow rate of 300 nL/min with a linear gradient from 25% (v/v) acetonitrile to 55% (with 0.1% formic acid) in 45 min. The nanoelectrospray source was powered in the range of 1.6–1.8 kV. The scan cycle was performed in top‐speed mode within a cycle time of 3 s, which can intelligently schedule MS and data‐dependent MS/MS scans. The full‐scan MS (m/z 300–1,500) experiment was performed in the Orbitrap at a resolution of 120 K and an automatic gain control (AGC) target value of 4 × 1018, while the data‐dependent MS/MS experiments was conducted in the C‐trap using higher energy collisional dissociation (HCD) with 15% normalized collision energy at a resolution of 15 K and an AGC value of 5 × 108.
To process MS data for the structural characterization of O‐glycans from sVASN, we used the X‐tract program to conduct peak deconvolution and output the peaklist with deconvoluted m/z and intensity using Xcalibur software from the raw MS data files. We next loaded the peaklist into the analytic module of GlycoWorkbench to annotate peak with glycan structure from the database of CFG according to the MS correlation with 5 ppm tolerance.18 We manually examined the MS/MS spectra of matched O‐glycan structures and annotated the carbohydrate fragmentation based on the nomenclature proposed by Domon and Costello.19 We also manually calculated the peak area in corresponding monoisotopic peak XICs to obtain the abundance of each glycoform. The relative abundance of individual glycoform was calculated based on the percentage (%) of sum integrated peak areas from all of the identified O‐glycoforms shown in Supporting Information Table S1. The results were conducted from two technical repeats.
Endothelial cell tube formation
The endothelial cell tube formation assay was performed as previously described.20 Matrigel (BD, Bedford, MA) was added to 48‐well plates in a volume of 150 μL and allowed to solidify at 37 °C for 30 min. HUVECs (15 K cells/well) were suspended in 50% CM and 50% regular medium (EGM‐2, Lonza, Walkersville, MD) before addition to Matrigel‐precoated plates. After 14 h, the formation of the capillary‐like tube network by HUVECs was imaged using phrase‐contrast microscopy, and the junctions and meshes of the network were quantified using ImageJ software.
Immunohistochemistry for CD31
Xenografted tumors were first fixed with zinc solution (550,523, BD Pharmingen) for 24 h. Tumors were embedded and dissected into 5 μm thick sections, followed by de‐waxing and blocking with 3% H2O2. The blood vessels in tumor sections were stained by purified rat antimouse CD3121 antibody (1:20, 550,274, BD Pharmingen) overnight at 4 °C. The detection was followed by polymer‐HRP IHC system and sections were counterstained with methyl green (1%). Digital images were captured by an Aperio ScanScope XT Slide Scanner (Aperio Technologies, Vista, CA) under 20‐fold magnification. The DAB‐positive areas were quantified using MetaMorph 4.6 software (Puchheim, Germany).22
Quantitative real‐time PCR reaction
Total RNA of breast cancer tissue was isolated using TRIzol reagent (Invitrogen), and cDNA was generated from 1,000 ng total RNA, using a High Capacity cDNA Reverse Transcription Kit (Applied BioSystems/ABI, Warrington, UK). Real‐time PCR assays were performed on an ABI 7500 Detection System (ABI) using SYBR Green MasterMix (ABI). For accurate normalization of quantitative data, multiple housekeeping genes, including GAPDH, GUSB (glucuronidase‐beta) and UBC (polyubiquitin) were assayed. Primer sequences are listed in Supporting Information Table S3.
Luminex‐based protein–protein interaction assays
Luminex immunosandwich assay was performed according to the manufacturer's manual (The xMAP Cookbook, 3rd Edition). Briefly, recombinant VASN was coupled on beads and underwent on‐bead removal of N‐glycans and/or de‐sialylation by incubating with the enzyme PNGase F (P0704; New England Biolabs, Ipswich, MA) at 37 °C for 1 h, or α2‐3 Neuraminidase (P0728, New England Biolabs) at 37 °C for 1 h. After removal of enzymes by washing 3 times with washing buffer (PBS‐BSA 1%), beads were divided equally into 96‐well plates for different reactions. In binding assay, serial dilutions of recombinant TGF‐β1 (in PBS‐BSA 1%, 1 h) were used for the interaction, and biotinylated anti‐TGF‐β1 antibody (1:100 in PBS‐BSA 1%) were used for detection. In control assays, biotinylated VASN antibody (1:100 in PBS‐BSA 1%, 1 h) was used to determine the amount of coated VASN on the beads. To confirm the removal of sialic acid from the recombinant coating on beads, PNA binding studies (1:1000 in PBS‐BSA 1%, 1 h) were performed. For detection, beads were incubated with Streptavidin‐phycoerythrin diluted 1:50 in PBS‐BSA 1% (SAPE, Millipore Corporation, Bedford) and shaken for 30 min. After extensive washing with PBS/BSA 1%, beads were analyzed with the Luminex 200 instrument.
Statistical analyses
All means data values were derived from three or more independent experiments and mean (SD) are presented. Differences between means were analyzed using Student's t‐test. p‐Values of <0.05 were considered statistically significant.
Results
Effects of altered ST3Gal1 expression on tumor growth and vascularity
Although overexpression of ST3Gal1 has been associated with tumoigenesis,23 however, the role of ST3Gal1 in breast cancer progression remains poorly understood. Therefore, we selected MCF7 to generate ST3Gal1‐silenced stable clone (MCF7/shST3Gal1) and control (MCF7/pVoid) because it expressed the highest level of ST3Gal1 amount 10 tested breast cancer cell (Supporting Information Fig. S1a). The transfection with ST3Gal1 shRNA greatly reduced the protein level of ST3Gal1 in MCF7/shST3Gal1 compare to MCF7/pVoid (Fig. 1 a). These stable clones were then implanted into the mammary fat pads of NSG mice, and the tumor growth was measured weekly. ST3Gal1 silencing significantly suppressed the rate of growth (Fig. 1 b) and reduced the size of the engrafted tumors (Fig. 1 c). The average weight of the MCF7/shST3Gal1 group (505 ± 223 mg) was twice smaller than the MCF7/pVoid group (1,102 ± 313 mg) (Fig. 1 d). In addition, gross appearance showed less vasculature surrounding tumors derived from MCF7/shST3Gal1 cells than tumors from MCF7/pVoid (Supporting information Fig. S2a). Therefore, we further evaluated tumor vascularity by immunohistochemistry (IHC) staining with anti‐CD31 antibodies (Fig. 1 e). The vascular density was quantified and the result shows that vascularity of MCF7/shST3Gal1 tumors (0.769% vessel area/field) was significantly reduced as compared to the MCF7/pVoid group (2.06% vessel area/field) (Fig. 1 e upper panel).
Figure 1.
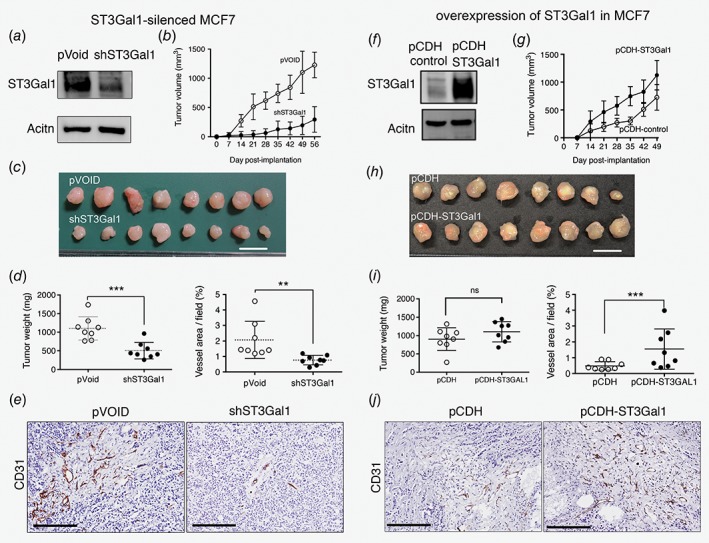
Effects of ST3Gal1 on tumor growth and vascular density in MCF7 engrafted tumors. (a‐e) MCF7 with knockdown of ST3Gal1 and control; (a) Western blotting (WB) analysis of ST3Gal1 in stable clones MCF7/pVoid and MCF7/shST3Gal1. (b) Tumor growth of MCF7/pVoid and MCF7/shST3Gal1. 1 × 1018 cells of each clone were injected into the mammary fat pads of female NSG mice (n = 8) and tumor volume was measured weekly. (c) Gross appearance and (d) tumor weight of xenografted tumors collected on days 56 after injection. (e) Blood vessels density in the harvested tumors, as identified by IHC staining with antimouse CD31 antibody (left panel), Bar: 200 μm. The DAB‐positive areas were quantified using MetaMorph 4.6 software22 (upper panel). (f‐j) MCF7 with overexpression of ST3Gal1 and control, (f) WB analysis of ST3Gal1 in stable clones of MCF7 transfected with pCDH‐, and pCDH‐ST3Gal1. The stable clones were used to generate xenograft tumor (n = 8 for each clone) and their (g) growth curve, (h) xenografted tumors, (i) tumor weight, (J) anti CD‐31 staining and quantification of positive areas were shown. Data are represented as mean (SD). Data are analyzed by Student's t‐test. *p < 0.05; **p < 0.01, *** p < 0.001.
In contrast, overexpression of ST3Gal1 in MCF7 cells (Fig. 1 f) increased the rate of growth (Fig. 1 g) and tended to enhance the size of the MCF7 engrafted tumors tumor (Fig. 2 h). Although the tumor weight at 49 days was not significantly different between the two groups (Fig. 1 i). However, overexpression of ST3Gal1 significantly increased vascularity in the engrafted tumors (pCDH control: pCDH‐ST3Gal1, 0.486%: 1.70% vessel area/field) (Fig. 1 j upper panel). In addition, ASB‐634, which is derived from breast cancer patient‐derived xenograft, was used to generate ST3Gal1 silenced stable clones for in vivo engraftment experiment. The engrafted tumors from ASB‐634/shST3Gal1 displayed a paler gross appearance and less vascular density (Supporting Information Fig. S2b‐d) than the tumors derived from ASB‐634/pVoid control. Together with the observation from MCF7, we suggested ST3Gal1 mediated‐tumor progression, at least in part through increased tumor angiogenesis.
Figure 2.
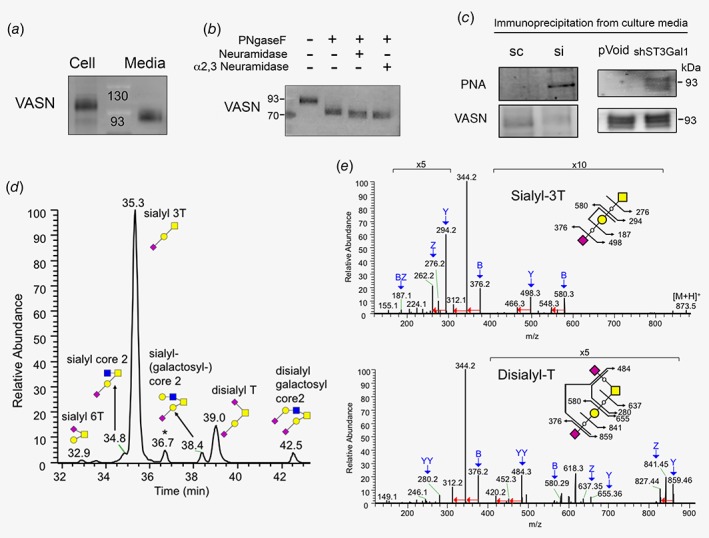
Identification of VASN as a substrate of ST3Gal1, and NanoLC‐MS/MS analyses of O‐glycan profiles on sVASN purified from MCF7 cells. (a) Detection of VASN in MCF7 cell extracts (110 kDa) and culture medium (sVASN, 93 kDa) by WB. (b) WB analysis of sVASN after treated with the indicated glycosidases. (c) WB analysis of PNA lectin or VASN antibody for anti‐VASN immunoprecipitated sVASN from CM of MCF7 using siRNA‐ (left panel) or shRNA‐ (right panel) mediated ST3Gal1 silencing. (d) The merged extracted ion chromatograms are resulted from all of their corresponding monoisotopic precursor ions (with 5 ppm tolerance) listed in Supporting Information Table S1. Note that three very minor levels of O‐glycoforms [galactosyl‐core 2, sialyl‐galactosyl‐core 2, and sialyl elongated core 1 (peaks at 32.6, 38.9 and 39.3 min, respectively)] are not indicated. (e) The precursor ions of sialyl‐3 T and disialyl‐T used for MS/MS analysis are 873.48 and 617.832+. Peak assignment for the fragment ions are highlighted with blue arrows and the neural loss of CH3OH (−32) from specific fragment ions are indicated with red arrow. PNA: peanut lectin agglutinin. sc: scramble siRNA control; si: ST3Gal1 siRNA.
VASN is a protein substrate of ST3Gal1
Peanut lectin agglutinin (PNA) recognizes the nonsialylated Galβ1–3GalNAc‐Ser/Thr core 1 O‐glycan.24 To identify ST3Gal1 substrate proteins, cell lysates from scramble control‐ or ST3Gal1 siRNA‐transfected cells were subjected to PNA precipitation. The differentially PNA‐binding proteins were identified using LTQ‐FT MS. VASN was identified as one of the ST3Gal1 substrate proteins. VASN was detected in total lysates of MCF7 cells at 110 kDa and sVASN was detected in culture media at 93 kDa due to the cleavage in the extracellular juxtamembrane region9 (Fig. 2 a). Treatment of sVASN with PNGase F reduced the molecular weight of VASN to slightly higher than the predicted value (71.72 kDa), and the combination of neuraminidases further reduced it to about 71 kDa, indicating that VASN is highly glycosylated (Fig. 2 b). Moreover, VASN immunoprecipitated from culture media derived from MCF7/siRNA (Fig. 2 c, left panel), and MCF7/shST3Gal1 (right panel) exhibited greater PNA staining than VASN from control cells indicating the exposure of Gal‐β‐(1‐3) GalNAc on VASN caused by siST3Gal1 or shST3Gal1. The mRNA level of siST3Gal1 and shST3Gal1 were reduced to 37% and 38%, respectively, of the controls (100%) (Supporting Information Fig. S1b). These results confirm that VASN is a substrate protein of ST3Gal1.
LC–MS/MS analysis identified α2,3 sialylation to be dominant O‐glycan structure of VASN
To delineate the O‐glycosylation of VASN, the sVASN secreted into MCF7‐conditioned medium was immunopurified using anti‐VASN antibody and separated by SDS‐PAGE, followed by staining with Coomassie Brilliant Blue. The protein band corresponding to the sVASN corresponding band was excised and subjected to in‐gel enzymatic digestion with PNGase F to remove the N‐glycans, followed by releasing the O‐glycan using alkaline β‐elimination.25 To stabilize the sialic acid residues, enhance MS detection sensitivity, enable glycan separation using reverse phase LC, and support detailed structural characterization using MS/MS, we conducted permethylation26 of O‐glycans and then analyzed the O‐glycoforms using LC–MS/MS. Table S1 in Supporting information summarizes all of the identified O‐glycoforms from sVASN, which includes the retention times on LC/MS, mass to charge ratios (m/z) of O‐glycan ions, structural information, and relative abundance based on the methodology described in materials and methods. The extracted ion chromatograms of all O‐glycan ions for the measurement of relative abundance are merged and shown in Figure 2 d. Notably, around 80% of sVASN O‐glycosylation are α2,3 sialylated, with that the majority are cancer relevant sialyl‐3 T (67.44%; with a peak of LC–MS trace at 35.3 min) and disialyl‐T (7.67%; peak at 39.0 min). Both are the O‐glycoforms catalyzed by ST3Gal1.
The resulting MS/MS peak assignment spectra of fragment ions for sialyl‐3 T and disialyl‐T are shown in Figure 2 e. Both MS/MS spectra show predominant dissociation of the sialyl linkage that results in the significant B fragment ion at m/z 376 and the subsequent abundant fragment ions at m/z 344 and 312 due to single and double neutral loss of CH3OH (−32; indicated with red arrow), respectively. In the left panel, the additional two pairs of B/Y ions (m/z 376/498 and 294/580), BZ ions (187), and Z ions (276) together suggest the linear structure of sialyl‐3 T. In the right panel, the two YY ions of di‐substituted HexNAc alditol (m/z 280) and Hex‐HexNAc alditol (484) are crucial diagnostic ions indicating a tetra‐saccharide precursor with a branching structure at the reducing end. Together with the other peak assignments, the spectra in the right panel suggest the structure of disialyl‐T. For the rest of O‐glycans listed in Supporting Information Table S1, their structural characterization by MS/MS spectra with peak assignment (Supporting Information Fig. S3) also suggested that most of the O‐glycan are α2,3 sialylated. These results confirm ST3Gal1 is the key enzyme to mediate the O‐glycoforms of VASN in MCF7.
Effects of α2,3 sialylated VASN on HUVEC tube formation and TGF‐β1 signaling
To examined whether the ST3Gal1 mediated VASN involved in angiogenesis. First, we assessed the effects of conditioned media (CM) from MCF7/pVoid, and MCF/shST3Gal1 on HUVEC tube formation. HUVEC cells (15 K cells/well) were suspended in 50% CM and 50% regular medium (EGM‐2) before addition to Matrigel precoated 48 well plates. After 14 h, CM from MCF7/shST3Gal1 significant decrease the HUVEC tube formation ability as shown by 23% decrease of meshes as compared to those in CM from MCF7/pVoid cells (value set at 100%). However, the addition of VASN antibody to CM partially prevented the CM‐caused decreasing in the formation of meshes (Fig. 3 a). The prevention of VASN bind to TGF‐β1 by anti‐VASN was verified and shown in Supporting Information Figure S5. On the other hand, TGF‐β1 induced the expression of several genes relevant to angiogenesis in HUVEC, including FGF1, FGF13, MMP2, and MMP9 (Supporting Information Fig. S6). The induction of these genes was suppressed by the addition of CM from MCF7/shST3Gal1 (Fig. 3 b). These results suggested that ST3Gal1 silencing and ST3Gal1‐modified VASN involved in the inhibition of angiogenesis. The effect of ST3Gal1 on VASN was further evaluated by overexpressed VASN in MCF7/pVoid and MCF7/shST3Gal1 (Fig. 3 c). CM from MCF7/shST3Gal1/VASN markedly decreased the activation of Smad2 to 35% (Fig. 3 d) and Smad3 to 61% (Supporting Information Fig. S7) of the control (100%) in HUVEC, respectively, indicating the desialylated‐VASN inhibited TGF‐β1/Smad signaling. Moreover, the proliferation rate of MCF7/shST3Gal1 was decreased as compared to MCF7/pVoid and it was further reduced by VASN overexpression in MCF7/shST3Gal1 (Supporting information Fig. S4), these results support the notion that desialylated VASN affected TGF‐β1 signaling.
Figure 3.
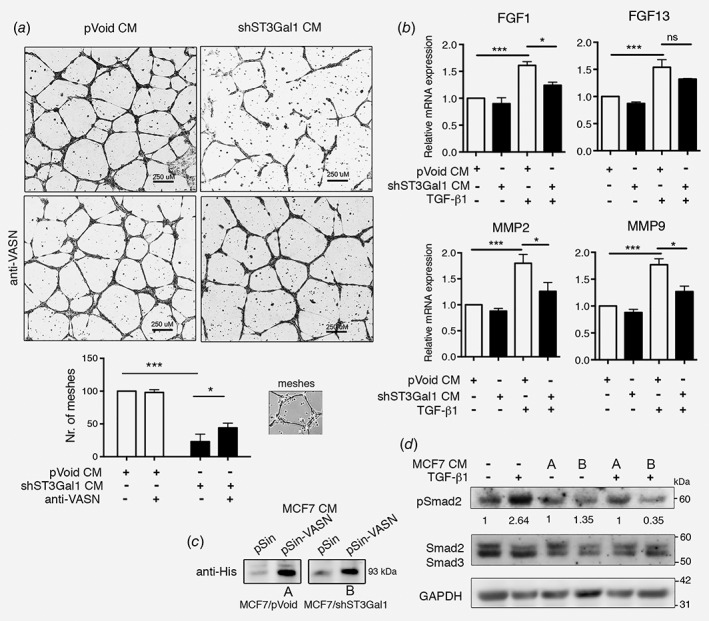
Effects of α2,3 sialylated VASN on HUVEC tube formation, and TGF‐β1 mediated angiogenesis and signaling pathway. (a) Representative phase contrast images of tube formation by HUVECs suspended in 50% CM from MCF7/shST3Gal1 or MCF7/pVoid stable clones, with or without anti‐VASN (upper panel). Scale bar = 250 μm. Numbers of meshes (as illustrated in inserts) were quantified using ImageJ software (lower panel; n = 3). Results are represented as means (SD). Data are analyzed by Student's t‐test (*p < 0.05, ***p < 0.001). (b) RT‐PCR analysis of angiogenesis related genes in HUVEC after treated with TGF‐β1 and CM from MCF7 stable clones. (c) WB analysis of VASN in CM after transfection of the an empty vector or pin‐VASN‐His into MCF7/pVOID (A) and MCF7/shST3Gal1 (B). (d) WB analysis of the protein extracts of HUVEC treated with CM from VASN over‐expression in MCF7/pVoid (A) and MCF7/shST3Gal1 (B) in the present of TGF‐β1 for 1 h, GAPDH was used as control. In c‐d, western blotting was performed with the indicated antibodies. WB, western blot; CM, conditioned media.
Effects of α2,3‐sialylation on VASN and TGF‐β1 interaction
It has been known that secreted VASN binds to TGF‐β1 and blocks its signaling, but whether α2,3‐sialylation of VASN affects its binding to TGF‐β1 has yet to be deciphered. A Luminex platform, as shown in the schematic illustration in Figure 4 a, was set up to determine the effect of ST3Gal1‐mediated sialylation on the binding of VASN to TGF‐β1. Briefly, recombinant VASN was coupled on beads and underwent on‐bead removal of N‐glycans and/or α2,3‐sialic acid before binding studies with recombinant TGF‐β1. As shown in Figure 4 b (left panel), TGF‐β1 binding to VASN‐beads was enhanced by 1.8‐fold when α2,3 sialic acid was removed by treatment with α2,3 neuraminidase. In order to rule out an effect of N‐linked α2,3 sialic acid, VASN‐beads were treated with PNGase F to remove N‐glycans. Treatment with PNGase F plus α2,3‐specific neuraminidases further increased the binding to 2.2‐fold. These data indicate that α2,3 sialylation on O‐linked VASN plays a major role in its binding to TGF‐β1. Meanwhile, the PNA and anti‐VASN bound to the VASN‐beads after treatment with glycosidase were also performed, indicating the successful removal of sialic acid from VASN coated bead (Fig. 4 b, middle panel), and it did not affect the VASN amount on beads (Fig. 4 b, right panel). Similar studies were performed using neuraminidase (N2876, SIGMA) purchase from another source, which catalyzes the α2,3‐linked sialic acid more efficient than α2,6‐ or α2,8‐linked sialic acid. As shown in Figure 4 c (left panel), neuraminidase treatment enhanced the binding of TGF‐β1 to VASN by 3.2‐fold. After treatment with neuraminidase plus PNGase F, the binding remained at 2.4‐fold of untreated VASN. The slight reduction of TGF‐β1 binding in the latter, suggest that α2,3 sialylation on N‐glycan may also affect the TGF‐β1 binding to VASN. The PNA (Fig. 4 c, middle panel) and anti‐VASN binding (Fig. 4 c, right panel) to the VASN‐beads after treatment with glycosidase also performed. On the other hand, as demonstrated by western blotting, TGF‐β1‐coated beads depleted recombinant VASN more efficiently when α2–3 sialylation was removed (Supporting Information Fig. S8). This result again indicates that removal of α2,3 sialic acid on VASN facilities it binding to TGF‐β1.
Figure 4.
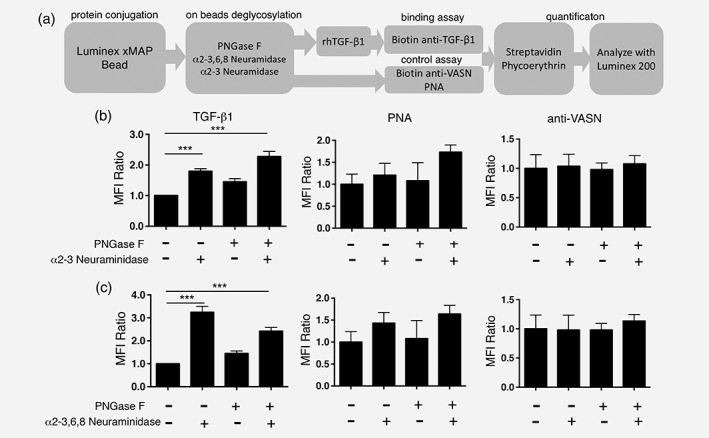
Removal of α2–3 sialic acid enhanced VASN binding to TGF‐β1. (a) Flow chart of Luminex immunosandwich assay. All values (Luminex fluorescence intensities) were normalized to wild‐type control. (b) Bar chart presenting the binding fold of TGF‐β1 to wild type (1×), VASN without α2,3‐sialic acid (1.8×), VASN without N‐glycan (1.4×), and VASN without N‐glycan and α2,3‐sialic acid (2.2×). Binding fold of PNA (middle panel) and anti‐VASN to VASN‐Bead (right panel) after treatment of glycosidase. (c) Bar chart presenting the binding fold of TGF‐β1 to wild type (1×), VASN without α2,3‐sialic acid (3.2×), VASN without N‐glycan (1.45×), and VASN without N‐glycans or α2,3‐sialic acid (2.42×). Binding fold of PNA (middle panel) and anti‐VASN to VASN‐Bead after treatment with glycosidase.
Expression levels of ST3GAL1, TGFB1 and VASN in primary breast cancer samples and their correlation
We next examined the clinical relevance of the expression levels of ST3GAl1, VASN, and TGFB1 in 114 pairs of primary breast cancer and their adjacent normal tissue by qPCR. Their clinical features of these patients were described by Fan et al.14 As shown in Figure 5 a, higher expression levels of ST3GAL1 (p = 0.001) and TGFB1 (p < 0.0001) were detected in the tumors than their counterpart, and the expression of these two genes showed a positive correlation in a Pearson Two‐dimensional analysis (Fig. 5 b upper panel, r = 0.322, p < 0.001). In addition, patients with grade 3 tumor expressed higher levels of ST3GAL1 than grade 1&2 (Supporting Information Fig. S9). In contrast, a lower expression level of VASN was observed in tumors than the normal tissue (p < 0.0001), and the expression level of VASN was negatively correlated to TGFB1 (Fig. 5 b lower panel, r = −0.193, p < 0.05). The correlation between gene expression level and relapse‐free survival (RFS) of 114 patients were further evaluated. With a median follow up of 93.73 (range 0.43–137.77) months, 37 of 114 patients (32.46%) had disease recurrence. Based on ROC curves, the best cutoff values for stratifying into high and low gene expression levels were selected for VASN (−2.88), TGFB1 (4.30), and ST3GAL1 (1.027). As shown in Figure 5 c, patients with low levels of VASN showed significantly shorter RFS than those with high levels of VASN (p = 0.029, HR = 2, 95% CI = 1.087–4.651). The adverse impact of low VASN expression was even more significant in the patients with grade 3 tumor (p = 0.017, HR = 2.6, 95% CI = 1.225–7.885) (right panel). Moreover, the association of low VASN to short RFS was evident only in patients with low expression of TGFB1 (p = 0.049, HR = 2.26, 95% CI = 1.004–7.195) (Fig. 5 d, right panel) but not those with high expression of TGFB1 (Fig. 5 d, left panel). Since ST3Gal1 is known to be transcriptionally activated by TGF‐β1,3 our findings suggest the presence of a feedback regulatory loop in which TGF‐β1 upregulates ST3Gal1 to circumvent the negative impact of VASN.
Figure 5.
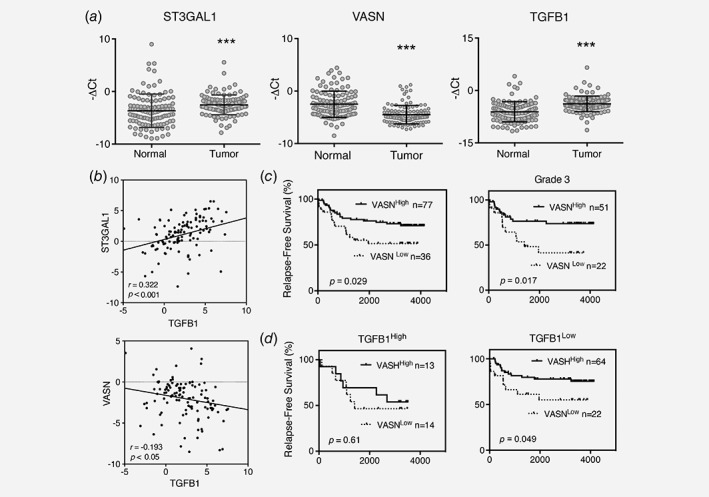
Expression levels of ST3GAL1, TGFB1 and VASN in 114 primary breast cancer samples and their correlation. (a) Expression levels of ST3GAL1, VASN and TGFB1 in tumor and adjacent normal tissue. (b) Pearson's correlation analysis for TGFB1 and ST3GAL1 (upper panel), or TGFB1 and VASN (lower panel). (c) RFS Kaplan–Meier plots of low and high VASN expression in overall study population (left panel) and in patients with grade 3 tumor (right panel). (d) Negative effect of VASN associated to high expression of TGFB1 high (left panel) and low expression of TGFB1 (right panel). Gene expression level was determined by RT‐PCR. Each scatter dot represents the value of minus delta threshold cycle (‐ΔCt) normalized to the average of three internal controls: GAPDH, GUSB and UBC. Expression level of each gene is stratified to high and low, according to ROC analysis. HR, Hazard Ratio. Statistical significance is determined by student's t‐test. ***p < 0.001.
Combination of expression levels of ST3GAL1 with VASN, or TGFB1 correlated with poor clinical outcome
The combined effects of ST3GAL1 and VASN, or ST3GAl1 and TGFB1 on RFS were also evaluated. As shown in upper panel of Figure 6 a, patients with ST3GAL1 High VASN Low had a significantly shorter RFS than those with ST3GAL1 Low VASN High (p = 0.025, HR = 2.69, 95% CI = 1.146–7.678). On the other hand, although there was no significant difference in RFS between patients with ST3Gal1 High TGFB1 High and those with ST3Gal1 Low TGFB1 Low (p = 0.093, HR = 2.08, 95% CI = 0.876–5.69, Fig. 6 b, upper panel). However, among patients with grade III tumors which expressed higher levels of ST3GAL1 than grade 1&2 (Supporting Information Fig. S9, p = 0.05), significantly shorter RFS was noted in patients with ST3GAL1 High VASN Low (p = 0.01, HR = 4.20, 95% CI = 1.457–16.39) or ST3Gal1 High TGFB1 High (p = 0.07, HR = 2.93, 95% CI = 0.906–10.43) than their counterparts (lower panels in Figs. 6 a and 6 b).
Figure 6.
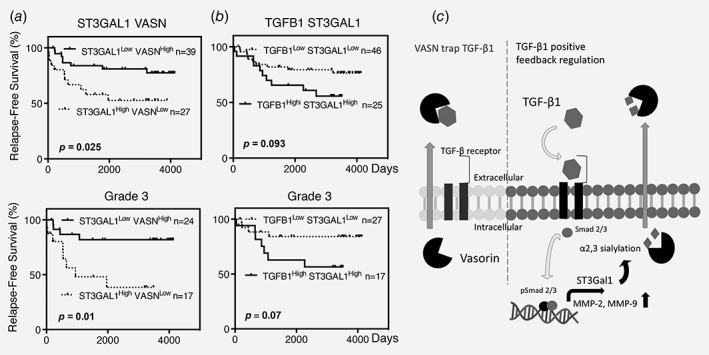
Correlation of combination of ST3GAL1/VASN and ST3GAL1/TGFB1 with RFS in overall study population (upper panel of a, b) and in patients with grade 3 tumor (lower panel of a, b). (a) Combination of ST3GAL1 and VASN. (b) Combination of TGFB1 and ST3GAL1. (c) Schematic illustration showing that α2,3 sialylation of VASN by ST3Gal1 reduces the ability of secreted VASN to trap TGF‐β1. HR, Hazard Ratio.
Furthermore, the association of these three genes with clinical‐pathological parameters in 114 patients was analyzed. As shown in Supporting Information Table S2, low expression of VASN was associated with a hazard ratio (HR) of 2.16 for tumor recurrence (p = 0.019, 95% CI: 1.13–4.14). High expressions of ST3GAL1 and TGFB1 individually showed a higher risk for recurrence (HR = 2.28), albeit the difference did not reach statistical significance (p = 0.059, 95% CI: 0.97–5.38). More importantly, multivariate Cox Proportional‐Hazards Regression survival analysis revealed that low VASN expression was an independent risk factor for relapse, with a HR of 2.06 (p = 0.003, 95% CI: 1.07–3.92).
Discussion
Overexpression of ST3Gal1 has been associated with mammary tumorigenesis.23 In noncancer cells, ST3Gal1 mediated‐sialylation of CD43 was found associated with anti‐CD43‐induced CD8+ T Lymphocyte apoptosis,27 and decreased CD8αβ‐MHCI binding avidity.28 However, the substrate of ST3Gal1 and mechanism that help to promote tumor progression is so far ill defined. By combining PNA‐precipitation with LTQ‐FT MS, a set of ST3Gal1 substrates have been identified and being investigated. Among them, ST3Gal1‐mediated sialylation of GFRA1 and CD55 was found to promote tumor growth, and evade immune surveillance, respectively. In our study, we demonstrated that the ST3Gal1 mediated‐O‐linked sialylation of VASN diminished its binding to TGF‐β1. Five N‐glycosylation sites and four O‐glycosylation sites (318, 487, 516, 567) were identified in the sequence.29 However, more than 30 potential O‐glycosylation sites on VASN were predicted by the NetOGLyc 4.0. The predicted sites were distributed throughout the protein sequence with many crowded near the EGF‐like domain, making it very challenging to decipher the precise O‐linked glycan site of VASN that modulates its interaction with TGF‐β1. Nonetheless, our demonstration of the importance of ST3Gal1‐mediated α2,3‐sialylation of VASN in modulating tumor angiogenesis illustrates the importance of delineating the involved O‐linked glycosylation sites on VASN in future studies.
TGF‐β is a key cytokine in maintaining tissue homeostasis.30, 31 However, accumulating evidence indicates that it also augments tumor growth, enhances invasion and metastasis32, 33 by modulating angiogenesis,34 promotes epithelial to mesenchymal transition,35 suppresses immune responses36 and enhances ECM production. High levels of circulating TGF‐β1 was detected in the plasma of patients with cancer,37, 38 which correlated with poor prognosis with increased probability of relapse.39, 40 Moreover, upregulation of TGF‐β1 has been noted in breast cancer specimens41 and increase the risk of advanced breast cancer39 with the promotion of angiogenesis.42 Among breast cancer cell lines, MCF7 cells expressed higher levels of TGF‐β1.43 A clone derived from MCF7 cells and secreted greater than 10‐fold of TGF‐β1 was more tumorigenic than the parental line, and tumor formation was abrogated by the administration of a TGF‐β1 neutralizing antibody.41 Gene knocks out of TGF‐β or its receptors and downstream signaling proteins led to embryonic lethality due to vascular defects.44 The correlation of TGFB1 and ST3GAL was reported in glioblastoma by Chong et al., and they demonstrated that ST3Gal1 was transcriptionally activated by TGF‐β1 using chromatin immunoprecipitation combined with anti‐pSmad2 antibody.3, 45 In our study, we further demonstrated that TGF‐β1 upregulates ST3Gal1 to circumvent the negative impact of VASN, which in turn trigger MMPs, FGFs expression to support angiogenesis in tumor. This finding reveals a noval glycosylation‐mediated feedback loop to the known regulatory mechanisms of the TGF‐β/Smad signaling system.46
Breast cancer is the most common cancer and the leading cause of death in women worldwide. Although the diagnosis and treatment of primary tumors have markedly improved, metastasis and/or recurrence of breast cancer remains the major cause of death.47 Thus, it is crucial to identify novel biomarkers for prognostication and therapy. In 114 breast cancer patients, we found that VASN was significantly downregulated in tumor compared to adjacent normal tissue, while ST3GAL1 and TGFB1 highly expressed in breast cancer. Kaplan–Meier analysis for RFS showed that lower VASN expression significantly correlated with a higher risk of breast cancer recurrence, suggesting that VASN probably acts as a tumor suppressor in breast cancer. Multivariant analyses indicated that VASN was an independent risk factor for breast cancer recurrence. On the other hand, there was a trend for an association of poorer RFS with higher expression of either ST3Gal1 or TGF‐β1. Importantly, the combination of low VASN and high ST3Gal1 expression increased the risk of the recurrence of disease, as demonstrated by a higher HR.
In summary, we have demonstrated for the first time a molecular mechanism underlying angiogenesis modulated by ST3Gal1‐mediated sialylation of VASN, supporting the notion that inhibition of ST3Gal1 is a promising new strategy for cancer therapy.
Supporting information
Figure S1: Western blot and RT‐PCR analysis of expression level of ST3Gal1. (a) Western blot analysis of ST3Gal1 expression level in several breast cancer cell lines. (b) RT‐QPCR analysis of mRNA levels of ST3Gal1 upon stable overexpression of shST3Gal1 (Left panel) and transiently transfected siRNA of ST3Gal1 in MCF7.
Figure S2: Effects of ST3Gal1 silencing on MCF7 and ASB‐634 on xenograft tumor. (a) Gross appearance of MCF7/pVoid, MCF7/shST3Gal1 xenograft tumor and surrounding tissues, (b) ASB‐634/pVoid, ASB634/shST3Gal1 xenograft tumor profile, (c) cross‐section and (d) blood vessels density in the harvested tumor, as identified by IHC staining with anti‐mouse CD31 antibody. The DAB‐positive areas were quantified using MetaMorph 4.6 software (lower panel).
Figure S3: Tandem mass spectra of indicated sialylated O‐glycans from sVASN. The m/z of precursor ions used for generation of MS/MS spectra are shown in the upper right corner of each sub‐figure. Peaks assignment for the fragment ions are highlighted with blue arrows and the neural loss of CH3OH (−32) from specific fragment ions are indicated with red arrows.
Figure S4: Proliferation of MCF/pVoid and MCF/shST3Gal1 with or without overexpression of VASN were monitored using the xCELLigence Real Time Cell analysis instrument.
Figure S5: Luminex immunosandwish assay of TGF‐β1 binding to VASN coated bead, with or without the presence of anti‐VASN. Data are represented as mean (SD). Data are analyzed by Student's t‐test. *** p < 0.001
Figure S6: RT‐PCR analysis of angiogenesis related genes from HUVEC incubated for 3 h with 1 ng/ml TGF‐β1. Data are represented as mean (SD). Data are analyzed by Student's t‐test. *** p < 0.001
Figure S7: Western blot analysis the treatment of HUVEC with CM from VASN expression in MCF7/pVoid and MCF7/shST3Gal1 in the presence of TGF‐β1 for 1 h, GAPDH was used as control.
Figure S8: VASN were treated with glycosidase and incubated with TGF‐β1 coated bead for 1 h. After removed of TGF‐β1 coated bead, the remaining VASN was analysis by Western blot analysis.
Figure S9: Expression levels of ST3GAL1 in different grade. Gene expression level was determined by RT‐PCR. Each scatter dot represents the value of minus delta threshold cycle (‐ΔCt) normalized to the average of 3 internal controls: GAPDH, GUSB and UBC.
Supplementary Table S1: Summary of identified O‐glycoforms from sVASN using LC–MS/MS analysis.
Supplementary Table S2. Univariate and multivariable Cox proportional regression analysis of mRNA expression and breast cancer recurrence.
Supplementary Table S3: qPCR primers
Acknowledgements
We thank Ms. Lo Fei‐Yun for excellent technical support. This work was supported by the grants from Ministry of Science and Technology and Chang Gung Medical Foundation in Taiwan for Alice L. Yu (MOST 103‐2321‐B‐182A‐005, MOST 104‐2321‐B‐182A‐003, MOST 105‐2321‐B‐182A‐001, OMRPG3C0014); John Yu (MOST 106‐3114‐B‐182A‐001).
Conflict of interest: The authors declare no conflict of interest.
Contributor Information
John Yu, Email: johnyu@gate.sinica.edu.tw.
Alice Lin‐Tsing Yu, Email: a1yu@ucsd.edu.
References
- 1. Dalziel M, Whitehouse C, McFarlane I, et al. The relative activities of the C2GnT1 and ST3Gal‐I glycosyltransferases determine O‐glycan structure and expression of a tumor‐associated epitope on MUC1. J Biol Chem 2001;276:11007–15. [DOI] [PubMed] [Google Scholar]
- 2. Tsuji S. Molecular cloning and functional analysis of sialyltransferases. J Biochem 1996;120:1–13. [DOI] [PubMed] [Google Scholar]
- 3. Chong YK, Sandanaraj E, Koh LW, et al. ST3GAL1‐associated transcriptomic program in glioblastoma tumor growth, invasion, and prognosis. J Natl Cancer Inst 2016;108:djv326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu H, Shi XL, Zhang HJ, et al. Overexpression of ST3Gal‐I promotes migration and invasion of HCCLM3 in vitro and poor prognosis in human hepatocellular carcinoma. Onco Targets Ther 2016;9:2227–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kudo T, Ikehara Y, Togayachi A, et al. Up‐regulation of a set of glycosyltransferase genes in human colorectal cancer. Lab Invest 1998;78:797–811. [PubMed] [Google Scholar]
- 6. Videira PA, Correia M, Malagolini N, et al. ST3Gal.I sialyltransferase relevance in bladder cancer tissues and cell lines. BMC Cancer 2009;9:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hakomori S. Aberrant glycosylation in tumors and tumor‐associated carbohydrate antigens. Adv Cancer Res 1989;52:257–331. [DOI] [PubMed] [Google Scholar]
- 8. Burchell J, Poulsom R, Hanby A, et al. An alpha2,3 sialyltransferase (ST3Gal I) is elevated in primary breast carcinomas. Glycobiology 1999;9:1307–11. [DOI] [PubMed] [Google Scholar]
- 9. Malapeira J, Esselens C, Bech‐Serra JJ, et al. ADAM17 (TACE) regulates TGFβ signaling through the cleavage of vasorin. Oncogene 2011;30:1912–22. [DOI] [PubMed] [Google Scholar]
- 10. Ikeda Y, Imai Y, Kumagai H, et al. Vasorin, a transforming growth factor β‐binding protein expressed in vascular smooth muscle cells, modulates the arterial response to injury in vivo . Proc Natl Acad Sci USA 2004;101:10732–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li S, Li H, Yang X, et al. Vasorin is a potential serum biomarker and drug target of hepatocarcinoma screened by subtractive‐EMSA‐SELEX to clinic patient serum. Oncotarget 2015;6:10045–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang A, Dong J, Li S, et al. Exosomal transfer of vasorin expressed in hepatocellular carcinoma cells promotes migration of human umbilical vein endothelial cells. Int J Biol Sci 2015;11:961–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Man J, Yu X, Huang H, et al. Hypoxic induction of Vasorin regulates Notch1 turnover to maintain glioma stem‐like cells. Cell Stem Cell 2018;22:104–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fan TC, Yeo HL, Hsu HM, et al. Reciprocal feedback regulation of ST3GAL1 and GFRA1 signaling in breast cancer cells. Cancer Lett 2018;434:184–95. [DOI] [PubMed] [Google Scholar]
- 15. Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat Protoc 2006;1:241–5. [DOI] [PubMed] [Google Scholar]
- 16. Morelle W, Michalski JC. Analysis of protein glycosylation by mass spectrometry. Nat Protoc 2007;2:1585–602. [DOI] [PubMed] [Google Scholar]
- 17. Kumagai T, Katoh T, Nix DB, et al. In‐gel beta‐elimination and aqueous‐organic partition for improved O‐ and sulfoglycomics. Anal Chem 2013;85:8692–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ceroni A, Maass K, Geyer H, et al. GlycoWorkbench: a tool for the computer‐assisted annotation of mass spectra of glycans. J Proteome Res 2008;7:1650–9. [DOI] [PubMed] [Google Scholar]
- 19. Domon B, Costello CE. A systematic nomenclature for carbohydrate fragmentations in FAB‐MS/MS spectra of glycoconjugates. Glycoconjugate Journal 1988;5:397–409. [Google Scholar]
- 20. Arnaoutova I, Kleinman HK. In vitro angiogenesis: endothelial cell tube formation on gelled basement membrane extract. Nat Protoc 2010;5:628–35. [DOI] [PubMed] [Google Scholar]
- 21. Delisser HM, Christofidou‐Solomidou M, Strieter RM, et al. Involvement of endothelial PECAM‐1/CD31 in angiogenesis. Am J Pathol 1997;151:671–7. [PMC free article] [PubMed] [Google Scholar]
- 22. Chantrain CF, Declerck YA, Groshen S, et al. Computerized quantification of tissue vascularization using high‐resolution slide scanning of whole tumor sections. J Histochem Cytochem 2003;51:151–8. [DOI] [PubMed] [Google Scholar]
- 23. Picco G, Julien S, Brockhausen I, et al. Over‐expression of ST3Gal‐I promotes mammary tumorigenesis. Glycobiology 2010;20:1241–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pereira ME, Kabat EA, Lotan R, et al. Immunochemical studies on the specificity of the peanut (Arachis hypogaea) agglutinin. Carbohydr Res 1976;51:107–18. [DOI] [PubMed] [Google Scholar]
- 25. Karlsson NG, Packer NH. Analysis of O‐linked reducing oligosaccharides released by an in‐line flow system. Anal Biochem 2002;305:173–85. [DOI] [PubMed] [Google Scholar]
- 26. Ruhaak LR, Zauner G, Huhn C, et al. Glycan labeling strategies and their use in identification and quantification. Anal Bioanal Chem 2010;397:3457–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Priatel JJ, Chui D, Hiraoka N, et al. The ST3Gal‐I sialyltransferase controls CD8(+) T lymphocyte homeostasis by modulating O‐glycan biosynthesis. Immunity 2000;12:273–83. [DOI] [PubMed] [Google Scholar]
- 28. Moody AM, Chui D, Reche PA, et al. Developmentally regulated glycosylation of the CD8αβ coreceptor stalk modulates ligand binding. Cell 2001;107:501–12. [DOI] [PubMed] [Google Scholar]
- 29. Steentoft C, Vakhrushev SY, Joshi HJ, et al. Precision mapping of the human O‐GalNAc glycoproteome through SimpleCell technology. EMBO J 2013;32:1478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Massague J. TGFβ in cancer. Cell 2008;134:215–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moses H, Barcellos‐Hoff MH. TGF‐β biology in mammary development and breast cancer. Cold Spring Harb Perspect Biol 2011;3:a003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Muraoka‐Cook RS, Kurokawa H, Koh Y, et al. Conditional overexpression of active transforming growth factor β1 in vivo accelerates metastases of transgenic mammary tumors. Cancer Res 2004;64:9002–11. [DOI] [PubMed] [Google Scholar]
- 33. Muraoka‐Cook RS, Shin I, Yi JY, et al. Activated type I TGFβ receptor kinase enhances the survival of mammary epithelial cells and accelerates tumor progression. Oncogene 2006;25:3408–23. [DOI] [PubMed] [Google Scholar]
- 34. Hasegawa Y, Takanashi S, Kanehira Y, et al. Transforming growth factor‐β1 level correlates with angiogenesis, tumor progression, and prognosis in patients with nonsmall cell lung carcinoma. Cancer 2001;91:964–71. [PubMed] [Google Scholar]
- 35. Cui W, Fowlis DJ, Bryson S, et al. TGFβ1 inhibits the formation of benign skin tumors, but enhances progression to invasive spindle carcinomas in transgenic mice. Cell 1996;86:531–42. [DOI] [PubMed] [Google Scholar]
- 36. Wan YY, Flavell RA. 'Yin‐Yang' functions of transforming growth factor‐β and T regulatory cells in immune regulation. Immunol Rev 2007;220:199–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Anscher MS, Peters WP, Reisenbichler H, et al. Transforming growth β as a predictor of liver and lung fibrosis after autologous bone marrow transplantation for advanced breast cancer. N Engl J Med 1993;328:1592–8. [DOI] [PubMed] [Google Scholar]
- 38. Levy L, Hill CS. Alterations in components of the TGF‐beta superfamily signaling pathways in human cancer. Cytokine Growth Factor Rev 2006;17:41–58. [DOI] [PubMed] [Google Scholar]
- 39. Bierie B, Chung CH, Parker JS, et al. Abrogation of TGF‐β signaling enhances chemokine production and correlates with prognosis in human breast cancer. J Clin Invest 2009;119:1571–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Padua D, Zhang XH, Wang Q, et al. TGFβ primes breast tumors for lung metastasis seeding through angiopoietin‐like 4. Cell 2008;133:66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Teicher BA. Malignant cells, directors of the malignant process: role of transforming growth factor‐beta. Cancer Metastasis Rev 2001;20:133–43. [DOI] [PubMed] [Google Scholar]
- 42. Vrana JA, Stang MT, Grande JP, et al. Expression of tissue factor in tumor stroma correlates with progression to invasive human breast cancer: paracrine regulation by carcinoma cell‐derived members of the transforming growth factor β family. Cancer Res 1996;56:5063–70. [PubMed] [Google Scholar]
- 43. Zhao Y, Hu J, Li R, et al. Enhanced NK cell adoptive antitumor effects against breast cancer in vitro via blockade of the transforming growth factor‐β signaling pathway. Onco Targets Ther 2015;8:1553–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lebrin F, Deckers M, Bertolino P, et al. TGF‐beta receptor function in the endothelium. Cardiovasc Res 2005;65:599–608. [DOI] [PubMed] [Google Scholar]
- 45. Son SW, Song KH, Kwon HY, et al. Transcriptional activation of pig Galbeta1,3GalNAc alpha2,3‐sialyltransferase (pST3Gal I) gene by TGF‐β1 in porcine kidney PK‐15 cells. Biochem Biophys Res Commun 2011;414:159–64. [DOI] [PubMed] [Google Scholar]
- 46. Yan X, Xiong X, Chen YG. Feedback regulation of TGF‐beta signaling. Acta Biochim Biophys Sin (Shanghai) 2018;50:37–50. [DOI] [PubMed] [Google Scholar]
- 47. Redig AJ, McAllister SS. Breast cancer as a systemic disease: a view of metastasis. J Intern Med 2013;274:113–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Western blot and RT‐PCR analysis of expression level of ST3Gal1. (a) Western blot analysis of ST3Gal1 expression level in several breast cancer cell lines. (b) RT‐QPCR analysis of mRNA levels of ST3Gal1 upon stable overexpression of shST3Gal1 (Left panel) and transiently transfected siRNA of ST3Gal1 in MCF7.
Figure S2: Effects of ST3Gal1 silencing on MCF7 and ASB‐634 on xenograft tumor. (a) Gross appearance of MCF7/pVoid, MCF7/shST3Gal1 xenograft tumor and surrounding tissues, (b) ASB‐634/pVoid, ASB634/shST3Gal1 xenograft tumor profile, (c) cross‐section and (d) blood vessels density in the harvested tumor, as identified by IHC staining with anti‐mouse CD31 antibody. The DAB‐positive areas were quantified using MetaMorph 4.6 software (lower panel).
Figure S3: Tandem mass spectra of indicated sialylated O‐glycans from sVASN. The m/z of precursor ions used for generation of MS/MS spectra are shown in the upper right corner of each sub‐figure. Peaks assignment for the fragment ions are highlighted with blue arrows and the neural loss of CH3OH (−32) from specific fragment ions are indicated with red arrows.
Figure S4: Proliferation of MCF/pVoid and MCF/shST3Gal1 with or without overexpression of VASN were monitored using the xCELLigence Real Time Cell analysis instrument.
Figure S5: Luminex immunosandwish assay of TGF‐β1 binding to VASN coated bead, with or without the presence of anti‐VASN. Data are represented as mean (SD). Data are analyzed by Student's t‐test. *** p < 0.001
Figure S6: RT‐PCR analysis of angiogenesis related genes from HUVEC incubated for 3 h with 1 ng/ml TGF‐β1. Data are represented as mean (SD). Data are analyzed by Student's t‐test. *** p < 0.001
Figure S7: Western blot analysis the treatment of HUVEC with CM from VASN expression in MCF7/pVoid and MCF7/shST3Gal1 in the presence of TGF‐β1 for 1 h, GAPDH was used as control.
Figure S8: VASN were treated with glycosidase and incubated with TGF‐β1 coated bead for 1 h. After removed of TGF‐β1 coated bead, the remaining VASN was analysis by Western blot analysis.
Figure S9: Expression levels of ST3GAL1 in different grade. Gene expression level was determined by RT‐PCR. Each scatter dot represents the value of minus delta threshold cycle (‐ΔCt) normalized to the average of 3 internal controls: GAPDH, GUSB and UBC.
Supplementary Table S1: Summary of identified O‐glycoforms from sVASN using LC–MS/MS analysis.
Supplementary Table S2. Univariate and multivariable Cox proportional regression analysis of mRNA expression and breast cancer recurrence.
Supplementary Table S3: qPCR primers


