Abstract
Kinase domain duplications of the epidermal growth factor receptor (EGFR‐KDD) have been identified and implicated to be oncogenic in nonsmall cell lung cancers (NSCLCs). However, its prevalence and clinical contributions in lung cancer are largely unknown. Here, we conducted a multicenter record review of 10,759 NSCLC patients who underwent genetic testing using next‐generation sequencing (NGS) targeting EGFR exons and the introns involved in EGFR‐KDD rearrangements. EGFR‐KDDs were identified in a total of 13 patients, which is approximately 0.12% of the total population reviewed, and also consisted of 0.24% (13/5394) of EGFR mutation‐positive patients. A total of 85% of patients (11/13) were identified with the canonical EGFR‐KDD duplication of exons 18–25, while the remaining two cases harbored duplications of EGFR exons 14–26 and exons 17–25, which have not been previously described. Importantly, none of the 13 patients had other coexisting driver mutations, highlighting the potential oncogenic role of this type of alteration. Three out of five patients who had exon 18–25 duplications showed partial antitumor responses to targeted therapies, while the other two patients demonstrated no clinical improvement. Furthermore, our data suggested that the EGFR T790 M mutation and EGFR amplification may represent the major resistance mechanisms against targeted therapies in tumors bearing EGFR‐KDD. In summary, our findings provide valuable insight into the prevalence of EGFR‐KDDs in NSCLCs and their clinical outcomes to targeted therapies.
Keywords: EGFR, kinase domain duplication, NSCLC, tyrosine kinase inhibitor, targeted therapy
Short abstract
What's new?
A rare class of mutation could provide a therapeutic target for lung cancer. Many NSCLCs arise with mutations in the EGFR tyrosine kinase domain, and treatment with EGFR tyrosine kinase inhibitors often boosts survival in these patients. Usually, these are deletions or point mutations, but sometimes the mutation is a kinase domain duplication (KDD). These authors investigated how frequently EGFR‐KDDs occur, and whether tyrosine kinase inhibitors work against them. By reviewing records from thousands of NSCLC patients, they discovered 13 EGFR‐KDDs. Tyrosine kinase inhibitors did suppress some of the tumors, although resistance arose in the event of additional EGFR mutations.
Introduction
The prospective identification and rational therapeutic targeting of tumor genomic alterations have revolutionized the care of patients with lung cancer. However, lung cancer remains one of the leading causes of cancer‐related deaths worldwide. Oncogenic mutations of the epidermal growth factor receptor (EGFR) tyrosine kinase domain are identified in a substantial subset of nonsmall cell lung cancers (NSCLCs), and patients with EGFR‐mutant lung cancers generally achieve better progression‐free survivals (PFS) when treated with EGFR tyrosine kinase inhibitors (TKIs), compared to standard chemotherapies.1, 2, 3 Such mutations, which most commonly occur as either small in‐frame deletions in exon 19 (Ex19del) or point mutations in exon 21 (L858R), confer constitutive activity of the EGFR tyrosine kinase and sensitivity to EGFR TKIs.4 Uncommon EGFR alterations, including rare point mutations and gene rearrangements, such as kinase domain duplications (KDDs) and gene fusions, have also been identified in previous studies.5, 6, 7
EGFR‐KDDs typically result from in‐frame tandem duplication of EGFR exons 18–25 and constitutively activate EGFR signaling by forming an intramolecule dimer. Such a recurrent alteration is often observed in lung, brain, and soft tissue cancers.6 A few reports showed that patients with lung adenocarcinomas harboring the EGFR‐KDD alteration had significant antitumor responses to EGFR TKI therapies, including gefitinib, erlotinib, and afatinib.5, 6 Thus, those findings suggested that EGFR‐KDD represents a driver aberration and a therapeutically actionable target in NSCLC patients. Herein, we interrogated next‐generation sequencing (NGS)‐based genetic testing data of 10,759 NSCLC cases from a multicenter database with known EGFR mutations, which provided the opportunity to determine the prevalence of EGFR‐KDD alterations in a large sample set. We also evaluated the clinical significance of EGFR‐KDDs, aiming to provide valuable insights into the clinical efficacy of TKIs that target common EGFR‐sensitizing mutations against this alteration, and the potential drug‐resistance mechanisms arising during disease progression.
Methods
We retrospectively reviewed records of 10,759 East Asian NSCLC patients who underwent genetic testing using capture‐based targeted NGS between August 2016 and April 2018 at hospitals across China, including the First Affiliated Hospital of Dalian Medical University (Liaoning, China), the First Affiliated Hospital of Zhengzhou University (Henan, China), the Beijing Tuberculosis and Thoracic Tumor Research Institute (Beijing, China), the First Affiliated Hospital of Nanjing Medical University (Jiangsu, China), the Affiliated Cancer Hospital & Institute of Guangzhou Medical University (Guangdong, China), and the Sun Yat‐sen University Cancer Centre (Guangdong, China). Written informed consent was collected from each patient upon sample collection according to the protocols approved by the ethical committee of each hospital. We identified patients with EGFR‐KDD alterations using a natural language search of the Laboratory Information Management System (LIMS) database for every EGFR mutation analysis performed on patients. Relevant demographic and clinical data were abstracted from the database for those cases, including age, gender, date of diagnosis, histology type, pathological stage, and evaluation of response after treatment with TKIs according to Response Evaluation Criteria In Solid Tumors (RECIST, version 1.1).8 Genomic profiling was performed by targeted sequencing of pan‐cancer relevant genes on post‐treatment samples from patients receiving TKI therapies.
Results
A multi‐institutional database revealed a total of 10,759 patients diagnosed with NSCLCs between August 2016 and April 2018. Approximately 90.2% (9,704/10,759) of all patients were lung adenocarcinomas, and approximately 50.1% (5,394/10,759) of harbored EGFR mutations. EGFR‐KDD was identified in 13 patients (Table 1), giving rise to the frequency of about 0.12% of the total NSCLCs and 0.24% of the EGFR mutation‐positive population. A total of 545 patients were identified with EGFR mutations other than EGFR L858R, ex19del, G719X, or L861Q, resulting in a frequency of 2.4% of EGFR‐KDD in the cohort bearing EGFR rare mutations.
Table 1.
Clinical information for NSCLC patients with EGFR‐KDD
| Patient ID | Age at diagnosis | Gender | Histology | Stage | Type of EGFR‐KDD | Sample type (Pre‐/Post‐ TKI) | EGFR amplification status |
|---|---|---|---|---|---|---|---|
| GS‐1 | 41 | Female | Lung adenocarcinoma | IV | exon 18–25 | P, T/ NA | no |
| GS‐2 | 48 | Male | Lung adenocarcinoma | NA | exon 18–25 | T/ NA | EGFR amp (pre) |
| GS‐3 | 61 | Male | Lung adenocarcinoma | IV | exon 18–25 | P, T/ P | EGFR amp (post) |
| GS‐4 | 63 | Male | Lung adenocarcinoma | IV | exon 18–25 | P/ NA | no |
| GS‐5 | 60 | Female | Lung adenocarcinoma | IV | exon 18–25 | NA/ P | EGFR amp (post) |
| GS‐6 | 67 | Male | Lung adenocarcinoma | IV | exon 18–25 | P, T/ NA | no |
| GS‐7 | 43 | Female | Lung adenocarcinoma | IV | exon 18–25 | NA/ P | EGFR amp (post) |
| GS‐8 | 52 | Male | Lung adenocarcinoma | IV | exon 17–25 | P, T/ NA | no |
| GS‐9 | NA | Male | Lung adenocarcinoma | NA | exon 18–25 | T/ NA | no |
| GS‐10 | 55 | Female | Lung adenocarcinoma | IV | exon 18–25 | P, T/ NA | no |
| GS‐11 | 87 | Male | Lung squamous cell carcinoma | IIA | exon 14–26 | P, T/ NA | no |
| GS‐12 | 84 | Male | Lung adenocarcinoma | IV | exon 18–25 | P/ NA | no |
| GS‐13 | 74 | Female | Lung adenocarcinoma | NA | exon 18–25 | T/ NA | no |
NA, not available; P, plasma; T, tumor tissue.
The median age of EGFR‐KDD patients at primary diagnosis was 60 years (range: 41–87 years) (Table 1). The male to female ratio of this cohort was 1.6:1. The stage of disease at diagnosis was 69.2% stage IV (9/13), 7.7% stage II (1/13), and the remaining three cases were undetermined. With the exception of one lung squamous cell carcinoma, all specimens were diagnosed with lung adenocarcinomas. Eight cases had an isolated EGFR‐KDD alteration, while three cases had a concurrent EGFR amplification. Eleven out of thirteen cases were identified with the previously described EGFR‐KDD that occurs in exons 18–25. However, the genomic breakpoints occurring in introns 17 and 25 varied between cases. Figure 1 a is representative of one example of such a case, while the other two patients were found to have atypical KDD events occurring in exons 17–25 and exons 14–26, respectively (Fig. 1 b and c). Furthermore, these EGFR‐KDD alterations were detected in plasma circulating cell‐free DNA (cfDNA) (Table 1).
Figure 1.
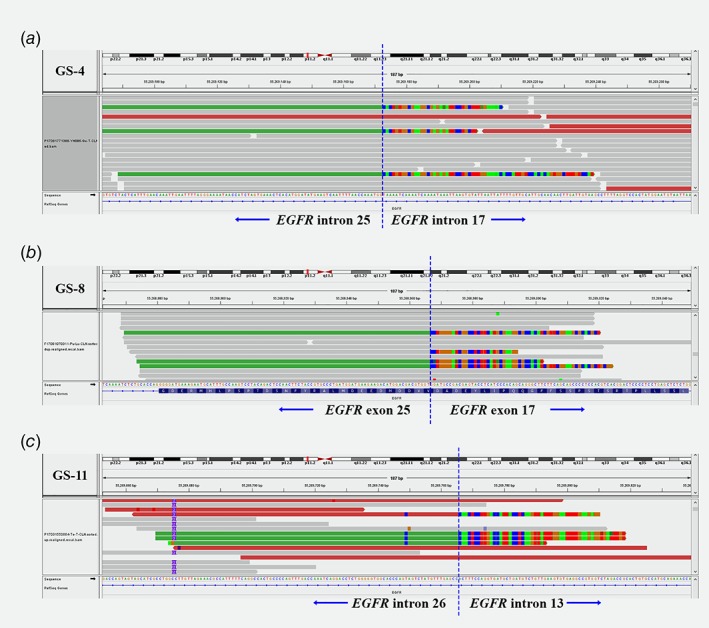
Visualization of EGFR‐KDD variants using the IGV Browser. The bottom of each panel consists of the EGFR reference sequence with nucleotides shown. Blue vertical dashed lines indicate where the breakpoints were localized. KDD events include canonical exon 18–25 duplications in GS‐4 (a), and uncommon ones, such as exons 17–25 in GS‐8 (b) and exons 14–26 in GS‐11 (c).
Patients’ treatment histories and their tumor responses to TKIs from our study and the literature are summarized in Table 2. In our study, patients with EGFR‐KDD‐driven lung adenocarcinomas (n = 5) showed variable antitumor responses to EGFR TKIs. Two patients (GS‐3 and GS‐7) progressed shortly after undergoing therapies on EGFR TKIs, including gefitinib, erlotinib and osimertinib with a PFS of less than 3 months. The other two patients exhibited partial responses to TKI treatments. One patient's (GS‐4) tumor responded to osimertinib after a relapse on afatinib, while the other patient (GS‐5) remained stable on gefitinib therapy with a PFS of 11 months. A fifth case was observed with a partial antitumor response to the VEGFR inhibitor, apatinib, combined with the first‐generation EGFR TKI, Icotinib,9 and is currently free of progressive disease. Targeted sequencing of post‐TKI samples revealed potential resistance mechanisms, including EGFR T790 M mutations (GS‐5), EGFR amplifications (GS‐3, GS‐5, and GS‐7), and missense mutations in PIK3R1 (GS‐3) and MAP2K4 (GS‐7); however, the clinical significance of such mutations remains unknown.
Table 2.
Summary of the responses of EGFR‐KDDs (exon 18–25) to targeted therapies in patients with the alteration
| Treatment and response | |||||||
|---|---|---|---|---|---|---|---|
| Database (EGFR‐KDD exon 18–25) | Treatment history | Best response to TKI | TKI and PFS (mo: month, yr: year) | Pre_TKI_alteration (Sample type) | Post_TKI_alteration (Sample type) | Potential AR | |
| our study | GS‐3 | Chemotherapy, targeted therapy (TKI, monoclonal antibody) | PD | (erlotinib, PD, 2 mo), (osimertinib, PD, 2 mo) | EGFR‐KDD, TP53 R280G (plasma and tissue) | EGFR‐KDD, EGFR amp (1.6‐fold), PIK3R1 Y463F (plasma) | EGFR amp, PIK3R1 Y463F |
| GS‐4 | Targeted therapy (TKI) | PR | (gefitinib, PR, 5 m0), (afatinib, PD, 2 mo), (osimertinib, PR, 4 mo [Not reached]) | EGFR‐KDD, ERBB2 amp (plasma) | NA | NA | |
| GS‐5 | Chemotherapy, targeted therapy (TKI) | SD | (gefitinib, SD, 11 mo) | NA | EGFR‐KDD, EGFR T790 M, EGFR amp (1.8‐fold) (plasma) | EGFR T790 M, EGFR amp | |
| GS‐6 | Chemotherapy, targeted therapy (TKI) | PR | (apatinib + icotinib, PR, 4 mo [Not reached]) | EGFR‐KDD, TP53 Y220C, PIK3CA E81G (plasma and tissue) | NA | NA | |
| GS‐7 | Chemotherapy, targeted therapy (TKI) | PD | (gefitinib, PD, 3 mo), (erlotinib, PD, 5 mo) | NA | EGFR‐KDD, EGFR amp (2.0‐fold), MAP2K4 S12 fs (plasma) | EGFR amp, MAP2K4 S12 fs | |
| Baik et al 5 | Targeted therapy (TKI) | PR | (gefitinib, PR, 6 yr), (erlotinib, PR, 3 yr) | not detected (tissue) | EGFR T790 M, CTNNB1 S37C (tissue) | EGFR T790 M, CTNNB1 S37C | |
| Gallant et al 6 | Chemotherapy, targeted therapy (TKI) | PR | (afatinib, PR, 7 cycles, about 10 mo) | EGFR amp not detected (tissue) | EGFR amp (tissue) | EGFR amp | |
| Wiest et al 11 | Chemotherapy, targeted therapy (TKI) | PR | (afatinib, PR, NA) | NA | NA | NA | |
PD, progressive disease; PR, partial response; SD, stable disease; NA, not available; AR, acquired resistance; amp, amplification.
Discussion
In our study, we reported that EGFR‐KDDs comprised 0.12% of all NSCLCs and 0.24% of all EGFR aberrations, including the canonical KDD rearrangements involving exons 18–25 and uncommon rearrangements, such as duplications of exons 14–26 and exons 17–25 that have not previously been described. The frequencies of such rearrangements were slightly higher than the previously reported statistics of 0.07% of lung cancers (N = 7,200) reported by Gallant et al.6 and 0.2% of the EGFR‐positive cohort (N = 1,510) reported by Costa et al.10. The higher frequency observed in our study is likely because EGFR mutations are more common in NSCLCs of ethnic Chinese individuals, compared to Caucasians.
EGFR‐KDD of exons 18–25 is the most frequent kinase domain duplication variant. Baik and colleagues reported the first case of this kind in a 45‐year‐old woman who was diagnosed with a nonmucinous bronchoalveolar carcinoma.5 The patient demonstrated a partial response to the first‐generation EGFR TKIs, including gefitinib and erlotinib, while resistance subsequently developed with the emergence of EGFR T790 M and CTNNB1 S37F missense mutations (Table 2). Another case study also reported a EGFR‐KDD of exons 18–25 in a 33‐year‐old male, which showed a preferable antitumor response to the second generation EGFR TKI, afatinib.6 Furthermore, the effect of afatinib in suppressing tumors harboring EGFR‐KDD of exons 18–25 was also observed in a 72‐year‐old patient reported by Wiest and colleagues.11 Here, we observed variable antitumor responses in a total of five patients who received targeted therapies, including gefitinib, erlotinib, osimertinib, and a VEGFR inhibitor, apatinib, revealing the therapeutic complexities and challenges associated with the treatment of patients with EGFR‐KDD aberrations. The best response was observed in patient GS‐5 who experienced a controlled disease while on gefitinib with a PFS of 11 months. Such stability was similar to the PFS seen in patients diagnosed with EGFR ex19del or L858R mutations,12, 13 further indicating that EGFR‐KDDs, although rare, are targetable tumor driver mutations. Subsequently, however, the EGFR T790 M mutation and EGFR amplification were detected in this patient's plasma ctDNA sample, and may explain the acquired resistance.
Kinase domain duplications are not only observed in EGFR as common genomic mechanisms underlying molecular adaption to the highly selective pressures and resistance to antibiotics and cancer therapies. In fact, BRAF‐KDDs have also been reported in gliomas.14 More recently, the analysis of 50,000 samples spanning multiple tumor types found that BRAF‐KDDs comprised 0.5% of all BRAF alterations identified.15 Additionally, KDD events were identified in other genes, including MET, FGFR1, FGFR3, RET, ERBB2, ALK, and NTRK in different types of tumors as potential driver mutations.16 It is therefore important to increase the panel coverage of intronic regions of such kinases for comprehensive NGS analyses in the future.
In summary, our study provided valuable insight into the prevalence and clinical outcomes of EGFR‐KDDs in NSCLCs. Our findings suggested that such rearrangements are clinically important genomic alterations and provide therapeutic targets for TKI treatments. Thus, the clinical incorporation of tumor‐profiling technologies is likely to further refine the clinicopathologic features of EGFR‐KDDs, and provide avenues for more effective therapeutic strategies.
Disclosures
Qiuxiang Ou, Xue Wu and Yang W. Shao are employees or shareholders of Geneseeq Technology Inc. Xiaonan Wang and Ming You are shareholders or employees of Nanjing Geneseeq Technology Inc. The remaining authors have no conflicts of interest to disclose.
Acknowledgements
We would like to thank the patients and their family members who consented to having their data presented in our study. We also thank the investigators and research staff at all hospitals and research sites involved.
[Correction added on January 7, 2019, after first online publication: December 16, 2018. Shucai Zhang's address has been updated]
Contributor Information
Jinguang Wang, Email: dlwangjg@163.com.
Zhihong Zhang, Email: zhangzhih2001@aliyun.com.
Shucai Zhang, Email: sczhang6304@163.com.
References
- 1. Giaccone G, Gallegos Ruiz M, Le Chevalier T, et al. Erlotinib for frontline treatment of advanced non‐small cell lung cancer: a phase II study. Clin Cancer Res 2006;12:6049–55. [DOI] [PubMed] [Google Scholar]
- 2. Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in untreated EGFR‐mutated advanced non‐small‐cell lung cancer. N Engl J Med 2018;378:113–25. [DOI] [PubMed] [Google Scholar]
- 3. Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin‐paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947–57. [DOI] [PubMed] [Google Scholar]
- 4. da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol 2011;6:49–69. [DOI] [PubMed] [Google Scholar]
- 5. Baik CS, Wu D, Smith C, et al. Durable response to tyrosine kinase inhibitor therapy in a lung cancer patient harboring epidermal growth factor receptor tandem kinase domain duplication. J Thorac Oncol 2015;10:e97–9. [DOI] [PubMed] [Google Scholar]
- 6. Gallant JN, Sheehan JH, Shaver TM, et al. EGFR kinase domain duplication (EGFR‐KDD) is a novel oncogenic driver in lung cancer that is clinically responsive to Afatinib. Cancer Discov 2015;5:1155–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Krawczyk P, Reszka K, Ramlau R, et al. Prevalence of rare EGFR gene mutations in nonsmall‐cell lung cancer: a multicenter study on 3856 polish Caucasian patients. Ann Oncol 2016;27:358–9. [DOI] [PubMed] [Google Scholar]
- 8. Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228–47. [DOI] [PubMed] [Google Scholar]
- 9. Shi Y, Zhang L, Liu X, et al. Icotinib versus gefitinib in previously treated advanced non‐small‐cell lung cancer (ICOGEN): a randomised, double‐blind phase 3 non‐inferiority trial. Lancet Oncol 2013;14:953–61. [DOI] [PubMed] [Google Scholar]
- 10. Costa DB. Kinase inhibitor‐responsive genotypes in EGFR mutated lung adenocarcinomas: moving past common point mutations or indels into uncommon kinase domain duplications and rearrangements. Transl Lung Cancer Res 2016;5:331–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wiest G, Kohlhäufel MMJ, Lakis S., et al. Detection of an EGFR kinase domain duplication in a lung adenocarcinoma patient by liquid biopsy using hybrid capture‐based next generation sequencing. New Oncol Abstract #291 2016. http://www.newoncology.com/fileadmin/content/Download_pdfs/Wiest_et_al_DGHO-2016-Abstract_final.pdf [Google Scholar]
- 12. Inoue A, Suzuki T, Fukuhara T, et al. Prospective phase II study of gefitinib for chemotherapy‐naive patients with advanced non‐small‐cell lung cancer with epidermal growth factor receptor gene mutations. J Clin Oncol 2006;24:3340–6. [DOI] [PubMed] [Google Scholar]
- 13. Sequist LV, Martins RG, Spigel D, et al. First‐line gefitinib in patients with advanced non‐small‐cell lung cancer harboring somatic EGFR mutations. J Clin Oncol 2008;26:2442–9. [DOI] [PubMed] [Google Scholar]
- 14. Rodriguez FJ, Ligon AH, Horkayne‐Szakaly I, et al. BRAF duplications and MAPK pathway activation are frequent in gliomas of the optic nerve proper. J Neuropathol Exp Neurol 2012;71:789–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Klempner SJ, Bordoni R, Gowen K, et al. Identification of BRAF kinase domain duplications across multiple tumor types and response to RAF inhibitor therapy. JAMA Oncol 2016;2:272–4. [DOI] [PubMed] [Google Scholar]
- 16. Ikeda S, Lg D, Pavlick JC, et al. Comprehensive genomic profiling (CGP) of 114,200 advanced cancers identifies recurrent kinase domain duplications (KDD) and novel oncogenic fusions in diverse tumor types. Ann Oncol 2017;28:x1–x6. doi: 10.1093/annonc/mdx652 [DOI] [Google Scholar]


