Abstract
Methods for 3D‐imaging of biological samples are experiencing unprecedented development, with tools such as X‐ray micro‐computed tomography (μCT) becoming more accessible to biologists. These techniques are inherently suited to small subjects and can simultaneously image both external and internal morphology, thus offering considerable benefits for invertebrate research. However, methods for visualising 3D‐data are trailing behind the development of tools for generating such data. Our aim in this article is to make the processing, visualisation and presentation of 3D‐data easier, thereby encouraging more researchers to utilise 3D‐imaging. Here, we present a comprehensive workflow for manipulating and visualising 3D‐data, including basic and advanced options for producing images, videos and interactive 3D‐PDFs, from both volume and surface‐mesh renderings. We discuss the importance of visualisation for quantitative analysis of invertebrate morphology from 3D‐data, and provide example figures illustrating the different options for generating 3D‐figures for publication. As more biology journals adopt 3D‐PDFs as a standard option, research on microscopic invertebrates and other organisms can be presented in high‐resolution 3D‐figures, enhancing the way we communicate science.
Keywords: Blender, computed tomography, Drishti, Meshlab, PDF
1. INTRODUCTION
1.1. 3D‐imaging of invertebrates
High‐resolution 3D‐imaging tools are providing new ways of looking at the external and internal morphology of organisms in the millimetre to micrometre size range (Friedrich & Beutel, 2008). Modern, non‐destructive imaging methods such as X‐ray micro‐computed tomography (μCT) are particularly well suited to invertebrate research, mostly overcoming the traditional challenges associated with the small size and fragility of these organisms. Indeed, 3D‐imaging has great potential for invertebrate research by improving morphological study through 3D‐geometric morphometrics, expediting taxonomy by alleviating the need for physical viewing of holotypes, and enhancing our understanding of evolution by allowing high‐resolution visualisation of external and internal anatomy.
Invertebrate studies utilising 3D‐imaging tools now encompass a broad range of topics, including neuro‐anatomy (Greco, Tong, Soleimani, Bell, & Schäfer, 2012; Ribi, Senden, Sakellariou, Limaye, & Zhang, 2008), developmental biology (Lowe, Garwood, Simonsen, Bradley, & Withers, 2013; Martin‐Vega, Simonsen, & Hall, 2017; Richards et al., 2012), descriptions of extant (Akkari, Enghoff, & Metscher, 2015) and extinct fossilized invertebrates (Barden, Herhold, & Grimaldi, 2017; Garwood & Sutton, 2010; van de Kamp et al., 2018), morphology and evolution of arthropod genitalia (McPeek, Shen, Torrey, & Farid, 2008; Schmitt & Uhl, 2015; Simonsen & Kitching, 2014; Tatarnic & Cassis, 2013; Wojcieszek, Austin, Harvey, Simmons, & Hayssen, 2012; Woller & Song, 2017; Wulff, Lehmann, Hipsley, & Lehmann, 2015), and 3D‐anatomical atlases (Bicknell, Klinkhamer, Flavel, Wroe, & Paterson, 2018). To highlight the growth of this field, the number of published studies has almost doubled in the last 5 years (Web of Science search for Topics including ‘micro‐CT’, ‘nano‐CT’ or ‘computed tomography’ in Biology: 393 from 2013 to 2017 compared to 201 from 2008 to 2012, accessed June 2018). However, methods for visualising 3D‐data have fallen behind the rapid technological development of tools for gathering such data, and thus the potential of the field is yet to be fully realised.
1.2. Visualisation of 3D‐data
As 3D‐data generation increases and scientific journals become primarily electronic, new presentation tools can be adopted that compliment the electronic format and allow us to more efficiently and effectively communicate science. The three primary modes of 3D‐data visualisation currently available for scientific publication are still images, videos, and interactive 3D‐files (most commonly 3D‐PDFs). Presently, still images are the most commonly used format, despite providing the most limited representation of 3D‐data. There appear to be two main reasons for this: relative ease of preparation, and inertia to incorporate multimedia figures in main texts, even from digital‐only journals. Unfortunately, most 3D‐figures are currently published in supplementary files, which require separate downloads and thus often go unseen. Although preparation of videos and 3D‐PDFs requires a set of skills and software that would be unfamiliar to many authors, these formats can more effectively convey 3D‐data. Structures of interest can still be annotated and highlighted in the same way as traditional 2D‐figures, and if prepared well, such figures can provide accurate representations of the subject's three‐dimensional form. The PDF format provides a standardised platform for dissemination of these data, as it allows embedding of all three formats (images, videos and 3D‐files), and provides a fully‐interactive environment for manipulating 3D‐data with Adobe Reader (freely available). Below we have summarised some of the recent studies (published since the review by Lautenschlager (2013)) that make use of 3D‐data in PDFs, and have highlighted the variety of ways this format can be used.
An excellent example of the utility of 3D‐PDFs is seen in van de Kamp, dos Santos Rolo, Vagovic, Baumbach, and Riedel (2014). They included a sophisticated animation of a Trigonopterus weevil moving into its ‘defence position’, demonstrating the advanced functionality possible with the PDF format, and exemplifying the untapped potential for scientific publication. Seidel and Lüter (2014) presented 3D‐PDFs of a brachiopod imaged with X‐ray CT, highlighting the benefits of these tools for examining the internal anatomy of animals without destructive dissection. Garcia, Fischer, Liu, Audisio, and Economo (2017) incorporated simple videos into PDF figures, providing excellent overviews of the 3D‐shape of each species to compliment the series of taxonomic images. Woller and Song (2017) and Bicknell et al. (2018) both provided high quality 3D‐PDF figures of CT reconstructed grasshopper reproductive structures and the horseshoe crab Limulus polyphemus respectively, demonstrating the utility of this medium for anatomical illustration and education. Additional details of these studies are available in Supporting Information Table 1, including the software used, its accessibility and functionality, the journal in which it was published, and whether the 3D‐figure was included in the main text or Supporting Information files.
Table 1.
Glossary of terms
| 3D‐reconstruction | The process of converting raw image projections (e.g., from X‐ray computed tomography) into cross‐sectional stacks of images, which resemble traditional tomographic sections. |
| 3D‐surface mesh | A series of 2D polygons (typically triangles or quadrangles) linked together to recreate the surface of a 3D‐object. This format is required for creating the interactive 3D‐PDFs described in this article, and for 3D‐printing. |
| Computed tomography | Commonly known as CT or ‘CAT’ scanning, a process for virtually recreating a three‐dimensional object from a series of sequential, cross‐sectional image slices, traditionally with microtome sectioning, but now more commonly with X‐ray imaging. |
| Projections | The raw images recovered from X‐ray CT imaging as the X‐ray source and camera rotate 360° around the subject (or the subject rotates as in μCT scanners). A higher number of projections results in a smaller angle between each projection, and therefore less noise in the reconstructed 3D‐image (but also a longer acquisition time). |
| Rendering | The process of adding colours, textures and lighting to a 3D‐object, which then determine the appearance of the final image or ‘render’. When imaging biological specimens, lighting is particularly important for visualising the true textures (e.g., from X‐ray CT) and colours (e.g., from photogrammetry) of the subject. |
| Rigging | The process of adding a virtual ‘skeleton’ to a 3D‐surface mesh in order to articulate joints and move or animate sections of the mesh independently. Used in this article to virtually reposition the mandibles of a scanned insect. |
| Segmentation (of 3D‐data) | The process of separating different regions of volumetric or surface mesh‐based 3D‐data. Typically used to aid visual differentiation, or for animation, but now also particularly useful for virtual dissections of internal morphology of invertebrates, which cannot easily be isolated by adjusting the visible range of densities (as one would for a vertebrate skeleton). |
| Volumetric 3D‐data | A cloud of ‘voxels’ (three‐dimensional pixels) that make up a virtually reconstructed 3D‐object generated by X‐ray CT scanning or similar, where each voxel contains information about the opacity of the original material (e.g., X‐ray absorption), thus providing measurable volumetric data of any part of the object. |
1.3. Challenges and directions for 3D‐visualisation
Despite the increasing number of studies presenting 3D‐data in the PDF format, we found that there was still a need for methods that did not rely on expensive licensed software. Here, we present a versatile framework for producing high‐quality figures for scientific publication (Figure 1), with the goal of addressing some of the current shortfalls of existing methods. This framework is largely based around free software: Drishti (Limaye, 2012), MeshLab (www.meshlab.net) and Blender (www.blender.org). Although many other packages have overlapping functionality (some free and some licensed), we found that these programs provided a good balance between ease of use, breadth of functionality and quality of output. As there are so few programs capable of generating 3D‐PDFs, our workflow is still reliant on the licensed Adobe Acrobat Pro for this final step. However, users can opt out of this final PDF‐creation step, and still generate still images, videos or 3D‐files in other formats (e.g., .ply or .obj), using only the free software included in our workflow.
Figure 1.
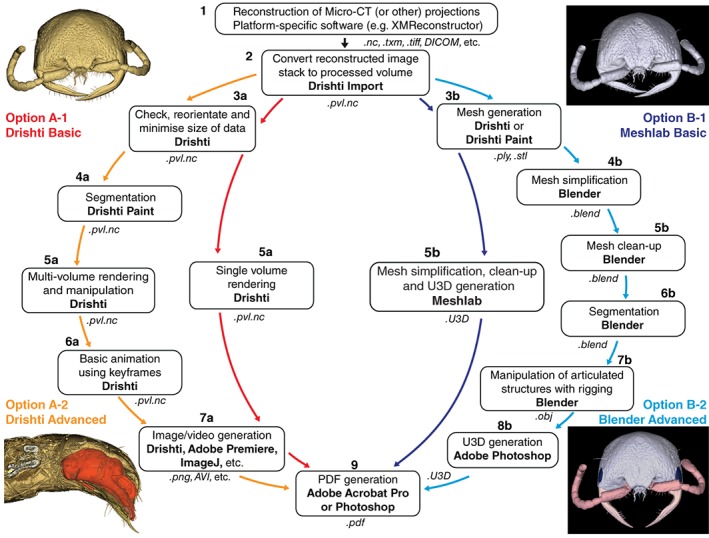
Framework for 3D‐data manipulation and visualisation, including four workflows (two basic and two advanced) for creating PDF figures from 3D‐data using the programs Drishti, MeshLab and Blender. The four images provide examples of the figures achievable from each of the corresponding workflows. Software used at each step is highlighted in bold, and file formats in italics
As with most new technologies, one of the main challenges associated with 3D‐data is the time investment required to familiarise oneself with the relevant software. This is further complicated by the fact that there is a myriad of different ways to prepare and display 3D‐data. To address this, we have provided four step‐by‐step workflows (Supporting Information Files 1–4); basic and advanced options for generating images and videos of volumetric data, and for creating interactive PDFs of surface‐mesh data. This is intended to allow complete beginners to produce 3D‐figures from μCT‐based (or other) 3D‐data, and to facilitate progression to more advanced manipulation of 3D‐data, producing segmented and animated 3D‐figures. As well as producing high‐quality figures for dissemination of 3D‐data, the two advanced workflows described here are intended to allow users to accurately visualise any component of an invertebrate specimen, external or internal. This is a fundamental requirement for morphometric analyses and can be particularly challenging in invertebrates, in contrast to hiding the low‐density soft‐tissues in order to examine vertebrate skeletons.
To facilitate the uptake of these methods we have provided four example figures to compare some of the ways 3D‐data can be visualised and prepared as PDF‐embedded figures. Figure 2 presents a volume rendering of a wasp head, made using the Basic Drishti workflow. The video in Figure 3 demonstrates virtual dissection of the terminalia of a wasp metasoma (abdomen), including depiction of the exertion of genitalia prior to mating, made using the Advanced Drishti workflow. Figure 4 is a 3D‐PDF of the same wasp head as Figure 2, but as a surface mesh made using the Basic MeshLab workflow. The 3D‐PDF in Figure 5 contains a segmented and “rigged” version of the same wasp head, this time prepared using the Advanced Blender workflow.
Figure 2.
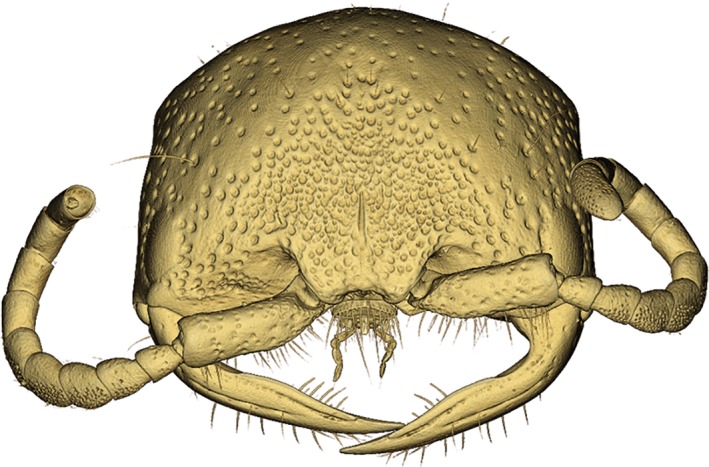
3D‐volume rendering of a female thynnine wasp head (Hymenoptera: Thynninae: Ariphron sp.) produced using the Basic Drishti workflow. This image exemplifies the detail available in μCT scan data, with fine setae and antennal sensory pores clearly visible
Figure 3.
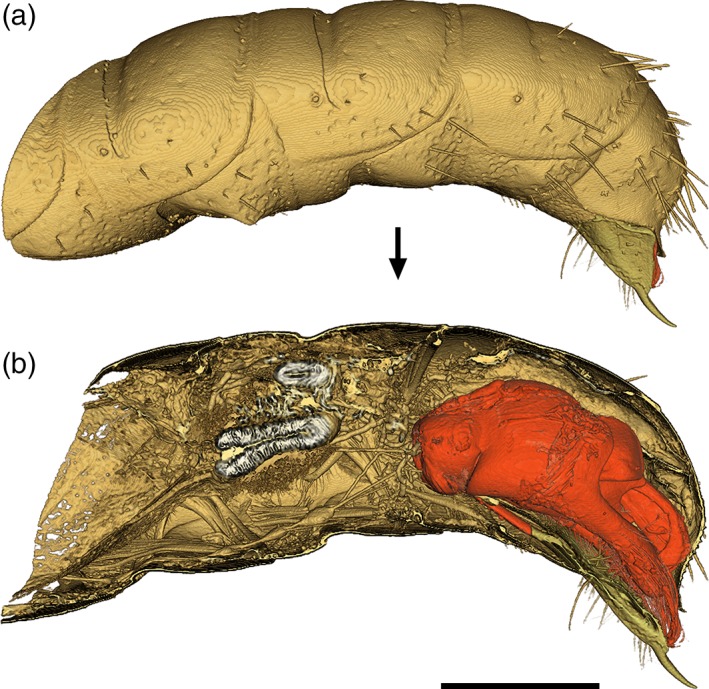
Note: This video can be viewed in the Supporting Information online .
(a) 3D‐rendering of the terminal abdominal segments of a male thynnine wasp (Hymenoptera: Thynninae: Catocheilus sp.). (b) Virtual dissection and segmentation of the male genitalia (red), generated using the Advanced Drishti workflow. Scale bar = 1 mm
Figure 4.
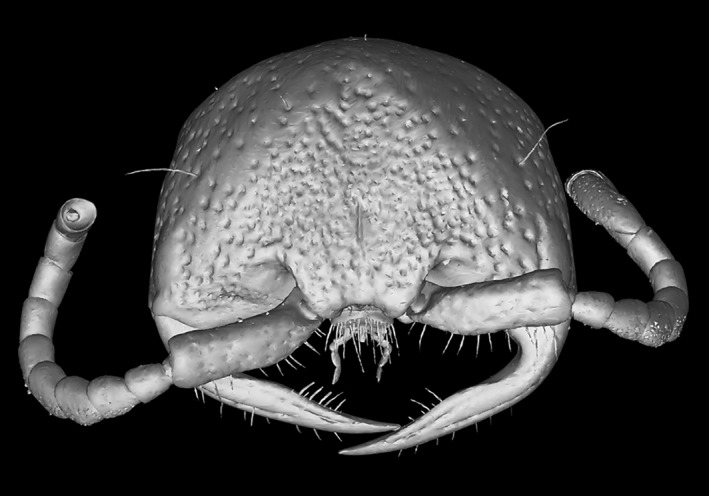
Note: To enable the interactive function of this figure, open the PDF in Adobe Reader program or web plug‐in.
Interactive 3D‐PDF of a female thynnine wasp head (Thynninae: Ariphron sp.) generated using the basic MeshLab workflow. This 3D‐mesh has been annotated to provide a self‐contained resource for science communication and education. A key benefit of presenting such data from primary research in the PDF format, is that those images can subsequently be used in education or for science communication without specialised software
Figure 5.
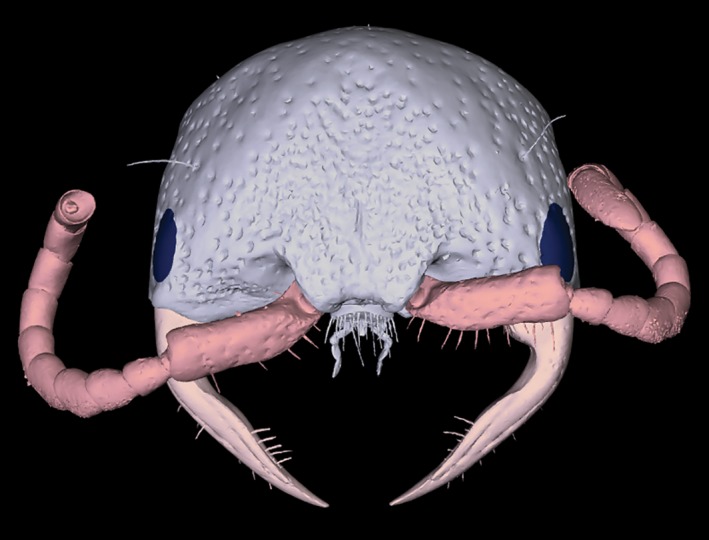
Note: To enable the interactive function of this figure, open the PDF in Adobe Reader program or web plug‐in.
Interactive 3D‐PDF of a female thynnine wasp head (Thynninae: Ariphron sp.) generated using the advanced blender workflow. This 3D‐mesh has been segmented and labeled to provide a highly informative scientific figure. Here, we also provide a basic example of how ‘rigging’ can be used to virtually reposition limbs or other components (in this case the mandibles of the wasp head). This can be thought of as ‘virtual insect pinning’, providing a standardized layout to facilitate examination, with the same reasoning as traditional insect pinning
2. MATERIALS AND METHODS
2.1. 3D‐imaging method
Our 3D‐data sets were obtained via μCT using a Zeiss Versa XRM‐520 X‐ray microscope at the Centre for Microscopy, Characterisation and Analysis at the University of Western Australia. Although this workflow was designed for CT‐based 3D‐data, the latter sections on generation of 3D‐PDFs can also be used with other types of 3D‐data obtained through techniques such as photogrammetry. The wasp head used in Figures 2, 4, and 5 belongs to an unidentified species of thynnine wasp from the genus Ariphron Erichson (Hymenoptera: Tiphiidae: Thynninae), and the metasoma used in Figure 3 is from an undescribed species in the genus Catocheilus Guérin‐Méneville (Thynninae; note that many thynnine wasps are undescribed, or not reliably identifiable with existing keys). Complete μCT imaging parameters and specimen details are available in Supporting Information Tables 2 and 3, respectively. The scan data used here is available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.vn6n74n.
2.2. Overview of workflow and software used
For all of our example data sets, μCT‐scans were reconstructed using XMReconstructor (Carl Zeiss Microscopy GmbH). For subsequent processing of reconstructed 3D‐data, we used Drishti v2.64 (Limaye, 2012), which includes three standalone programs: Drishti, Drishti Import and Drishti Paint. We used Drishti Import to convert our reconstructed .txm files to processed volume .pvl.nc files, then Drishti for reorientating and cropping volumes. Segmentation was carried out either in Drishti Paint, or in Blender v2.78 (www.blender.org), depending on the intended final figure type (explained in detail below). For video generation, we used Drishti to animate volumes with keyframe sequences, then either the free program ImageJ (Rasband, 1997–2016) or Adobe Premiere Pro for converting image sequences into movies. For creating 3D‐PDFs, surface meshes were generated as .ply or .stl files from Drishti or Drishti Paint, then simplified, segmented and exported as .obj or .stl files using either MeshLab v2016.12 (www.meshlab.net) or Blender. Note that the aforementioned 3D‐files can alternatively be used ‘as is’, and viewed with free 3D software such as MeshLab, Mac Preview or Windows 3D Builder, thereby removing the need for licenced Adobe software. For PDF generation, we used Adobe Photoshop (tested on CC versions 2014 and 2018) to convert .obj and .stl files to .u3d files, with the final .pdf files then created and annotated with Adobe Acrobat Pro (tested on versions XI and DC). We tested this workflow on both MacOS v10.12.6 and Windows 10 operating systems.
In an effort to provide a user‐friendly resource for 3D‐data manipulation and figure generation, we have presented the workflows in three complimentary formats. The first is a flowchart (Figure 1), which provides an overview of the different pathways and major steps, as well as the programs and file formats used at each step. The second is the methods text that follows below, in which we outline the methods we used to generate the example figures (Figures 2, 3, 4, 5). The third is a series of four comprehensive, step‐by‐step instructional guides (Supporting Information Files 1–4), which provide users with all of the information needed to take reconstructed 3D‐data through to both basic and advanced 3D‐figures.
2.3. Workflow
When using this workflow, the numbered steps provide a link between the workflow diagram (Figure 1), this methods text, and the step‐by‐step instructional guides (Supporting Information Files 1–4).
2.3.1. Reconstruction of μCT (or other) projections
Reconstruction of our μCT .txrm projections was done using XMReconstructor (which accompanies Zeiss Xradia X‐ray CT scanners), producing .txm image stack files. The workflow described here is compatible with most types of reconstructed 3D‐data, but given the platform‐specific nature of reconstruction software, we recommend following the software instructions or the guidance of your imaging facility for this initial step.
2.3.2. Convert reconstructed image stack to processed volume
After loading our .txm file into Drishti Import (various other image stack formats compatible), we selected the usable signal from the scan in the histogram provided, to exclude the majority of the background noise. Next, we defined the physical bounds of the volume by setting the limiting sliders for the Z‐axis and resizing the white bounding box for the X‐ and Y‐axes. With the desired range of densities and physical bounds of the volume selected, the file was saved in the .pvl.nc format for subsequent loading into Drishti or Drishti Paint.
2.3.3. Check, reorientate and minimise size of data
Our first checks of scan quality were done by loading the .pvl.nc file into Drishti. At this stage, we also re‐orientated the volume so that the subject was aligned with all three major axes, which greatly simplifies downstream processes such as segmentation. To do this, we first adjusted the transfer function (which allows users to define which voxels are visualised, based on the opacity values collected with the original scan data) to remove the ‘noise’ around the subject. Once the subject was visible, we orientated it with the major axes, set the transfer function to include all densities, then saved the data as a .pvl.nc file in the new orientation using the ‘reslice’ function. Next, we loaded the new ‘resliced’ .pvl.nc file into Drishti, and repeated this process, but this time with the bounding box adjusted to tightly crop the subject in its new orientation. After this second reslice operation the data is often reduced by up to half the size of the original .pvl.nc file.
2.3.4. Segmentation in Drishti paint
In Figure 3, segmentation of components was carried out in Drishti Paint by loading our resliced .pvl.nc file, then ‘tagging’ individual components in the 3D preview window. This method of segmentation allows users to retain all of the volumetric data information, so that structures can still be quantified after segmentation (as opposed to post‐meshing segmentation, which results in a reduced representation of the original data). After tagging, the data was exported in one of two ways: by extracting new .pvl.nc files for each segmented component (for animation and video generation in Drishti), or by generating .ply mesh files (for importing those components into Blender). The first of these two options flows on to step 5a, and the second to Step 5b.
2.3.5. Volume rendering and manipulation in Drishti
We used Drishti for rendering single volumes for image generation, manipulating multiple volumes in relation to each other, and as a tool for virtual dissection. After loading one or more .pvl.nc volume files (maximum of four) into Drishti, we used the ‘Bricks Editor’ to transform (scale, rotate and translate) individual components, while adjusting the transfer functions, lighting and shadows to achieve differential rendering for each component and aid visualisation.
2.3.6. Basic animation using keyframes in Drishti
To create basic animations and image sequences, we saved snapshots of the manipulated volumes in a logical sequence of states or positions with the Keyframe Editor. Drishti then interpolates the intermediate images between the user‐defined keyframes. Thus, a higher number of frames between each keyframe results in smoother movies.
2.3.7. Image and video generation from Drishti volume renderings
After rendering and manipulating volumes, we exported still images either as single images of the current rendered screen view, or by saving image sequences from Keyframes (from which any single image can also be used on its own). For generation of movies, we used either ImageJ or Adobe Premiere Pro to compile those image sequences generated in Drishti. We found ImageJ had an easier learning curve than Adobe Premiere Pro, but was limited in the file formats available for export. In the Windows version of Drishti, keyframe sequences can be exported directly as .wmv movie files, but exporting as image sequences provides more flexibility of file format and image size, and allows parameters such as frame rate to be specified when compiling the movie.
2.3.8. Mesh generation
Surface meshes can be generated in both Drishti and Drishti Paint. However, we used Drishti Paint exclusively, and highly recommend this option over the ‘Mesh Generator’ plugin in Drishti. We found that it consistently produced higher quality meshes, and usually smaller file sizes. If segmenting in Drishti Paint, individual components can be meshed based on the tag number used. Even if segmenting in Blender, the tag and ‘fill’ functions can be useful to select only the subject itself, excluding any ‘scan debris’ that might be in the volume as well. During mesh generation, we found that we could maintain greater detail by leaving smoothing and hole‐filling operations for post mesh‐generation steps.
2.3.9. Mesh simplification
To produce high‐quality 3D‐PDFs of manageable file sizes, it is important to simplify and clean meshes. This can be done either in Blender or in MeshLab (we use MeshLab for the basic workflow and Blender for the Advanced). To streamline this process for beginners, we created a Python script to automate the simplification step in Blender. Step 4b can either be followed manually, or automated in the Python Interactive Console within Blender, using the script provided in Supporting Information File 5.
For the Advanced Blender workflow, we imported and then resized and centred our meshes. We found that our Zeiss μCT‐data meshed in Drishti was usually several orders of magnitude bigger or smaller than the default field of view in Blender, and sometimes not centred, and thus not visible. Following this, we reduced the number of vertices (or faces/polygons) in the mesh with the ‘decimate’ tool, to between 10 and 50% of the initial number (depending on complexity), thereby removing unnecessarily complex geometry in the mesh, and reducing the file to a workable size.
An additional method for reducing the vertex count and file size is to hollow out objects where possible, which although both time‐consuming and requiring manual input, is a very effective way to reduce file sizes. This can be particularly useful with μCT‐data sets that contain detailed information on internal anatomy, but are often used to examine external features only. We did this by selecting the vertices visible on the external surface of the object, then inverting the selection and deleting the internal vertices. This step is essentially the same between the MeshLab and Blender workflows, but is carried out slightly differently (refer to workflows for more details).
2.3.10. Mesh clean‐up in blender
As general good practice, and of particular importance for 3D‐printing applications, surface meshes should undergo a ‘clean‐up’ step to remove artefacts produced during the meshing process. These steps include removal of holes where the volume might have had low‐opacity voxels, removal of duplicated edges and vertices (doubles), and resolving non‐manifold geometry. We performed all of these clean‐up operations in Blender, typically iterating through each of them several times to achieve the desired result, often with liberal use of the undo command. Following these steps, we performed a second decimation to reduce final vertex counts to between 20,000 and 200,000, depending on complexity.
2.3.11. Mesh simplification, clean‐up and U3D generation in MeshLab
In the Basic MeshLab workflow, we performed a simplified version of the Blender clean‐up process, as this was intended to be a quick and easy workflow. We first hollowed the mesh using the Ambient Occlusion tool to select external vertices, before inverting the selection and deleting. We then performed a decimation step to further simplify the mesh, followed by removal of duplicated vertices and removal of disconnected geometry. The resultant mesh was then exported directly as a .U3D file, to be annotated and saved as a .PDF in Adobe Acrobat.
2.3.12. Segmentation in blender
An alternative to the Drishti Paint segmentation method described above is to generate a mesh of the whole subject, then use Blender to perform segmentation after mesh simplification and clean‐up. We performed this segmentation by creating ‘vertex groups’ for each component, and assigning different materials to each for differentiation. This allows the entire mesh to be manipulated as a whole ‘body’ in the following Step 7b. However, in order for the segments to be recognised as individual objects in the final PDF, these vertex groups must be separated before exporting the completed mesh from Blender (described in more detail in the Advanced Blender workflow).
2.3.13. Manipulation of articulated structures with rigging in blender
For the wasp head in Figure 5 we used the rigging tools in Blender to re‐position the mandibles, as a virtual emulation of traditional insect setting and pinning. This tool is useful for providing an overview image of the subject, as it would appear in life, or for rearranging body parts obscuring a structure of interest. However, it is difficult to fully control the way Blender deforms the mesh around articulation points, and so this tool should be used with caution for anatomical or taxonomic images. First, we added armatures to the limbs, and then used the existing vertex groups for assignment of vertices to each armature, before positioning the limbs in ‘Pose Mode’, that is, manually re‐positioning each group of vertices around its new articulation point. After setting pose positions, vertex assignments can be refined either with the “weight paint” tool or by amending the assignments of each vertex group.
2.3.14. U3D conversion
Creating interactive 3D‐PDFs in Adobe Acrobat Pro requires files to be in the .U3D format. For the Advanced Blender workflow, we used Adobe Photoshop to convert meshes from Blender to .U3D files. Although the free program MeshLab can create .U3D files with simple meshes (as in our Basic workflow), we found that segmented components and associated materials could not be preserved through the file conversion with this program. By contrast, Photoshop is capable of converting various file types to .U3D while preserving materials and components, and we also found it useful for final colour adjustment to ensure consistency among figures. While we would prefer to utilise a free option for this step, we were unable to find one at the time of writing this manuscript.
2.3.15. 3D‐PDF generation
We used Adobe Acrobat Pro to create all of our final PDFs. This included still images exported from Drishti, videos compiled from Drishti image sequences, and .U3D files exported from Photoshop. For the interactive 3D‐PDFs, we used Acrobat Pro to set placeholder images (so that users not viewing the PDF in its interactive mode are still presented with a representative figure), create ‘views’ to guide users to important aspects of the subject with different rendering settings (e.g., opacity or lighting), and to label components. We found that the 3D Comment tool in Acrobat Pro provided a simple solution for labelling components in 3D‐PDFs, in such a way that the label is always visible when rotating the model (not a trivial task with other software).
3. DISCUSSION
3.1. Future directions and challenges with 3D‐imaging
Although 3D‐imaging in itself is not novel, its accessibility to research biologists has improved dramatically in recent years, and the community is currently still in the early stages of exploring its potential uses, particularly for invertebrates and other small organisms. Thus, it is an important time to explore different methods, but also to start establishing standards. For example, while it is possible to view some 3D‐file types (e.g., .stl and .obj) with native MacOS and Windows programs (‘Preview’ and ‘3D Builder’, respectively), we believe that using the well‐established and cross‐platform PDF format will provide more stability and consistency for the long‐term.
Here, we have provided examples of different 3D‐visualisation formats, with figures including still images, videos and interactive 3D‐PDFs. These figures can all be embedded in the main PDF files of papers (as they are here), thereby improving their visibility and facilitating easier dissemination. While there are advantages to each format, it would usually not be possible or practical to use all three in a single scientific publication, mostly due to file size restrictions. Interactive 3D‐PDFs are the only format that provides viewers complete control over how subjects are viewed. Users can view, manipulate and annotate these 3D‐figures with the free Adobe Reader program or web browser plug‐in, on any desktop computer.
Aside from the viewing platform itself, the manipulation and generation of 3D‐figures presents its own challenges. Most of the programs available for manipulating 3D‐data were designed for modelling in architecture, animated movies and computer games. Thus, while there is a plethora of programs available, it is difficult for new users to determine which are the most appropriate. We chose to use Drishti, MeshLab and Blender primarily because they are free and open‐source, and provide nearly unlimited options for expansion beyond the basic and advanced outputs presented here. We recognise that these programs can be time‐consuming to learn and implement, and so have provided a series of user‐friendly, step‐by‐step guides for visualising and manipulating 3D‐data (Supporting Information Files 1–4).
An additional justification for incorporating Blender into our workflow was its integration with the Python programming language, which allows users to automate functions and processes without extensive programming experience. This enabled us to automate the initial simplification steps in the Advanced Blender workflow, and choose from three different output meshes of varying simplification levels. As well as removing the manual input typically required in this process, this automation alleviates one of the most common challenges of working with large 3D‐volume files, which is the need for more graphical processing power than most standard desktop computers possess. Although the process might still take a long time, it can run unattended, while still allowing users to choose from various outputs on completion. As more and more 3D‐data are generated, we foresee an increased need for automation, so that processing tasks can run in the background and utilise specialised computing resources (just as bioinformatics is now largely outsourced to cluster‐based computing systems).
In addition to the visualisation tools we have described here, we envision virtual reality or augmented reality tools, along with 3D‐printing, becoming increasingly common and useful in science education. Even as this manuscript was being prepared, a significant new tool became available for testing: Drishti in Virtual Reality (https://github.com/nci/drishti/releases). The ability to explore in virtual reality the external and internal anatomy of microscopic organisms, and to handle enlarged yet accurate physical models of those same specimens, could significantly improve teaching quality, and potentially replace (or at least compliment) traditional expensive and destructive whole‐class collection and dissection exercises.
3.2. The art in the science of 3D‐imagery—Rendering and segmentation
One of the most important aspects of visualising 3D‐data is the segmentation and differentiation of structures of interest. Segmentation is a critical step required to perform quantitative shape analyses of invertebrate morphology. For example, to perform any sort of geometric morphometric analysis on insect genitalia, the structures of interest must first be revealed using a segmentation method such as that described in our advanced Drishti workflow. The majority of existing studies using geometric morphometrics have looked at either external morphology, or skeletons of vertebrates (which can easily be revealed by narrowing the range of visible densities from a CT‐scan; Adams, Rohlf, & Slice, 2013). Geometric morphometrics can and should be utilised more often in studies on invertebrate morphology, and simply require a few extra steps to process the raw 3D‐data, as described here.
Further to the analytical requirement of segmentation described above, differentiation of structures is important simply for accurate visualisation. Without appropriate rendering with lighting, colour and textures, 3D‐data can be difficult or impossible to interpret, especially for someone who is not familiar with the subject being displayed. The workflows provided here offer comprehensive step‐by‐step instructions to help users improve the interpretability of their 3D‐figures. Although this is not strictly necessary to achieve an interactive 3D‐figure for publication, we consider it an important component of the 3D‐data visualisation process. Each of the advanced workflows comprises segmentation and rendering options: the advanced Drishti pathway includes segmentation carried out in Drishti Paint specifically, with resultant components rendered in Drishti and most suited to creating still images or videos. The advanced Blender pathway involves segmentation in Blender after mesh generation, with rendering carried out either in Blender, Photoshop or Acrobat Pro (prior to saving the final 3D‐PDF).
For complex subjects with internal details that are desired in the final figure (e.g., μCT‐scan data), we recommend segmentation in Drishti Paint, as it allows complete preservation of external and internal structures. For the most accurate portrayal of fine details, this method should be used in conjunction with videos or still images, because the generation of surface meshes inherently reduces the detail of complex objects (unless a very high number of polygons is used, which in turn leads to unwieldy file sizes). Figures 2 and 4 demonstrate the difference in detail between volume and surface‐mesh renderings, as seen in the reduction of setae visibility, and obscuring of antennal sensory pores in the meshed example. For simple objects, or when fine details are not important, mesh segmentation in Blender is relatively fast and effective (demonstrated in Figure 5). When coupled with the 3D‐PDF format, segmented meshes can be used to communicate a huge amount of information about numerous aspects of a single subject, all in one interactive figure. We envision users progressing from the basic to the advanced workflows presented here, and then gaining the confidence to utilise the extensive additional tools available in Drishti, MeshLab and Blender.
As 3D‐imaging techniques become more numerous and accessible to biologists, there is exciting potential for further development of innovative ways to present the data. Already, 3D‐PDFs can be included alongside traditional illustrations and figures in electronic formats, with summarising placeholder images for printing. Unfortunately, there are very few journals that currently offer this direct embedding of 3D or other multimedia figures in the main PDF of publications. For example, the literature review of Newe and Becker (2018) revealed that only 34 out of 156 relevant publications had their 3D‐PDFs embedded in the main text. We hope that more journals and their publishers will adopt 3D‐PDFs as a standard option for figures. This will be particularly valuable for microscopic invertebrates and other organisms that can now be imaged and visualised in high‐resolution 3D for the first time, and enhance the way we communicate science.
Supporting information
Drishti basic workflow
Drishti advanced workflow
Meshlab basic workflow
Blender advanced workflow
Mesh simplification script
Supplementary tables
Supplementary Video
ACKNOWLEDGMENTS
This research was carried out primarily at The Australian National University, supported by an Australian Government Research Training Program (RTP) Scholarship, and was additionally funded by a John Noble Award for Invertebrate Research from the Linnean Society of NSW, both to TLS. The authors acknowledge the facilities, and the scientific and technical assistance of the Australian Microscopy & Microanalysis Research Facility at the Centre for Microscopy, Characterisation & Analysis, The University of Western Australia, a facility funded by the University, State and Commonwealth Governments. Special thanks to A. Limaye for his ongoing support with Drishti, G. Brown for specimen identifications, and the Australian National Insect Collection for specimen loans. Also thanks to M. Vidal‐Garcia, M. Cosgrove and two anonymous reviewers for comments that greatly improved the manuscript.
Semple TL, Peakall R, Tatarnic NJ. A comprehensive and user‐friendly framework for 3D‐data visualisation in invertebrates and other organisms. Journal of Morphology. 2019;280:223–231. 10.1002/jmor.20938
Funding information Australian Government, Research Training Program (RTP) Scholarship; Linnean Society of NSW, John Noble Award for Invertebrate Research; Australian National University
REFERENCES
- Adams, D. C. , Rohlf, F. J. , & Slice, D. E. (2013). A field comes of age: Geometric morphometrics in the 21st century. Hystrix, the Italian Journal of Mammalogy, 24(1), 7–14. 10.4404/hystrix-24.1-6283 [DOI] [Google Scholar]
- Akkari, N. , Enghoff, H. , & Metscher, B. D. (2015). A new dimension in documenting new species: High‐detail imaging for Myriapod taxonomy and first 3D cybertype of a new millipede species (Diplopoda, Julida, Julidae). PLoS One, 10(8), e0135243 10.1371/journal.pone.0135243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barden, P. , Herhold, H. W. , & Grimaldi, D. A. (2017). A new genus of hell ants from the cretaceous (Hymenoptera: Formicidae: Haidomyrmecini) with a novel head structure. Systematic Entomology, 42, 837–846. [Google Scholar]
- Bicknell, R. D. C. , Klinkhamer, A. J. , Flavel, R. J. , Wroe, S. , & Paterson, J. R. (2018). A 3D anatomical atlas of appendage musculature in the chelicerate arthropod Limulus polyphemus . PLoS One, 13(2), e0191400 10.1371/journal.pone.0191400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich, F. , & Beutel, R. G. (2008). Micro‐computer tomography and a renaissance of insect morphology. Paper presented at the Proc. SPIE 7078, Developments in X‐Ray Tomography VI, San Diego, California.
- Garcia, F. H. , Fischer, G. , Liu, C. , Audisio, T. L. , & Economo, E. P. (2017). Next‐generation morphological character discovery and evaluation: An X‐ray micro‐CT enhanced revision of the ant genus Zasphinctus wheeler (Hymenoptera, Formicidae, Dorylinae) in the Afrotropics. ZooKeys, 693, 33–93. 10.3897/zookeys.693.13012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garwood, R. , & Sutton, M. (2010). X‐ray micro‐tomography of carboniferous stem‐Dictyoptera: New insights into early insects. Biology Letters, 6(5), 699–702. 10.1098/rsbl.2010.0199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco, M. K. , Tong, J. , Soleimani, M. , Bell, D. , & Schäfer, M. O. (2012). Imaging live bee brains using minimally‐invasive diagnostic radioentomology. Journal of Insect Science, 12(89), 1–7. 10.1673/031.012.8901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautenschlager, S. (2013). Palaeontology in the third dimension: A comprehensive guide for the integration of three‐dimensional content in publications. Paläontologische Zeitschrift, 88(1), 111–121. 10.1007/s12542-013-0184-2 [DOI] [Google Scholar]
- Limaye, A. (2012). Drishti: a volume exploration and presentation tool. Proc. SPIE 8506, Developments in X‐Ray Tomography VIII, 85060X.
- Lowe, T. , Garwood, R. J. , Simonsen, T. J. , Bradley, R. S. , & Withers, P. J. (2013). Metamorphosis revealed: Time‐lapse three‐dimensional imaging inside a living chrysalis. Journal of the Royal Society: Interface, 10(84), 20130304 10.1098/rsif.2013.0304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin‐Vega, D. , Simonsen, T. J. , & Hall, M. J. R. (2017). Looking into the puparium: Micro‐CT visualization of the internal morphological changes during metamorphosis of the blow fly, Calliphora vicina, with the first quantitative analysis of organ development in cyclorrhaphous dipterans. Journal of Morphology, 278(5), 629–651. 10.1002/jmor.20660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPeek, M. A. , Shen, L. , Torrey, J. Z. , & Farid, H. (2008). The tempo and mode of three‐dimensional morphological evolution in male reproductive structures. The American Naturalist, 171(5), E158–E178. 10.1086/587076 [DOI] [PubMed] [Google Scholar]
- Newe, A. , & Becker, L. (2018). Three‐dimensional portable document format (3D PDF) in clinical communication and biomedical sciences: Systematic review of applications, tools, and protocols. JMIR Medical Informatics, 6(3), e10295 10.2196/10295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband, W. S. (1997. –2016). ImageJ. Bethesda, MD: U.S. National Institutes of Health. Retrieved from https://imagej.nih.gov/ij/
- Ribi, W. , Senden, T. J. , Sakellariou, A. , Limaye, A. , & Zhang, S. (2008). Imaging honey bee brain anatomy with micro‐X‐ray‐computed tomography. Journal of Neuroscience Methods, 171(1), 93–97. 10.1016/j.jneumeth.2008.02.010 [DOI] [PubMed] [Google Scholar]
- Richards, C. S. , Simonsen, T. J. , Abel, R. L. , Hall, M. J. , Schwyn, D. A. , & Wicklein, M. (2012). Virtual forensic entomology: Improving estimates of minimum post‐mortem interval with 3D micro‐computed tomography. Forensic Science International, 220(1–3), 251–264. 10.1016/j.forsciint.2012.03.012 [DOI] [PubMed] [Google Scholar]
- Schmitt, M. , & Uhl, G. (2015). Functional morphology of the copulatory organs of a reed beetle and a shining leaf beetle (Coleoptera: Chrysomelidae: Donaciinae, Criocerinae) using X‐ray micro‐computed tomography. Zookeys, 547, 193–203. 10.3897/zookeys.547.7143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel, R. , & Lüter, C. (2014). Overcoming the fragility ‐ X‐ray computed micro‐tomography elucidates brachiopod endoskeletons. Frontiers in Zoology, 11(1), 65 10.1186/s12983-014-0065-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen, T. J. , & Kitching, I. J. (2014). Virtual dissections through micro‐CT scanning: A method for non‐destructive genitalia ‘dissections’ of valuable Lepidoptera material. Systematic Entomology, 39(3), 606–618. 10.1111/syen.12067 [DOI] [Google Scholar]
- Tatarnic, N. J. , & Cassis, G. (2013). Surviving in sympatry: Paragenital divergence and sexual mimicry between a pair of traumatically inseminating plant bugs. The American Naturalist, 182(4), 542–551. 10.1086/671931 [DOI] [PubMed] [Google Scholar]
- van de Kamp, T. , dos Santos Rolo, T. , Vagovic, P. , Baumbach, T. , & Riedel, A. (2014). Three‐dimensional reconstructions come to life‐interactive 3D PDF animations in functional morphology. PLoS One, 9(7), e102355 10.1371/journal.pone.0102355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Kamp, T. , Schwermann, A. H. , Dos Santos Rolo, T. , Losel, P. D. , Engler, T. , Etter, W. , … Krogmann, L. (2018). Parasitoid biology preserved in mineralized fossils. Nature Communications , 9(1), 3325. doi: 10.1038/s41467-018-05654-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcieszek, J. M. , Austin, P. , Harvey, M. S. , Simmons, L. W. , & Hayssen, V. (2012). Micro‐CT scanning provides insight into the functional morphology of millipede genitalia. Journal of Zoology, 287(2), 91–95. 10.1111/j.1469-7998.2011.00892.x [DOI] [Google Scholar]
- Woller, D. A. , & Song, H. (2017). Investigating the functional morphology of genitalia during copulation in the grasshopper Melanoplus rotundipennis (Scudder, 1878) via correlative microscopy. Journal of Morphology, 278(3), 334–359. 10.1002/jmor.20642 [DOI] [PubMed] [Google Scholar]
- Wulff, N. C. , Lehmann, A. W. , Hipsley, C. A. , & Lehmann, G. U. (2015). Copulatory courtship by bushcricket genital titillators revealed by functional morphology, μCT scanning for 3D reconstruction and female sense structures. Arthropod Structure and Development, 44(4), 388–397. 10.1016/j.asd.2015.05.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Drishti basic workflow
Drishti advanced workflow
Meshlab basic workflow
Blender advanced workflow
Mesh simplification script
Supplementary tables
Supplementary Video


