Abstract
The H9N2 subtype of avian influenza A viruses (AIV) has spread among domestic poultry and wild birds worldwide. H9N2 AIV is sporadically transmitted to humans from avian species. A total of 42 laboratory‐confirmed cases of non‐fatal human infection with the Eurasian Y280 and G1 lineages have been reported in China, Hong Kong, Bangladesh and Egypt since 1997. H9N2 AIV infections in poultry have become endemic in Asia and the Middle East and are a major source of viral internal genes for other AIV subtypes, such that continuous monitoring of H9N2 AIV is recommended. In this study, a new, one‐step, real‐time RT‐PCR assay was developed to detect two major Eurasian H9 lineages of AIV capable of causing human infection. The sensitivity of this assay was determined using in vitro‐transcribed RNA, and the detection limit was approximately 3 copies/reaction. In this assay, no cross‐reactivity was observed against RNA from H1–15 subtypes of influenza A viruses, influenza B viruses and other viral respiratory pathogens. In addition, this assay could detect the H9 hemagglutinin (HA) gene from artificially reconstituted clinical samples spiked with H9N2 virus without any non‐specific reactions. Therefore, this assay is highly sensitive and specific for H9 HA detection. The assay is useful both for diagnostic purposes in cases of suspected human infection with influenza H9N2 viruses and for the surveillance of both avian and human influenza viruses.
Keywords: avian influenza, diagnosis, H9N2, influenza, real‐time RT‐PCR
Abbreviations
- AIV
avian influenza A virus
- Cp
crossing point
- Ct
threshold cycle
- GISAID
Global Initiative on Sharing All Influenza Data
- HA
hemagglutinin
- NA
neuraminidase
- rRT‐PCR
real‐time RT‐PCR
1. INTRODUCTION
Influenza A viruses are single‐stranded, negative‐sense RNA viruses belonging to the Orthomyxoviridae family. The natural host of influenza A viruses are wild aquatic birds, with 16 hemagglutinin (HA) and nine neuraminidase (NA) subtypes identified in avian species 1. Avian influenza viruses (AIV) of the H9N2 subtype circulate primarily among wild birds and domestic poultry, but the viruses can infect swine and humans as well. A total of 42 cases of non‐fatal human infection were reported in Asia and the Middle East as of March 2018 (http://www.who.int/influenza/human_animal_interface/HAI_Risk_Assessment/en/) 2, 3, 4, 5, 6, 7, 8, 9.
H9N2 viruses have been widely and consistently isolated worldwide since their first isolation from turkeys in Wisconsin, USA, in 1966 10. H9N2 viruses are divided into a North American lineage and a Eurasian lineage 11. The North American lineage H9N2 viruses are typically detected in shorebirds and wild ducks, and no cases of human infection have been reported to date 12. The Eurasian lineage of H9N2 AIV circulating in Asia, the Middle East and Europe have been classified into two major lineages, Y280 and G1, and one minor Korean lineage. Since 1997, sporadic laboratory‐confirmed cases of avian‐to‐human transmission of Y280‐lineage viruses in China and G1‐lineage viruses in China, Hong Kong, Bangladesh and Egypt have been reported (http://www.who.int/influenza/human_animal_interface/HAI_Risk_Assessment/en/) 2, 3, 4, 5, 6, 7, 8, 9. However, the results of serologic studies in Asia and the Middle East suggest that the number of humans infected by H9N2 AIV is much greater than the number of laboratory‐confirmed cases 13, 14, 15, 16, 17, 18. It is thus important to monitor H9N2 AIV in wild birds and poultry in order to assess the risk for human infection.
Molecular diagnostic techniques such as the PCR method can be used as diagnostic tools for virus identification and assessing viral infection. In particular, real‐time RT‐PCR (rRT‐PCR) is one of the most widely used methods for detecting viral genes, and rRT‐PCR assays for detecting H9 viruses have been reported 19, 20, 21, 22. However, the sequences of probes and primers used in rRT‐PCR in previous studies were designed for detecting viruses of the North American lineage or past circulated G1‐lineage H9 AIV. These methods did not use minor groove binder (MGB) probes, resulting in different conditions for these assays compared with the assay used in Japan for detecting other influenza viruses. Therefore, our newly developed, one‐step rRT‐PCR assay was designed to detect both recent Y280‐ and G1‐lineage H9 AIV, including those causing human infection.
2. MATERIALS AND METHODS
2.1. Primer and probe design
The nucleotide sequences of the HA genes of H9 subtype AIV were aligned using ClustalX 2.1 software with the sequences of all human viruses registered to date and avian viruses in Asia and the Middle East registered after 2014 in the GenBank/EMBL/DDBJ (https://www.ncbi.nlm.nih.gov/genbank/) and Global Initiative on Sharing All Influenza Data (GISAID) (http://www.gisaid.org) databases 23, 24. On the basis of highly conserved sequences in the HA1 region, primers were designed to detect as many human and avian viruses as possible by rRT‐PCR using the MGB TaqMan® probe (Thermo Fisher Scientific, Waltham, MA, USA). The sequences and positions of the primers and probes are listed in Table 1.
Table 1.
Primers and probes used in the H9 real‐time RT‐PCR assay
| Name | Sequence (5–3′) † | Position ‡ | Product size (bp) |
|---|---|---|---|
| NIID‐H9 TMPrimer‐F1 | AATGTYCCTGTGACACATGCCAAAGA | 121–146 | |
| NIID‐H9 TMPrimer‐R1 | AGRTCACAAGAAGGRTTGCCATA | 238–260 | 140 |
| NIID‐H9 Probe1 | (FAM)CATYCCATTRTGCTCTGTGTGGAG(MGB) | 151–174 |
Probe was labeled with FAM at the 5′‐end and minor groove binder at the 3′‐end.
Nucleotide numbering is based on the HA gene CDS of A/Hong Kong/308/2014 (H9N2).
2.2. One‐step rRT‐PCR assay
The reaction was performed using AgPath‐ID™ one‐step RT‐PCR reagents (Thermo Fisher Scientific) in a 25 μL reaction mixture containing 12.5 μL of 2× RT‐PCR buffer, 1 μL of 25× RT‐PCR enzyme mix, 0.1 μL (20 U) of RNase inhibitor (Thermo Fisher Scientific), 600 nM each forward and reverse primer, 100 nM TaqMan MGB probe and 5 μL of RNA template. The rRT‐PCR assays were carried out using a LightCycler® 480 II (Roche, Basel, Switzerland) under the following conditions: 50°C for 10 min, 95°C for 10 min, and 45 cycles of 15 s at 95°C, 30 s at 56°C and 15 s at 72°C. Amplification data were collected at 56°C (annealing step) and analyzed according to the second derivative maximum method in the LightCycler® 480 SW1.5 software.
2.3. Viruses and viral RNA extraction
H1–15 subtypes of influenza A viruses, influenza B viruses and 19 viral respiratory pathogens stored in our laboratory were used in this study (Tables 2, 3, 4). Viral RNA was extracted from 140 μL cultures of each virus propagated in embryonated chicken eggs to 60 μL of AVE (elution buffer supplied with the kit) using a QIAamp® viral RNA mini kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. In this study, the copy number of the M gene of the 18 H9 viruses was determined quantitatively as previously described 25 using an influenza A (type A) rRT‐PCR assay targeted to the universal M gene of all influenza A viruses 26. The threshold cycle (Ct) values of viral RNA extracted from 19 viral respiratory pathogens were determined using the multiplex real‐time PCR assay described previously, with minor modifications 27.
Table 2.
Detection limit of the new H9 rRT‐PCR assay in comparison with the type A rRT‐PCR assay
| Type A rRT‐PCR assay | H9 rRT‐PCR assay | |||||
|---|---|---|---|---|---|---|
| Lineage | Virus strain | M gene copy number † | No. of positive results | Cp value ‡ (average ± SD) | No. of positive results | Cp value ‡ (average ± SD) |
| Y280 | A/chicken/Hong Kong/G9/1997 (H9N2) | 5 | 3/3 | 37.54 ± 0.58 | 3/3 | 36.33 ± 0.44 |
| A/Hong Kong/308/2014 (H9N2) | 5 | 2/3 | 38.6 | 3/3 | 37.88 ± 1.01 | |
| A/swine/Hong Kong/9/98 (H9N2) | 5 | 3/3 | 37.82 ± 0.54 | 3/3 | 37.82 ± 0.54 | |
| A/Hunan/44558/2015 (H9N2) § | 5 ¶ | Not available | 3/3 | 38.77 ± 1.19 | ||
| A/duck/Japan/AQ‐HE5/2015 (H9N2) | 5 | 3/3 | 37.56 ± 0.55 | 3/3 | 37.51 ± 0.49 | |
| A/chicken/Japan/AQ‐HE14/2015 (H9N2) | 5 | 3/3 | 37.23 ± 0.54 | 3/3 | 39.12 ± 0.76 | |
| A/duck/Japan/AQ‐HE28/2015 (H9N2) | 5 | 3/3 | 38.04 ± 0.82 | 3/3 | 38.49 ± 1.45 | |
| A/chicken/Japan/AQ‐HE61/2015 (H9N2) | 5 | 3/3 | 38.83 ± 1.05 | 3/3 | 37.37 ± 0.39 | |
| A/chicken/Japan/AQ‐HE28‐28/2016 (H9N2) | 5 | 3/3 | 38.19 ± 1.03 | 3/3 | 37.86 ± 0.11 | |
| A/chicken/Japan/AQ‐HE28‐50/2016 (H9N2) | 5 | 3/3 | 38.59 ± 1.23 | 3/3 | 37.45 ± 0.14 | |
| A/chicken/Japan/AQ‐HE28‐57/2016 (H9N2) | 5 | 3/3 | 38.25 ± 0.23 | 3/3 | 38.13 ± 0.54 | |
| G1 | A/Hong Kong/1073/99 (H9N2) | 5 | 3/3 | 38.72 ± 1.13 | 3/3 | 38.30 ± 0.83 |
| A/chicken/Bangladesh/28182/2016 (H9N2) § | 5 ¶ | Not available | 2/3 | 38.88 | ||
| A/chicken/Egypt/F12173D/2016 (H9N2) § | 5 ¶ | Not available | 3/3 | 36.94 ± 0.19 | ||
| A/parakeet/Chiba/1/97 (H9N2) | 5 | 3/3 | 38.45 ± 0.42 | 3/3 | 36.10 ± 0.39 | |
| A/parakeet/Narita/92a/98 (H9N2) | 5 | 3/3 | 38.09 ± 1.24 | 3/3 | 36.40 ± 0.60 | |
| Korean | A/duck/Hong Kong/448/78 (H9N2) | 5 | 3/3 | 38.21 ± 0.06 | 3/3 | 37.50 ± 0.56 |
| A/duck/Hong Kong/702/79 (H9N5) | 5 | 3/3 | 37.83 ± 1.09 | 3/3 | 37.94 ± 1.79 | |
| A/duck/Hokkaido/31/97 (H9N2) | 500 | 3/3 | 31.16 ± 0.04 | 2/3 | 40.00 | |
| 50 | 3/3 | 34.46 ± 0.53 | 0/3 | ‐ | ||
| 5 | 3/3 | 38.54 ± 1.41 | 0/3 | ‐ | ||
| A/duck/Fukui/3/2005 (H9N1) | 5 | 3/3 | 38.61 ± 1.21 | 2/3 | 40.00 | |
| North American | A/turkey/Wisconsin/1/66 (H9N2) | 500 | 3/3 | 31.07 ± 0.09 | 3/3 | 38.39 ± 0.47 |
| 50 | 3/3 | 34.45 ± 0.22 | 1/3 | 40.00 | ||
| 5 | 2/3 | 38.47 | 0/3 | ‐ | ||
Copy number of the M gene corresponding to the detection limit of the H9 HA gene (copies/reaction).
Crossing point (Cp) values were analyzed according to the second derivative maximum method in the Light Cycler® 480 SW1.5 software. The Cp value of 40.00 was detectable.
In vitro‐transcribed RNA was used for the HA gene of each isolate.
Copy number of the H9 HA gene (copies/reaction).
rRT‐PCR, real‐time RT‐PCR
Table 3.
Panel of non‐H9 influenza viruses used in the H9 rRT‐PCR assay
| Sample | Strain or sample name | Type or subtype | Type A or B/NS rRT‐PCR assays † | H9 rRT‐PCR assay† |
|---|---|---|---|---|
| Virus isolate | A/duck/Alberta/35/76 | H1N1 | 18.84 | N.D. |
| A/Brisbane/59/2007 | H1N1 | 25.45 | N.D. | |
| A/Narita/1/2009 | H1N1pdm09 | 23.91 | N.D. | |
| A/duck/Germany/1215/73 | H2N3 | 19.82 | N.D. | |
| A/duck/Ukraine/1/63 | H3N8 | 18.11 | N.D. | |
| A/Uruguay/716/2007 | H3N2 | 25.20 | N.D. | |
| A/Indiana/12/2012 | H3N2v | 21.75 | N.D. | |
| A/duck/Czechoslovakia/56 | H4N6 | 19.60 | N.D. | |
| A/duck/Hyogo/1/2010 | H4N6 | 23.64 | N.D. | |
| A/blow fly/Kyoto/93/2004 | H5N1 | 26.24 | N.D. | |
| A/chicken/Ibaraki/1/2005 | H5N2 | 18.17 | N.D. | |
| A/white swan/Hokkaido/4/2011 | H5N1 | 18.66 | N.D. | |
| A/turkey/Massachusetts/3740/65 | H6N2 | 23.29 | N.D. | |
| A/duck/Hong Kong/301/78 | H7N1 | 21.24 | N.D. | |
| A/duck/Fukui/1/2004 | H7N7 | 24.65 | N.D. | |
| A/Anhui/1/2013 | H7N9 | 24.62 | N.D. | |
| A/turkey/Ontario/6118/68 | H8N4 | 22.57 | N.D. | |
| A/duck/Shizuoka/45/2011 | H8N4 | 21.84 | N.D. | |
| A/chicken/Germany/N/49 | H10N7 | 17.86 | N.D. | |
| A/duck/England/56 | H11N6 | 19.00 | N.D. | |
| A/duck/Alberta/60/76 | H12N5 | 19.39 | N.D. | |
| A/gull/Maryland/704/77 | H13N6 | 19.60 | N.D. | |
| A/mallard/Gurjev/263/82 | H14N5 | 18.92 | N.D. | |
| A/duck/Australia/341/83 | H15N8 | 19.20 | N.D. | |
| B/Florida/04/2006 | Type B | 25.40 | N.D. | |
| B/Brisbane/60/2008 | Type B | 29.61 | N.D. | |
| B/Massachusetts/2/2012 | Type B | 30.07 | N.D. |
Crossing point values were determined using the second derivative maximum method in Light Cycler® 480 SW1.5 software.
N.D., not detected; rRT‐PCR, real‐time RT‐PCR.
Table 4.
Panel of non‐influenza respiratory pathogens used in the H9 rRT‐PCR assay
| Respiratory pathogen | Other PCR assay † | H9 rRT‐PCR assay ‡ |
|---|---|---|
| Respiratory syncytial virus A | 24.1 | N.D. |
| Respiratory syncytial virus B | 26.0 | N.D. |
| Human parainfluenza virus type 1 (strain C35) | 17.5 | N.D. |
| Human parainfluenza virus type 2 (strain GREER) | 18.5 | N.D. |
| Human parainfluenza virus type 3 (strain Washington/1957 C243) | 18.5 | N.D. |
| Human parainfluenza virus type 4a (strain M‐25) | 22.1 | N.D. |
| Human parainfluenza virus type 4b (strain CH19503) | 20.0 | N.D. |
| Human rhinovirus type A | 30.9 | N.D. |
| Human rhinovirus type B | 28.6 | N.D. |
| Human metapneumovirus type A1 | 26.1 | N.D. |
| Human metapneumovirus type B2 | 25.7 | N.D. |
| Human coronavirus OC43 | 26.8 | N.D. |
| Human coronavirus 229E | 25.9 | N.D. |
| Human coronavirus NL63 | 27.0 | N.D. |
| Human coronavirus HKU1 | 25.0 | N.D. |
| Human bocavirus | 24.2 | N.D. |
| Human enterovirus | 28.9 | N.D. |
| Human adenovirus 2 | 27.0 | N.D. |
| Human adenovirus 4 | 30.0 | N.D. |
These results were obtained by multiplex real‐time PCR assay as described in the main text. Threshold cycle values were determined using 7500 software, version 2.3.
Crossing point values were determined using the second derivative maximum method in Light Cycler® 480 SW1.5 software.
N.D., not detected; rRT‐PCR, real‐time RT‐PCR.
2.4. Clinical specimens and identification of seasonal influenza viruses
Nasal swabs or aspirate samples collected from patients with influenza‐like illness were collected at Showa General Hospital, Japan, between 2014 and 2016 and suspended using a UTM 360 C kit (Copan, Brescia, Italy). The study protocol was approved by the ethics committees of both the National Institute of Infectious Diseases and Showa General Hospital, and the study was performed in compliance with the Declaration of Helsinki. Informed consent was obtained from all patients. Total RNA was extracted using the QIAamp viral RNA mini kit (Qiagen) according to the manufacturer's instructions. The type and subtype of influenza viruses in each clinical sample were determined using rRT‐PCR as previously described 26, 28.
2.5. Phylogenetic analysis of the H9 HA gene
Phylogenetic analysis of the H9 HA gene was performed using Molecular Evolutionary Genetics Analysis software (MEGA, version 7.0) 29. Evolutionary history was inferred using the neighbor‐joining method 30. Evolutionary distances were computed using Kimura's two‐parameter method 31. Bootstrap values of the HA genes were calculated from 1000 replicates 32.
2.6. Preparation of in vitro‐transcribed RNA
To evaluate the sensitivity of the H9 rRT‐PCR assay, three in vitro‐transcribed full‐length H9 HA gene RNA were used. RNA transcripts of the full‐length H9 HA gene were synthesized from artificial DNA (Eurofins Genomics, Tokyo, Japan) of A/Hunan/44558/2015 (H9N2) (GISAID accession no. EPI680526), A/chicken/Bangladesh/28182/2016 (H9N2) (WSS1378750) and A/chicken/Egypt/F12173D/2016 (H9N2) (EPI953355) using the following procedure. The H9 HA artificial DNA were amplified by PCR using Phusion high‐fidelity DNA polymerase (New England BioLabs, Ipswich, MA, USA) with the paired primers T7 + Stop‐R (5′‐TAATACGACTCACTATAGGGTTA‐3′) and Hunan‐F (5′‐ATGGAGACAGTATCACTAATAACTA‐3′), Bangladesh‐F (5′‐ATGGAAACAGTAACACTGTTGAC‐3′) or Egypt‐F (5′‐ATGGAAATAATACCACTGATG‐3′) (underline indicates the T7 promoter sequence) according to the manufacturer's instructions. RNA were transcribed using the T7 RiboMAX™ Express large‐scale RNA production system (Promega, Madison, WI, USA) and treated with TURBO® DNase (Thermo Fisher Scientific) to degrade the template DNA according to the manufacturer's instructions. dNTP and NTP were removed using MicroSpin G‐25 columns (GE Healthcare, Piscataway, NJ, USA) according to the manufacturer's instructions. Transcribed RNA were quantified using a NanoDrop™ spectrometer (Thermo Fisher Scientific), and the copy number was then calculated. The integrity of transcribed RNA was assessed using a 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA, USA). Transcript dilutions were performed in nuclease‐free water containing 10 ng/μL of carrier RNA (Qiagen).
2.7. Validation and evaluation of the H9 rRT‐PCR assay
The analytical sensitivity of the assay was assessed by testing serial dilutions of two quantified in vitro‐transcribed RNA from A/Hunan/44558/2015 (H9N2) of the Y280 lineage and A/chicken/Egypt/F12173D/2016 (H9N2) of the G1 lineage using six replicates of each concentration. The limit of detection was calculated using StatPlus® Professional Version 2009 for Windows (Build 5.8.4.3) by probit regression analysis 33 with a 95% probability end‐point.
For evaluation of the H9 rRT‐PCR assay, extracted RNA were prepared at 1, 10 and 100 copies/μL based on the number of M genes determined using type A rRT‐PCR for 10 Y280 viruses, three G1 viruses, four Korean viruses and one North American virus, and three synthetic RNA were also prepared at 1, 10 and 100 copies/μL. The H9 rRT‐PCR assay was performed in triplicate for each dilution. Results were considered to be positive when the crossing point (Cp) value was given by the second derivative maximum method in the Light Cycler® 480 SW1.5 software. For positive samples with Cp > 40 (flagged as “late Cp call” [last five cycles] with high uncertainty by the LightCycler 480 software), the Light Cycler 480 software applied 40.00 to the sample. The number of positive results per test number and Cp values are shown in Table 2.
The specificity of the H9 rRT‐PCR assay was validated using RNA extracted from 24 representative subtype viruses except for the H9 subtype of influenza A and three influenza B viruses, and 19 viral respiratory pathogens (Tables 3 and 4).
2.8. Evaluation of the H9 rRT‐PCR assay using artificially reconstituted clinical samples
Artificially reconstituted clinical samples spiked with H9N2 virus (130 μL of H1N1pdm09, H3N2 or type B positive clinical specimens, or influenza A and B viruses negative clinical specimens + 10 μL of A/Hong Kong/308/2014 [H9N2]), those not spiked with H9N2 virus (130 μL of H1N1pdm09, H3N2 or type B positive clinical specimens, or influenza A and B viruses negative clinical specimens + 10 μL of PBS) and diluted H9N2 virus (130 μL of PBS + 10 μL of A/Hong Kong/308/2014 [H9N2]) were prepared. Total RNA was extracted using the QIAampviral RNA mini kit (Qiagen) according to the manufacturer's instructions. Simultaneous with the H9 rRT‐PCR assay, type A, B/NS, H1pdm and H3 rRT‐PCR assays were performed using primer and probe sets previously described under the same conditions used for the H9 rRT‐PCR assay 26, 28.
3. RESULTS
The phylogenetic tree based on the H9 HA gene, including the 21 H9 AIV examined in this study and 44 representative H9 AIV from the GenBank/EMBL/DDBJ and GISAID databases, is shown in Figure 1. The detection limit of the H9 rRT‐PCR assay was determined by testing six replicates each of 10‐fold serial dilutions of 5.0 × 107 copies/reaction of in vitro‐transcribed full‐length H9 HA RNA derived from isolate A/Hunan/44558/2015 (H9N2) of the Y280 lineage and isolate A/chicken/Egypt/F12173D/2016 (H9N2) of the G1 lineage (Figure 1). An amplification plot for A/Hunan/44558/2015 (H9N2) obtained as raw data is shown in Figure S1. Probit regression analysis of the data from the two viruses tested showed detection limits of 3.1 and 1.8 copies/reaction, respectively. The efficiency of the assay for the two viruses tested was 96.3% and 93.8%, respectively; the R 2 value was 0.99 for each virus, and the slope of the standard curve was −3.41 and −3.48 in the range between 5.0 and 5.0 × 107 copies/reaction, respectively (Figure 2).
Figure 1.
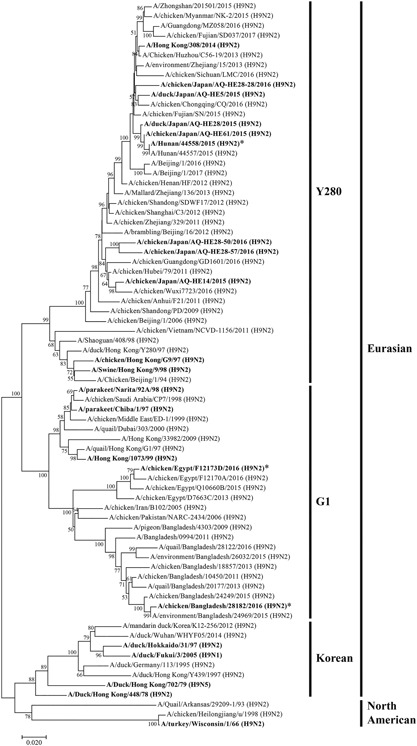
Phylogenetic tree for the H9 HA genes. The tree was constructed using the neighbor‐joining method with MEGA7 software. Evolutionary distances were computed using the Kimura two‐parameter method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) and values of more than 50% are shown next to the branches. The viruses used in this study are shown in bold. *Viruses for which in vitro‐transcribed RNA was used for rRT‐PCR
Figure 2.
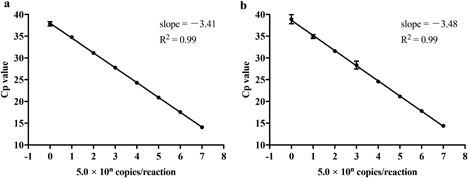
Dynamic range of the H9 rRT‐PCR assay. Standard curve (crossing point [Cp] value vs log10 concentration) for serial dilutions of in vitro‐transcribed RNA of the HA gene of (a) A/Hunan/44558/2015 (H9N2), Y280 lineage and (b) A/chicken/Egypt/F12173D/2016 (H9N2), G1 lineage. The standard curve was generated using the average Cp values obtained from six replicates. The correlation coefficient (R 2) and slope of the standard curve are shown in each graph
The H9 AIV examined in this study were classified into four lineages (Figure 1). The AIV of 11 belonged to Y280, five AIV belonged to G1, four AIV belonged to the Korean lineage and one AIV belonged to the North American lineage; all were detected by the H9 rRT‐PCR assay (Table 2). Phylogenetically, H9N2 viruses isolated from smuggled meats illegally imported into Japan, including A/duck/Japan/AQ‐HE5/2015 (H9N2), A/chicken/Japan/AQ‐HE14/2015 (H9N2), A/duck/Japan/AQ‐HE28/2015 (H9N2), A/chicken/Japan/AQ‐HE61/2015 (H9N2), A/chicken/Japan/AQ‐HE28‐28/2016 (H9N2), A/chicken/Japan/AQ‐HE28‐50/2016 (H9N2) and A/chicken/Japan/AQ‐HE28‐57/2016 (H9N2), were classified into the Y280 lineage along with closely related AIV recently isolated from poultry in China (Figure 1). All RNA extracted from the AIV (except for A/duck/Hokkaido/31/97 [H9N2], belonging to the Korean lineage, and A/turkey/Wisconsin/1/66 [H9N2], belonging to the North American lineage) and both G1‐ and one of the Y280‐lineage virus in vitro‐transcribed H9 HA RNA could be detected at concentrations corresponding to 5, 50 and 500 copies of the M gene RNA per reaction and 5, 50 and 500 copies of H9 HA gene per reaction using the newly established H9 rRT‐PCR assay (Table 2 and data not shown). For these AIV, the H9 rRT‐PCR assay exhibited amplification plots similar to the type A rRT‐PCR assay (Figure S2a). A/duck/Hokkaido/31/97 (Korean lineage) and A/turkey/Wisconsin/1/66 (North American lineage), which were isolated over 20 years ago, could also be detected using the H9 rRT‐PCR assay at RNA concentrations corresponding to a minimum of 500 and 50 copies/reaction of M gene RNA in viral extracted RNA, respectively (Table 2). However, both viruses could be detected at 5, 50 and 500 copies of M gene RNA per reaction using the type A rRT‐PCR assay, suggesting that the H9 rRT‐PCR assay is less sensitive against these past prevalent viruses (Figure S2b).
The cross‐reactivity of the H9 rRT‐PCR assay was evaluated against other HA subtypes, H1 through H15 (except for H9) influenza A viruses, influenza B viruses and 19 other respiratory viruses (Tables 3 and 4). No cross‐reactivity was observed against RNA derived from these isolates, and no non‐specific reactions were observed (Tables 3 and 4).
To demonstrate the robustness of the H9 rRT‐PCR assay, we evaluated artificially reconstituted clinical specimens with or without seasonal influenza viruses and spiked or not spiked with the H9N2 virus. The H9 rRT‐PCR assays could detect the HA gene of the H9N2 virus from all 12 artificially reconstituted samples spiked with the H9N2 virus, and no non‐specific reactions were observed in the 12 clinical samples not spiked with the H9N2 virus (Table 5). The Cp values for all 12 artificially reconstituted samples in the H9 rRT‐PCR assay were almost the same as the Cp value for PBS spiked with the same amount of the H9N2 virus.
Table 5.
Detection of the H9 HA gene from artificially reconstituted clinical samples spiked with H9N2 virus
| Sample name | Type or subtype | Virus‐spike | Type A rRT‐PCR assay † | H9 rRT‐PCR assay † | Other type/subtype rRT‐PCR assays † , ‡ |
|---|---|---|---|---|---|
| F16‐9 | H1N1pdm09 | + | 21.05 | 31.56 | 20.59 |
| − | 21.06 | N.D. | 20.58 | ||
| F16‐26 | H1N1pdm09 | + | 20.62 | 31.62 | 20.74 |
| − | 20.64 | N.D. | 20.78 | ||
| F16‐61 | H1N1pdm09 | + | 23.88 | 31.21 | 23.25 |
| − | 23.01 | N.D. | 23.06 | ||
| F14‐53 | H3N2 | + | 19.13 | 31.68 | 18.54 |
| − | 19.06 | N.D. | 18.48 | ||
| F15‐7 | H3N2 | + | 18.30 | 31.84 | 17.84 |
| − | 19.31 | N.D. | 17.99 | ||
| F16‐17 | H3N2 | + | 16.73 | 31.72 | 16.31 |
| − | 17.45 | N.D. | 16.48 | ||
| F15‐15 | Type B | + | 32.61 | 32.42 | 23.12 |
| − | N.D. | N.D. | 22.77 | ||
| F16‐44 | Type B | + | 32.31 | 32.02 | 21.52 |
| − | N.D. | N.D. | 21.16 | ||
| F16‐56 | Type B | + | 33.32 | 31.35 | 18.75 |
| − | N.D. | N.D. | 18.90 | ||
| F16‐52 | − | + | 32.35 | 31.57 | N.T. |
| − | N.D. | N.D. | N.T. | ||
| F16‐68 | − | + | 32.64 | 31.70 | N.T. |
| − | N.D. | N.D. | N.T. | ||
| F16‐76 | − | + | 32.12 | 31.67 | N.T. |
| − | N.D. | N.D. | N.T. | ||
| PBS | − | + | 31.51 | 31.43 | N.T. |
| − | N.D. | N.D. | N.T. |
Crossing point values were determined using the second derivative maximum method in Light Cycler® 480 SW1.5 software.
Other assays were as follows: H1pdm rRT‐PCR assay for F16‐9, F16‐26, and F16‐61; H3 rRT‐PCR assay for F14‐53, F15‐7, and F16‐17; B/NS rRT‐PCR assay for F15‐15, F16‐44, and F16‐56.
N.D., not detected; N.T., not tested; rRT‐PCR, real‐time RT‐PCR.
4. DISCUSSION
Among H9N2 viruses, those of the Y280 lineage are spreading primarily in poultry in China, and this has resulted in occasional cases of human infection. H9N2 viruses of the G1 lineage are more prevalent in poultry in Asia and the Middle East, with recent reports of sporadic transmission to humans in China, Hong Kong, Bangladesh and Egypt. We therefore developed a new H9 rRT‐PCR assay that is highly sensitive and specific for viruses of the Y280 and G1 lineages. Our H9 rRT‐PCR assay also exhibited good linearity (R 2 = 0.99) and high sensitivity for detecting in vitro‐transcribed HA gene RNA from the Y280‐lineage virus A/Hunan/44558/2015 (H9N2) and the G1‐lineage virus A/chicken/Egypt/F12173D/2016 (H9N2) (Figure 2). The H9 HA gene RNA was detected for all isolates of both lineages examined in this study at the equivalent of a minimum of 5 copies of the M gene per reaction (Table 2). Therefore, the sensitivity of the H9 rRT‐PCR assay is comparable to that of our previously developed rRT‐PCR assay for the universal detection of M genes of all influenza A viruses 26. The sensitivity of the assay was at least 10‐ and 100‐fold lower for A/duck/Hokkaido/31/97 (Korean lineage) and A/turkey/Wisconsin/1/66 (North American lineage), respectively, compared with other Y280‐ or G1‐lineage viruses. The A/duck/Hokkaido/31/97 virus had one mismatch in the forward primer region and four mismatches in the reverse primer region, whereas the A/turkey/Wisconsin/1/66 virus had four mismatches in the forward primer region, three mismatches in the reverse primer region and one mismatch in the probe region. As these viruses had many more mismatches in the primer/probe regions compared with the other viruses examined, the decrease in sensitivity was attributed to these mismatches. However, the mismatches in the primer and probe sequences are not conserved in recently described viruses of the Y280 and G1 lineages (data not shown). Moreover, there are no reports to date of human infections caused by viruses of the Korean and North American lineages derived from poultry. Reports of Y280‐ and G1‐lineage viruses with an HA‐Q226L substitution are increasing 7, 8, 34. The adaptation of Y280 and G1 lineages to humans may be dependent on HA‐L226, which prefers α‐2,6‐linked sialic acids 35. These results suggest that the H9 rRT‐PCR assay described here is highly sensitive for current epidemic strains of the Y280 and G1 lineages in poultry that are transmissible to humans.
The viral load of clinical specimens from patients infected with AIV is often very low compared with specimens from seasonal influenza cases 36. Therefore, a highly sensitive detection system is needed to diagnose cases of human infection with AIV. An evaluation of the H9 rRT‐PCR assay using artificially reconstituted clinical samples showed that the assay could detect a small viral load (Cp > 30) of H9N2 when the specimen contained a viral load of seasonal influenza A virus higher (Cp < 23.25) than the amount of spiked H9N2 virus (Table 5). In all artificially reconstituted clinical specimens containing influenza B virus and non‐influenza viruses, the Cp values for the H9 and type A rRT‐PCR assays were almost the same as that for diluted H9N2 virus in PBS. These results suggested that the sensitivity of the H9 rRT‐PCR assay is sufficient and that there are no non‐specific reactions or interference in analyses of clinical specimens with and without seasonal influenza virus.
Even though H9N2 avian influenza viruses circulate worldwide, only 42 cases of human infection were confirmed between 1997 and May 2018. However, the results of serologic studies in Asia and the Middle East suggest that the actual number of humans infected with H9N2 AIV is much greater than the number of confirmed cases 13, 14, 15, 16, 17, 18. For example, the seroprevalence among avian‐exposed humans in Egypt is reportedly between 5.6% and 7.5% 16. Indeed, H9 HA was shown to have a human influenza virus‐like binding property 37. In addition, H9N2 viruses have been identified as a major source of six internal genes in H5N1 38, H7N9 39 and H10N8 40 viruses. Therefore, given the pandemic potential of H9N2 viruses, continuous monitoring and surveillance of these viruses using the highly sensitive rRT‐PCR assay is needed.
At present, the risk of H9N2 exposure may be limited to countries in which the viruses are endemic in poultry, such as China, Egypt and Bangladesh. However, the virus can be transferred to other countries through AIV‐infected migratory birds, illegal importation of raw poultry products from birds infected with AIV 41, or travel by persons infected with an AIV, such as the case of an H7N9 HP AIV human infection in Taiwan in 2017 (http://www.who.int/csr/don/22-february-2017-ah7n9-china/en/). These dangers highlight the need for a highly sensitive and specific system for detecting H9N2 viruses even in countries with a low risk of infection.
In our previous study, we developed rRT‐PCR assays to detect types of influenza A and B viruses, determine subtypes of H1pdm09, former H1 (Russian flu), H3, H5 and H7 influenza A viruses, and discriminate between the Victoria and Yamagata lineages of influenza B viruses. These assays can be performed under the same conditions as the H9 assay described in the present report (http://www.who.int/influenza/gisrs_laboratory/molecular_diagnosis/en/) 25, 26, 28. Hence, by combining these methods, influenza viruses can be easily and simultaneously identified with respect to type and subtype or lineage with high sensitivity for diagnostic and monitoring purposes.
In summary, our newly developed rRT‐PCR assay is capable of detecting two major Eurasian H9 lineages of AIV known to cause human infection. This assay can serve as a useful tool for not only highly sensitive and specific laboratory diagnostic testing for H9 infections in humans but also surveillance and monitoring of the spread of H9 AIV, including those that circulate among avian species and infect humans.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest regarding this manuscript.
Supporting information
Additional Supporting Information may be found online in the supporting information tab for this article.
Figure S1. Amplification plot of in vitro‐transcribed RNA in the H9 rRT‐PCR assay. Amplification plot for serial dilutions of in vitro‐transcribed RNA of the HA gene of A/Hunan/44558/2015 (H9N2), Y280 lineage. Shown are results for six replicates in each copy number of concentration (copies/reaction). rRT‐PCR, real‐time RT‐PCR.
Figure S2. Comparison of amplification plots for the H9 and type A rRT‐PCR assays. Amplification plot for serial dilutions of virus RNA of (a) A/chicken/Japan/AQ‐HE28‐50/2016 (H9N2) (Y280 lineage), and (b) A/turkey/Wisconsin/1/66 (H9N2) (North American lineage), for the H9 and type A rRT‐PCR assays. Shown are results for three replicates in each copy number of concentration (copies/reaction) for each assay. rRT‐PCR, real‐time RT‐PCR.
ACKNOWLEDGMENTS
The authors thank the St Jude Children's Research Hospital, USA, for providing the A/Hong Kong/308/2014 (H9N2) isolate. The authors also thank the National Institute for Biological Standards and Control, a Centre of the Medicines and Healthcare Products Regulatory Agency, UK, for providing the A/chicken/Hong Kong/G9/1997 (H9N2) and A/Hong Kong/1073/99 (H9N2) isolates; Animal Quarantine Service, Ministry of Agriculture, Forestry and Fisheries, Japan, for providing the A/duck/Japan/AQ‐HE5/2015 (H9N2), A/chicken/Japan/AQ‐HE14/2015 (H9N2), A/duck/Japan/AQ‐HE28/2015 (H9N2), A/chicken/Japan/AQ‐HE61/2015 (H9N2), A/chicken/Japan/AQ‐HE28‐28/2016 (H9N2), A/chicken/Japan/AQ‐HE28‐50/2016 (H9N2) and A/chicken/Japan/AQ‐HE28‐57/2016 (H9N2) isolates; the authors, originating and submitting laboratories of the sequences from GISAID's EpiFlu™ Database, on which this research is based; and Drs Hideyuki Kubo and Atsushi Kaida, Osaka City Institute of Public Health and Environmental Sciences, for providing DNA/RNA of viral respiratory pathogens. This research was supported by AMED under grant number JP18fk0108030.
Saito S, Takayama I, Nakauchi M, et al. Development and evaluation of a new real‐time RT‐PCR assay for detecting the latest H9N2 influenza viruses capable of causing human infection. Microbiol Immunol. 2019;63:21–31. 10.1111/1348-0421.12666
REFERENCES
- 1. Wright P.F., Neumann G., Kawaoka Y. (2013) Orthomyxoviriuses. In Knipe D.M., Howley P.M. eds. Fields Virology. 6th edn. Philadelphia, PA: Wolters Kluwer Health/Lippincott Williams & Wilkins. [Google Scholar]
- 2. Dalby A.R., Iqbal M. (2014) A global phylogenetic analysis in order to determine the host species and geography dependent features present in the evolution of avian H9N2 influenza hemagglutinin. PeerJ 2: e655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Peiris M., Yuen K.Y., Leung C.W., Chan K.H., Ip P.L., Lai R.W., Orr W.K., Shortridge K.F. (1999) Human infection with influenza H9N2. Lancet 354: 916–7. [DOI] [PubMed] [Google Scholar]
- 4. Butt A.M., Siddique S., Idrees M., Tong Y. (2010) Avian influenza A (H9N2): computational molecular analysis and phylogenetic characterization of viral surface proteins isolated between 1997 and 2009 from the human population. Virol J 7: 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cheng V.C., Chan J.F., Wen X., Wu W.L., Que T.L., Chen H., Chan K.H., Yuen K.Y. (2011) Infection of immunocompromised patients by avian H9N2 influenza A virus. J Infect 62: 394–9. [DOI] [PubMed] [Google Scholar]
- 6. Shanmuganatham K., Feeroz M.M., Jones‐Engel L., Smith G.J., Fourment M., Walker D., Mcclenaghan L., Alam S.M., Hasan M.K., Seiler P., Franks J., Danner A., Barman S., Mckenzie P., Krauss S., Webby R.J., Webster R.G. (2013) Antigenic and molecular characterization of avian influenza A(H9N2) viruses, Bangladesh. Emerg Infect Dis 19: 1393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shanmuganatham K., Feeroz M.M., Jones‐Engel L., Walker D., Alam S., Hasan M., Mckenzie P., Krauss S., Webby R.J., Webster, R.G. (2014) Genesis of avian influenza H9N2 in Bangladesh. Emerg Microbes Infect 3: e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sun Y., Liu J. (2015) H9N2 influenza virus in China: A cause of concern. Protein Cell 6: 18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chakraborty A., Arifeen S.E., Streafield P.K. (2011) Outbreak of mild respiratory disease caused by H5N1 and H9N2 infections among young children in Dhaka, Bangladesh, 2011. HSB (Health Sci Bull), 9: 5–12 (En), 5–12 (Bengali). [Google Scholar]
- 10. Homme P.J., Easterday B.C. (1970) Avian influenza virus infections. I. Characteristics of influenza A‐turkey‐Wisconsin‐1966 virus. Avian Dis 14: 66–74. [PubMed] [Google Scholar]
- 11. Guo Y.J., Krauss S., Senne D.A., Mo I.P., Lo K.S., Xiong X.P., Norwood M., Shortridge K.F., Webster R.G., Guan Y. (2000) Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology 267: 279–88. [DOI] [PubMed] [Google Scholar]
- 12. Jackwood M.W., Stallknecht D.E. (2007) Molecular epidemiologic studies on North American H9 avian influenza virus isolates from waterfowl and shorebirds. Avian Dis 51: 448–50. [DOI] [PubMed] [Google Scholar]
- 13. Uyeki T.M., Nguyen D.C., Rowe T., Lu X., Hu‐Primmer J., Huynh L.P., Hang N.L., Katz J.M. (2012) Seroprevalence of antibodies to avian influenza A (H5) and A (H9) viruses among market poultry workers, Hanoi, Vietnam, 2001. PLoS ONE 7: e43948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang R., Wang A.R., Liu Z.H., Liang W., Li X.X., Tang Y.J., Miao Z.M., Chai T.J. (2013) Seroprevalence of avian influenza H9N2 among poultry workers in Shandong Province, China. Eur J Clin Microbiol Infect Dis 32: 1347–51. [DOI] [PubMed] [Google Scholar]
- 15. Yu Q., Liu L., Pu J., Zhao J., Sun Y., Shen G., Wei H., Zhu J., Zheng R., Xiong D., Liu X., Liu J. (2013) Risk perceptions for avian influenza virus infection among poultry workers, China. Emerg Infect Dis 19: 313–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gomaa M.R., Kayed A.S., Elabd M.A., Zeid D.A., Zaki S.A., El Rifay A.S., Sherif L.S., Mckenzie P.P., Webster R.G., Webby R.J., Ali M.A., Kayali G. (2015) Avian influenza A(H5N1) and A(H9N2) seroprevalence and risk factors for infection among Egyptians: a prospective, controlled seroepidemiological study. J Infect Dis 211: 1399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Heidari A., Mancin M., Nili H., Pourghanbari G.H., Lankarani K.B., Leardini S., Cattoli G., Monne I., Piccirillo A. (2016) Serological evidence of H9N2 avian influenza virus exposure among poultry workers from Fars province of Iran. Virol J 13: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li X., Tian B., Jianfang Z., Yongkun C., Xiaodan L., Wenfei Z., Yan L., Jing T., Junfeng G., Tao C., Rongbao G., Dayan W., Shu Y. (2017) A comprehensive retrospective study of the seroprevalence of H9N2 avian influenza viruses in occupationally exposed populations in China. PLoS ONE 12: e 0178328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Das A., Suarez D.L. (2007) Development and bench validation of real‐time reverse transcription polymerase chain reaction protocols for rapid detection of the subtypes H6, H9, and H11 of avian influenza viruses in experimental samples. J Vet Diagn Invest 19: 625–34. [DOI] [PubMed] [Google Scholar]
- 20. Monne I., Ormelli S., Salviato A., De Battisti C., Bettini F., Salomoni A., Drago A., Zecchin B., Capua I., Cattoli G. (2008) Development and validation of a one‐step real‐time PCR assay for simultaneous detection of subtype H5, H7, and H9 avian influenza viruses. J Clin Microbiol 46: 1769–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ben Shabat M., Meir R., Haddas R., Lapin E., Shkoda I., Raibstein I., Perk S., Davidson I. (2010) Development of a real‐time TaqMan RT‐PCR assay for the detection of H9N2 avian influenza viruses. J Virol Methods 168: 72–7. [DOI] [PubMed] [Google Scholar]
- 22. Slomka M.J., Hanna A., Mahmood S., Govil J., Krill D., Manvell R.J., Shell W., Arnold M.E., Banks J., Brown I.H. (2013) Phylogenetic and molecular characteristics of Eurasian H9 avian influenza viruses and their detection by two different H9‐specific RealTime reverse transcriptase polymerase chain reaction tests. Vet Microbiol 162: 530–– 42. [DOI] [PubMed] [Google Scholar]
- 23. Larkin M.A., Blackshields G., Brown N.P., Chenna R., Mcgettigan P.A., Mcwilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–8. [DOI] [PubMed] [Google Scholar]
- 24. Shu Y., Mccauley J. (2017) GISAID: Global initiative on sharing all influenza data − from vision to reality. Euro Surveill 22: 30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Takayama I., Takahashi H., Nakauchi M., Nagata S., Tashiro M., Kageyama T. (2015) Development of a diagnostic system for novel influenza A(H7N9) virus using a real‐time RT‐PCR assay in Japan. Jpn J Infect Dis 68: 113–8. [DOI] [PubMed] [Google Scholar]
- 26. Nakauchi M., Yasui Y., Miyoshi T., Minagawa H., Tanaka T., Tashiro M., Kageyama T. (2011) One‐step real‐time reverse transcription‐PCR assays for detecting and subtyping pandemic influenza A/H1N1 2009, seasonal influenza A/H1N1, and seasonal influenza A/H3N2 viruses. J Virol Methods 171: 156–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kaida A., Kubo H., Takakura K., Sekiguchi J., Yamamoto S.P., Kohdera U., Togawa M., Amo K., Shiomi M., Ohyama M., Goto K., Hase A., Kageyama T., Iritani N. (2014) Associations between co‐detected respiratory viruses in children with acute respiratory infections. Jpn J Infect Dis 67: 469–75. [DOI] [PubMed] [Google Scholar]
- 28. Nakauchi M., Takayama I., Takahashi H., Oba K., Kubo H., Kaida A., Tashiro M., Kageyama T. (2014) Real‐time RT‐PCR assays for discriminating influenza B virus Yamagata and Victoria lineages. J Virol Methods 205: 110–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kumar S., Stecher G., Tamura K. (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saitou N., Nei M. (1987) The neighbor‐joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–25. [DOI] [PubMed] [Google Scholar]
- 31. Kimura M. (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–20. [DOI] [PubMed] [Google Scholar]
- 32. Felsenstein J. (1985) Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783–91. [DOI] [PubMed] [Google Scholar]
- 33. Finney D.J. (1971) Probit Analysis, 3rd edn. Cambridge: Cambridge University Press. [Google Scholar]
- 34. Fusaro A., Monne I., Salviato A., Valastro V., Schivo A., Amarin N.M., Gonzalez C., Ismail M.M., Al‐Ankari A.R., Al‐Blowi M.H., Khan O.A., Maken Ali A.S., Hedayati A., Garcia Garcia J., Ziay G.M., Shoushtari A., Al Qahtani K.N., Capua I., Holmes E.C., Cattoli G. (2011) Phylogeography and evolutionary history of reassortant H9N2 viruses with potential human health implications. J Virol 85: 8413–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wan H., Perez D.R. (2007) Amino acid 226 in the hemagglutinin of H9N2 influenza viruses determines cell tropism and replication in human airway epithelial cells. J Virol 81: 5181–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yu L., Wang Z., Chen Y., Ding W., Jia H., Chan J.F., To K.K., Chen H., Yang Y., Liang W., Zheng S., Yao H., Yang S., Cao H., Dai X., Zhao H., Li J., Bao Q., Chen P., Hou X., Li L., Yuen K.Y. (2013) Clinical, virological, and histopathological manifestations of fatal human infections by avian influenza A(H7N9) virus. Clin Infect Dis 57: 1449–57. [DOI] [PubMed] [Google Scholar]
- 37. Shelton H., Ayora‐Talavera G., Ren J., Loureiro S., Pickles R.J., Barclay W.S., Jones I.M. (2011) Receptor binding profiles of avian influenza virus hemagglutinin subtypes on human cells as a predictor of pandemic potential. J Virol 85: 1875–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guan Y., Shortridge K.F., Krauss S., Webster R.G. (1999) Molecular characterization of H9N2 influenza viruses: Were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc Natl Acad Sci USA 96: 9363–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu D., Shi W., Shi Y., Wang D., Xiao H., Li W., Bi Y., Wu Y., Li X., Yan J., Liu W., Zhao G., Yang W., Wang Y., Ma J., Shu Y., Lei F., Gao G.F. (2013) Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: Phylogenetic, structural, and coalescent analyses. Lancet 381: 1926–32. [DOI] [PubMed] [Google Scholar]
- 40. Chen H., Yuan H., Gao R., Zhang J., Wang D., Xiong Y., Fan G., Yang F., Li X., Zhou J., Zou S., Yang L., Chen T., Dong L., Bo H., Zhao X., Zhang Y., Lan Y., Bai T., Dong J., Li Q., Wang S., Zhang Y., Li H., Gong T., Shi Y., Ni X., Li J., Zhou J., Fan J., Wu J., Zhou X., Hu M., Wan J., Yang W., Li D., Wu G., Feng Z., Gao G.F., Wang Y., Jin Q., Liu M., Shu Y. (2014) Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: A descriptive study. Lancet 383: 714–21. [DOI] [PubMed] [Google Scholar]
- 41. Shibata A., Hiono T., Fukuhara H., Sumiyoshi R., Ohkawara A., Matsuno K., Okamatsu M., Osaka H., Sakoda Y. (2018) Isolation and characterization of avian influenza viruses from raw poultry products illegally imported to Japan by international flight passengers. Transbound Emerg Dis 65: 465–75. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found online in the supporting information tab for this article.
Figure S1. Amplification plot of in vitro‐transcribed RNA in the H9 rRT‐PCR assay. Amplification plot for serial dilutions of in vitro‐transcribed RNA of the HA gene of A/Hunan/44558/2015 (H9N2), Y280 lineage. Shown are results for six replicates in each copy number of concentration (copies/reaction). rRT‐PCR, real‐time RT‐PCR.
Figure S2. Comparison of amplification plots for the H9 and type A rRT‐PCR assays. Amplification plot for serial dilutions of virus RNA of (a) A/chicken/Japan/AQ‐HE28‐50/2016 (H9N2) (Y280 lineage), and (b) A/turkey/Wisconsin/1/66 (H9N2) (North American lineage), for the H9 and type A rRT‐PCR assays. Shown are results for three replicates in each copy number of concentration (copies/reaction) for each assay. rRT‐PCR, real‐time RT‐PCR.


