Abstract
Objective
To investigate the genetic background influencing the development of cardiovascular (CV) disease in patients with rheumatoid arthritis (RA).
Methods
We performed a genome‐wide association study (GWAS) in which, after quality control and imputation, a total of 6,308,944 polymorphisms across the whole genome were analyzed in 2,989 RA patients of European origin. Data on subclinical atherosclerosis, obtained through assessment of carotid intima‐media thickness (CIMT) and presence/absence of carotid plaques by carotid ultrasonography, were available for 1,355 individuals.
Results
A genetic variant of the RARB gene (rs116199914) was associated with CIMT values at the genome‐wide level of significance (minor allele [G] β coefficient 0.142, P = 1.86 × 10−8). Interestingly, rs116199914 overlapped with regulatory elements in tissues related to CV pathophysiology and immune cells. In addition, biologic pathway enrichment and predictive protein–protein relationship analyses, including suggestive GWAS signals of potential relevance, revealed a functional enrichment of the collagen biosynthesis network related to the presence/absence of carotid plaques (Gene Ontology no. 0032964; false discovery rate–adjusted P = 4.01 × 10−3). Furthermore, our data suggest potential influences of the previously described candidate CV risk loci NFKB1,MSRA, and ZC3HC1 (P = 8.12 × 10−4, P = 5.94 × 10−4, and P = 2.46 × 10−4, respectively).
Conclusion
The present findings strongly suggest that genetic variation within RARB contributes to the development of subclinical atherosclerosis in patients with RA.
Introduction
Cardiovascular (CV) disease is the most common cause of morbidity and mortality in patients with rheumatoid arthritis (RA) 1, 2, 3. In RA patients, CV disease may develop as a result of an accelerated atherosclerotic process 4. Surrogate markers for subclinical atherosclerosis, i.e., increased carotid intima‐media thickness (CIMT) and presence of carotid plaques 5, 6, are excellent predictors of future CV events. Traditional CV risk factors and chronic inflammation do not fully explain the increased CV predisposition observed in patients with RA, accounting for only ~70% of the population‐attributable risk for CV disease outcomes 7. Cumulative knowledge clearly suggests that genetic factors may play a relevant role in this phenomenon 8, but the specific genetic component of CV disease in RA remains elusive.
Genome‐wide association studies (GWAS) constitute a hypothesis‐free approach in which millions of common genetic variations across the whole genome are interrogated 9. This strategy has been of great help in elucidating relevant inroads into the genetics of several complex human diseases 10. The use of this technology has substantially increased the number of established RA susceptibility loci from 3 to >100 during the last decade 11. Nevertheless, there are currently no available GWAS data specifically focused on CV disease in patients with RA.
Taking into account all of these considerations, we undertook the first GWAS on the development of CV disease in RA. This multicenter study included a large number of patients with RA, in whom the presence/absence of CV events and subclinical atherosclerosis were evaluated.
Patients and methods
Study population
A total of 3,433 unrelated Spanish patients of European ancestry, all of whom had RA according to the 2010 American College of Rheumatology/European League Against Rheumatism classification criteria 12, were enrolled in the study. Centers involved in patient recruitment included Hospital Universitario Lucus Augusti, Hospital Universitario Marqués de Valdecilla, Hospital Universitario de Basurto, Hospital Universitario Central de Asturias, Hospital Clínico Universitario de Santiago, Hospital Universitario de Bellvitge, Hospital Universitario San Cecilio, Hospital Universitario Reina Sofía, Hospital Universitario de Canarias, Hospital Universitario Doctor Peset, Hospital General Universitario de Ciudad Real, Hospital Clínico San Carlos, Hospital Universitario La Paz, Hospital Universitario de la Princesa, Hospital General Universitario Gregorio Marañón, and Hospital Universitario 12 de Octubre. Before being included in the study, all patients provided written informed consent according to the Declaration of Helsinki. The procedures followed were in accordance with the standards and requirements of the human experimentation ethics committees at all participating centers.
Genotyping and quality control
Genomic DNA was extracted from peripheral blood using standard procedures. Genotyping was conducted at the Human Genotyping Unit of the National Genotyping Center in Spain, using the GWAS platform Infinium HumanCore BeadChip in an iScan system, according to the protocol recommended by the manufacturer (Illumina). Single‐nucleotide polymorphisms (SNPs) with a cluster separation of <0.4 were removed after the calling.
Raw data were subjected to stringent quality control filters using the software Plink (version 1.07) 13. Polymorphisms with call rates of <0.98 and minor allele frequencies of <0.01, as well as those that deviated from Hardy‐Weinberg equilibrium (P < 0.001), were filtered out. Similarly, samples with <95% successfully called polymorphisms, and 1 subject per pair of first‐degree relatives (identity by descent >0.4), were removed. Sex chromosomes were also excluded from the analysis.
To ensure reliability of the results, the associated SNP described below was re‐genotyped using a predesigned TaqMan 5′ SNP genotyping assay (C_154503570_10) in a 7900HT Fast Real‐Time PCR System (Applied Biosystems), and the TaqMan types were compared with the corresponding imputed data.
Imputation methods
After application of the quality control filters, whole‐genome SNP genotype imputation in autosomal chromosomes was carried out in the Michigan Imputation Server (MIS) 14, using ShapeIT16 software (version v2.r790) for haplotype reconstruction and the updated Haplotype Reference Consortium data (version r1.1) as a reference panel, which combine sequencing data from a total of 32,470 individuals from multiple studies (including the 1000 Genomes Project) 15. The quality control filters mentioned above were also applied to the imputed data using Plink. In addition, singletons (r2 ≤ 0.2) were excluded. Finally, possible population substratification was controlled by principal components (PC) analysis using Plink and gcta64 and R‐base software under GNU Public license v2. The first 10 PCs for each individual were calculated and plotted to identify outliers, and those deviating from the cluster centroid by >4 SD were excluded.
After quality control, 6,308,944 SNPs and 2,989 RA patients remained for analysis in the final data set. Data on demographic, RA clinical, and CV disease–related characteristics are shown in Table 1. Information related to CV events was obtained from the medical records of each patient, with traditional CV risk factors and CV events defined as previously described 3, 6. Briefly, individuals were considered to have ischemic heart disease (IHD) if any of the following criteria were satisfied: a recorded diagnosis of ischemic cardiopathy due to an acute coronary syndrome (acute myocardial infarction or unstable angina), abnormal Q waves seen on electrocardiography, and/or >50% stenosis of at least 1 coronary vessel seen on coronary images. A patient was considered to have heart failure based on the Framingham criteria. Cerebrovascular accident was recorded if patients had a stroke and/or transient ischemic attacks (TIAs). Strokes were classified according to their clinical features and were confirmed by computed tomography and/or magnetic resonance imaging. TIAs were diagnosed if the symptoms were self‐limited in <24 hours, without residual neurologic damage. Finally, peripheral arterial disease was considered to be present if confirmed by Doppler imaging and arteriography 3, 6.
Table 1.
Demographic, clinical, and CV disease–related characteristics of the 2,989 RA patients whose samples were included in the filtered data seta
| Demographic and RA characteristics | |
| Age at the time of disease onset, mean ± SD years | 49.8 ± 14.9 |
| Follow‐up time, mean ± SD years | 11.7 ± 9.1 |
| Women, % | 74.7 |
| RF positiveb | 1,585/2,432 (65.2) |
| ACPA positive | 1,365/2,286 (59.7) |
| Erosions | 1,125/2,148 (52.4) |
| Extraarticular manifestationsc | 575/1,994 (28.8) |
| Traditional CV risk factors | |
| Hypertension | 1,018/2,585 (39.4) |
| Diabetes mellitus | 318/2,585 (12.3) |
| Dyslipidemia | 1,122/2,585 (43.4) |
| Obesity | 605/2,585 (23.4) |
| Smoking | 957/2,585 (37.0) |
| CV events | 467/2,989 (15.6) |
| Ischemic heart disease | 224/2,989 (7.5) |
| Heart failure | 146/2,989 (4.9) |
| Cerebrovascular accident | 125/2,989 (4.2) |
| Peripheral artery disease | 60/2,989 (2.0) |
Except where indicated otherwise, values are the number of patients/number assessed (%). CV = cardiovascular; RA = rheumatoid arthritis; RF = rheumatoid factor; ACPA = anti–citrullinated protein antibody.
At least 2 determinations at different times were required for analysis of this result.
Patients were considered to have extraarticular manifestations if they experienced at least 1 of the following: nodular disease, Felty's syndrome, pulmonary fibrosis, rheumatoid vasculitis, or secondary Sjögren's syndrome 3.
Subclinical atherosclerosis examination
Information on subclinical atherosclerosis was available for 1,355 RA patients from the filtered data sets. Subclinical atherosclerosis examination was assessed with a carotid ultrasound technique (evaluation of CIMT and presence/absence of carotid plaques). At the hospitals in Santander, Bilbao, Granada, Córdoba, Tenerife, Valencia, Ciudad Real, and Madrid, the ultrasound examination was performed using a commercial scanner (16,17). Patients from Lugo were assessed by high‐resolution B‐mode ultrasound 18. CIMT was measured at the far wall of the right and left common carotid arteries over the proximal 15‐mm–long segment. CIMT was determined as the average of 3 measurements in each common carotid artery. Consistency of results between these 2 ultrasound methods was previously reported 19, supporting the fact that the use of 2 different instruments to collect CIMT data did not influence the results derived from this analysis. In addition, these studies were performed by experts with high intra‐ and interobserver reliability, who have collaborated closely in the assessment of subclinical atherosclerosis in RA. Criteria for determining the presence of plaque in the accessible extracranial carotid tree were defined as described by Touboul et al 20.
Statistical analysis
Estimations for statistical power were obtained with CaTS Power Calculator for Genetic Studies software, which implements the methods described by Skol et al 21 (Supplementary Tables 1–4, on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.40734/abstract). All statistical analyses were conducted with Plink. First, we compared the genotype frequencies of all SNPs according to a continuous CV disease outcome variable (CIMT values) by linear regression assuming an additive model. The first 10 PCs, age at the time of the carotid ultrasound examination, and sex were included in the model as covariates. Subsequently, we compared the genotype frequencies of all SNPs according to binary CV disease outcome variables (presence/absence of CV events, IHD, and carotid plaques) by logistic regression on the best‐guess genotypes assuming an additive model. The first 10 PCs, age at the time of RA diagnosis, and sex were included as covariates for the presence/absence of CV events and IHD analyses, and the first 10 PCs, age at the time of carotid ultrasound examination, and sex were included as covariates for the presence/absence of carotid plaque analysis. Finally, P values, beta coefficients, standard errors, odds ratios, and 95% confidence intervals were calculated. The statistical threshold was set at the genome‐wide level of significance (P < 5 × 10−8).
Performance of functional annotations of the associated variants
In a further step, we evaluated the putative functional implications of the identified CV risk signals by integrating our data with functional annotation data available in public databases, using different bioinformatics approaches. For this purpose, we first identified all of the potential polymorphisms in high linkage disequilibrium (LD; r2 > 0.8) of the associated signals of our GWAS, using the European populations from the 1000 Genomes Project and Plink. All of those potential polymorphism taggers would be considered equally as candidates for prioritizing causality or hypothesizing possible molecular causes of the observed associations in the subsequent bioinformatic approaches. Then, the online tools RegulomeDB 22, HaploReg (version 4.1) 23, and Capture HiC Plotter (CHi‐CP) 24 were used to evaluate the possible regulatory effect of the associated signals and their possible implications in the clinical phenotypes analyzed.
Candidate genomic regions and pathway enrichment analysis
Finally, we assessed the statistical significance in our GWAS of previously described CV risk–associated genomic regions (±100 kbp 3′ and 5′ of the reported gene) through candidate gene studies 8 and a recently published meta‐analysis of ImmunoChip data 25. Regarding the HLA region, a more comprehensive analysis was conducted. We extracted the extended HLA region (29,000,000–34,000,000 bp in chromosome 6) and imputed SNPs, classic HLA alleles at 2‐ and 4‐digits, and polymorphic amino acid positions, as previously described 26, 27, 28.
Additionally, a biologic pathway enrichment analysis involving genes that showed suggestive P values in our study (P < 1 × 10−4) was performed by using the tool for that purpose from the Gene Ontology (GO) reference genome project 29, 30, powered by the Protein Analysis Through Evolutionary Relationships Classification System 31. Moreover, we conducted a predictive protein–protein interaction analysis among these same markers, using the Search Tool for the Retrieval of Interacting Genes/Proteins database 32. P values less than 0.05 after correction for multiple testing were considered significant.
Results
Testing for association with CV disease outcomes
Figure 1 and Supplementary Figure 1 (available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.40734/abstract) summarize the overall results obtained for each CV disease outcome analysis performed. Interestingly, a statistically significant signal at the genome‐wide level of significance was associated with CIMT values (Figure 1). This signal corresponded with the genetic variant rs116199914, which maps to the 3′‐untranslated region (3′‐UTR) of the retinoic acid receptor β gene (RARB) (Table 2). The minor allele (G) of this SNP was significantly related to increased CIMT values (β = 0.142, P = 1.86 × 10−8) (Table 2). To rule out the possibility that bias due to incorrect genotyping or imputation could have affected these results, we obtained direct genotypes using TaqMan probes for rs116199914. The overall concordance reached after comparing TaqMan types with the corresponding imputed data was 99.94%. Based on previous studies that demonstrated association between anti–citrullinated protein antibody (ACPA) positivity and CV disease in RA 33, 34, we evaluated the potential association between the genetic variant rs116199914 and ACPA status. However, no statistically significant results were observed (data not shown). Several suggestive associations with CIMT values were also detected, although none of them reached the genome‐wide level of statistical significance (Figure 1). Among them, intronic variants of both RARB and the positive regulatory domain zinc‐finger protein 10 gene (PRDM10), as well as a disequilibrium block of intergenic polymorphisms at chromosome 12, had the most suggestive P values.
Figure 1.
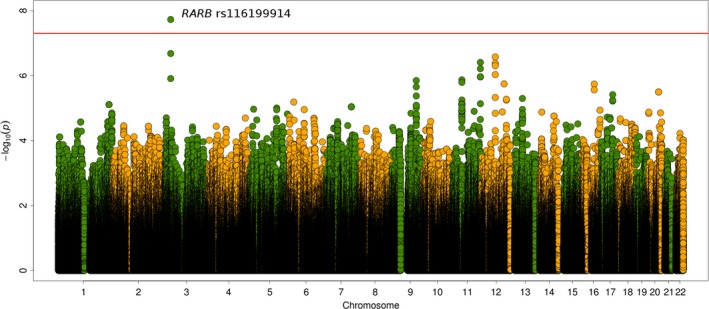
Manhattan plot representation of the analysis of carotid intima‐media thickness values as the cardiovascular disease outcome. The −log10 P values are plotted against their physical chromosomal position. The red line represents the genome‐wide level of significance (P < 5 × 10−8).
Table 2.
Index signals showing the lowest P values according to the different CV disease outcomesa
| CV disease outcome, Chr. | Position in Chr. (GRCh37) | SNP ID | GENCODE gene | Change | Minor allele | MAF | P | β [SE] or OR (95% CI) |
|---|---|---|---|---|---|---|---|---|
| CIMT values | ||||||||
| 3 | 25.638.355 | rs116199914 | RARB (3′‐UTR) | G<A | G | 0.012 | 1.86 × 10−8 b | 0.142 [0.025] |
| 3 | 25.622.694 | rs77388418 | RARB (intronic) | C<T | C | 0.014 | 2.07 × 10−7 | 0.124 [0.024] |
| 12 | 63.337.536 | rs1695024 | 8 kb 3′ of Y_RNA | A<G | A | 0.230 | 2.64 × 10−7 | 0.031 [0.006] |
| 11 | 129.852.180 | rs111703287 | PRDM10 (intronic) | T<C | T | 0.014 | 3.90 × 10−7 | 0.119 [0.023] |
| CV events | ||||||||
| 1 | 166.485.891 | rs6684311 |
27 kb 5′ of RP11‐ 276E17.2 |
G<C | G | 0.189 | 2.85 × 10−7 | 1.68 (1.38–2.05) |
| IHD | ||||||||
| 1 | 245.338.976 | rs112844193 | KIF26B (intronic) | T<C | T | 0.054 | 1.35 × 10−7 | 2.67 (1.85–3.85) |
| 7 | 120.966.790 | rs3779381 | WNT16 (intronic) | G<A | G | 0.283 | 2.09 × 10−7 | 1.77 (1.23–2.19) |
| 1 | 156.057.417 | rs112941217 | LMNA (intronic) | C<T | C | 0.030 | 4.67 × 10−7 | 4.81 (2.61–8.87) |
| Carotid plaques | ||||||||
| 17 | 15.008.430 | rs8066891 |
123 bp 3′ of RP11‐ 924A14.1 |
G<A | G | 0.171 | 4.47 × 10−6 | 0.58 (0.46–0.73) |
| 9 | 29.148.449 | rs12683261 | 259 kb 5′ of MIR873 | A<G | A | 0.031 | 4.57 × 10−6 | 0.25 (0.14–0.45) |
| 1 | 240.599.906 | rs9727451 | FMN2 (intronic) | A<G | A | 0.087 | 4.69 × 10−6 | 2.13 (1.54–2.95) |
| 4 | 166.579.647 | rs2611206 | 26 kb 5′ of RP11‐340B18.1 | A<G | A | 0.126 | 4.84 × 10−6 | 0.53 (0.41–0.69) |
CV = cardiovascular; Chr. = chromosome; SNP = single‐nucleotide polymorphism; MAF = minor allele frequency; OR = odds ratio; 95% CI = 95% confidence interval; CIMT = carotid intima‐media thickness; 3′‐UTR = 3′‐untranslated region; IHD = ischemic heart disease.
Statistically significant at the genome‐wide level of significance.
Likewise, several trends of association were observed when the presence/absence of CV events, IHD, and carotid plaques were analyzed (Table 2 and Supplementary Figure 1, http://onlinelibrary.wiley.com/doi/10.1002/art.40734/abstract). Regarding the presence/absence of CV events, 2 intergenic variants in high LD at chromosome 1 exhibited the lowest P values. According to the presence/absence of IHD, an intronic variant of the kinesin family member 26B gene (KIF26B) and 2 disequilibrium blocks of polymorphisms at chromosomes 1 and 7 represented the strongest signals. Similarly, an intronic variant of the formin 2 gene (FMN2) and intergenic polymorphisms located at chromosomes 4, 9, and 17 exhibited the lowest P values regarding the presence/absence of carotid plaques.
Similar results were obtained when the analyses were also performed with traditional CV risk factors (smoking, diabetes mellitus, hypertension, obesity, and dyslipidemia) as covariates. In this regard, a statistically significant signal at the genome‐wide level of significance that corresponded to RARB rs116199914 was associated with CIMT values (minor allele β = 0.137, P = 4.35 × 10−8). In addition, trends of association were again observed when the presence/absence of CV events, IHD, and carotid plaques were analyzed (data not shown).
Functional annotations of the associated variants
We evaluated the possible functional implications of the associated genetic variant rs116199914 by integrating our data with data from public databases. First, we searched for proxies (r2 > 0.8) of rs116199914 in the 5 populations of European origin in the 1000 Genomes Project (Iberian population in Spain, Utah residents of North and Western European ancestry, British in England and Scotland, Toscani in Italy, and Finnish in Finland). Since no proxies were identified, we functionally annotated just the rs116199914 polymorphism. As this SNP is located in the 3′‐UTR of the RARB gene, we used bioinformatic tools aimed at exploring annotations of the noncoding genome with putative regulatory effects on gene expression (including effect on regulatory motifs, chromatin state, and protein binding, as well as expression from expression quantitative trait locus studies) in GEO, ENCODE, Roadmap Epigenomics, and promoter CHi‐C data sets, and published literature.
Interestingly, RegulomeDB results suggested that rs116199914 may represent a DNA element with relevant regulatory effects (score 6). Additional functional implications were suggested with both HaploReg version 4.1. and CHi‐C. In particular, overlapping with histone marks in tissues related to CV pathophysiology and cells of the immune system was observed (Figure 2). Specifically, rs116199914 was described to overlap with the enhancer histone mark H3K4me1 and the promoter histone mark H3K9ac in fetal heart, and with histone marks enriched at promoters and enhancers in immune cells 23. Furthermore, as derived from the CHi‐C data sets, rs116199914 was reported to interact with, among others, NF‐κB inhibitor–interacting Ras‐like 1 gene (NKIRAS1) in total CD4 Mycosis fungoides cells and total CD8 cells 35 (Supplementary Figure 2, on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.40734/abstract). In addition, rs116199914 was described to affect the sequence‐specific binding for NFAT 23.
Figure 2.
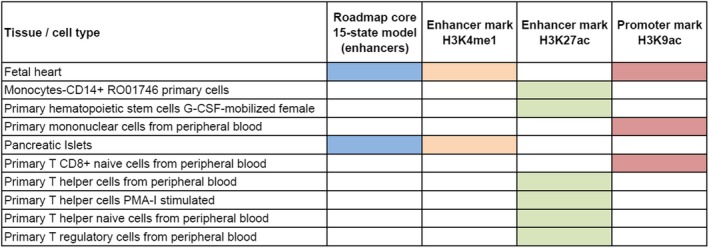
Regulatory chromatin annotations of RARB rs116199914 in tissues related to cardiovascular pathology and cells of the immune system, according to ENCODE data. The chromatin 15‐state model was developed using 5 marks and 127 epigenomes from the Roadmap Epigenomics Project. G‐CSF = granulocyte colony‐stimulating factor; PMA‐I = phorbol myristate acetate/ionomycin.
Candidate genes and pathway analysis
We also determined the statistical significance, in our GWAS, of previously described CV risk genes by candidate studies 8 and a recently published meta‐analysis of ImmunoChip data 25. P values of <0.05 were observed across most of the evaluated loci (Supplementary Table 5, http://onlinelibrary.wiley.com/doi/10.1002/art.40734/abstract). Among them, the lowest P values were detected for associations of the NFKB1 and methionine sulfoxide reductase A gene (MSRA) regions with the presence of CV events (P = 8.12 × 10−4 and P = 5.94 × 10−4, respectively), as well as the zinc‐finger C3HC‐type containing 1 gene (ZC3HC1) region with CIMT values (P = 2.46 × 10−4). The association between NFKB1 and CV events remained statistically significant after correction for multiple testing (rs227361, false discovery rate–adjusted P = 4.50 × 10−2). Regarding the HLA system, no statistically significant results were observed across this genomic region (Supplementary Figure 3, http://onlinelibrary.wiley.com/doi/10.1002/art.40734/abstract).
In addition, analysis of possible biologic pathway enrichments and predictive protein–protein relationships was performed for the gene products of loci that showed P values of potential relevance in our study (P < 1 × 10−4). In this regard, the molecular network of the selected proteins related to the presence/absence of carotid plaques had significantly more interactions than expected (number of nodes 51, number of edges 8, average node degree 0.314, clustering coefficient 0.235; expected number of edges 3, protein–protein interaction enrichment P = 1.68 × 10−2) (Figure 3). In accordance with the functional enrichments of the network, the most significantly associated GO term corresponded to “collagen biosynthetic process” (GO number 0032964) (false discovery rate–adjusted P = 4.01 × 10−3). No statistically significant results were obtained when these analyses were performed according to CIMT values, presence/absence of CV events, or IHD.
Figure 3.
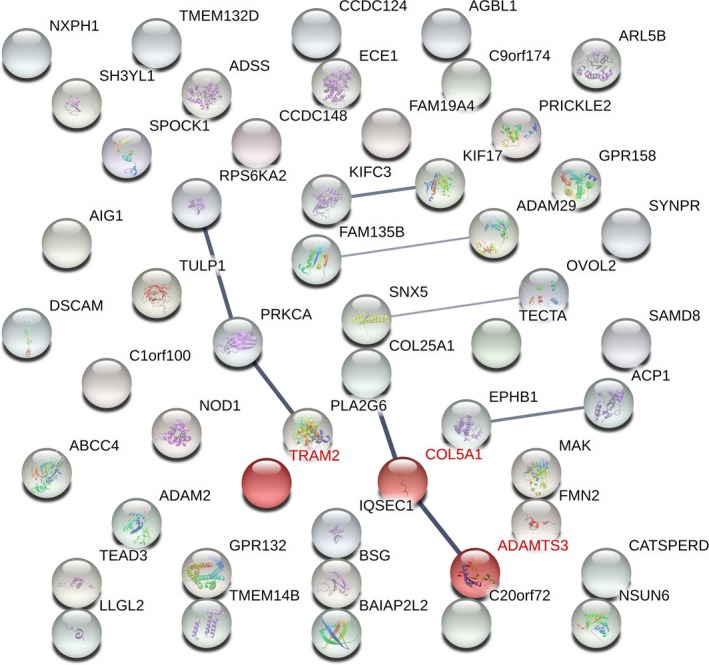
Interaction network formed by the encoded proteins of genes showing P values of potential relevance in our study (P < 1 × 10−4), according to the presence/absence of carotid plaques. The width of the gray lines indicates the reliability of each interaction. Proteins of the collagen biosynthetic process pathway (GO number 0032964) are highlighted in red.
Discussion
During the last decade, the genetic basis of the increased predisposition to CV disease observed in RA patients has been comprehensively investigated using a candidate gene strategy 8. However, not until the present study have GWAS data been generated and analyzed. Therefore, the results presented here may represent a turning point for better understanding of the pathogenic mechanisms underlying this severe complication of RA.
A genetic marker of the RARB gene (rs116199914) was associated, at the genome‐wide level of significance, with subclinical atherosclerosis, assessed by CIMT. Interestingly, this signal overlaps with promoter and enhancer histone marks in fetal heart and immune cells. In addition, rs116199914 has been described to interact with the gene NKIRAS1. These data are striking, as NKIRAS1 encodes a crucial protein for the inhibition of NF‐κB 36, 37, which is one of the most relevant molecules involved in inflammation processes 38 and is considered to be a key regulator of several atherosclerosis genes 39. A previous candidate gene study demonstrated the influence of a promoter genetic variant in the NF‐κB coding gene (NFKB1) on the risk of developing CV events among patients with RA 39. Additionally, the use of drugs that block cytokines of the NF‐κB signaling pathway has been described as a promising therapeutic strategy to attenuate the heightened CV risk in patients with RA 40, 41 and to provide a beneficial effect on surrogate CV disease markers in those patients 42, 43.
In addition, the associated variant identified in our study was shown to affect sequence‐specific binding of NFAT, which regulates inducible gene transcription during the immune response 44, 45, 46. Originally, NFAT was described as being mainly expressed in activated T cells 46 and other immune cells 45. Currently, its regulatory roles in blood vessels and heart tissue are well established 47, 48, 49. Furthermore, a role of this molecule in angiogenic processes has been confirmed 48. Consistent with this, cumulative knowledge clearly demonstrates that the chronic inflammation observed in patients with RA, critical for the development of atherosclerosis, is often accompanied by imbalanced angiogenesis 50. In accordance with that, increased serum levels of the angiogenic molecule angiopoietin 2 have been found to correlate with the development of CV events in patients with RA 50.
Our results suggest a functional impact of the genetic variant RARB rs116199914. In this regard, it could be speculated that the interaction between this polymorphism and NKIRAS1 modulates the expression of the latter, affecting the inhibition of NF‐κB. This may trigger the regulation of genes encoding proinflammatory cytokines, adhesion molecules, chemokines, and inducible nitric oxide synthase, thus contributing to endothelial damage and subsequently to CV disease. Similarly, since RARB rs116199914 has been described to affect the sequence‐specific binding of NFAT as noted above, it may be reasonable to consider that this phenomenon modulates the expression of genes related to angiogenic processes in atherosclerosis.
Additional suggestive signals of potential relevance were observed when both the presence/absence of CV events (including IHD) and subclinical atherosclerosis were tested. However, those signals did not reach the genome‐wide level of significance, probably due to insufficient statistical power to detect risk variants with low‐to‐moderate effects. Biologic pathway enrichment and protein–protein interaction analyses revealed a functional enrichment of the collagen biosynthesis network according to the presence/absence of carotid plaques. This result is consistent with the fact that collagen constitutes the main component of the fibrous cap of the carotid plaque and contributes to its structural integrity and vulnerability 51. Indeed, a recent Metabochip analysis performed in American patients with RA revealed a suggestive association between a genetic variant in the Colα1(IV) gene (COL4A1) and carotid plaques 52.
Finally, our results support the implication of the previously reported candidate CV risk gene NFKB1 and suggest a potential influence of both MSRA and ZC3HC1 in the development of CV disease in RA. In contrast, a relevant influence of the HLA region in this process, though suggested previously by others 8, was not supported by our data.
There is evidence that current CV risk screening and management strategies underestimate the actual degree of predisposition to CV in patients with RA. In this context, genetic markers related to the development of CV disease in patients with RA may be used as additional tools to identify those patients at high CV risk, who may definitively benefit from active therapy to prevent CV events. Accordingly, the results of our study may help in the design of efficient tools to identify RA patients who are more likely to develop CV disease based on their genetic background.
A potential major limitation of the present study is the lack of replication of the discovery findings in an independent cohort of patients with RA. In addition, the study could have been underpowered to detect associations with small effect size. Further investigations to confirm our results are needed. Interestingly, Karpouzas et al reported that the frequency of unstable, noncalcified plaques is increased among patients with RA 53. Unstable plaques are very dangerous since they are particularly susceptible to disruption. Vulnerable plaques are generally characterized as those having a thin inflamed fibrous cap over a very large lipid core. Since the conventional carotid ultrasound technique performed in our study did not allow us to identify the presence of unstable plaques, we believe further investigations aimed at identifying a potential role of the genetic variant RARB rs116199914 in the risk of unstable plaques should be conducted.
In conclusion, through a whole‐genome screening of common genetic variation, we have identified RARB rs116199914 as the main genetic variant associated with CIMT values in patients with RA. This finding could potentially lead to an improved ability to predict and screen for this condition and initiate treatment to prevent life‐threatening CV events in RA patients.
Author Contributions
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. González‐Gay had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design
López‐Mejías, Carmona, Mijares, Lera‐Gómez, Martín, Llorca, González‐Gay.
Acquisition of data
González‐Juanatey, Corrales, Vicente, Miranda‐Filloy, Ramírez Huaranga, Blanco, Robustillo‐Villarino, Rodríguez‐Carrio, Alperi‐López, Alegre‐Sancho, Pérez‐Pampín, González, Ortega‐Castro, López‐Pedrera, García Vivar, Gómez‐Arango, Raya, Narvaez, Balsa, López‐Longo, Carreira, González‐Álvaro, Rodríguez‐Rodríguez, Fernández‐Gutiérrez, Ferraz‐Amaro, Gualillo, Castañeda, González‐Gay.
Analysis and interpretation of data
López‐Mejías, Carmona, Genre, Remuzgo‐Martínez, Pulito‐Cueto, Martín, Llorca, González‐Gay.
Supporting information
Supported in part by the European Union FEDER fund and the Instituto de Salud Carlos III (ISCIII) (Fondo de Investigación Sanitaria grants PI06/0024, PS09/00748, PI12/00060, PI15/00525, and CP16/00033), the ISCIII RETICS programs (RD12/0009 and RD16/0012), the European IMI BTCure Program. Dr. López‐Mejías is recipient of the Miguel Servet type I Program Fellowship from the ISCIII and the European Social Fund (ESF) (grant CP16/00033). Dr. Carmona's work was supported by the Spanish Ministry of Economy and Competitiveness (Ramón y Cajal Program grant RYC‐2014‐16458). Dr. Genre is recipient of a Sara Borrell postdoctoral fellowship from the ISCIII and ESF (grant CD15/00095). Dr. Remuzgo‐Martínez’ work was supported by the ISCIII and European Regional Development Fund (ERDF) (RETICS Program RD16/0012/0009). Ms Pulito‐Cueto's work was supported by a predoctoral grant from the Instituto de Investigación Sanitaria Valdecilla (PREVAL 18/01). Ms Mijares’ work was supported by the Miguel Servet type I Program from the ISCIII and ESF (grant CP16/00033). Dr. Gualillo is staff personnel of Xunta de Galicia (Servizo Galego de Saude [SERGAS]) through a research‐staff stabilization contract (ISCIII/SERGAS) and his work is funded by ISCIII and FEDER (grants RD16/0012/0014 [RIER] and PI17/00409). He is beneficiary of project funds from the Research Executive Agency (REA) of the European Union in the framework of MSCA‐RISE Action of the H2020 Programme, project 734899—Olive‐Net.
Drs. López‐Mejías and Carmona contributed equally to this work. Drs. Martín, Llorca, and González‐Gay contributed equally to this work.
References
- 1. Aviña‐Zubieta JA, Choi HK, Sadatsafavi M, Etminan M, Esdaile JM, Lacaille D. Risk of cardiovascular mortality in patients with rheumatoid arthritis: a meta‐analysis of observational studies. Arthritis Rheum 2008;59:1690–7. [DOI] [PubMed] [Google Scholar]
- 2. Castañeda S, Nurmohamed MT, Gonzalez‐Gay MA. Cardiovascular disease in inflammatory rheumatic diseases. Best Pract Res Clin Rheumatol 2016;30:851–69. [DOI] [PubMed] [Google Scholar]
- 3. Gonzalez‐Gay MA, Gonzalez‐Juanatey C, Lopez‐Diaz MJ, Piñeiro A, Garcia‐Porrua C, Miranda‐Filloy JA, et al. HLA–DRB1 and persistent chronic inflammation contribute to cardiovascular events and cardiovascular mortality in patients with rheumatoid arthritis. Arthritis Rheum 2007;57:125–32. [DOI] [PubMed] [Google Scholar]
- 4. Gonzalez‐Gay MA, Gonzalez‐Juanatey C, Martin J. Rheumatoid arthritis: a disease associated with accelerated atherogenesis. Semin Arthritis Rheum 2005;35:8–17. [DOI] [PubMed] [Google Scholar]
- 5. Evans MR, Escalante A, Battafarano DF, Freeman GL, O'Leary DH, del Rincón I. Carotid atherosclerosis predicts incident acute coronary syndromes in rheumatoid arthritis. Arthritis Rheum 2011;63:1211–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gonzalez‐Juanatey C, Llorca J, Martin J, Gonzalez‐Gay MA. Carotid intima‐media thickness predicts the development of cardiovascular events in patients with rheumatoid arthritis. Semin Arthritis Rheum 2009;38:366–71. [DOI] [PubMed] [Google Scholar]
- 7. Crowson CS, Rollefstad S, Ikdahl E, Kitas GD, van Riel PL, Gabriel SE, et al. Impact of risk factors associated with cardiovascular outcomes in patients with rheumatoid arthritis. Ann Rheum Dis 2018;77:48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. López‐Mejías R, Castañeda S, González‐Juanatey C, Corrales A, Ferraz‐Amaro I, Genre F, et al. Cardiovascular risk assessment in patients with rheumatoid arthritis: the relevance of clinical, genetic and serological markers. Autoimmun Rev 2016;15:1013–30. [DOI] [PubMed] [Google Scholar]
- 9. McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, et al. Genome‐wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 2008;9:356–69. [DOI] [PubMed] [Google Scholar]
- 10. López‐Mejías R, Carmona FD, Castañeda S, Genre F, Remuzgo‐Martínez S, Sevilla‐Perez B, et al. A genome‐wide association study suggests the HLA class II region as the major susceptibility locus for IgA vasculitis. Sci Rep 2017;7:5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 2014;506:376–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO III, et al. 2010 rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum 2010;62:2569–81. [DOI] [PubMed] [Google Scholar]
- 13. Purcell S, Neale B, Todd‐Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole‐genome association and population‐based linkage analyses. Am J Hum Genet 2007;81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, et al. Next‐generation genotype imputation service and methods. Nat Genet 2016;48:1284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 2016;48:1279–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Corrales A, González‐Juanatey C, Peiró ME, Blanco R, Llorca J, González‐Gay MA. Carotid ultrasound is useful for the cardiovascular risk stratification of patients with rheumatoid arthritis: results of a population‐based study. Ann Rheum Dis 2014;73:722–7. [DOI] [PubMed] [Google Scholar]
- 17. Corrales A, Parra JA, González‐Juanatey C, Rueda‐Gotor J, Blanco R, Llorca J, et al. Cardiovascular risk stratification in rheumatic diseases: carotid ultrasound is more sensitive than Coronary Artery Calcification Score to detect subclinical atherosclerosis in patients with rheumatoid arthritis. Ann Rheum Dis 2013;72:1764–70. [DOI] [PubMed] [Google Scholar]
- 18. Gonzalez‐Juanatey C, Llorca J, Garcia‐Porrua C, Martin J, Gonzalez‐Gay MA. Effect of anti–tumor necrosis factor α therapy on the progression of subclinical atherosclerosis in severe rheumatoid arthritis. Arthritis Rheum 2006;55:150–3. [DOI] [PubMed] [Google Scholar]
- 19. Naredo E, Möller I, Gutiérrez M, Bong DA, Cobo T, Corominas H, et al. Multi‐examiner reliability of automated radio frequency‐based ultrasound measurements of common carotid intima‐media thickness in rheumatoid arthritis. Rheumatology (Oxford) 2011;50:1860–4. [DOI] [PubMed] [Google Scholar]
- 20. Touboul PJ, Hennerici MG, Meairs S, Adams H, Amarenco P, Bornstein N, et al. Mannheim carotid intima‐media thickness consensus (2004‐2006): an update on behalf of the Advisory Board of the 3rd and 4th Watching the Risk Symposium, 13th and 15th European Stroke Conferences, Mannheim, Germany, 2004, and Brussels, Belgium, 2006. Cerebrovasc Dis 2007;23:75–80. [DOI] [PubMed] [Google Scholar]
- 21. Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication‐based analysis for two‐stage genome‐wide association studies. Nat Genet 2006;38:209–13. [DOI] [PubMed] [Google Scholar]
- 22. Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 2012;22:1790–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. 1000 Genomes Project Consortium , Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schofield EC, Carver T, Achuthan P, Freire‐Pritchett P, Spivakov M, Todd JA, et al. CHiCP: a web‐based tool for the integrative and interactive visualization of promoter capture Hi‐C datasets. Bioinformatics 2016;32:2511–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Leonard D, Svenungsson E, Dahlqvist J, Alexsson A, Ärlestig L, Taylor KE, et al. Novel gene variants associated with cardiovascular disease in systemic lupus erythematosus and rheumatoid arthritis. Ann Rheum Dis 2018;77:1063–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Carmona FD, Mackie SL, Martín JE, Taylor JC, Vaglio A, Eyre S, et al. A large‐scale genetic analysis reveals a strong contribution of the HLA class II region to giant cell arteritis susceptibility. Am J Hum Genet 2015;96:565–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mayes MD, Bossini‐Castillo L, Gorlova O, Martin JE, Zhou X, Chen WV, et al. Immunochip analysis identifies multiple susceptibility loci for systemic sclerosis. Am J Hum Genet 2014;94:47–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Raychaudhuri S, Sandor C, Stahl EA, Freudenberg J, Lee HS, Jia X, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet 2012;44:291–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gene Ontology Consortium . Gene Ontology Consortium: going forward. Nucleic Acids Res 2015;43:D1049–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. The Gene Ontology Consortium , Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. Gene ontology: tool for the unification of biology. Nat Genet 2000;25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res 2003;13:2129–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta‐Cepas J, et al. STRING v10: protein‐protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sokolove J, Brennan MJ, Sharpe O, Lahey LJ, Kao AH, Krishnan E, et al. Brief report: citrullination within the atherosclerotic plaque: a potential target for the anti‐citrullinated protein antibody response in rheumatoid arthritis. Arthritis Rheum 2013;65:1719–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mackey RH, Kuller LH, Deane KD, Walitt BT, Chang YF, Holers VM, et al. Rheumatoid arthritis, anti‐CCP positivity, and cardiovascular disease risk in the Women's Health Initiative. Arthritis Rheumatol 2015;67:2311–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Javierre BM, Burren OS, Wilder SP, Kreuzhuber R, Hill SM, Sewitz S, et al. Lineage‐specific genome architecture links enhancers and non‐coding disease variants to target gene promoters. Cell 2016;167:1369–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dieguez‐Gonzalez R, Akar S, Calaza M, Perez‐Pampin E, Costas J, Torres M, et al. Genetic variation in the nuclear factor κB pathway in relation to susceptibility to rheumatoid arthritis. Ann Rheum Dis 2009;68:579–83. [DOI] [PubMed] [Google Scholar]
- 37. Fenwick C, Na SY, Voll RE, Zhong H, Im SY, Lee JW, et al. A subclass of Ras proteins that regulate the degradation of IκB. Science 2000;287:869–73. [DOI] [PubMed] [Google Scholar]
- 38. Baldwin AS. Control of oncogenesis and cancer therapy resistance by the transcription factor NF‐κB. J Clin Invest 2001;107:241–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. López‐Mejías R, García‐Bermúdez M, González‐Juanatey C, Castañeda S, Miranda‐Filloy JA, Gómez‐Vaquero C, et al. NFKB1‐94ATTG ins/del polymorphism (rs28362491) is associated with cardiovascular disease in patients with rheumatoid arthritis. Atherosclerosis 2012;224:426–9. [DOI] [PubMed] [Google Scholar]
- 40. Barnabe C, Martin BJ, Ghali WA. Systematic review and meta‐analysis: anti–tumor necrosis factor α therapy and cardiovascular events in rheumatoid arthritis. Arthritis Care Res (Hoboken) 2011;63:522–9. [DOI] [PubMed] [Google Scholar]
- 41. Greenberg JD, Kremer JM, Curtis JR, Hochberg MC, Reed G, Tsao P, et al. Tumour necrosis factor antagonist use and associated risk reduction of cardiovascular events among patients with rheumatoid arthritis. Ann Rheum Dis 2011;70:576–82. [DOI] [PubMed] [Google Scholar]
- 42. Hürlimann D, Forster A, Noll G, Enseleit F, Chenevard R, Distler O, et al. Anti‐tumor necrosis factor‐α treatment improves endothelial function in patients with rheumatoid arthritis. Circulation 2002;106:2184–7. [DOI] [PubMed] [Google Scholar]
- 43. Mäki‐Petäjä KM, Hall FC, Booth AD, Wallace SM, Yasmin, Bearcroft PW, et al. Rheumatoid arthritis is associated with increased aortic pulse‐wave velocity, which is reduced by anti‐tumor necrosis factor‐α therapy. Circulation 2006;114:1185–92. [DOI] [PubMed] [Google Scholar]
- 44. Macian F. NFAT proteins: key regulators of T‐cell development and function. Nat Rev Immunol 2005;5:472–84. [DOI] [PubMed] [Google Scholar]
- 45. Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol 1997;15:707–47. [DOI] [PubMed] [Google Scholar]
- 46. Shaw JP, Utz PJ, Durand DB, Toole JJ, Emmel EA, Crabtree GR. Identification of a putative regulator of early T cell activation genes. Science 1988;241:202–5. [DOI] [PubMed] [Google Scholar]
- 47. Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells. Cell 2002;109 Suppl:S67–79. [DOI] [PubMed] [Google Scholar]
- 48. Graef IA, Chen F, Crabtree GR. NFAT signaling in vertebrate development. Curr Opin Genet Dev 2001;11:505–12. [DOI] [PubMed] [Google Scholar]
- 49. Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev 2003;17:2205–32. [DOI] [PubMed] [Google Scholar]
- 50. López‐Mejías R, Corrales A, Genre F, Hernández JL, Ochoa R, Blanco R, et al. Angiopoietin‐2 serum levels correlate with severity, early onset and cardiovascular disease in patients with rheumatoid arthritis. Clin Exp Rheumatol 2013;31:761–6. [PubMed] [Google Scholar]
- 51. Ross R. Atherosclerosis: an inflammatory disease. N Engl J Med 1999;340:115–26. [DOI] [PubMed] [Google Scholar]
- 52. Arya R, Escalante A, Farook VS, Restrepo JF, Battafarano DF, Almeida M, et al. A genetic association study of carotid intima‐media thickness (CIMT) and plaque in Mexican Americans and European Americans with rheumatoid arthritis. Atherosclerosis 2018;271:92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Karpouzas GA, Malpeso J, Choi TY, Li D, Munoz S, Budoff MJ. Prevalence, extent and composition of coronary plaque in patients with rheumatoid arthritis without symptoms or prior diagnosis of coronary artery disease. Ann Rheum Dis 2014;73:1797–804. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


