Abstract
Background
Treatment with apremilast has recently demonstrated clinically meaningful improvement in moderate hidradenitis suppurativa (HS).
Objective
To evaluate the change in expression of inflammatory markers in lesional skin of HS patients receiving apremilast 30 mg twice daily (n = 15) for 16 weeks compared with placebo (n = 5).
Methods
At baseline, 5‐mm punch biopsies were obtained from an index lesion (HSL) and non‐lesional (HSN) skin in the same anatomical area. Subsequent HSL samples were taken as close as possible to the previously biopsied site at week 4 and week 16. After sampling, biopsies were split; one half was processed for in vivo mRNA analysis using real‐time quantitative PCR; the other half was cultured for ex vivo protein analysis using a proximity extension assay (Olink). Linear mixed effects models were calculated to compare the levels of inflammatory markers in HSL skin between apremilast and placebo over time.
Results
At baseline, 17 proteins with a fold change >2 in HSL vs. HSN skin were identified in 20 patients. The top five were IL‐17A (5), S100A12, CST5, IL‐12/23p40, CD6 (1) with fold changes ranging from 6.6 to 1638, respectively (FDR <0.044). Linear mixed effects models for 75 assays were calculated. Protein levels of S100A12 decreased during treatment in the apremilast group compared with the placebo group (p = 0.014, FDR = 0.186). None of the 14 genes exhibited significant changes in expression over time. However, an evident downward trend in relative mRNA expression of IL‐17A and IL‐17F was demonstrated in patients receiving apremilast.
Conclusion
We did not detect statistically significant changes in inflammatory markers in HSL skin of HS patients receiving apremilast compared with placebo, despite clinical improvement in the apremilast group. Nonetheless, S100A12 and IL‐17A were significantly elevated in HSL skin and showed a decrease in response to apremilast. The translational model in clinical trials involving HS clearly needs further improvement.
Introduction
Hidradenitis suppurativa (HS) is a chronic, recurrent, auto‐inflammatory skin disorder with limited effective treatment options.1 Recently, treatment with apremilast for 16 weeks has demonstrated clinically meaningful improvement in moderate HS.2 In this study (NCT03238469; EudraCT 2016‐000859‐27), twenty patients with moderate HS (HS‐PGA score of 3) were randomized in a 3 : 1 ratio to receive blinded treatment of apremilast 30 mg twice daily or placebo, respectively. Demographic and disease characteristics were similar between treatment arms. The mean age of the study population was 35 years (range: 21–64), most patients were female (17/20) and current smoker (16/20), the mean BMI was 32.7 ± standard deviation (SD) 6.2, and the mean abscess and inflammatory nodule (AN) count was 6.0 ± 1.8. The objective of this translational study was to evaluate the change in expression of inflammatory markers in lesional skin of HS patients receiving apremilast 30 mg twice daily for 16 weeks compared with placebo.
Material and Methods
Ethical statement
This investigator‐initiated trial was approved by the medical ethics committee of the Erasmus University Medical Center Rotterdam, The Netherlands (NL.57003.078.16), and conducted according to the Declaration of Helsinki principles. All patients provided written informed consent for study participation and use of their skin biopsies.
Biopsy procedures and sample processing
At baseline, two 5‐mm punch biopsies were taken: one was obtained from an erythematous, indurated, inflamed lesion, i.e. the index lesion (HSL), and one from the normal‐appearing unaffected non‐lesional (HSN) skin in the same anatomical area (Fig. 1). Subsequent HSL skin samples were taken as close as possible to the previously biopsied index site at week 4 (early response) and week 16 (end of study). After sampling, biopsies were split whereby one half was processed for mRNA analysis of the biopsy, representing the in vivo gene expression, and the other half was cultured for ex vivo protein analysis. The supernatants and the biopsies in 250 μL lysis buffer containing 1% ß‐mercaptoethanol were transferred to a polypropylene tube and stored at −20°C until further analysis.
Figure 1.
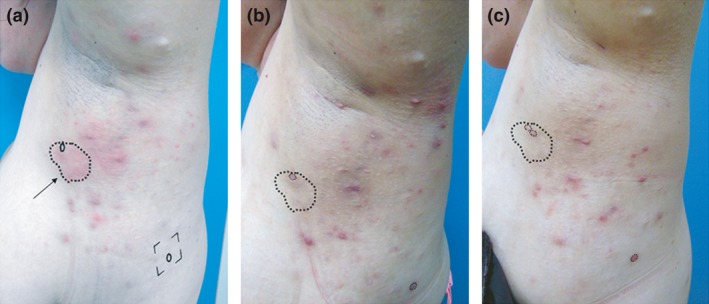
Biopsy procedure in a patient with a good clinical response to apremilast. Note that all deep‐seated lesions have resolved. Erythematous lesions seen at week 16 are scars and superficial folliculitis. Panel (a; left) Baseline. A 5‐mm punch biopsy was obtained from an erythematous, indurated, inflamed lesion (arrow pointing to index lesion). An additional 5‐mm punch biopsy of the non‐lesional skin was taken from the same anatomical area to serve as control (circle within the square). Panel (b; middle) Week 4 and Panel (c; right) Week 16. Subsequent skin samples were taken as close as possible to the previously biopsied index site (small circles within dashed line).
Ex vivo skin culture and protein quantification
After sampling, half of the 5‐mm biopsy was cultured for 24 h at 37°C in an atmosphere of 5% CO2 and 98% humidity in a transwell system (Netwell; Costar, Cambridge, MA, USA) as previously described.3, 4 In short, samples were placed in a 12‐well plate with the epidermis exposed to the air and the dermis immersed in 1 mL Iscove's modified Dulbecco's medium (Gibco, Paisley, UK) containing 0.5% human AB serum, penicillin (100 U/mL) and streptomycin (100 U/mL). A broad spectrum of 92 inflammatory markers was measured in the supernatant using the Olink Proseek Multiplex Inflammation panel (Olink Proteomics, Uppsala, Sweden).
In vivo gene expression
Total RNA was isolated using the GenElute Mammalian Total RNA Miniprep Kit (Sigma‐Aldrich, St. Louis, MA, USA), treated with 0.1 U/μL DNAse (Invitrogen, Carlsbad, CA, USA) and cDNA was synthesized with SuperScript II reverse transcriptase, random hexamer primers (Invitrogen) and oligo(dT)15 (Promega, Madison, WI, USA). Primers and probes were designed and chosen using ProbeFinder Software and the Universal Probe Library (Roche Applied Science, Indianapolis, IN, USA). ABL1 was used as housekeeping gene. Real‐time quantitative PCR for 14 genes (IFN‐ɣ, TNF‐α, IL‐1ß, IL‐6, IL‐8, IL‐10, IL‐23p19, IL‐12/23p40, IL‐17A, IL‐17F, IL‐31, CXCL‐10, CCL‐5 and MPO) was performed with the ViiA7 System and using the QuantStudio Real‐Time PCR Software version 1.3 (Applied Biosystems, Waltham, MA, USA).
Statistical analysis
Protein data are presented as normalized protein expression (NPX), which are log2‐transformed arbitrary units. Relative mRNA expression levels for week 4 and week 16 were calculated using the 2−ΔΔCT method using a baseline value of 1 for each individual sample.5
First, differentially expressed proteins at baseline were explored by calculating a fold change with the median NPX of HSL skin divided by the median NPX of HSN skin. If the fold change was more than 2, a Wilcoxon signed rank test was subsequently used to pairwise compare protein levels between HSL and HSN skin. Second, linear mixed effects models and associated ANOVA for repeated measures were calculated to compare inflammatory protein and mRNA expression levels between apremilast and placebo over time. Fixed effects were treatment, time and treatment × time. Third, the Spearman's rho was used to assess translational linkage, i.e. correlation between the levels of inflammation and the AN count.
A two‐sided P‐value below 0.05 was considered significant. P‐values were adjusted for multiple testing within each test using the Benjamini‐Hochberg approach. Statistical analyses were conducted in R Statistical Software version 3.5 (R Foundation for Statistical Computing, Vienna, Austria) using the lmerTest package.
Results
At baseline, 17 proteins with a fold change >2 were identified (Table 1). The top‐five were IL‐17A (5), S100A12, CST5, IL‐12p40, CD6 (1) with fold changes ranging from 6.6 to 1638, respectively (FDR ≤0.044). Out of 92 Olink assays, 75 were included in the linear mixed effects models. The assays removed had less than 25% detectability including TNF‐α and IFN‐γ. Only the protein levels of S100A12 decreased over time in the apremilast vs. placebo group, statistically non‐significant after correction for multiple testing (P = 0.014, FDR = 0.186; Fig. 2).
Table 1.
Proteins with a fold change >2 in HSL vs. HSN skin at baseline
| # | Protein | Fold change HSL/HSN | NPX HSL n = 20 | NPX HSN n = 20 | Unadj. P‐value | Adj. P‐value | Missing data | ||
|---|---|---|---|---|---|---|---|---|---|
| Median | IQR | Median | IQR | FDR | % | ||||
| 1 | CD6 | 1638 | 3.5 | 2.0–4.1 | 0.002 | 0.0–0.2 | 3.4 e‐07 | 0.018 | None |
| 2 | IL‐12p40 | 38 | 0.9 | 0.5–1.8 | 0.024 | 0.0–0.1 | 7.2 e‐06 | 0.038 | 7.5 |
| 3 | CST5 | 23 | 0.9 | 0.6–1.1 | 0.041 | 0.0–0.1 | 6.1 e‐07 | 0.024 | 1.1 |
| 4 | S100A12 | 11 | 4.5 | 3.1–6.3 | 0.4 | 0.4–0.8 | 2.7 e‐05 | 0.041 | 20.0 |
| 5 | IL‐17A | 6.6 | 3.1 | 1.8–3.8 | 0.5 | 0.5–0.6 | 2.9 e‐05 | 0.044 | 26.3 |
| 6 | TNFSF14 | 4.5 | 3.4 | 2.5–4.1 | 0.8 | 0.4–1.0 | 1.0 e‐06 | 0.029 | 6.3 |
| 7 | AXIN1 | 4.4 | 1.9 | 1.7–2.1 | 0.4 | 0.4–0.5 | 8.8 e‐07 | 0.026 | 8.8 |
| 8 | β‐NGF | 3.2 | 1.1 | 0.6–1.4 | 0.3 | 0.3–0.6 | 5.8 e‐04 | 0.047 | 15.0 |
| 9 | TNFSF11 | 3.1 | 2.2 | 1.2–3.3 | 0.7 | 0.7–0.7 | 4.6 e‐07 | 0.021 | 28.8 |
| 10 | CD244 | 3.0 | 2.7 | 2.3–3.1 | 0.9 | 0.9–0.9 | 1.1 e‐07 | 0.012 | 23.8 |
| 11 | CD5 | 3.0 | 7.3 | 5.9–7.6 | 2.5 | 2.2–2.6 | 1.3 e‐08 | 0.003 | None |
| 12 | TNFRSF9 | 2.9 | 6.4 | 5.2–7.6 | 2.2 | 1.8–2.7 | 1.3 e‐08 | 0.006 | 1.3 |
| 13 | OSM | 2.9 | 6.4 | 6.0–7.8 | 2.2 | 1.3–3.4 | 4.0 e‐06 | 0.035 | 3.8 |
| 14 | SIRT2 | 2.8 | 5.0 | 4.5–5.4 | 1.8 | 1.5–1.9 | 1.9 e‐07 | 0.015 | 5.0 |
| 15 | GDNF | 2.7 | 2.9 | 2.0–3.8 | 1.1 | 0.8–1.2 | 1.6 e‐06 | 0.032 | 5.0 |
| 16 | IL‐18R1 | 2.3 | 4.2 | 3.6–5.2 | 1.8 | 1.7–2.0 | 1.3 e‐08 | 0.009 | None |
| 17 | ST1A1 | 2.1 | 1.5 | 0.8–1.8 | 0.7 | 0.5–1.2 | 1.3 e‐02 | 0.050 | 10.0 |
Missing data: samples with NPX‐values below background level throughout all samples, i.e. HSN and HSL baseline, HSL week 4 and HSL week 16. NPX: a high NPX value corresponds with a high protein concentration. Unadj. P‐value: unadjusted P‐value by Wilcoxon signed rank, NPX HSL vs. NPX HSN. Adj. P‐value (FDR): adjusted P‐value Benjamini Hochberg test, NPX HSL vs. NPX HSN.
FDR: false discovery rate; HS: hidradenitis suppurativa; HSL: HS lesional; HSN: HS non‐lesional; IQR: interquartile range; NPX: normalized protein expression.
Figure 2.
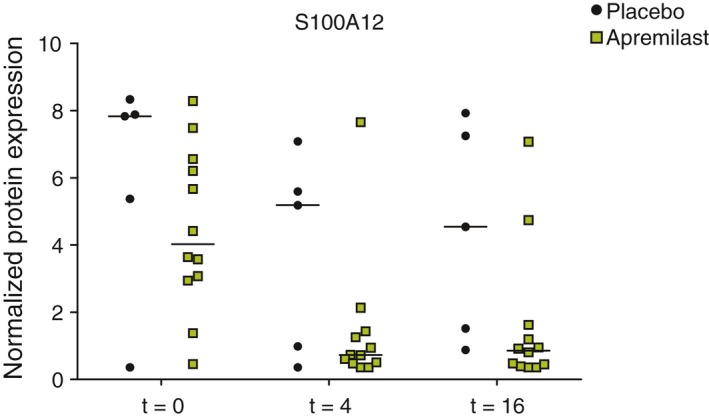
Protein levels of S100A12 in patients receiving apremilast (n = 15) and placebo (n = 5), linear mixed effect model: P = 0.014, FDR = 0.186. t: week. Data at week 16 for two patients receiving apremilast are missing as these two patients discontinued after respectively week 4 and week 8.
None of the 14 genes analysed by linear mixed effects models yielded significant differences. Only the relative mRNA expression of IL‐17A and IL‐17F demonstrated an evident but non‐significant downward trend in patients receiving apremilast vs. placebo (P > 0.05; Fig. 3). In addition, there was no correlation between S100A12 protein levels or IL‐17A/F mRNA expression and the favourable clinical response in the apremilast group as measured by the AN count (data not shown).
Figure 3.
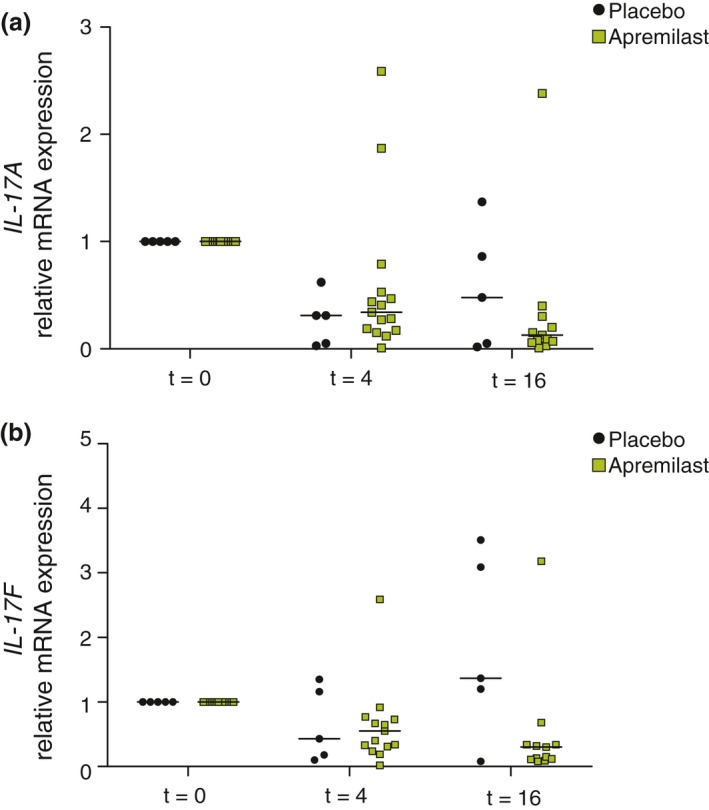
Relative mRNA expression levels of IL‐17A and IL‐17F in patients receiving apremilast (n = 15) and placebo (n = 5). IL: interleukin; t: week. Panel (a; above) IL‐17A. At t = 16 in the apremilast group: one data point is out of the y‐axis range. Linear mixed effect model: P > 0.05. Panel (b; below). IL‐17F. At both t = 4 and t = 16 in the apremilast group, one data point is out of the y‐axis range. Linear mixed effect model: P > 0.05. Data at week 16 for two patients receiving apremilast are missing as these two patients discontinued after respectively week 4 and week 8.
Discussion
This translational study aimed to identify inflammatory skin markers in response to apremilast 30 mg twice daily for 16 weeks in patients with moderate HS, which showed positive clinical results and have been published previously.2 However, S100A12 protein levels and IL‐17A and IL‐17F mRNA expression showed a clear but non‐significant decrease in patients receiving apremilast compared with placebo.
S100A12 (calgranulin C, EN‐RAGE) is a pro‐inflammatory, calcium‐binding antimicrobial protein that interacts with the receptor for advanced glycation end products (RAGE) and is overexpressed in various inflammatory skin diseases, e.g. psoriasis.6, 7 Previously, S100A8 and S100A9 (calprotectin or calgranulin A/B) have been demonstrated to be elevated in the serum and lesional skin of HS patients.8, 9 It is known that IL‐17 is a potent inducer of S100A8 and S100A9 in keratinocytes.10, 11 The other way around, S100A12 stimulates T cells to produce IL‐17A.12 Thus, elevated levels of IL‐17A and S100A12 in HSL skin at baseline and the responsiveness of these inflammatory markers to apremilast treatment is not surprising. Moreover, apremilast's impact on the Th17 pathway was recently demonstrated by the significant reduction of IL‐17A and IL‐17F protein levels in the plasma of psoriasis patients.13
The non‐significant findings using a broad panel of inflammatory markers may be explained by the regression to the mean phenomenon, particularly at the individual level.13 The highly inflammatory index lesions could have spontaneously improved over time in the context of the fluctuating nature of the disease, especially in the placebo group. The effect of regression to the mean increases with larger measurement variability.14 Expanding the number of biopsies from index lesions, representing several degrees of inflammation, would theoretically be a solution. However, taking more biopsies would not be feasible for ethical reasons.
Another explanation for the decreased levels of inflammation over time in both groups could be that the inflammatory infiltrate of the index lesion was gradually reduced by successively taking biopsies from the same nodule. Moreover, the substrate for the foreign body inflammation such as hair fragments and keratin fibres, may have also been (partially) removed by the biopsy procedure.15 In addition, this exploratory study might be underpowered which may also have resulted in non‐significant results.
Previously, two prospective uncontrolled studies investigating infliximab and ustekinumab in HS were also unable to link translational data to the clinical response.16, 17 However, in these studies, only inflammatory markers in serum were investigated. Strengths of our study include the placebo‐controlled trial design, the use of skin (target organ) instead of serum or plasma, and the standardized biopsy procedure of an index lesion. A limitation is the small study population.
In conclusion, this translational study investigating apremilast in moderate HS did not detect statistically significant changes in important inflammatory markers in lesional skin compared with placebo over 16 weeks of treatment, despite positive clinical findings. Nonetheless, related S100A12 and IL‐17A were significantly elevated in HSL skin and showed a decline only in the apremilast group. In addition, our findings highlight the challenge of assessing pharmacodynamics in the skin in a highly fluctuating inflammatory disease. A better translational model in clinical trials involving HS has yet to be developed.
Acknowledgements
We thank Eddy Florencia and Patrick Asmawidjaja for providing technical assistance in processing the skin samples.
Conflicts of interest
A.R.J.V.V., N.D., A.M.C.M.: no conflicts of interest to declare. H.H.v.d.Z.: advisory board member for AbbVie, InflaRX and Galderma. M.B.A.v.D.: consultant, speaker, principal investigator or received grants from Abbvie, Celgene, Eli Lilly, Janssen‐Cilag, Novartis, Pfizer, Cutanea Life Sciences, Idera Pharmaceuticals. E.P.P.: consultant, speaker, principal investigator or received grants from Abbvie, Amgen, Biogen, Celgene, Eli Lilly, Janssen‐Cilag, Novartis, Pfizer and UCB.
Funding sources
This study was sponsored by Celgene (Summit, NJ, USA).
Ethical review board approval: IRB of the Erasmus University Medical Center (NL.57003.078.16, MEC‐2016‐377)
Trial registration number: NCT03049267 on ClinicalTrials.gov, registered 25 October 2016.
References
- 1. Zouboulis CC, Bechara FG, Dickinson‐Blok JL et al Hidradenitis suppurativa/acne inversa: a practical framework for treatment optimization ‐ systematic review and recommendations from the HS ALLIANCE working group. J Eur Acad Dermatol Venereol 2018. 10.1111/jdv.15233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vossen ARJV, van Doorn MBA, van der Zee HH, Prens EP. Apremilast for moderate hidradenitis suppurativa: results of a randomized controlled trial. J Am Acad Dermatol 2018. pii: S0190‐9622(18)32202–3. [DOI] [PubMed] [Google Scholar]
- 3. Wei L, Debets R, Hegmans JJ et al IL‐1 beta and IFN‐gamma induce the regenerative epidermal phenotype of psoriasis in the transwell skin organ culture system. IFN‐gamma up‐regulates the expression of keratin 17 and keratinocyte transglutaminase via endogenous IL‐1 production. J Pathol 1999; 187: 358–364. [DOI] [PubMed] [Google Scholar]
- 4. Companjen AR, van der Wel LI, Wei L et al A modified ex vivo skin organ culture system for functional studies. Arch Dermatol Res 2001; 293: 184–190. [DOI] [PubMed] [Google Scholar]
- 5. Rao X, Huang X, Zhou Z et al An improvement of the 2^(‐delta delta CT) method for quantitative real‐time polymerase chain reaction data analysis. Biostat Bioinforma Biomath 2013; 3: 71–85. [PMC free article] [PubMed] [Google Scholar]
- 6. Eckert RL, Broome AM, Ruse M et al S100 proteins in the epidermis. J Invest Dermatol 2004; 123: 23–33. [DOI] [PubMed] [Google Scholar]
- 7. Wilsmann‐Theis D, Wagenpfeil J, Holzinger D et al Among the S100 proteins, S100A12 is the most significant marker for psoriasis disease activity. J Eur Acad Dermatol Venereol 2016; 30: 1165–1170. [DOI] [PubMed] [Google Scholar]
- 8. Lima AL, Karl I, Giner T et al Keratinocytes and neutrophils are important sources of proinflammatory molecules in hidradenitis suppurativa. Br J Dermatol 2016; 174: 514–521. [DOI] [PubMed] [Google Scholar]
- 9. Wieland CW, Vogl T, Ordelman A et al Myeloid marker S100A8/A9 and lymphocyte marker, soluble interleukin 2 receptor: biomarkers of hidradenitis suppurativa disease activity? Br J Dermatol 2013; 168: 1252–1258. [DOI] [PubMed] [Google Scholar]
- 10. Cho KA, Suh JW, Lee KH et al IL‐17 and IL‐22 enhance skin inflammation by stimulating the secretion of IL‐1beta by keratinocytes via the ROS‐NLRP3‐caspase‐1 pathway. Int Immunol 2012; 24: 147–158. [DOI] [PubMed] [Google Scholar]
- 11. Bai B, Yamamoto K, Sato H et al Complex regulation of S100A8 by IL‐17, dexamethasone, IL‐4 and IL‐13 in HaCat cells (human keratinocyte cell line). J Dermatol Sci 2007; 47: 259–262. [DOI] [PubMed] [Google Scholar]
- 12. Kessel C, Lippitz K, Weinhage T et al Proinflammatory cytokine environments can drive interleukin‐17 overexpression by gamma/delta T cells in systemic juvenile idiopathic arthritis. Arthritis Rheumatol 2017; 69: 1480–1494. [DOI] [PubMed] [Google Scholar]
- 13. Garcet S, Nograles K, Correa da Rosa J et al Synergistic cytokine effects as apremilast response predictors in patients with psoriasis. J Allergy Clin Immunol 2018; 142: 1010–1013.e6. [DOI] [PubMed] [Google Scholar]
- 14. Barnett AG, van der Pols JC, Dobson AJ. Regression to the mean: what it is and how to deal with it. Int J Epidemiol 2005; 34: 215–220. [DOI] [PubMed] [Google Scholar]
- 15. van der Zee HH, de Ruiter L, Boer J et al Alterations in leucocyte subsets and histomorphology in normal‐appearing perilesional skin and early and chronic hidradenitis suppurativa lesions. Br J Dermatol 2012; 166: 98–106. [DOI] [PubMed] [Google Scholar]
- 16. Blok JL, Li K, Brodmerkel C et al Ustekinumab in hidradenitis suppurativa: clinical results and a search for potential biomarkers in serum. Br J Dermatol 2016; 174: 839–846. [DOI] [PubMed] [Google Scholar]
- 17. Montaudie H, Seitz‐Polski B, Cornille A et al Interleukin 6 and high‐sensitivity C‐reactive protein are potential predictive markers of response to infliximab in hidradenitis suppurativa. J Am Acad Dermatol 2017; 76: 156–158. [DOI] [PubMed] [Google Scholar]


