Abstract
Objectives
The pathogenesis of Graves’ disease (GD) remains unclear. In terms of environmental factors, GD development may be associated with chronic inflammation caused by alteration of the intestinal flora. This study explored the association of intestinal flora alteration with the development of GD among the Han population in southwest China.
Design and methods
Fifteen GD patients at the Affiliated Hospital of Zunyi Medical College between March 2016 and March 2017 were randomly enrolled. Additionally, 15 sex- and age-matched healthy volunteers were selected as the control group during the same period. Fresh stool samples were collected, and bacterial 16S RNA was extracted and amplified for gene sequencing with the Illumina MiSeq platform. The sequencing results were subjected to operational taxonomic unit-based classification, classification verification, alpha diversity analysis, taxonomic composition analysis and partial least squares-discriminant analysis (PLS-DA).
Results
The diversity indices for the GD group were lower than those for the control group. The GD group showed significantly higher abundances of Firmicutes, Proteobacteria and Actinobacillus and a higher Firmicutes/Bacteroidetes ratio than the control group. PLS-DA suggested the satisfactory classification of the flora between the GD group and the control group. The abundances of the genera Oribacterium, Mogibacterium, Lactobacillus, Aggregatibacter and Mogibacterium were significantly higher in the GD group than in the control group (P < 0.05).
Conclusions
The intestinal flora of GD patients was significantly different from that of the healthy population. Thus, alteration of intestinal flora may be associated with the development of GD.
Keywords: autoimmune thyroid disease, Graves’ disease, gut microbiota, 16S RNA
Introduction
Graves’ disease (GD) is an organ-specific autoimmune thyroid disease (AITD) (1, 2) and the most common cause of hyperthyroidism in iodine-sufficient geographical areas (3). According to the literature, the prevalence of autoimmune thyroiditis diseases varies according to ethnicity (4). GD has a prevalence rate of approximately 1.2% (0.5% overt and 0.7% subclinical) (3) and an annual incidence of 20–50 cases per 100,000 persons in the USA (5), and the incidence among the population in China is approximately 1.2% (6). GD can lead to damage in various systems, such as the nervous system, somatosensory system and cardiovascular system and can even induce life-threatening thyroid crisis.
Risk factors for GD include genetic factors, environmental factors and immune factors (7, 8). However, the exact mechanisms underlying the development of GD remain unclear. Segmented filamentous bacteria (SFB) may be effective inducers of Th17 cells in the intestinal mucosal lamina propria (9), whereas Clostridium IV and XIVa can induce colonic Treg cells (10), indicating that alteration of the intestinal flora may lead to a Th17/Treg imbalance in AITD. Scholars have proposed a classic molecular simulation mechanism: infectious agents have a molecular structure similar to that of the antigenic determinants of thyroid-stimulating hormone (TSH) receptors, resulting in cross-reactivity of antibodies at the level of the TSH receptor (e.g., TSH receptor-like molecules in Yersinia enterocolitica can increase the risk of GD) (11).
The intestinal flora has been called the second human genome; its alteration is associated with abnormal development and metabolic disorders (12). The different bacteria that compose the intestinal flora play active roles in metabolism, immune development and host defense against pathogens. In addition, these microorganisms participate in intestinal mucosal immunity and autoimmunity, function as key regulators of the immune system, play roles in the differentiation of different effector and memory T cell populations and are necessary for the production of proinflammatory (e.g., IL-17) and anti-inflammatory molecules (e.g., IL-10); therefore, the intestinal microflora are associated with a variety of autoimmune and inflammatory diseases via interactions between the innate and adaptive immune systems (13, 14, 15). The thyroid gland is the largest endocrine-regulating organ in humans. However, studies on the potential association and interactions between the thyroid gland and the intestinal flora have been reported only rarely. Although Zhou et al. found that alterations in the diversity and similarity of the gut microbiota occurred in hyperthyroid patients compared with healthy individuals (16), their results need to be further validated and more systemic studies need to be conducted.
To clarify this concept, the current study investigated the possible differences in the structure and composition of the intestinal flora between GD patients and healthy populations. All the participants were from the Han population in southwest China. The results of this study may provide a reference for future research on the association between intestinal flora and thyroid diseases.
Materials and methods
Study subjects
Fifteen patients who were newly diagnosed with GD at the Affiliated Hospital of Zunyi Medical College between March 2016 and March 2017 were randomly selected; these patients constituted the GD group. The patients’ ages ranged from 46 to 55 years. In addition, 15 healthy age- and sex-matched volunteers who underwent medical examination during the same period constituted the control group. All the participants were from the Han population. GD diagnosis was in accordance with the diagnostic criteria described in the Guidelines for the Diagnosis and Treatment of Thyroid Diseases in China (2008) (17). The control group participants had no medical or family history of thyroid diseases and had normal thyroid function and negative autoantibody responses. They had no inflammatory diseases or hypersensitivity. In addition, in each group, participants who met any of the following criteria were excluded from this study: (1) the presence of osteoporosis, pregnancy or autoimmune diseases, such as type 1 diabetes, systemic lupus erythematosus, rheumatoid arthritis and autoimmune hepatitis; (2) the presence of infectious disease in the digestive tract, oral cavity, respiratory tract or urinary tract; (3) recent use of antibiotics, probiotics or other medicine (such as metformin, acarbose or herbal preparations) and food that could possibly influence the intestinal flora; (4) the use of any anti-thyroid gland medicine before recruitment and (5) the existence of current stress.
This study was approved by the Ethics Committee of the Affiliated Hospital of Zunyi Medical College. Informed written consent was obtained from each participant.
Methods
Sampling
Fresh stool was sampled in the morning on the second day after recruitment. Approximately 2 g was removed from the interior of the middle segment of the stool. The samples were stored at −80°C until use in the following experiments.
Stool flora DNA extraction
DNA of the stool flora was extracted with an OMEGA Soil DNA kit (Omega Bio-Tek, Norcross, GA, USA) in strict accordance with the kit instructions.
Polymerase chain reaction (PCR)
The sequences of microbial rRNA that can reflect the flora composition and diversity were used as targets, and sample-specific barcode sequences were used for PCR amplification of the variable regions of the 16S rRNA gene (Q5 HiFi DNA polymerase; NEB, Ipswich, MA, USA). The amplification products were subjected to 2% agarose gel electrophoresis, and the target segments were recovered with a gel recovery kit (Axygen, Union City, CA, USA). Fluorescence quantitation was performed with a Quant-iT PicoGreen dsDNA assay kit (Thermo Fisher). Sequencing libraries were prepared with a TruSeq Nano DNA LT Library Prep kit (Illumina).
Sequencing
Sequencing was performed with the Illumina MiSeq platform. Qualified sequencing libraries underwent 2 × 300 bp double-end sequencing with a MiSeq Reagent Kit V3 as the corresponding reagent (600 cycles). For rRNAs that contained multiple conserved regions and hypervariable regions, primers were designed based on the conserved regions to amplify a single variable region or multiple variable regions of the rRNA gene. Then, sequencing was performed to analyze microbial diversity.
Statistical analysis
The original high-flux sequencing data were initially screened according to the sequence quality. The sequences were identified and then assigned to corresponding samples according to the primer and barcode information. Operational taxonomic unit (OTU)-based classification was performed on the obtained sequences; the representative sequence of each OTU was used for taxonomic identification. The diversity of each sample was evaluated based on the abundances of the OTUs in the sample. The absolute composition of each sample at different classification levels was analyzed, and significant differences were tested. Measurement data between groups were compared using t tests. P < 0.05 was considered indicative of significance.
On the basis of the outcomes of statistical analysis, the differences in the flora structure and species between samples were further assessed. A sequence similarity of 97% was the threshold set for OTU classification, corresponding to the allowable sequence difference at the taxonomic species level (18). Alpha diversity was assessed based on the Chao1, abundance-based coverage estimator (ACE), Shannon and Simpson indices (18). Partial least squares-discriminant analysis (PLS-DA) was performed on the basis of the species abundance matrix and the maximum covariance of the assigned classification information, and the samples were resequenced in a low-dimensional coordinate system. R software was used to construct the PLS-DA model based on the species abundance matrix and the classification data for the samples.
Results
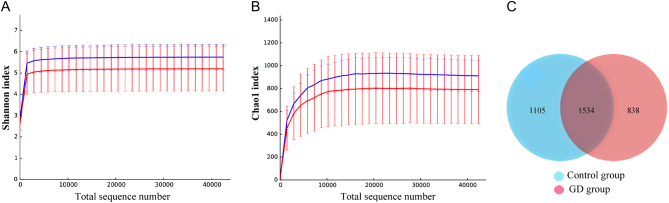
A total of 658,223 high-quality sequences were obtained via Illumina sequencing, with an average of 65,822 ± 3817 valid sequences. A total of 329,741 sequences were obtained from the control group, and 328,482 sequences were obtained from the GD group for phylogenetic analysis. The Chao1, ACE, Shannon and Simpson indices of the GD group showed lower values than those of the control group, although the differences were not significant (P > 0.05; Table 1). The curves showed that the control group had a greater abundance of species in the intestinal flora than did the GD group, although the difference was not significant (Fig. 1A and B). Based on the OTU abundance matrix, the number of OTUs common to both groups was calculated with R software, and a Venn diagram was constructed (Fig. 1C). In total, 3477 OTUs are shown in the diagram: 1534 were shared by both groups, 838 belonged only to the GD group and 1105 were unique to the control group.
Table 1.
Alpha diversity based on operational taxonomic units (OTUs).
| Group | Number of valid sequences | Number of OTUs attributable to known classification units (%) | Alpha diversity | |||
|---|---|---|---|---|---|---|
| ACE index | Chao1 index | Shannon index | Simpson index | |||
| Control | 329741 | 100 | 917.08 | 911.31 | 5.75 | 0.927 |
| GD | 328482 | 100 | 792.10 | 790.26 | 5.20 | 0.889 |
GD, Graves’ disease.
Figure 1.
Rarefaction curves and a Venn diagram of the number of OTUs. (A) Shannon curves for the control (blue) and GD (red) groups. Control group: Ave 5.753 and Err 0.599. GD group: Ave 5.209 and Err 1.054. (B) Chao1 curves for the control (blue) and GD (red) groups: new species were observable with increased sequencing depth. For a total sequence number of 42379, 902.460 and 780.380 sequences were observed in the control and GD groups, respectively. (C) Venn diagram for the OTUs. The overlapping region indicates the number of OTUs shared by both groups, and the nonoverlapping regions indicate the number of OTUs belonging to only one group.
A PLS-DA model was constructed based on the species abundance matrix and the grouped sample data, and the typical outcomes are shown in Fig. 2 (the results of the remaining 10 GD and ten control samples are provided in Supplementary Fig. 1, see section on supplementary data given at the end of this article). As shown in Fig. 2, the grouped samples were separated, which should be attributed to the presence of different sets, thus indicating that the classification model in this study had satisfactory performance and could differentiate the GD group from the control group.
Figure 2.
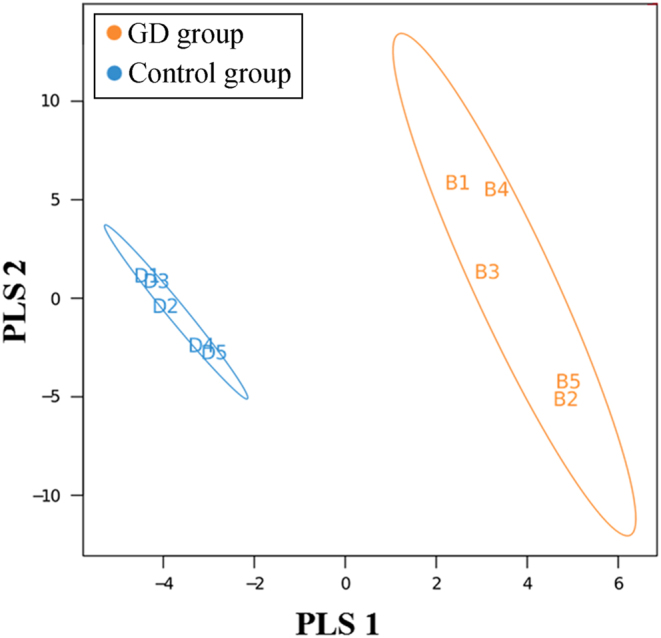
The PLS-DA model diagram of five samples from the GD group and five from the control group. Each point in the diagram represents one sample. Points of the same color belong to the same group, and points in the same group are enclosed in an ellipse. The distance between the two ellipses indicates that the model achieved satisfactory classification performance.
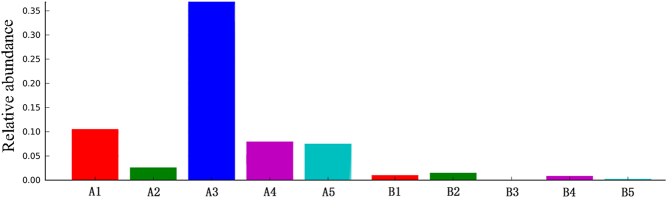
To analyze bacterial 16S rRNA genes, the Greengenes database was used as the template sequence for the OTUs to describe the taxonomic composition of the intestinal flora. The analysis identified a total of nine phyla, namely, Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, Fusobacteria, Verrucomicrobia, Cyanobacteria, TM7 bacteria and Tenericutes. Among these bacterial phyla, Firmicutes accounted for the highest proportion and was found in up to 72.0% of the sequences; Firmicutes, Bacteroidetes and Proteobacteria were found in 91.3% of the sequences. The GD group showed a significantly higher proportion of Firmicutes and a significantly lower proportion of Bacteroidetes than the control group (Fig. 3 and Supplementary Fig. 2). The number of sequences of each classification unit at the phylum and genus levels were compared between the groups based on the Metastats statistical algorithm (19) with Mothur software. The GD group and the control group did not show significant differences at the phylum level (P > 0.05). However, at the genus level, the GD group had significantly higher abundances of Oribacterium, Mogibacterium, Lactobacillus, and Aggregatibacter than the control group (P < 0.05). Linear discriminant analysis effect size (LEfSe) was performed, and according to the screening outcomes, Lactobacillus was the critical flora member. The bar chart shows that the abundance of Lactobacillus was significantly higher in the 15 stool samples from the GD group than in the samples from the control group (P < 0.05; Fig. 4 and Supplementary Fig. 3). The relative abundances of Prevotella in the GD group and the control group were 2.08 and 0.14%, respectively, although the difference was not significant.
Figure 3.
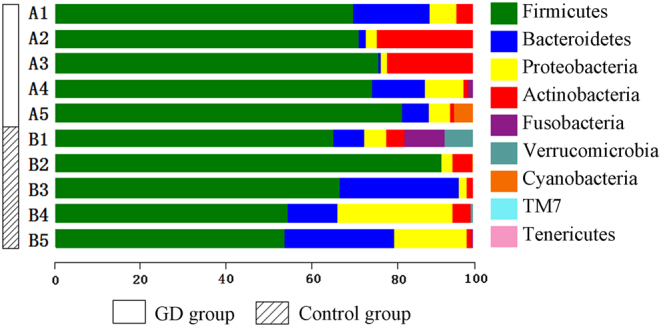
The composition and abundance distribution of the community classification at the phylum level based on the analysis of the five samples from the GD group and five from the control group. The horizontal axis indicates the relative abundance of each phylum. A longer bar of a certain color indicates a higher relative abundance of that phylum in the corresponding sample. Each horizontal bar represents a sample, and the phyla are differentiated by different colors. GD, Graves’ disease.
Figure 4.
The relative abundance of Lactobacillus in five samples from the GD group and five from the control group. (A) GD group. (B) The control group.
Discussion
To date, the pathogenesis of GD remains unclear. In recent years, numerous studies have proved that the interactions of epigenetic, environmental and immune factors play an important role in the development of GD (20, 21, 22). Among these factors, the infection factor has attracted increasing research attention. Yersinia infections are associated with the development of GD (12), and cross-reactions can occur between anti-Yersinia antibodies and the TSH receptor in GD patients (21, 22).
Recent studies have found that environmental factors promoting the development of GD are associated with chronic inflammation caused by variations in the intestinal flora (16, 23). Zhou et al. found that compared with healthy individuals, in patients with hyperthyroidism, the abundance of the intestinal microbial flora increases, the proportions of Bifidobacterium and Lactobacillus decrease, and the proportion of Enterococci increases (16). However, our findings contradicted these findings, possibly because of the following reasons: (1) the patients enrolled in their study were from different regions than those enrolled in our study; and (2) the severity of disease between their study and our study was different.
In this study, we used the 454 pyrosequencing-based technique to detect variations in flora in stool samples from GD patients; our findings may elucidate the pathogenesis of GD and treatment methods for GD. Compared with traditional microbial techniques, next-generation high-flux sequencing technology allows innovative studies on microorganisms and overcomes the limitations of traditional technologies for bacterial research, thus improving the ability to identify important intestinal microorganisms that cause GD and thereby providing a more comprehensive profile of the stool flora associated with GD. In this study, the structure and diversity of the intestinal flora were explored according to the abundance and uniformity of OTUs based on 16S rRNA gene sequences. We found that the species abundance in the control group was significantly higher than that in the GD group, which allowed the two groups to be clearly separated. This result indicates that the flora structure is significantly different between GD patients and healthy individuals. The proportion of Firmicutes was significantly lower in the control group than in the GD group, whereas the proportion of Bacteroidetes was significantly higher in the control group than in the GD group. Furthermore, the ratio of Firmicutes to Bacteroidetes in the control group was significantly lower than that in the GD group. In addition, our study showed that at the genus level, the abundances of Oribacterium, Mogibacterium, Lactobacillus and Aggregatibacter in the GD group were significantly higher than those in the control group (P < 0.05). Although no significant difference in the abundance of Prevotella was observed, the relative abundance of Prevotella in the GD group was higher than that in the control group, and this finding was consistent with that reported in the literature (24). Lactobacilli have been proved to be beneficial bacteria that help protect Th17 cells and support the barrier function of the intestinal mucosa by secreting IL-17 and IL-22. A recent study of macaques found that Lactobacillus regulated the Th17 axis by inhibiting the activity of indoleamine 2,3-dioxygenase 1 (25). A Th17/Treg imbalance causes inflammatory disorders, indicating that the imbalance of the immune-inflammation axis participates in inflammatory reactions and target organ damage. Numerous studies have proved the involvement of Th17 cells in autoimmune diseases, such as inflammatory bowel disease, rheumatoid arthritis, psoriasis and asthma (26, 27, 28, 29). In this study, the abundance of Lactobacillus in the GD group was higher than that in the control group, suggesting that the development of GD may be associated with Th17 cells. However, an association between intestinal flora and Th17 cells in GD patients remains to be confirmed. In new GD patients, the number of Th17 cells among PBMCs, the level of IL-17 secreted by PBMCs and the expression of ROR-γt mRNA were significantly increased, and these increases were associated with the severity of the disease (30, 31, 32). IL-17+ cell infiltration is found in the thyroid gland of GD patients, further suggesting the possible involvement of Th17 cells in the development of GD (33, 34, 35).
Hyperthyroidism is a common condition that is difficult to cure completely. Although the development of modern medical science has resulted in great changes in the diagnosis and treatment of autoimmune diseases, including GD, the pathogeneses of these diseases have not been fully elucidated, and abnormal thyroid function-related indices tend to occur repeatedly during treatment. Furthermore, although current treatment approaches for treating GD can achieve reliable curative effect, there are still concerns on the part of clinicians with regard to the choice of the treatment protocol due to safety considerations. Therefore, more safe and effective treatment methods for GD are needed. Recent studies have evidenced the possible association of alterations of the intestinal flora with the development of GD (11, 20). This association is also supported by the results of this study, which may provide new strategies for exploration into the treatment modalities of GD in clinical practice.
Although some novel and unambiguous findings were noted, this study had some limitations. First, this study suffered from a relatively small sample size due to relatively strict exclusion criteria. To overcome this limitation, multicenter studies with a larger sample size need to be conducted in the future. Second, this study was preliminary, and a negative control group with other AITDS, a control group with a different cause of hyperthyroidism (such as autonomous adenoma and iatrogenesis) and a control group with another endocrine-related, rather than the thyroid gland, autoimmune disease were not included. Therefore, our results remain to be validated by enrolling these groups in future studies.
In conclusion, the abundances of Firmicutes, Proteobacteria and Actinobacillus, as well as the Firmicutes/Bacteroidetes ratio, are increased in GD patients. At the genus level, the abundances of Oribacterium, Mogibacterium, Lactobacillus, Aggregatibacter and Mogibacterium are significantly increased. Thus, GD patients are subject to alterations of the intestinal flora. The results of this study may provide a theoretical reference in the search for novel microbial biomarkers and augment the understanding of the influence of the intestinal flora on GD.
Supplementary Material
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
This work was supported by the National Natural Science Foundation of China (grant number 81560147) and the Science and Technology Support Program of Guizhou Province (contract number: CK-1127). Part of the work was done in the Key Laboratory of Basic Pharmacology of the Ministry of Education and the Joint International Research Laboratory of Ethnomedicine of the Ministry of Education at Zunyi Medical University.
References
- 1.Kahaly GJ, Bartalena L, Hegedüs L. The American Thyroid Association/American Association of Clinical Endocrinologists guidelines for hyperthyroidism and other causes of thyrotoxicosis: an European perspective. Thyroid 2011. 21 . ( 10.1089/thy.2011.2106.ed3) [DOI] [PubMed] [Google Scholar]
- 2.Zheng HJ, Wei P, Wei JP. New research on Graves’ disease. Chinese Journal of Immunology 2017. 33 . ( 10.3969/j.issn.1000-484X.2017.04.030) [DOI] [Google Scholar]
- 3.Hollowell JG, Staehling NW, Flanders WD, Hannon WH, Gunter EW, Spencer CA, Braverman LE, Serum TSH. T4, and thyroid antibodies in the United States population (1988 to 1994): national health and nutrition examination survey (NHANES III). Journal of Clinical Endocrinology and Metabolism 2002. 87 . ( 10.1210/jcem.87.2.8182) [DOI] [PubMed] [Google Scholar]
- 4.McLeod DS, Caturegli P, Cooper DS, Matos PG, Hutfless S. Variation in rates of autoimmune thyroid disease by race/ethnicity in US military personnel. JAMA 2014. 311 . ( 10.1001/jama.2013.285606) [DOI] [PubMed] [Google Scholar]
- 5.Zimmermann MB, Boelaert K. Iodine deficiency and thyroid disorders. Lancet: Diabetes and Endocrinology 2015. 3 . ( 10.1016/S2213-8587(14)70225-6) [DOI] [PubMed] [Google Scholar]
- 6.Lu ZY, Zhong NS. Internal Medicine, pp . Beijing, China: People’s Medical Publishing House, 2008. [Google Scholar]
- 7.Lee HJ, Li CW, Hammerstad SS, Stefan M, Tomer Y. Immunogenetics of autoimmune thyroid diseases: a comprehensive review. Journal of Autoimmunity 2015. 64 . ( 10.1016/j.jaut.2015.07.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiersinga WM. Clinical relevance of environmental factors in the pathogenesis of autoimmune thyroid disease. Endocrinology and Metabolism 2016. 31 . ( 10.3803/EnM.2016.31.2.213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaboriau-Routhiau V, Rakotobe S, Lécuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, et al The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 2009. 31 . ( 10.1016/j.immuni.2009.08.020) [DOI] [PubMed] [Google Scholar]
- 10.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011. 331 . ( 10.1126/science.1198469) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heyma P, Harrison LC, Robins-Browne R. Thyrotrophin (TSH) binding sites on Yersinia enterocolitica recognized by immunoglobulins from humans with Graves’ disease. Clinical and Experimental Immunology 1986. 64 . [PMC free article] [PubMed] [Google Scholar]
- 12.Grice EA, Segre JA. The human microbiome: our second genome. Annual Review of Genomics and Human Genetics 2012. 13 . ( 10.1146/annurev-genom-090711-163814) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X, Zou Q, Zeng B, Fang Y, Wei H. Analysis of fecal Lactobacillus community structure in patients with early rheumatoid arthritis. Current Microbiology 2013. 67 . ( 10.1007/s00284-013-0338-1) [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, Wu X, Li J, Tang L, Li Y, et al The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nature Medicine 2015. 21 . ( 10.1038/nm.3914) [DOI] [PubMed] [Google Scholar]
- 15.Coretti L, Natale A, Cuomo M, Florio E, Keller S, Lembo F, Chiariotti L, Pero R. The interplay between defensins and microbiota in Crohn’s disease. Mediators of Inflammation 2017. 2017 8392523 ( 10.1155/2017/8392523) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou L, Li XL, Ahme A, Wu D, Liu L, Qiu J, Yan Y, Jin M, Xin Y. Gut microbe analysis between hyperthyroid and healthy individuals. Current Microbiology 2014. 69 . ( 10.1007/s00284-014-0640-6) [DOI] [PubMed] [Google Scholar]
- 17.Chinese Society of Endocrinology. Guidelines for Diagnosis and Treatment of Thyroid Diseases in China, p 47 Eds Teng WP, Zeng ZP, Li GW. & Ning G. Beijing, China: Chinese Society of Endocrinology, 2008. [Google Scholar]
- 18.Blaxter M, Mann J, Chapman T, Thomas F, Whitton C, Floyd R, Abebe E. Defining operational taxonomic units using DNA barcode data. Philosophical Transactions of the Royal Society of London: Series B, Biological Sciences 2005. 360 . ( 10.1098/rstb.2005.1725) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White JR, Ngarajan N, Pop M. Statistical methods for detecting differential abundant features in clinical metagenomic samples. PLoS Computational Biology 2009. 5 e1000352 ( 10.1371/journal.pcbi.1000352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen CR, Zhao JJ, Xu MY, Tang JF, Li FY, Chen JL. Studies on the relationship between Yersinia infection and the pathogenesis of Graves’ disease. Chinese Journal of Endocrinology and Metabolism 1998. 14 . ( 10.3760/j.issn:1000-6699.1998.03.005) [DOI] [Google Scholar]
- 21.Kalmykova EN, Dergunova ES, Ermoleava TN, Gorshkova RP, Komandrova NA. Piezoquartz immunosensors for assessing the interactions between Yersinia enterocolitica, lipopolysaccharides and antibodies to them. Prikladnaia Biokhimiia i Mikrobiologiia 2008. 44 . ( 10.1134/S0003683808040066) [DOI] [PubMed] [Google Scholar]
- 22.Hochel I, Skvor J. Characterization of rabbit antibodies for immunochemical detection of Yersinia enterocolitica. Folia Microbiologica 2007. 52 . ( 10.1007/BF02932112) [DOI] [PubMed] [Google Scholar]
- 23.Bongers G, Pacer ME, Geraldino TH, Chen L, He Z, Hashimoto D, Furtado GC, Ochando J, Kelley KA, Clemente JC, et al Interplay of host microbiota, genetic perturbations, and inflammation promotes local development of intestinal neoplasms in mice. Journal of Experimental Medicine 2014. 211 . ( 10.1084/jem.20131587) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han L. Detection the Frequency of Th17 Cell and the Special Bacteria in Intestinal Flora of Graves Disease Patients. Dissertation, Lanzhou University, 2017. [Google Scholar]
- 25.Vujkovic-Cvijin I, Swainson LA, Chu SN, Ortiz AM, Santee CA, Petriello A, Dunham RM, Fadrosh DW, Lin DL, Faruqi AA, et al Gut-resident lactobacillus abundance associates with IDO1 inhibition and Th17 dynamics in SIV-infected macaques. Cell Reports 2015. 13 . ( 10.1016/j.celrep.2015.10.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitra A, Raychaudhuri SK, Raychaudhuri SP. IL-17 and IL-17: an auspicious therapeutic target for psoriatic disease. Actas Dermo-Sifiliográficas 2014. 105 . [DOI] [PubMed] [Google Scholar]
- 27.Sato KTh. Cells and rheumatoid arthritis-from the stand-point of osteoclast differentiation. Allergology International 2008. 57 . ( 10.2332/allergolint.R-07-158) [DOI] [PubMed] [Google Scholar]
- 28.Hundorfean G, Neurath MF, Mudter J. Functional relevance of Thelper 17 (Th17) cells and the IL-17 cytokine family in inflammatory bowel disease. Inflammatory Bowel Diseases 2012. 18 . ( 10.1002/ibd.21677) [DOI] [PubMed] [Google Scholar]
- 29.Cosmi L, Liotta F, Maggi E, Romagnani S, Annunziato F. Th17 cells: new players in asthma pathogenesis. Allergy 2011. 66 . ( 10.1111/j.1398-9995.2011.02576.x) [DOI] [PubMed] [Google Scholar]
- 30.Horie I, Abiru N, Saitoh O, Ichikawa T, Iwakura Y, Eguchi K, Nagayama Y. Distinct role of T helper Type 17 immune response for Graves’ hyperthyroidism in mice with different genetic backgrounds. Autoimmunity 2011. 44 . ( 10.3109/08916931003777247) [DOI] [PubMed] [Google Scholar]
- 31.Bossowski A, Moniuszko M, Idźkowska E, Grubczak K, Singh P, Bossowska A, Diana T, Kahaly GJ. Decreased proportions of CD4 + IL17+/CD4 + CD25 + CD127− and CD4 + IL17+/CD4 + CD25 + CD127 − FoxP3+ T cells in children with autoimmune thyroid diseases. Autoimmunity 2016. 49 320328 ( 10.1080/08916934.2016.1183654) [DOI] [PubMed] [Google Scholar]
- 32.Li C, Yuan J, Zhu YF, Yang XJ, Wang Q, Xu J, He ST, Zhang JA. Imbalance of Th17/Treg in different subtypes of autoimmune thyroid diseases. Cellular Physiology and Biochemistry 2016. 40 . ( 10.1159/000452541) [DOI] [PubMed] [Google Scholar]
- 33.Zhang YN, Zhou XN, An J, Zhu TN, Zhao RJ. [The change of Th17 in patients with Graves’ disease]. Chinese Journal of Immunology 2012. 28 . ( 10.3969/j.issn.1000-484X.2012.07.014) [DOI] [Google Scholar]
- 34.Peng D, Xu B, Wang Y, Guo H, Jiang Y. A high frequency of circulating Th22 and Th17 cells in patients with new onset Graves’ disease. PLoS ONE 2013. 8 e68446 ( 10.1371/journal.pone.0068446) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin J, Zhou J, Fan C, Zhao N, Liu Y, Wang S, Cui X, Huang M, Guan H, Li Y, et al Increased circulating Th17 but decreased CD4+Foxp3+Treg and CD19+CD1dhiCD5+ Breg subsets in new-onset Graves’ disease. BioMed Research International 2017. 2017 8431838 ( 10.1155/2017/8431838) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



 This work is licensed under a
This work is licensed under a