Abstract
Receptor occupancy, the ratio between amount of drug bound and amount of total receptor on single cells, is a biomarker for treatment response to therapeutic monoclonal antibodies. Receptor occupancy is traditionally measured by flow cytometry. However, spectral overlap in flow cytometry limits the number of markers that can be measured simultaneously. This restricts receptor occupancy assays to the analysis of major cell types, although rare cell populations are of potential therapeutic relevance. We therefore developed a receptor occupancy assay suitable for mass cytometry. Measuring more markers than currently available in flow cytometry allows simultaneous receptor occupancy assessment and high‐parameter immune phenotyping in whole blood, which should yield new insights into disease activity and therapeutic effects. However, varying sensitivity across the mass cytometer detection range may lead to misinterpretation of the receptor occupancy when drug and receptor are detected in different channels. In this report, we describe a method for optimization of mass cytometry receptor occupancy measurements by using antibody‐binding quantum simply cellular (QSC) beads for standardization across channels with different sensitivities. We evaluated the method in a mass cytometry‐based receptor occupancy assay for natalizumab, a therapeutic antibody used in multiple sclerosis treatment that binds to α4‐integrin, which is expressed on leukocyte cell surfaces. Peripheral blood leukocytes from a treated patient were stained with a panel containing metal‐conjugated antibodies for detection of natalizumab and α4‐integrin. QSC beads with known antibody binding capacity were stained with the same metal‐conjugated antibodies and were used to standardize the signal intensity in the leukocyte sample before calculating receptor occupancy. We found that QSC bead standardization across channels corrected for sensitivity differences for detection of drug and receptor and generated more accurate results than observed without standardization. © 2019 The Authors. Cytometry Part A published by Wiley Periodicals, Inc. on behalf of International Society for Advancement of Cytometry.
Keywords: receptor occupancy, biomarkers, QSC beads, CyTOF, standardization, optimization, multiple sclerosis, natalizumab, quantitative analysis, mass cytometry
Receptor occupancy (RO) by therapeutic monoclonal antibodies is a potential biomarker for therapeutic response and may support dose optimization in precision medicine 1, 2. RO assays generally involve measuring bound drug relative to total target receptor on single cells by flow cytometry. Mass cytometry has rapidly evolved to become a relevant tool in several fields of translational clinical research 3, 4, 5, 6. In mass cytometry, antibodies are conjugated to purified metal isotopes instead of fluorophores, which dramatically reduces signal overlap and allows simultaneous detection of more than 40 parameters in individual cells by inductively‐coupled plasma mass spectrometry 7. Mass cytometry permits measurement of RO in conjunction with more markers, and in more cell types of interest, than is currently possible by flow cytometry. In order to be useful, estimation of RO using mass cytometry must be reliable and reproducible. Mass cytometers have varying sensitivity over the detection range of metal isotopes (up to fivefold difference in CyTOF 1 and 2, lower in Helios), and each mass cytometer has its own sensitivity pattern 8, 9. In a RO assay, differences in detection sensitivity of anti‐drug and anti‐receptor antibodies will result in either over‐ or underestimation of the RO, depending on which antibody is detected in the most sensitive channel.
Quantum simply cellular (QSC) beads are cell‐sized particles with known antibody binding capacity that were developed for flow cytometry to enable determination of absolute numbers of cellular epitopes 10. We aimed to obtain more accurate RO estimation in mass cytometry by employing QSC beads for standardization across channels with varying detection sensitivity. We evaluated the applicability of QSC bead standardization in a mass cytometry‐based RO assay for natalizumab. Natalizumab is a humanized monoclonal IgG4 antibody that binds to the α4 subunit of surface integrins on leukocytes, thereby preventing leukocytes from entering the central nervous system over the blood‐brain‐barrier. Natalizumab is used in the treatment of multiple sclerosis (MS) 11, and the natalizumab RO has been suggested as a biomarker for monitoring response to therapy 12, 13. The assay used here was adapted from a natalizumab RO assay previously published for flow cytometry 14 in which bound natalizumab was detected by an anti‐IgG4 and total α4 integrin was detected by an anti‐α4 integrin antibody that binds to a different epitope of the α4 integrin than natalizumab. We demonstrated how the different detection sensitivities for natalizumab and α4 integrin influenced the mass cytometry‐based RO assay results and how accurate and reproducible RO determination was achieved by standardization with QSC beads.
Materials and Methods
Subjects and Samples
The study was approved by the Regional Ethics Committee (approval REK 2016/579), and samples were collected after written informed consent from one healthy donor and one MS patient receiving natalizumab therapy (4 weeks after the last infusion) at the Department of Neurology, Haukeland University Hospital. Whole blood was obtained in heparinized vacutainer tubes (Greiner Bio‐One GmbH, Kremsmünster, Austria), incubated with Proteomic stabilizer (Smart Tube, Inc., San Carlos, CA) for 10 min according to the manufacturer's protocol, and stored at −80°C.
Mass Cytometry Antibody Panel and Titration
The 34 marker mass cytometry antibody panel (Supporting Information Table S1a) consisted of 25 metal‐conjugated antibodies purchased pre‐conjugated from Fluidigm (South San Francisco, CA) and nine antibodies purchased from Biolegend (San Diego, CA), R&D Systems, (Minneapolis, MN) and Abcam (Cambridge, Great Britain) that were conjugated to metal isotopes with the Maxpar Antibody Labeling Kit (Fluidigm) according to the manufacturer's protocol. In the RO assay, bound natalizumab was measured with an anti‐IgG4 (conjugated to 169Tm) specific for the Fc portion of human IgG4 and total α4 integrin was measured with anti‐α4 integrin (conjugated to 141Pr) specific for a different epitope than natalizumab (Fig. 1). Antibody titrations were performed on the patient's peripheral blood leukocytes (PBLs) under the same conditions as the experiment (barcoded samples, staining volume 100 μl, 1.5 × 106 cells), and anti‐IgG4 (169Tm) and anti‐α4 integrin (141Pr) were titrated to saturating concentrations. An antibody cocktail containing the complete panel except anti‐IgG4 and anti‐α4 integrin was pre‐made and stored in Maxpar Cell Staining Buffer (CSB; Fluidigm) in aliquots at −80°C for up to 9 days during which time the three replicate experiments were performed. Anti‐IgG4 and anti‐α4 integrin were added to the antibody cocktail aliquot on the day of the experiments.
Figure 1.
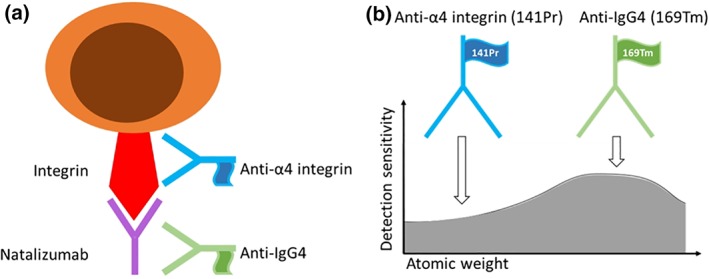
Natalizumab RO assay: (a) Natalizumab was detected with anti‐IgG4 (169Tm), and its receptor was detected with anti‐α4 integrin (141Pr). (b) Metal‐conjugated antibodies are detected with different sensitivity depending on the atomic weight of the metal tag (graph adapted from Tricot et al.). [Color figure can be viewed at wileyonlinelibrary.com]
Quality Control Experiments
Quality control (QC) experiments were performed on the same patient PBL sample under the same conditions as the main experiments. The following were analyzed:
Unstained samples
To examine whether PBLs contained any metals in the detection range of the mass cytometer (in MS patients gadolinium can originate from intravenous contrast used in MRI scans), an unstained aliquot of the barcoded PBL sample, with only the DNA intercalation reagent used for cell detection, was acquired.
Mass‐minus‐one controls
Two mass‐minus‐one (MMO) controls were performed to test for spillover from other markers in the panel into the channels used for detection of anti‐IgG4 and anti‐α4 integrin. Barcoded PBLs were stained with the complete panel except for either anti‐IgG4 or anti‐α4 integrin.
Biological negative control
PBLs from an untreated healthy donor were barcoded and pooled with patient PBLs to serve as a negative control for binding of anti‐IgG4 in the absence of natalizumab.
Positive control
Patient PBLs incubated with natalizumab, which were expected to have a RO of 100%, were used as a positive control. The same sample was also used as reference in some of the RO calculation methods.
Test of competitiveness between natalizumab and anti‐α4 integrin
To examine whether natalizumab and anti‐α4 integrin bound to different epitopes of α4 integrin without competition, detection of α4 integrin on PBLs from the healthy donor was compared with and without prior incubation with natalizumab.
Barcoding, Pooling, and Freezing of Aliquots
Whole blood samples stored in Proteomic stabilizer were thawed, and red blood cell lysis was performed with Thaw‐Lyse buffer 1 (Smart Tube, Inc.) according to the manufacturer's manual. A total of 6 × 106 PBLs from each sample were permeabilized and barcoded using the Cell‐ID 20‐Plex Pd Barcoding kit (Fluidigm) according to the manufacturer's protocol. The two samples were washed in Maxpar PBS (Fluidigm), pooled, and stored in three aliquots each containing 3 × 106 cells in Maxpar PBS with 10% DMSO (Dimethyl sulfoxide, Sigma‐Aldrich, Darmstadt, Germany) at −80°C for up to 9 days. All centrifugation steps were performed at room temperature at 800g.
In Vitro Incubation with Natalizumab and Antibody Staining
The in vitro incubation with natalizumab and antibody staining (Fig. 2a) was performed by the same operator in the same lab on three separate days. On each of the days, one aliquot of 3 × 106 barcoded and pooled PBLs was thawed and washed in Maxpar CSB. For optional incubation in vitro with natalizumab, the sample was split into two tubes, which were both incubated for 20 min at room temperature in Maxpar CSB with 100 U/ml heparin (LEO Pharma A/S, Ballerup, Denmark) to avoid nonspecific eosinophil antibody binding 15. To one of the tubes, natalizumab (Lot 28918, Biogen, Cambridge, Massachusetts) was added to a final concentration of 10 μg/ml. Both tubes were incubated for 30 min at room temperature with intermittent vortexing and washed thoroughly in Maxpar CSB. Prior to antibody staining, the number of cells in each tube was adjusted to 1 × 106, and cells were incubated again for 20 min at room temperature in Maxpar CSB with 100 U/ml heparin. Aliquots of the metal‐conjugated antibody cocktail were thawed, anti‐IgG4 and anti‐α4 integrin were added, and antibody staining was performed in a total volume of 100 μl. After 30‐min incubation at room temperature with intermittent vortexing, samples were washed with Maxpar CSB, and a 10‐min post‐staining fixation was performed in fresh 2% paraformaldehyde (Thermo Scientific, Waltham, MA) in Maxpar PBS at room temperature. Samples were washed with Maxpar PBS, resuspended in 1 ml of 125 nM Cell‐ID™ Intercalator‐Ir in Maxpar Fix and Perm Buffer (Fluidigm), and stored at 4°C for 3–4 h. Immediately prior to acquisition, PBL were washed in Maxpar PBS and Maxpar Water (Fluidigm), resuspended in 0.1× EQ Four Element Calibration Beads (Fluidigm) in Maxpar Water and filtered (Corning Falcon Test Tube with Cell Strainer Snap Cap, Fisher Scientific, Hampton, NN). All centrifugation steps were performed at room temperature at 800g.
Figure 2.
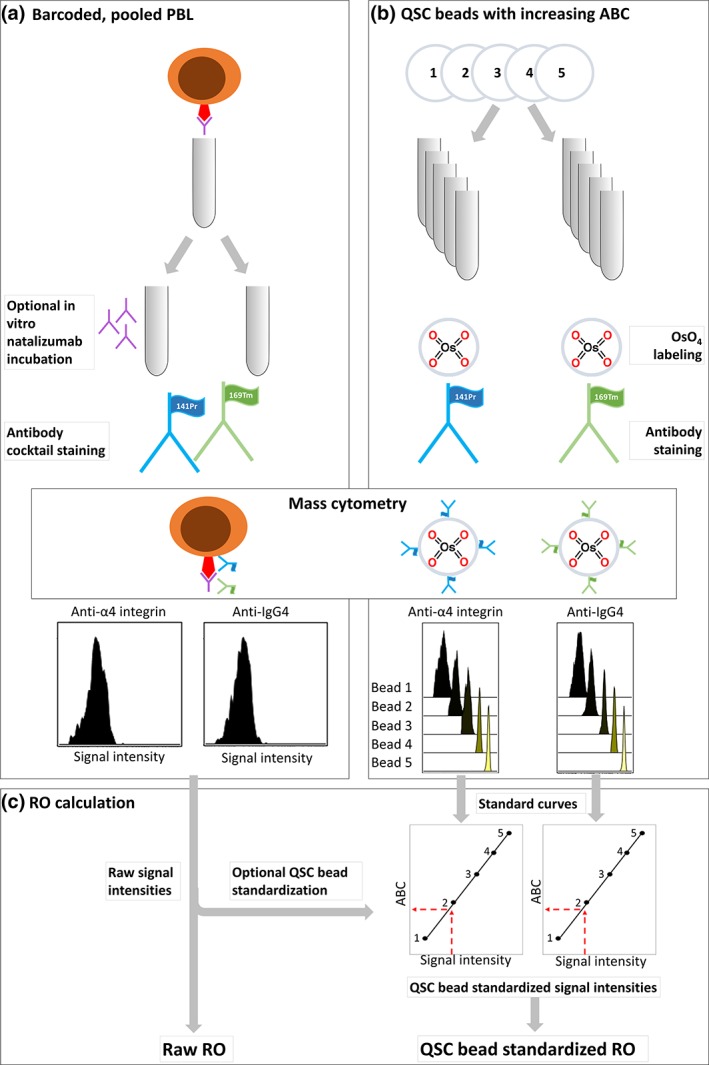
Experimental workflow: (a) peripheral blood leukocytes (PBLs) were split into two aliquots for optional in vitro incubation with natalizumab, stained with an antibody cocktail containing anti‐IgG4 and anti‐α4 integrin, and analyzed on a Helios mass cytometer. (b) Quantum simply cellular (QSC) beads with known antibody binding capacity (ABC) were labeled with OsO4, stained with anti‐IgG4 or anti‐α4 integrin, and acquired on the same mass cytometer on the same day. (c) Standard curves were created based on anti‐IgG4 and anti‐α4 integrin signal intensities from QSC beads with known ABC, and signal intensities of the same antibodies from the PBL samples were plotted into the standard curves for standardization before RO calculation. [Color figure can be viewed at wileyonlinelibrary.com]
Adaptation of QSC Bead Protocol for Mass Cytometry
QSC anti‐mouse beads (Cat code 815A, Bangs Laboratories Inc., Fishers, IN) with increasing antibody binding capacity (ABC) for mouse‐IgG were stained and acquired on the same days as PBL samples (Fig. 2b) and used to create standard curves of signal intensity (dual counts) from known numbers of anti‐IgG4 (169Tm) and anti‐α4 integrin (141Pr) (Fig. 2c). The QSC bead kit consisted of four bead populations with known ABC (12,319–814,348, lot 13,359). To cover the range of anti‐IgG4 (169Tm) and anti‐α4 integrin (141Pr) dual count values in our PBL samples, we purchased one additional QSC bead population with low ABC (2,685, lot 13,289), resulting in five QSC bead populations with ABC range of 2,685–814,548.
QSC beads are identified by forward and side scatter in flow cytometry, and they do not contain any metal in the detection range of the mass cytometer. To enable identification of QSC beads on the mass cytometer, the manufacturer's staining protocol was modified by addition of an osmium tetroxide (OsO4) labeling step prior to antibody staining. Four drops of each of the QSC bead populations were incubated separately with 0.01–0.001% OsO4 (CAS#20816‐12‐0, Electron Microscopy Sciences, Hatfield, PA) diluted in Maxpar PBS. After 30 min, beads were washed four times in Maxpar PBS, once in Maxpar CSB, and stained separately with 1 μg of either anti‐IgG4 (169Tm) or anti‐α4 integrin (141Pr) in a total volume of 100 μl Maxpar CSB for 30 min at room temperature. QSC beads were washed twice in Maxpar PBS and once in Maxpar Water, resuspended in 200 μl 0.1 × EQ Four Element Calibration Beads in Maxpar water, and filtered. The five QSC bead populations were kept separate through all staining and acquisition steps, and all centrifugation steps were performed at room temperature at 2500g.
A flow cytometry QC experiment was performed to examine whether OsO4 labeling affected the ABC of QSC beads. QSC beads with and without OsO4 labeling prior to antibody incubation with anti‐IgG‐PE (Abcam, Supporting Information Table S1b) were acquired on a flow cytometer (BD LSR Fortessa, BD Biosciences, Franklin Lakes, NJ). Apart from staining QSC beads with a fluorochrome labeled anti‐IgG4‐PE antibody, instead of a metal‐conjugated antibody, the QSC bead protocol described above was followed. To evaluate the correlation between QSC beads on mass and flow cytometry, QSC beads coated with anti‐IgG4 (169Tm) or anti‐IgG4‐PE were acquired by mass or flow cytometry, respectively, and signal intensities were compared.
Acquisition on the Helios® Instrument
In each of the three replicate experiments, freshly stained QSC beads and PBL samples were analyzed with the same standard settings on a Helios mass cytometer (Fluidigm). Before acquisition, tuning (CyTOF Tuning Solution, Fluidigm) and calibration (EQ Four Element Calibration Beads, Fluidigm) were performed according to the manufacture's guidelines. PBL sample acquisition was performed at a rate of 300–400 events per second.
Mass Cytometry Data Processing and Analysis
FCS files from analyses of QSC beads and PBL samples were normalized to the EQ beads using the Fluidigm normalizer (Fluidigm). Normalized QSC bead FCS files were exported to Cytobank software (Cytobank Inc., Santa Clara, CA) for gating and downstream analysis. Signal intensity (median dual counts) of QSC beads stained with anti‐IgG4 and anti‐α4 integrin, respectively, and the corresponding ABC values were plotted using QuickCal template (Bangs Laboratories) to create individual standard curves for the antibodies.
Normalized PBL sample FCS files were debarcoded (Fluidigm Debarcoder) and exported to Cytobank software for gating and downstream analysis. Cleanup gating was performed to obtain single PBL cells, and eight cell types of interest were identified by manual gating: memory B cells, monocytes, CD4 effector memory (TEM), central memory (TCM), effector memory RA (TEMRA) T cells, and CD8 TEM, TCM, and TEMRA cells.
In patient PBLs, 90th percentiles of anti‐IgG4 (169Tm) and anti‐α4 integrin (141Pr) dual counts in the eight cell types were exported for further calculations (Fig. 2c, left). Ninetieth percentiles were used instead of medians because of the bimodal distribution of α4 integrin and natalizumab in some cell types. For optional QSC bead standardization (Fig. 2c, right), these 90th percentiles were plotted against the respective standard curves in the QuickCal template and the corresponding ABC values, which will be referred to as QSC bead standardized signal intensities, were used in subsequent calculations. Untreated PBLs from the healthy donor were used as negative controls.
Calculation of Receptor Occupancy
In patient PBLs with and without in vitro natalizumab incubation, RO was calculated as a percent ratio between signal intensities of anti‐IgG4 (169Tm) and anti‐α4 integrin (141Pr) with and without QSC bead standardization (Fig. 2c):
-
i
Raw RO (ROraw) based on raw 90th percentile dual counts:
-
ii
QSC bead standardized RO (ROstandardized) based on QSC bead standardized signal intensities:
In patient PBLs not incubated with natalizumab, we calculated RO by an additional approach by determining the ratio between RO (as calculated above) in the sample and RO in the in vitro natalizumab saturated aliquot of the same sample:
-
ii
ROraw in the sample relative to ROraw in the 100% saturated aliquot:
-
ii
ROstandardized in the sample relative to ROstandardized in the 100% saturated aliquot:
Statistics
In the sample incubated in vitro with natalizumab, RO results were compared to 100% using a one‐sample t test. Otherwise, results from different RO calculation methods were compared using a paired t test. Statistical significance was defined as P < 0.05. We used R version 3.4.3 16 for statistical analysis and Inkspace (Free Software Foundation, Inc., Boston, MA) for illustrations.
Results
Quantification of Natalizumab and α4 Integrin in PBL Cell Subtypes
Memory B cells, monocytes, CD4 TEM, TCM, and TEMRA cells, and CD8 TEM, TCM, and TEMRA cells were gated in PBL samples as illustrated in Supporting Information Figure S1a. The 90th percentile dual counts of anti‐IgG4 (169Tm) and anti‐α4 integrin (141Pr) of these cell types in the patient PBL samples with and without natalizumab incubation in vitro (Supporting Information Fig. S1b) were exported for RO calculations.
QSC Beads Were Used for Standardization of Signal Intensities
QSC beads were gated as illustrated in Supporting Information Figure S2a. Median dual counts of anti‐IgG4 (169Tm) and anti‐α4 integrin (141Pr) in the five QSC bead populations were determined (Fig. 3; Supporting Information Fig. S2b) and used to create standard curves for each of the antibodies in the QuickCal Template. QSC bead standardization of the PBL samples was performed by plotting raw signal intensities (90th percentile dual counts) of anti‐IgG4 (169Tm) and anti‐α4 integrin (141Pr) in the cell types against the respective standard curves, as shown in detail in Supporting Information Figure S2c. OsO4 labeling of the QSC beads prior to antibody staining did not alter antibody binding capacity (Supporting Information Fig. S3a). Moreover, there was a linear correlation between ABC of the same QSC beads acquired by mass cytometry and by flow cytometry (Supporting Information Fig. S3b).
Figure 3.
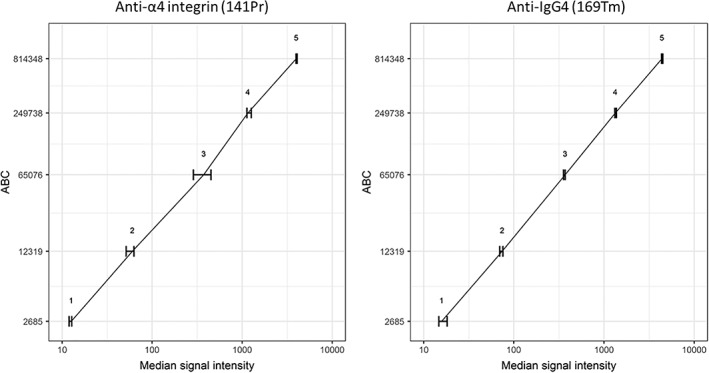
Median signal intensity of anti‐α4 integrin (141Pr) and anti‐IgG4 (169Tm) on QSC beads with known antibody binding capacity (ABC). Error bars show the range of measured signal intensities in three replicate experiments. The same data for each of the experiments are shown in Supporting Information Figure. S2b.
Bead Standardization Compensated for Overestimation of RO
Table 1 shows RO values from different calculation methods based on data collected on patient PBLs with and without in vitro incubation with natalizumab. Samples incubated with natalizumab were expected to have ROs of 100% in all cell subtypes. We compared two calculation methods (Fig. 4aa): ROraw and ROstandardized. We found that ROraw was significantly different from the expected 100% in all eight cell subtypes identified (p < 0.0001, median 126%, IQR: 116–130%), whereas ROstandardized was not significantly different from 100% (p = 0.45, median 101%, IQR: 94–106%).
Table 1.
Receptor occupancy (RO) in eight cell types in three replicate experiments with the same patient PBL sample
| (a) ROraw and ROstandardized in patient PBLs saturated in vitro with natalizumab with expected RO of 100%. | ||||||
|---|---|---|---|---|---|---|
| RO raw | RO standard zed | |||||
| Day 1 | Day 2 | Day 3 | Day 1 | Day 2 | Day 3 | |
| Mem B cell | 120% | 129% | 132% | 98% | 105% | 111% |
| Monocyte | 114% | 108% | 113% | 94% | 89% | 95% |
| CD4 T CM | 115% | 110% | 122% | 92% | 86% | 101% |
| CD4 T EM | 114% | 116% | 126% | 92% | 94% | 106% |
| CD4 T EMRA | 116% | 130% | 129% | 93% | 102% | 107% |
| CD8 T CM | 129% | 127% | 148% | 103% | 101% | 123% |
| CD8 T EM | 123% | 125% | 131% | 99% | 100% | 110% |
| CD8 T EMRA | 130% | 132% | 147% | 105% | 106% | 123% |
| (b) ROraw, ROstandardized, ROvs100%raw, and ROvs100%standardized in patient PBL with unknown RO (i.e., not incubated with natalizumab in vitro). | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RO raw | RO standardized | RO vs. 100% raw | RO vs. 100% standardized | |||||||||
| Day 1 | Day 2 | Day 3 | Day 1 | Day 2 | Day 3 | Day 1 | Day 2 | Day 3 | Day 1 | Day 2 | Day 3 | |
| Mem B cell | 101% | 95% | 96% | 82% | 77% | 81% | 84% | 73% | 73% | 83% | 73% | 73% |
| Monocyte | 76% | 72% | 76% | 62% | 59% | 64% | 67% | 67% | 67% | 66% | 66% | 67% |
| CD4 T CM | 73% | 77% | 82% | 57% | 60% | 68% | 63% | 70% | 67% | 62% | 69% | 67% |
| CD4 T EM | 77% | 75% | 79% | 61% | 59% | 66% | 68% | 64% | 63% | 66% | 63% | 62% |
| CD4 T EMRA | 95% | 89% | 94% | 75% | 69% | 78% | 82% | 68% | 73% | 81% | 68% | 73% |
| CD8 T CM | 85% | 84% | 93% | 67% | 66% | 78% | 66% | 66% | 63% | 65% | 66% | 63% |
| CD8 T EM | 86% | 88% | 87% | 68% | 70% | 73% | 70% | 70% | 66% | 69% | 70% | 66% |
| CD8 T EMRA | 82% | 87% | 86% | 65% | 69% | 72% | 63% | 66% | 59% | 62% | 65% | 59% |
Figure 4a.
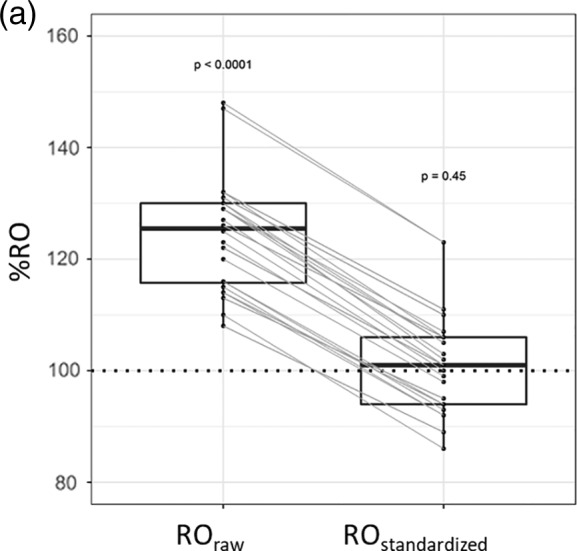
Receptor occupancy (RO) in three replicate experiments with patient PBL aliquots. RO raw and RO stand in the PBLs incubated in vitro with natalizumab (all cell types combined) with expected RO 100% (marked by a horizontal line). Each dot represents RO in one cell type, and lines connect RO values determined in the same measurement using the two calculation methods. P values for comparison of mean RO to the expected (100%) in a one‐sample t test.
In the samples not incubated with natalizumab, RO was unknown. Four different RO calculation methods were compared (Fig. 4b): In addition to ROraw and ROstandardized, we calculated ROvs100%raw and ROvs100%standardized based on RO in the sample relative to RO in the corresponding 100% saturated sample. As for the in vitro saturated samples, we found that ROraw was higher than ROstandardized in each of the eight cell subtypes (p values, medians and ranges are shown in Fig. 4b). Neither ROstandardized nor ROvs100%raw were significantly different from ROvs100%standardized in any of the subtypes (p values, medians and ranges are shown in Fig. 4b).
Figure 4b.
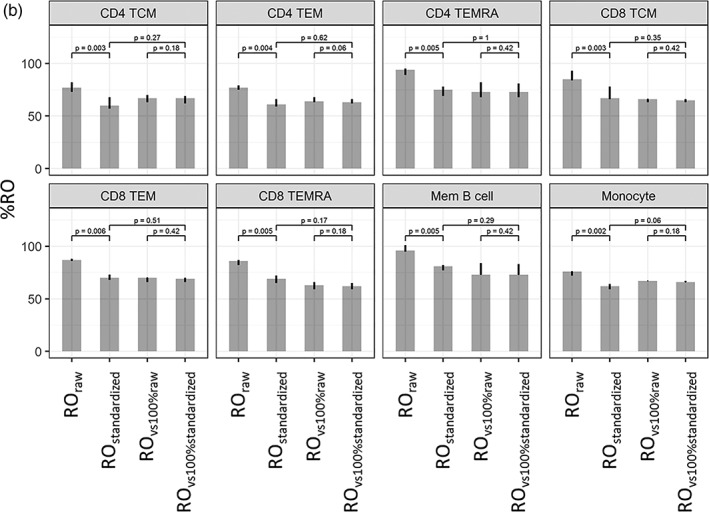
Receptor occupancy (RO) in three replicate experiments with patient PBL aliquots. ROraw, ROstandardized, ROvs. 100% raw, and ROvs. 100% standardized in PBL aliquots with unknown RO. Heights of the bars are median values, and the error bars indicate the range of measured values in three replicate experiments. P values for comparison of mean RO using a paired t test.
To determine whether the overestimation of RO could be caused by interfering factors, such as unwanted signal in the channels for detection of anti‐IgG4 (169Tm) and anti‐α4 integrin (141Pr), several control experiments were performed. First, unstained PBL samples did not contain any detectable metals (Supporting Information Fig. S4a). Second, MMO controls did not reveal spillover into either of the two channels (Supporting Information Fig. S4b). Third, there was minimal nonspecific binding of anti‐IgG4 in the absence of natalizumab in untreated PBLs from the healthy donor (Supporting Information Fig. S4c). Finally, detection of α4 integrin by the anti‐α4 integrin antibody was not reduced by bound natalizumab (Supporting Information Fig. S4d).
Discussion
Embedding RO assays into high‐parameter mass cytometry may be a valuable addition to therapy monitoring. However, to be useful, mass cytometry‐based RO assay results must be reliable and reproducible. In this article, we showed how direct comparison of drug and receptor detected in mass cytometer channels with different sensitivities led to misinterpretation of RO. We demonstrated that reliable results can be obtained by standardization across channels using QSC beads with known ABC. A QSC bead protocol was adapted from flow to mass cytometry. In the mass cytometry assay, signal intensities from metal‐conjugated antibodies in the sample were standardized with standard curves created from QSC beads coated with known numbers of the same metal‐conjugated antibodies.
We performed our mass cytometry RO assay for the therapeutic antibody natalizumab on PBLs from one treated patient with one healthy donor as a negative control in three replicate experiments. An in vitro natalizumab‐saturated aliquot with expected RO of 100% was used as a positive control. Natalizumab was detected with anti‐IgG4 (169Tm) and its target receptor was detected with anti‐α4 integrin (141Pr). Based on prior knowledge, 169 is a more sensitive channel of the mass cytometer than 141, and we therefore expected overestimation of the RO. Indeed, we observed a consistent overestimation of the ROraw in all cell types (median 126%) of the sample saturated in vitro with natalizumab with known RO of 100%. After QSC bead standardization of anti‐IgG4 (169Tm) and anti‐α4 integrin (141Pr) signal intensities, the resulting ROstandardized was no longer significantly different from the expected (median 101%). The same pattern was observed in the sample with unknown RO: ROraw was higher in all cell types compared to ROstandardized.
QC experiments did not reveal other explanations for the overestimation of RO: We did not detect preexisting (in vivo) metal in the sample, spillover from other markers in the panel into channels of interest, or nonspecific binding of anti‐IgG4, and bound natalizumab did not interfere with detection of integrin. Overall, our results indicate that deviance in channel sensitivity for anti‐IgG4 (169Tm) and anti‐α4 integrin (141Pr) was indeed the cause of the overestimation of RO and that this could be corrected by QSC bead standardization. In the sample with unknown RO, after correcting for RO in the corresponding 100% saturated sample, the effect of QSC bead standardization disappeared so that there was no significant difference between ROvs100%raw and ROvs100%standardized in any of the cell types. There was also no difference between ROstandardized and ROvs100%standardized. This indicates that using RO in a 100% saturated sample as a reference mitigates overestimation of RO.
The general effect of the deviance in detection sensitivity on RO results can be predicted by the relative location of the antibody metal tags in the detection spectrum 8: If the drug is detected in a more sensitive channel than the receptor, the RO will be overestimated and vice versa. However, the detection sensitivity pattern varies between machines and cannot be exactly predicted by existing tools. EQ calibration beads and tuning solution only contain certain metals, whereas QSC beads are stained with the actual metal‐conjugated antibodies used to stain the samples. The approach described here may also correct for differences in batch‐to‐batch variability in labeling efficiency (how many metal isotopes are conjugated to the antibody) of in‐house conjugated antibodies but that was not tested in our study.
As no studies of RO in mass cytometry have yet been published, the problem with diverging detection sensitivity has not yet been addressed, but a similar problem arises in flow cytometry where antibodies are conjugated to fluorophores with different brightness. Some have addressed this by using various antibody‐binding beads 1, 10. Others have avoided measuring receptors in various ways: for example, comparing bound drug in the sample to an in vitro drug‐saturated sample as an indirect measure of total receptor level 17 or comparing bound drug during treatment to a baseline before treatment 18. The latter method does not take into account changes in total receptor levels during treatment. Advantages of using QSC beads instead of staining several aliquots of the sample as a reference are that less sample is consumed and that acquisition time is decreased. Measuring drug and receptor on the same cells in the same run means that there is no batch‐to‐batch variability and takes into account varying receptor level during treatment. Labeling QSC beads with metal before antibody staining is time‐consuming, and commercial antibody capture beads pre‐labeled with metal in the detection range of mass cytometer would simplify the protocol.
Importantly, we here used QSC beads only for standardization and not for absolute quantitation of ABC. In mass cytometry, signal intensity is proportional to the amount of metal‐conjugated antibody bound per QSC bead or cell, but no actual counting of epitopes is performed by the instrument. Therefore, ABC values obtained by mass cytometry require careful interpretation, and we refer to the resulting semi‐quantitative value as QSC bead standardized signal intensity.
Despite efforts to reduce experimental variability by using frozen aliquots of one barcoded sample and one antibody cocktail, we observed some day‐to‐day variability in the three replicate experiments performed over a period of 9 days. This could be due to variations in sample handling, staining, cell numbers, and pipetting in the many steps of the protocol. A superior approach for isolating the effect of QSC bead standardization and eliminating other experimental variation would be to run the same stained beads and PBL samples on different mass cytometers simultaneously.
Reproducibility over time and between instruments is crucial in longitudinal and multicenter studies. In flow cytometry, QSC beads allow comparison of experiments over time and between different instruments 19, 20. In mass cytometers, individual detection sensitivity patterns for different machines and variations in machine performance over time may affect results in longitudinal and multicenter studies. Acquisition of data on samples and on QSC beads labeled with the same metal‐conjugated antibodies as used to stain the samples on the same mass cytometer on same day may correct for such variations.
In conclusion, our findings suggest that QSC bead standardization offers an effective means to standardize signal intensities across channels of different sensitivity resulting in reliable and accurate RO results. We demonstrated this in a natalizumab RO assay, but the approach is applicable for RO assays of any drug‐receptor pair or in other mass cytometry experiments involving comparison of abundance of two or more markers. QSC beads should cover the whole range of dual count values in the samples and may be labeled with any metal within the detection range of the mass cytometer, but alterations of bead ABC should be examined. There are certain factors that use of beads cannot correct. QC experiments should be performed to evaluate unspecific binding and spillover into the channels for detection of drug or receptor, and we suggest use of an in vitro drug‐saturated sample with known RO as a reference to validate the results. Future studies should evaluate whether QSC beads can, as in flow cytometry, be used for standardization of experiments performed on different mass cytometers and over time, which would be an important step toward applicability of mass cytometry in multicenter and longitudinal clinical studies.
Supporting information
Table S1: Antibodies used in the experiments.
Supplementary Figure 1: a) Gating strategy for PBL samples. Eight cell types of interest were identified: memory B cells, monocytes, CD4 effector memory (TEM), central memory (TCM), effector memory RA (TEMRA) T cells, and CD8 TEM, TCM, and TEMRA cells. b) Eight cell types in patient PBL samples without (‐nata) and with (+nata) in vitro natalizumab incubation. Shown are 90th percentile of anti‐IgG4 and anti‐α4 integrin dual counts in experiments conducted on three separate days.
Supplementary Figure 2: a) Gating strategy of QSC beads labeled with OsO4 prior to metal conjugated antibody. b) QSC bead standardization of patient PBL samples: Standard curves were created in QuickCal (Bangs Laboratories) based on median dual counts of anti‐IgG4 (169Tm) and anti‐α4 integrin (141Pr) (previous panel) in five QSC bead populations with increasing ABC. For bead standardization of patient PBL samples, raw signal intensities (dual count 90th percentiles, shown in supplementary Figure 1b) of the same antibodies in eight cell types were plotted into the correlating standard curves and translated into ABC values. Standard curves in the three replicate experiments (days 1, 2, and 3) are shown.
Supplementary Figure 3: a) QSC beads acquired on a Fortessa flow cytometer without (left) and with (right) OsO4 labeling prior to antibody staining stained with anti‐IgG4‐PE. Median fluorescence intensity (MFI) in the four standard beads are shown.b) Correlation of signal intensity of the same five QSC bead populations stained with either anti‐IgG4‐PE or anti‐IgG4 (169Tm) and acquired with flow or mass cytometry, respectively.
Supplementary Figure 4: Quality control experiments using PBL samples: a) Preexisting metal in the detection channels for natalizumab (IgG4 169Tm, left) or α4 integrin (CD49d 141Pr, right) in unstained PBL samples from the healthy donor (top) and the patient (bottom). Samples were barcoded and Ir‐intercalated for event detection. b) Mass‐minus‐one (MMO) controls in patient PBLs: 141 signal in sample stained with the panel minus anti‐α4 integrin (left) and 169 signal in sample stained with the panel minus anti‐IgG4 (right).c) Healthy donor PBLs not treated with natalizumab. Median dual counts of anti‐IgG4 in each of the eight cell types are shown. d) Healthy donor PBLs with (top) and without (bottom) in vitro incubation with natalizumab. The presence of drug (IgG4, left) did not affect detection of α4 integrin (CD49d, right), indicating non‐competitive binding of anti‐α4 integrin and natalizumab to different epitopes of α4 integrin. The 90th percentiles of anti‐IgG4 and anti‐ α4 integrin in non‐granulocytes are shown.
ACKNOWLEDGMENTS
The authors thank the Neurological Research Laboratory at Haukeland University Hospital for cooperation on sample collection, the Flow Cytometry Core at the University of Bergen for technical support and helpful discussions, and Novartis for financial research support.
Contributor Information
Gerd Haga Bringeland, Email: gerd.haga.bringeland@helse-bergen.no.
Sonia Gavasso, Email: sonia.gavasso@helse-bergen.no.
Literature Cited
- 1. Moulard M, Ozoux ML. How validated receptor occupancy flow cytometry assays can impact decisions and support drug development. Cytometry B Clin Cytom 2016;90(2):150–158. [DOI] [PubMed] [Google Scholar]
- 2. Stewart JJ, Green CL, Jones N, Liang M, Xu Y, Wilkins DEC, Moulard M, Czechowska K, Lanham D, McCloskey TW, et al. Role of receptor occupancy assays by flow cytometry in drug development. Cytometry B Clin Cytom 2016;90(2):110–116. [DOI] [PubMed] [Google Scholar]
- 3. Gaudillière B, Fragiadakis GK, Bruggner RV, Nicolau M, Finck R, Tingle M, Silva J, Ganio EA, Yeh CG, Maloney WJ, et al. Clinical recovery from surgery correlates with single‐cell immune signatures. Sci Transl Med 2014;6(255): 255ra131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gaudillière B, Ganio EA, Tingle M, Lancero HL, Fragiadakis GK, Baca QJ, Aghaeepour N, Wong RJ, Quaintance C, El‐Sayed YY, et al. Implementing mass Cytometry at the bedside to study the immunological basis of human diseases: Distinctive immune features in patients with a history of term or preterm birth. Cytometry A 2015;87(9):817–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nair N, Mei HE, Chen SY, Hale M, Nolan GP, Maecker HT, Genovese M, Fathman CG, Whiting CC. Mass cytometry as a platform for the discovery of cellular biomarkers to guide effective rheumatic disease therapy. Arthritis Res Ther 2015;17:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. O'Gorman WE, Hsieh EWY, Savig ES, Gherardini PF, Hernandez JD, Hansmann L, Balboni IM, Utz PJ, Bendall SC, Fantl WJ, et al. Single‐cell systems‐level analysis of human toll‐like receptor activation defines a chemokine signature in patients with systemic lupus erythematosus. J Allergy Clin Immunol 2015;136(5):1326–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, Pavlov S, Vorobiev S, Dick JE, Tanner SD. Mass cytometry: Technique for real time single cell multitarget immunoassay based on inductively coupled plasma time‐of‐flight mass spectrometry. Anal Chem 2009;81(16):6813–6822. [DOI] [PubMed] [Google Scholar]
- 8. Tricot S, Meyrand M, Sammicheli C, Elhmouzi‐Younes J, Corneau A, Bertholet S, Malissen M, le Grand R, Nuti S, Luche H, et al. Evaluating the efficiency of isotope transmission for improved panel design and a comparison of the detection sensitivities of mass cytometer instruments. Cytometry A 2015;87(4):357–368. [DOI] [PubMed] [Google Scholar]
- 9. Takahashi C, Au‐Yeung A, Fuh F, Ramirez‐Montagut T, Bolen C, Mathews W, O'Gorman WE. Mass cytometry panel optimization through the designed distribution of signal interference. Cytometry A 2017;91(1):39–47. [DOI] [PubMed] [Google Scholar]
- 10. Engelberts PJ, Badoil C, Beurskens FJ, Boulay‐Moine D, Grivel K, Parren PWHI, Moulard M. A quantitative flow cytometric assay for determining binding characteristics of chimeric, humanized and human antibodies in whole blood: Proof of principle with rituximab and ofatumumab. J Immunol Methods 2013;388(1–2):8–17. [DOI] [PubMed] [Google Scholar]
- 11. Hutchinson M. Natalizumab: A new treatment for relapsing remitting multiple sclerosis. Ther Clin Risk Manag 2007;3(2):259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wipfler P, Harrer A, Pilz G, Oppermann K, Afazel S, Haschke‐Becher E, Sellner J, Trinka E, Kraus J. Natalizumab saturation: Biomarker for individual treatment holiday after natalizumab withdrawal? Acta Neurol Scand 2014;129(3):e12–e15. [DOI] [PubMed] [Google Scholar]
- 13. Puñet‐Ortiz J, Hervás‐García JV, Teniente‐Serra A, Cano‐Orgaz A, Mansilla MJ, Quirant‐Sánchez B, Navarro‐Barriuso J, Fernández‐Sanmartín MA, Presas‐Rodríguez S, Ramo‐Tello C, et al. Monitoring CD49d receptor occupancy: A method to optimize and personalize natalizumab therapy in multiple sclerosis patients. Cytometry B Clin Cytom. 2018;94(2):327–333. [DOI] [PubMed] [Google Scholar]
- 14. Schneider‐Hohendorf T, Rossaint J, Mohan H, Böning D, Breuer J, Kuhlmann T, Gross CC, Flanagan K, Sorokin L, Vestweber D, et al. VLA‐4 blockade promotes differential routes into human CNS involving PSGL‐1 rolling of T cells and MCAM‐adhesion of TH17 cells. J Exp Med 2014;211(9):1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rahman AH, Tordesillas L, Berin MC. Heparin reduces nonspecific eosinophil staining artifacts in mass cytometry experiments. Cytometry A 2016;89(6):601–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. R core team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing, 2017. [Google Scholar]
- 17. Reilly M, Miller RM, Thomson MH, Patris V, Ryle P, McLoughlin L, Mutch P, Gilboy P, Miller C, Broekema M, et al. Randomized, double‐blind, placebo‐controlled, dose‐escalating phase I, healthy subjects study of intravenous OPN‐305, a humanized anti‐TLR2 antibody. Clin Pharmacol Ther 2013;94(5):593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bensinger W, Maziarz RT, Jagannath S, Spencer A, Durrant S, Becker PS, Ewald B, Bilic S, Rediske J, Baeck J, et al. A phase 1 study of lucatumumab, a fully human anti‐CD40 antagonist monoclonal antibody administered intravenously to patients with relapsed or refractory multiple myeloma. Br J Haematol 2012;159(1):58–66. [DOI] [PubMed] [Google Scholar]
- 19. Perfetto SP, Ambrozak D, Nguyen R, Chattopadhyay PK, Roederer M. Quality assurance for polychromatic flow cytometry using a suite of calibration beads. Nat Protoc 2012;7(12):2067–2079. [DOI] [PubMed] [Google Scholar]
- 20. Mizrahi O, Ish Shalom E, Baniyash M, Klieger Y. Quantitative flow cytometry: Concerns and recommendations in clinic and research. Cytometry B Clin Cytom 2018;94(2):211–218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Antibodies used in the experiments.
Supplementary Figure 1: a) Gating strategy for PBL samples. Eight cell types of interest were identified: memory B cells, monocytes, CD4 effector memory (TEM), central memory (TCM), effector memory RA (TEMRA) T cells, and CD8 TEM, TCM, and TEMRA cells. b) Eight cell types in patient PBL samples without (‐nata) and with (+nata) in vitro natalizumab incubation. Shown are 90th percentile of anti‐IgG4 and anti‐α4 integrin dual counts in experiments conducted on three separate days.
Supplementary Figure 2: a) Gating strategy of QSC beads labeled with OsO4 prior to metal conjugated antibody. b) QSC bead standardization of patient PBL samples: Standard curves were created in QuickCal (Bangs Laboratories) based on median dual counts of anti‐IgG4 (169Tm) and anti‐α4 integrin (141Pr) (previous panel) in five QSC bead populations with increasing ABC. For bead standardization of patient PBL samples, raw signal intensities (dual count 90th percentiles, shown in supplementary Figure 1b) of the same antibodies in eight cell types were plotted into the correlating standard curves and translated into ABC values. Standard curves in the three replicate experiments (days 1, 2, and 3) are shown.
Supplementary Figure 3: a) QSC beads acquired on a Fortessa flow cytometer without (left) and with (right) OsO4 labeling prior to antibody staining stained with anti‐IgG4‐PE. Median fluorescence intensity (MFI) in the four standard beads are shown.b) Correlation of signal intensity of the same five QSC bead populations stained with either anti‐IgG4‐PE or anti‐IgG4 (169Tm) and acquired with flow or mass cytometry, respectively.
Supplementary Figure 4: Quality control experiments using PBL samples: a) Preexisting metal in the detection channels for natalizumab (IgG4 169Tm, left) or α4 integrin (CD49d 141Pr, right) in unstained PBL samples from the healthy donor (top) and the patient (bottom). Samples were barcoded and Ir‐intercalated for event detection. b) Mass‐minus‐one (MMO) controls in patient PBLs: 141 signal in sample stained with the panel minus anti‐α4 integrin (left) and 169 signal in sample stained with the panel minus anti‐IgG4 (right).c) Healthy donor PBLs not treated with natalizumab. Median dual counts of anti‐IgG4 in each of the eight cell types are shown. d) Healthy donor PBLs with (top) and without (bottom) in vitro incubation with natalizumab. The presence of drug (IgG4, left) did not affect detection of α4 integrin (CD49d, right), indicating non‐competitive binding of anti‐α4 integrin and natalizumab to different epitopes of α4 integrin. The 90th percentiles of anti‐IgG4 and anti‐ α4 integrin in non‐granulocytes are shown.


