Abstract
Background
Nasopharyngeal carcinoma (NPC) is related to Epstein‐Barr virus (EBV) in endemic areas; however, the role of viruses in nonendemic countries is unclear. Our nationwide study investigated the prevalence and prognostic significance of EBV and human papillomaviruses (HPVs) in Finnish NPC tumors.
Methods
We analyzed samples from 150 patients diagnosed between 1990 and 2009. Viral status was determined using EBV and HPV RNA in situ hybridizations, and p16 immunohistochemistry. Patient and treatment characteristics were obtained from patient records.
Results
In our white patient cohort, 93 of 150 (62%) patients were EBV‐positive and 21/150 (14%) patients were HPV‐positive with no coinfections. Thirty‐six (24%) tumors were negative for both viruses. The 5‐year disease‐specific survival for patients with EBV‐positive, HPV‐positive, and EBV/HPV‐negative tumors was 69%, 63%, and 39%, respectively. In multivariable‐adjusted analysis, overall survival was better among patients with EBV‐positive (P = .005) and HPV‐positive (P = .03) tumors compared to patients with EBV/HPV‐negative tumors.
Conclusions
In our low‐incidence population, EBV and HPV are important prognostic factors for NPC.
Keywords: Epstein‐Barr virus, human papillomavirus, nasopharyngeal carcinoma, p16 immunohistochemistry, viral carcinogenesis
1. INTRODUCTION
The development of new immunomodulating therapies has increased interest in the infectious causes of cancer. Worldwide, it is estimated that 10% of newly diagnosed cancers are attributable to viral infections,1 and according to the International Agency for Research on Cancer (IARC), the Epstein‐Barr virus (EBV) and high‐risk human papillomaviruses (HPV) have been classified as group 1 (well‐established) carcinogenic agents in humans.2 EBV is etiologically linked to Burkitt's lymphoma, extranodal NK/T‐cell lymphoma (nasal type), Hodgkin lymphoma, nasopharyngeal carcinoma (NPC), and lymphoepithelioma‐like carcinoma.3 Presumably, a specific combination of an EBV variant and a particular human leukocyte antigen type is needed for proliferating epithelial cells to escape immune control.4, 5 This hypothesis may partially explain the remarkable geographical differences in the incidence of NPC. In endemic areas of South‐Eastern China, age‐adjusted incidence may reach more than 30 per 100000 persons per year,6 whereas in nonendemic Northern Europe, the incidence is only 0.4 per 100000 in males and 0.2 per 100000 in females.7 Distribution of the histological subtypes of NPC and supposed etiological factors also differ between geographical regions. According to the WHO classification of head and neck tumors, NPC is subdivided into 3 major types: keratinizing squamous cell carcinoma (KSCC), non‐keratinizing carcinoma (NKC), and basaloid squamous cell carcinoma.8 NKC can be further subdivided into differentiated and undifferentiated types.8 In high‐incidence endemic areas, up to 99.6% of NPC tumors display the NKC subtypes, which are more consistently associated with EBV positivity than KSCC.9, 10 By contrast, in low‐incidence nonendemic areas, the prevalence of the NKC subtypes is distinctly lower, whereas the prevalence of KSCC is higher than in endemic areas.10, 11
High‐risk HPV infection has been suggested to be one of the etiological factors causing NPC in whites.12 Studies from both endemic and nonendemic regions have reported the occurrence of HPV in NPC tumors, with or without the coexistence of EBV.13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31 Unlike in oropharyngeal carcinoma,32 no statistical significance for outcome has been attributed to HPV in NPC with 1 exception: Dogan et al. reported that patients with HPV‐positive tumor had, similarly to the patients with EBV‐positive tumor, significantly better overall survival (OS) than the patients with EBV/HPV‐negative tumor.13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31 In addition, Jiang et al. suggested that although overexpression of p16 is not a significant prognostic marker for OS in NPC, it correlates with better progression‐free survival and locoregional control in patients with EBV‐positive tumor.22
Previous studies on viral infections in NPC have mainly reported on small and heterogeneous series of patients treated at 1 or 2 institutions. Thus, data on the prognostic value of EBV and HPV in NPC are limited. The aim of this retrospective whole population‐based study was to describe the status of EBV and HPV in Finnish NPC cases and relate them to histopathological NPC subtypes and patient survival.
2. PATIENTS AND METHODS
2.1. Patients
In a nationwide search for the years 1990 to 2009, a total of 207 patients with newly diagnosed primary NPC were identified from the files of the Finnish Cancer Registry.33 All patients had biopsies taken from their tumor at the time of diagnosis, and histological slides stained with hematoxylin and eosin were available for 168 patients. These specimens were re‐reviewed by an experienced head and neck pathologist (I.L.) to reclassify the cases according to the fourth edition of the WHO histological classification.8 Formalin‐fixed paraffin‐embedded (FFPE) tissue samples were obtained for 150 patients of the study population. Clinical records were collected from 5 university hospitals and 3 other major hospitals in Finland for patient characteristics, presentation of the disease, treatment, and follow‐up. All tumors were confirmed to originate from the nasopharynx. The clinical stage of the disease was determined according to the International Union Against Cancer (UICC) staging system, seventh edition.34 Dates and causes of death were acquired from the Finnish Cancer Registry and Statistics Finland. The study was approved by the Research Ethics Committee of the Hospital District of Southwest Finland, National Institute for Health and Welfare (THL), and National Supervisory Authority for Welfare and Health (Valvira).
2.2. Tissue microarray construction
Tissue microarrays (TMAs) were constructed using an automated tissue microarrayer (TMA Grand Master; 3D Histech Ltd, Budapest, Hungary) to create 5 new paraffin blocks from representative 1 mm core samples (n = 324) taken from the original FFPE blocks of the tumors. Each patient was represented in the array by at least 1 core, usually 2. Scores from the duplicate cores were averaged to produce a single score. Five of the original tumor specimens were derived from neck metastases, whereas the remaining 145 were from the primary tumors. The TMAs also included control tissues from the liver and the placenta.
2.3. In situ hybridizations for EBV‐encoded RNA and HPV DNA
Sections with the thickness of 5 μm were cut from the TMA blocks with a microtome, transferred onto glass slides and incubated for 2 hours at 58°C. For EBV RNA and HPV DNA detection, chromogenic in situ hybridization (ISH) was performed using automated Benchmark XT system (Ventana/Roche Medical Systems Inc., Tucson, Arizona). The Ventana EBER probe detects early RNA transcripts of EBV. In the automated process, deparaffinization and proteolytic treatment with Protease 3 (28 minutes) were followed by hybridization with the EBV‐encoded RNA (EBER) probe at 57°C for 1 hour and counterstaining with red stain. Positive hybridization was defined as strong diffuse signals in the nucleus of nearly all (>90%) tumor cells. The Ventana HPV III Family 16 probe was used to detect high‐risk HPV genotypes 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, and 66. In the automated process, deparaffinization and proteolytic treatment with Protease 3 (20 minutes) were followed by hybridization with the HPV probe in 52°C for 2 hours and counterstaining with red stain. Positive staining for HPVs was recognized as dark blue granules in the nuclei of neoplastic cells. Stainings were interpreted by a pathologist (I.L.) blinded to clinical data.
2.4. ISH for high‐risk HPV E6/E7 mRNA
RNA ISH for high‐risk HPV E6/E7 mRNA was performed manually using the RNAscope 2.5 HD Reagent kit (Advanced Cell Diagnostics, Inc., Hayward, California) according to the manufacturer's protocol. FFPE TMA sections of 5 μm were incubated for 1 hour at 58°C. After deparaffinization, the sections were pretreated with hydrogen peroxidase for 10 minutes at room temperature. Target retrieval was performed for 15 minutes at 100°C. The sections underwent protease treatment (RNAscope Protease Plus) for 30 minutes at 40°C in a hybridization oven followed by hybridization with a high‐risk HPV 18 cocktail probe for genotypes 16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 73, and 82 for 2 hours at 40°C in a hybridization oven. Preamplifiers and amplifiers were hybridized consecutively, accompanied with chromogenic signal detection with diaminobenzidine (DAB). Finally, the slides were counterstained with hematoxylin. An endogenous housekeeping gene HS‐PPIB probe was used as a positive control and a bacterial gene DapB probe as a negative control. The staining was examined (by S.S.) using a qualitative scoring system: a positive staining was recognized as weak or strong intensity of dot‐like nuclear and cytoplasmic signals in at least 10 cells.
2.5. Immunohistochemistry for p16
Immunohistochemical (IHC) stains were performed on 3.5‐μm thick TMA sections using the automated Benchmark XT system (Ventana/Roche). For detection of p16 protein, after deparaffinization and epitope retrieval with CC1 buffer for 60 minutes, the sections were incubated for 16 minutes with a mouse monoclonal antibody against p16 protein (clone E6H4; Ventana/Roche). To visualize the p16 antibodies, a Ventana UltraView Universal DAB Detection Kit was used, and the sections were counterstained with hematoxylin and Bluing Reagent. Positive p16 expression was defined as strong diffuse nuclear and cytoplasmic staining in 75% or more of tumor cells.
2.6. HPV genotyping polymerase chain reaction
DNA was extracted from 3 to 5 sections cut from FFPE sections (total area of the sample was approximately 1 cm2) using high salt method as described previously.35 DNA was amplified with primer sets 1 and 2 from the Multiplex HPV Genotyping Kit (DiaMex GmbH, Heidelberg, Germany). Primer set 2 (DNA quality control primers) contains primers to amplify ß‐globin gene fragments to verify the amount and the quality of human genomic sample DNA. A negative control contained no genomic DNA to confirm the absence of contamination in the amplification reactions. The labeled hybrids were analyzed with a Luminex LX‐100 analyzer (Bio‐Plex 200 System; Bio‐Rad Laboratories, Hercules, California). The Multiplex HPV Genotyping Kit detects 24 low‐risk and high‐risk HPV genotypes.
2.7. Treatment
Patients were treated with radiotherapy (RT) with or without concurrent chemotherapy (CT) (Table 1). Treatment strategies were somewhat heterogeneous over time, as the study period covered the shift from definitive RT to chemoradiotherapy (CRT) protocols and from 3D conformal to intensity‐modulated radiotherapy techniques. However, the treatment parameters, such as the doses of RT and CT and the duration of treatment, were consistently similar among all histological tumor subgroups. The most common schedule was 2 Gy daily fractions 5 times per week; however, 8 patients (5%) received hyperfractionated accelerated RT with 1.6 Gy twice daily and a planned interim break of about 11 days.36 For all patients, the median dose was 70 Gy in the nasopharyngeal tumor area, 60 Gy in the involved lymph nodes, and 50 Gy in the elective neck area. The median treatment time was 7 weeks. Four patients (3%) were treated with additional intracavitary brachytherapy, and a second course of RT for locoregional recurrences was given to 7 (5%) patients. In total, 76 patients (51%) received concurrent CT usually with platinum‐based (87%) cytostatic drugs. Furthermore, neoadjuvant CT was given to 10 patients (7%) and adjuvant CT to 32 patients (21%). A neck dissection was performed in 17 patients (11%) as part of the primary treatment. Seven patients (5%) with a compromised general condition and/or distant metastases received only palliative treatment and were omitted from the survival analyses.
Table 1.
Patient characteristics stratified according to viral status
| Characteristics | No. of patients (%) | |||
|---|---|---|---|---|
| All | EBV‐positive | HPV‐positive | EBV/HPV‐negative | |
| Total number | 150 (100) | 93 (62) | 21 (14) | 36 (24) |
| Sex | ||||
| Male | 101 (67) | 69 (74) | 16 (76) | 16 (44) |
| Female | 49 (33) | 24 (26) | 5 (24) | 20 (56) |
| Age at diagnosis (y) | ||||
| Mean (SD) | 57.0 (15) | 54.5 (15) | 56.9 (13) | 63.5 (15) |
| Range | 12‐85 | 12‐82 | 30‐79 | 21‐85 |
| Ethnicity | ||||
| Finnish | 145 (97) | 88 (95) | 21 (100) | 36 (100) |
| Othera | 5 (3) | 5 (5) | 0 (0) | 0 (0) |
| Smoking | ||||
| Smoker or ex‐smoker | 71 (47) | 43 (46) | 12 (57) | 16 (45) |
| Nonsmoker | 36 (24) | 25 (27) | 3 (14) | 8 (22) |
| Not known | 43 (29) | 25 (27) | 6 (29) | 12 (33) |
| T classification | ||||
| T1 | 54 (36) | 40 (43) | 3 (14) | 11 (31) |
| T2 | 40 (27) | 28 (30) | 6 (29) | 6 (17) |
| T3 | 26 (17) | 11 (12) | 4 (19) | 11 (31) |
| T4 | 30 (20) | 14 (15) | 8 (38) | 8 (22) |
| N classification | ||||
| N0 | 53 (35) | 35 (38) | 4 (19) | 14 (39) |
| N1 | 33 (22) | 20 (22) | 8 (38) | 5 (14) |
| N2 | 51 (34) | 30 (32) | 8 (38) | 13 (36) |
| N3a | 10 (7) | 6 (6) | 1 (5) | 3 (8) |
| N3b | 3 (2) | 2 (2) | 0 (0) | 1 (3) |
| Overall stage | ||||
| I | 19 (13) | 15 (16) | 1 (5) | 3 (8) |
| II | 37 (25) | 27 (29) | 5 (24) | 5 (14) |
| III | 52 (35) | 30 (32) | 6 (28) | 16 (44) |
| IV | 42 (28) | 21 (23) | 9 (43) | 12 (33) |
| Treatment | ||||
| Radiotherapy | 67 (45) | 46 (49) | 7 (33) | 14 (39) |
| Chemoradiotherapy | 76 (51) | 45 (48) | 12 (57) | 19 (53) |
| Palliative | 7 (5) | 2 (2) | 2 (10) | 3 (8) |
| Irradiation technique | ||||
| 2‐dimensional radiotherapy | 15 (10) | 11 (12) | 1 (5) | 3 (8) |
| 3‐dimensional radiotherapy | 88 (60) | 55 (59) | 11 (52) | 22 (61) |
| Intensity‐modulated radiotherapy | 44 (30) | 26 (28) | 9 (43) | 9 (25) |
Abbreviations: EBV, Epstein–Barr virus; HPV, human papillomavirus.
Three from South‐East Asia, 1 from Africa, and 1 from Eastern Europe.
2.8. Statistical analysis
Statistical analyses were carried out using the SAS System for Windows, release 9.4 (SAS Institute Inc., Cary, North Carolina). Mean ages in the histological subgroups were compared with the 1‐way analysis of variance using the Tukey's method for pairwise comparisons. Categorical variables between patients with different viral status were compared with Pearson chi‐squared test or Fisher's exact test. Survival rates were calculated using the Kaplan‐Meier method. Follow‐up time was calculated from the end of the primary treatment, usually from the last day of RT, to the end of the follow‐up or the date of death. Age‐adjusted and multivariable‐adjusted Cox regression was used to test the association of viral status with disease‐specific survival (DSS) and OS. The multivariable Cox regression analysis was adjusted for the potential confounding factors such as age, sex, smoking, T class, N class, total RT dose, and treatment (RT vs CRT).33 To avoid multicollinearity problems, histology was excluded from the multivariable model due to the high correlation with viral status. The results are expressed using hazard ratios with 95% confidence intervals. P‐values of less than .05 were considered as statistically significant.
3. RESULTS
3.1. Overall characterization of the patients
Analyses were restricted to 150 patients with tissue specimens appropriate for TMA. The demographic and clinical characteristics are shown in Table 1. The mean age (SD) of patients was 57.0 (15) years. A total of 145 of 150 (97%) patients were whites of Finnish ethnic background, and 101 of 150 (67%) patients were men. Thirty‐three patients (22%) had KSCC, and 25 patients (17%) had non‐keratinizing differentiated carcinoma (NK‐D), whereas 92 patients (61%) had non‐keratinizing undifferentiated carcinoma (NK‐U) (Table 2). There were no basaloid SCCs. Smoking was frequent and two‐thirds of the patients (71/107, 66%) with known history were current or ex‐smokers. The percentages of known smokers were 68, 57, and 69 in KSCC, NK‐D, and NK‐U, respectively. Unfortunately, smoking history was unknown for almost one‐third of the study population (43/150, 29%). The majority of patients (63%) were seen with locoregionally advanced (stage III/IV) disease at the time of diagnosis.
Table 2.
Relationship between tumor histology according to the WHO classification and viral status
| WHO type | No. of patients (%) | |||
|---|---|---|---|---|
| All | EBV‐positive | HPV‐positive | EBV/HPV‐negative | |
| Keratinizing SCC | 33 | 2 (6) | 12 (36) | 19 (58) |
| Non‐keratinizing differentiated | 25 | 13 (52) | 3 (12) | 9 (36) |
| Non‐keratinizing undifferentiated | 92 | 78 (85) | 6 (6) | 8 (9) |
| Total | 150 | 93 (62) | 21 (14) | 36 (24) |
Abbreviations: EBV, Epstein–Barr virus; HPV, human papillomavirus; SCC, squamous cell carcinoma.
3.2. EBV and HPV status and disease characteristics
All tumors showing a positive EBER ISH (Figure 1F) reaction were regarded as EBV‐positive. For HPV positivity, we determined that the sample should be positive for both p16 IHC and HPV DNA/RNA ISH (Figure 1E, H, G) or/and HPV polymerase chain reaction (PCR). We performed HPV DNA and RNA ISH on all 150 samples but HPV genotyping only for p16‐positive samples. Among the 21 p16‐positive cases, 18 had sufficient FFPE sample for DNA extraction.
Figure 1.
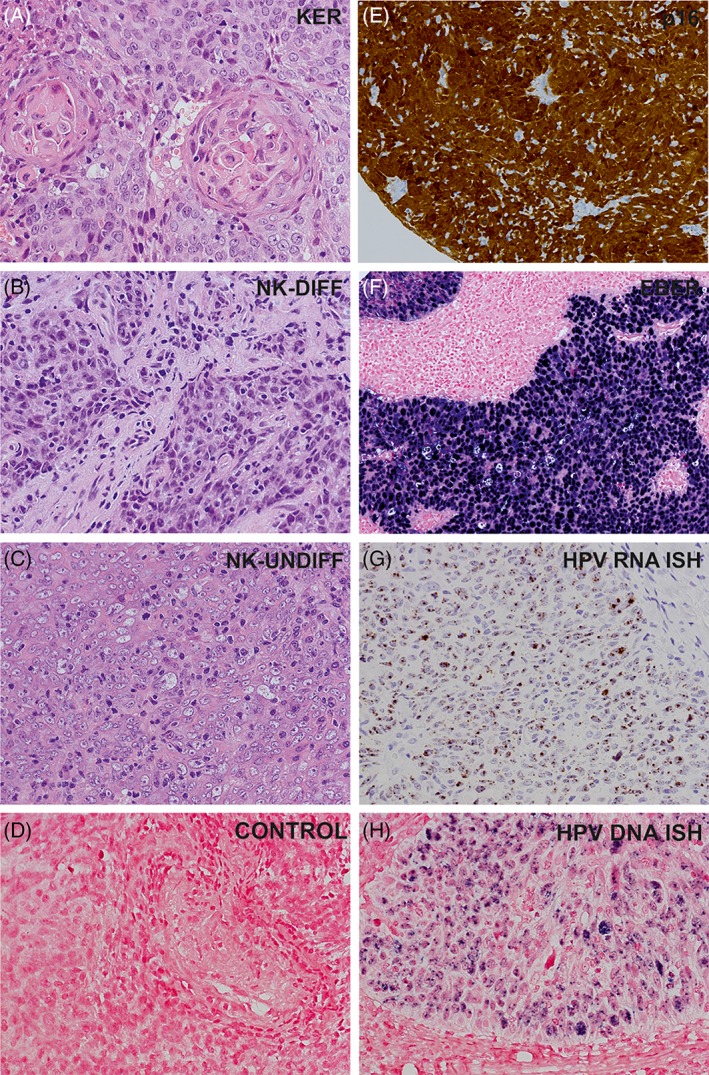
Histopathological subtypes of nasopharyngeal carcinoma observed in the study (A‐C). A, Keratinizing squamous cell carcinoma. B, Non‐keratinizing carcinoma, differentiated type. C, Nonkeratinizing carcinoma, undifferentiated type. D, Negative control for HPV DNA in situ hybridization. A‐H, Magnification ×250. EBV, Epstein‐Barr virus; HPV, human papillomavirus; E, Immunohistochemical staining for p16. F, EBV RNA in situ hybridization (EBER). G, HPV E6/E7 mRNA in situ hybridization. H, HPV DNA in situ hybridization
Overall, in 93 of 150 (62%) patients, the tumors were positive for EBV and negative for HPV (EBV‐positive group); in 21 of 150 (14%) patients, they were positive for HPV and negative for EBV (HPV‐positive group); and in 36 of 150 (24%) patients, they were negative for both EBV and HPV (EBV/HPV‐negative group) (Table 2). None of the patients had a coinfection with both EBV and HPV. Among the p16‐positive tumors, only 1 tumor remained negative for HPV DNA with both ISH and HPV genotyping methods (1/21, 5%). Interestingly, this case was confirmed HPV‐positive with the HPV RNA ISH method. In contrast, 16 of 21 (76%) p16‐positive tumors were positive in HPV DNA ISH; in 16 of 18 (89%) p16‐positive tumors available for HPV genotyping method, HPV DNA was amplified with PCR. There were 3 negative cases in HPV DNA ISH but positive in HPV DNA PCR, and 1 case that was negative in HPV DNA PCR but positive in HPV DNA ISH. All p16‐positive tumors showed positivity in HPV RNA ISH. HPV genotyping was performed for 18 p16‐positive samples: HPV16 was the most prevalent genotype (11/18, 61%) followed by HPV18 (2/18, 11%). In addition, HPV11, HPV33, and HPV59 were present in 1 sample each.
The incidences of EBV or HPV did not vary over the 20‐year period of the study. The patients with EBV‐positive or HPV‐positive tumors were younger than the patients with EBV/HPV‐negative tumors with mean (SD) ages of 54.5 (15) years for EBV‐positive, 56.9 (13) years for HPV‐positive, and 63.5 (15) years for patients with EBV/HPV‐negative tumors. The age difference was statistically significant in the patients with EBV‐positive tumors but not in the patients with HPV‐positive tumors compared to the patients with EBV/HPV‐negative tumors (P = .007 and P = .24, respectively). There were significantly more women in the EBV/HPV‐negative group (56%) compared to the EBV‐positive (26%, P = .001) and the HPV‐positive (24%, P = .020) groups. Table 2 shows the distribution of EBV‐ and HPV‐positive carcinomas in relation to histological subtypes (P < .0001). More than one‐third of KSCC tumors (12/33, 36%) were HPV‐positive, whereas HPV was only detected in 3 of 25 (12%) NK‐D tumors and in 6 of 92 (6%) NK‐U tumors. In contrast, the majority of non‐keratinizing tumors were EBV‐positive (91/117, 78%), whereas only 2 of 33 (6%) of KSCCs presented with EBV positivity. Considering the extent of the disease at the time of diagnosis, EBV‐positive tumors showed a significantly smaller T classification than the EBV/HPV‐negative tumors (P = .030). However, no statistically significant differences were found between EBV or HPV status and N classification, distant metastases, or overall stage.
3.3. Viral status and clinical outcomes
The median follow‐up time was 63 months for patients treated with curative intent (n = 143). Nearly all patients had a minimum of 5‐year follow‐up; only 4 survivors were observed after treatment for fewer than 60 months (49‐57 months). Altogether, 67 patients (47%) were seen with a treatment failure, residual or recurrence. The median latency for overall failure was 5 months (range, 0‐201 months). The patients with EBV‐positive tumors had significantly less local failures than those who had EBV/HPV‐negative tumors (P = .014). The rate of local failure was 25% in the patients with EBV‐positive, 32% in the patients with HPV‐positive, and 49% in the patients with EBV/HPV‐negative tumors, respectively. There were no statistical differences in nodal, distal, or overall failures; however, the patients with EBV/HPV‐negative tumors tended to have more failures than patients in other groups.
The 5‐year DSS for the patients with EBV‐positive tumors was 69%, whereas for the patients with HPV‐positive tumors and with EBV/HPV‐negative tumors, it was 63% and 39%, respectively (Figure 2B). In age‐adjusted Cox regression analysis, EBV positivity (P = .007) was a significant prognostic factor for better DSS compared to EBV/HPV negativity (Table 3).
Figure 2.
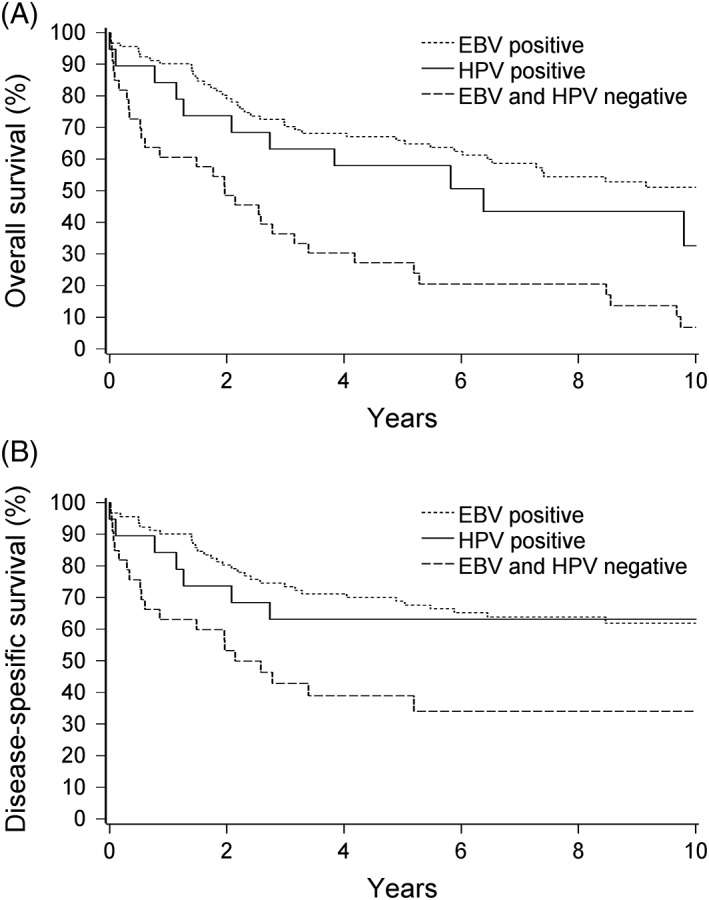
A, Overall survival and (B) disease‐free survival for different viral statuses. EBV, Epstein‐Barr virus; HPV, human papillomavirus
Table 3.
Age‐adjusted and multivariable Cox regression analysis of 143 patients relative to disease‐specific survival and overall survival
| Patient survival | Age‐adjusted | Multivariable | Adjusteda | |
|---|---|---|---|---|
| HR (95% CI) | P‐value | HR (95% CI) | P‐value | |
| Disease‐specific survival | ||||
| Viral status | ||||
| EBV‐positive vs EBV/HPV‐negative | 0.45 (0.25‐0.80) | .007 | 0.69 (0.33‐1.44) | .32b |
| HPV‐positive vs EBV/HPV‐negative | 0.50 (0.21‐1.20) | .12b | 0.44 (0.16‐1.17) | .10b |
| Overall survival | ||||
| Viral status | ||||
| EBV‐positive vs EBV/HPV‐negative | 0.38 (0.23‐0.61) | <.0001 | 0.44 (0.25‐0.78) | .005 |
| HPV‐positive vs EBV/HPV‐negative | 0.61 (0.31‐1.20) | .15b | 0.45 (0.21‐0.94) | .03 |
Abbreviations: CI, confidence interval; EBV, Epstein‐Barr virus; HPV, human papillomavirus; HR, hazard ratio.
Adjusted for age, sex, smoking, T classification, N classification, total radiotherapy dose, and treatment.
Nonsignificant P‐value.
The 5‐year OS was 66% for the patients with EBV‐positive tumors, 58% for the patients with HPV‐positive tumors, and 27% for the patients with EBV/HPV‐negative tumors (Figure 2A). In age‐adjusted Cox regression analysis, EBV positivity (P < .0001) was a significant prognostic factor for better OS compared to EBV/HPV negativity (Table 3). In multivariable‐adjusted Cox regression analysis, both EBV positivity (P = .005) and HPV positivity (P = .034) were significant prognostic factors for better OS (Table 3B).
All patients who were p16‐positive were also HPV‐positive, and thus their survival was similar as described previously for HPV positivity.
4. DISCUSSION
This study focused on the prognostic value of EBV and HPV status in the NPC tumors of 150 Finnish patients diagnosed during 1990 to 2009. To our knowledge, this is the first nationwide study and one of the largest studies published to date of EBV and/or HPV status and their impact on the outcome of NPC. It is widely accepted that EBV is etiologically associated with NPC in high‐incidence endemic regions, such as South‐Eastern China, but its significance in low‐incidence nonendemic countries has been equivocal.10 A recent meta‐analysis based on 8 studies of NPC in low‐incidence regions including the United States, the United Kingdom, Greece, and Denmark revealed that the prevalence of EBV positivity ranged from 0% to 83%, with an average of approximately 42% in solely white patients.31 In our nationwide series of Finnish patients with NPC, of which 97% were of white origin, EBV was detected in 62% of their tumors. Importantly, our study indicated that EBV is a significant favorable prognostic factor of NPC in Finland. We found that both 5‐year DSS and OS were significantly better in the EBV‐positive patient group compared to the EBV/HPV‐negative patient group (Figure 2). Our results are in line with other reports from low‐incidence areas indicating that patients with EBV‐positive NPC tumors fare significantly better than patients with EBV‐negative tumors.13, 28, 30
High prevalence of HPV positivity has been reported in NPC tumors in nonendemic areas, but its prognostic significance remains unclear. Recent studies from the United States and the United Kingdom examined tumor samples from 30 to 88 patients using HPV DNA ISH and/or PCR, and reported HPV positivity ranging from 6% to 30%.13, 25, 27, 28, 29 Only a few studies have assessed the association of HPV status and disease outcome, and mostly they have not found statistically significant differences in survival between patients with HPV‐positive and HPV‐negative tumors.13, 20, 27, 28, 29 In our study, HPV was detected in 14% of all cases. We used a HPV E6/E7 mRNA ISH method,37 detecting transcriptionally active high‐risk HPV mRNA, which has not been used in NPC studies before. In contrast to other studies,13, 20, 27, 29 the patients with HPV‐positive tumors of our cohort showed better survival than the patients with HPV‐negative tumors: in multivariable‐adjusted Cox regression analysis, the patients with HPV‐positive tumors showed significantly better OS compared to the patients with EBV/HPV‐negative tumors. Moreover, Kaplan‐Meier analysis indicated better DSS for the patients with HPV‐positive tumors compared to the patients with EBV/HPV‐negative tumors, although the difference was not statistically significant. We assume that this lack of statistical significance might be due to the low number of patients in the HPV‐positive group.
The absence of coinfections with EBV and HPV is interesting and might reflect mutually exclusive pathogenetic mechanisms. Histologically, our EBV‐positive tumors were almost exclusively NKCs in line with prior findings from endemic and nonendemic areas.10 In contrast, less than half of all HPV‐positive tumors (43%) had non‐keratinizing histology, whereas the majority were KSCCs. Due to our whole population‐based patient material, the latter result introduces new data on the histological distribution of HPV‐positive NPC. Additional studies are needed in both nonendemic and endemic regions to clarify the role of HPV in the pathogenesis of NPC.
HPV‐associated NPC bears some resemblance to oropharyngeal SCC (OPSCC) as patients with HPV‐associated OPSCC also have better survival than their HPV‐negative counterparts.38 However, the incidence of HPV‐associated OPSCC has increased since the early 1970s,39 whereas no rise in the incidence of NPC or HPV‐associated NPC was seen in our cohort during the 20‐year period of 1990‐2009. Due to the presence of high‐risk HPV in 14% of our cases, we look forward to learning if HPV vaccines show efficacy in preventing HPV‐positive NPC, as has been the case in cervical cancer. 40 Possible presence of HPV in NPC may challenge current diagnostic work‐ups, as one must consider NPC in addition to OPSCC as a possible primary tumor in cases of HPV‐positive neck metastasis. This may also have direct consequences to the irradiated volume in cases of HPV‐positive metastasis from an unknown primary.
We used a sophisticated HPV E6/E7 mRNA ISH method to detect transcriptionally active high‐risk HPV in NPC samples.37 The results were fully consistent with IHC detection of p16 overexpression, as all of the p16‐positive tumors in our study were also positive in HPV E6/E7 mRNA ISH. Conversely, high‐risk HPV mRNA was not identified in p16‐negative samples. This is an important finding, suggesting that p16 is a useful surrogate marker for identifying HPV in NPC. Nevertheless, due to high specificity, the HPV E6/E7 mRNA ISH is a recommended method for detecting high‐risk HPV.37 To identify HPV genotype distribution, we used PCR and multiplex genotyping. We found that HPV16 was the most prevalent genotype (61%) and HPV18 was the second most prevalent, in line with prior results on other HPV‐associated head and neck malignancies.41
This study carries the limitations of a retrospective study: for example, patient information on smoking habits and alcohol use was often incomplete or absent. This limits the potential to evaluate the outcome data. Furthermore, the treatment strategies of our patients have been somewhat inconsistent across the various hospitals without regard to viral status. However, the strengths of this study include a whole population‐based representation of patients with NPC nationwide in 1 country during a 20‐year study period. The Finnish Cancer Registry, population statistics, and the uniform nationwide health care system enabled nearly full coverage of patients treated for NPC.42 A high proportion of all diagnostic histopathological samples were retrieved and usable for TMA (150/207 samples). Some samples were discarded from TMA due to their inadequately small size.
Patients with EBV‐associated NPC are potential candidates for new immunotherapies. Indeed, recent clinical trials have studied therapeutic EBV vaccines and immune‐checkpoint inhibitors in patients with NPC with encouraging results for antitumor activity.43, 44, 45 EBV vaccines have mainly implied adoptive T‐cell therapy with EBV‐specific T cells prepared in vitro for infusion into patients. 43 Studies on immune‐checkpoint inhibitors pembrolizumab and nivolumab have been conducted mostly with patients with EBV‐positive tumors, and therefore more studies are needed to show possible differences in the effect of immunomodulators related to viral status.44, 45 The high proportion of patients with EBV‐positive NPC in our cohort emphasizes the value of EBER ISH in histopathological diagnosis of nasopharyngeal tumors and their metastases in low‐incidence areas. In addition, an analysis of cell‐free EBV DNA load in peripheral blood or in nasopharyngeal brushings could help predict recurrences after treatment.46, 47 One quarter of the tumor samples in this study did not reveal either EBV or HPV positivity, and these patients with virus‐negative NPC had the poorest outcome. It is tempting to speculate that these tumors might be genetically different from the virus‐positive similar to findings from other HPV‐negative head and neck cancers,48 and thus they might require more intensive therapy as in HPV‐negative oropharyngeal carcinomas.
In conclusion, we found that the majority of Finnish patients with NPC have viral etiology and almost two‐thirds of the cases associate with EBV and 1 out of 7 with HPV. The favorable prognosis for virus‐associated patient groups is in line with results from endemic regions. This highlights the role of radiosensitivity and immunological aspects in guiding therapeutic approaches. Patients with p16‐positive cervical lymph node metastasis from an unknown primary tumor may harbor NPC, although p16‐positive oropharyngeal cancer remains the most common alternative.
ACKNOWLEDGMENTS
We thank Prof. Reidar Grénman for the initiation of this project and Antti Mäkitie, Kauko Saarilahti, Tuija Wigren, Merja Korpela, and Leena Voutilainen for collecting patient data. Additionally, we thank Petri Koivunen, Sinikka Collanus, Heikki Peuravuori, Minnamaija Lintunen, and Auria Biobank for technical assistance. This study was funded by Tuulikki Edessalo Foundation (Turun Yliopisto), Turku University Hospital Research Funds (EVO), and the Finnish Otologic Research Funds.
Ruuskanen M, Irjala H, Minn H, et al. Epstein‐Barr virus and human papillomaviruses as favorable prognostic factors in nasopharyngeal carcinoma: A nationwide study in Finland. Head & Neck. 2019;41:349–357. 10.1002/hed.25450
Funding information Turku University Hospital Research Funds (EVO); University of Turku: Tuulikki Edessalo Foundation; The Finnish ORL‐HNS Foundation
REFERENCES
- 1. Schiller JT, Lowy DR. Virus infection and human cancer: an overview. Recent Results Cancer Res. 2014;193:1‐10. [DOI] [PubMed] [Google Scholar]
- 2. Bouvard V, Baan R, Straif K, et al. A review of human carcinogens—Part B: biological agents. Lancet Oncol. 2009;10:321‐322. [DOI] [PubMed] [Google Scholar]
- 3. Rickinson AB. Co‐infections, inflammation and oncogenesis: future directions for EBV research. Semin Cancer Biol. 2014;26:99‐115. [DOI] [PubMed] [Google Scholar]
- 4. Raab‐Traub N. Novel mechanisms of EBV‐induced oncogenesis. Curr Opin Virol. 2012;2:453‐458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tsai M, Raykova A, Klinke O, et al. Spontaneous lytic replication and epitheliotropism define an Epstein‐Barr virus strain found in carcinomas. Cell Rep. 2013;5:458‐470. [DOI] [PubMed] [Google Scholar]
- 6. Jia WH, Huang QH, Liao J, et al. Trends in incidence and mortality of nasopharyngeal carcinoma over a 20‐25 year period (1978/1983‐2002) in Sihui and Cangwu counties in southern China. BMC Cancer. 2006;6:178‐185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E386. [DOI] [PubMed] [Google Scholar]
- 8. El‐Naggar AK, Chan JKC, Grandis JR, Takata T, Slootweg PJ. WHO Classification of Head and Neck Tumours. Lyon, France: International Agency for Research on Cancer; 2017:64‐70. [Google Scholar]
- 9. Lee AW, Sze WM, Au JS, et al. Treatment results for nasopharyngeal carcinoma in the modern era: the Hong Kong experience. Int J Radiat Oncol Biol Phys. 2005;61:1107‐1116. [DOI] [PubMed] [Google Scholar]
- 10. Chang ET, Adami HO. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev. 2006;15:1765‐1777. [DOI] [PubMed] [Google Scholar]
- 11. Vaughan TL, Shapiro JA, Burt RD, et al. Nasopharyngeal cancer in a low‐risk population: defining risk factors by histological type. Cancer Epidemiol Biomarkers Prev. 1996;5:587‐593. [PubMed] [Google Scholar]
- 12. Chua MLK, Wee JTS, Hui EP, Chan ATC. Nasopharyngeal carcinoma. The Lancet. 2016;387:1012‐1024. [DOI] [PubMed] [Google Scholar]
- 13. Stenmark MH, Mchugh JB, Schipper M, et al. Nonendemic HPV‐positive nasopharyngeal carcinoma: association with poor prognosis. Int J Radiat Oncol Biol Phys. 2014;88:580‐588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tyan YS, Liu ST, Ong WR, Chen ML, Shu CH, Chang YS. Detection of Epstein‐Barr virus and human papillomavirus in head and neck tumors. J Clin Microbiol. 1993;31:53‐56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rassekh CH, Rady PL, Arany I, et al. Combined Epstein‐Barr virus and human papillomavirus infection in nasopharyngeal carcinoma. Laryngoscope. 1998;108:362‐367. [DOI] [PubMed] [Google Scholar]
- 16. Tung YC, Lin KH, Chu PY, Hsu CC, Kuo WR. Detection of human papilloma virus and Epstein‐Barr virus DNA in nasopharyngeal carcinoma by polymerase chain reaction. Kaohsiung J Med Sci. 1999;15:256‐262. [PubMed] [Google Scholar]
- 17. Punwaney R, Brandwein MS, Zhang DY, et al. Human papillomavirus may be common within nasopharyngeal carcinoma of Caucasian Americans: investigation of Epstein‐Barr virus and human papillomavirus in Eastern and Western nasopharyngeal carcinoma using ligation‐dependent polymerase chain reaction. Head Neck. 1999;21:21‐29. [DOI] [PubMed] [Google Scholar]
- 18. Mirzamani N, Salehian P, Farhadi M, Tehran EA. Detection of EBV and HPV in nasopharyngeal carcinoma by in situ hybridization. Exp Mol Pathol. 2006;81:231‐234. [DOI] [PubMed] [Google Scholar]
- 19. Laantri N, Attaleb M, Kandil M, et al. Human papillomavirus detection in moroccan patients with nasopharyngeal carcinoma. Infect Agents Cancer. 2011;6:3‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang C, Hsiao J, Yang M, et al. Human papilloma virus detection in neoplastic and non‐neoplastic nasopharyngeal tissues in Taiwan. J Clin Pathol. 2011;64:571‐577. [DOI] [PubMed] [Google Scholar]
- 21. Kano M, Kondo S, Wakisaka N, et al. The influence of human papillomavirus on nasopharyngeal carcinoma in Japan. Auris Nasus Larynx. 2017;44:327‐332. [DOI] [PubMed] [Google Scholar]
- 22. Jiang W, Chamberlain P, Garden AS, et al. Prognostic value of p16 expression in Epstein‐Barr virus positive nasopharyngeal carcinomas. Head Neck. 2016;38:E1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hørding U, Nielsen H, Daugaard S, Albeck H. Human papillomavirus types 11 and 16 detected in nasopharyngeal carcinomas by the polymerase chain reaction. Laryngoscope. 1994;104:99‐102. [DOI] [PubMed] [Google Scholar]
- 24. Giannoudis A, Ergazaki M, Segas J, et al. Detection of Epstein‐Barr virus and human papillomavirus in nasopharyngeal carcinoma by the polymerase chain reaction technique. Cancer Lett. 1995;89:177‐181. [DOI] [PubMed] [Google Scholar]
- 25. Lo EJ, Bell D, Woo JS, et al. Human papillomavirus and WHO type I nasopharyngeal carcinoma. Laryngoscope. 2010;120:1990‐1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maxwell JH, Kumar B, Feng FY, et al. HPV‐positive/p16‐positive/EBV‐negative nasopharyngeal carcinoma in white North Americans. Head Neck. 2010;32:562‐567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Robinson M, Suh Y, Paleri V, et al. Oncogenic human papillomavirus‐associated nasopharyngeal carcinoma: an observational study of correlation with ethnicity, histological subtype and outcome in a UKpopulation. Infect Agents Cancer. 2013;8:30‐36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dogan S, Hedberg ML, Ferris RL, Rath TJ, Assaad AM, Chiosea SI. Human papillomavirus and Epstein‐Barr virus in nasopharyngeal carcinoma in a low‐incidence population. Head Neck. 2013;36:511‐516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin Z, Khong B, Kwok S, et al. Human papillomavirus 16 detected in nasopharyngeal carcinomas in white Americans but not in endemic southern Chinese patients. Head Neck. 2014;36:709‐714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dogan HT, Kılıçarslan A, Doğan M, Süngü N, Güler Tezel G, Güler G. Retrospective analysis of oncogenic human papilloma virus and Epstein‐Barr virus prevalence in Turkish nasopharyngeal cancer patients. Pathol Res Pract. 2016;212:1021‐1026. [DOI] [PubMed] [Google Scholar]
- 31. Svajdler M, Kaspirkova J, Mezencev R, et al. Human papillomavirus and Epstein‐Barr virus in nasopharyngeal carcinoma in a non‐endemic eastern European population. Neoplasma. 2016;63:107‐113. [DOI] [PubMed] [Google Scholar]
- 32. Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24‐35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ruuskanen M, Grenman R, Leivo I, et al. Outcome of nasopharyngeal carcinoma in Finland: a nationwide study. Acta Oncol. 2018;57:251‐256. [DOI] [PubMed] [Google Scholar]
- 34. Sobin LH, Gospodarowicz MK, Wittekind C. TNM Classification of Malignant Tumours. Hoboken, United Kingdom: John Wiley & Sons, Incorporated; 2009:30‐38. [Google Scholar]
- 35. Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lindholm P, Valavaara R, Aitasalo K, et al. Preoperative hyperfractionated accelerated radiotherapy and radical surgery in advanced head and neck cancer: a prospective phase II study. Radiother Oncol. 2006;78:146‐151. [DOI] [PubMed] [Google Scholar]
- 37. Bishop JA, Ma XJ Wang H, et al. Detection of transcriptionally active high‐risk HPV in patients with head and neck squamous cell carcinoma as visualized by a novel E6/E7 mRNA in situ hybridization method. Am J Surg Pathol. 2012;36:1874‐1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Attner P, Du J, Näsman A, et al. Human papillomavirus and survival in patients with base of tongue cancer. Int J Cancer. 2011;128:2892‐2897. [DOI] [PubMed] [Google Scholar]
- 39. Näsman A, Attner P, Hammarstedt L, et al. Incidence of human papillomavirus (HPV) positive tonsillar carcinoma in Stockholm, Sweden: an epidemic of viral‐induced carcinoma? Int J Cancer. 2009;125:362‐366. [DOI] [PubMed] [Google Scholar]
- 40. Lehtinen M, Lagheden C, Luostarinen T, et al. Ten‐year follow‐up of human papillomavirus vaccine efficacy against the most stringent cervical neoplasia end‐point‐registry‐based follow‐up of three cohorts from randomized trials. BMJ Open. 2017;7:11‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kreimer AR, Clifford G, Boyle P, Franceschi S. Human papillomavirus types in head and neck squamous cell carcinomas worldwide: a systematic review. Cancer Epidemiol Biomarkers Prev. 2005;14:467‐475. [DOI] [PubMed] [Google Scholar]
- 42. Teppo L, Pukkala E, Lehtonen M. Data quality and quality control of a population‐based cancer registry. Experience in Finland. Acta Oncol. 1994;33:365‐369. [DOI] [PubMed] [Google Scholar]
- 43. Taylor GS, Steven NM. Therapeutic vaccination strategies to treat nasopharyngeal carcinoma. Chin Clin Oncol. 2016;5:23‐36. [DOI] [PubMed] [Google Scholar]
- 44. Hsu C, Lee S, Ejadi S, et al. Safety and antitumor activity of pembrolizumab in patients with programmed death‐ligand 1–positive nasopharyngeal carcinoma: results of the KEYNOTE‐028 study. J Clin Oncol. 2017;35:4050‐4056. [DOI] [PubMed] [Google Scholar]
- 45. Ma BB, Lim WT, Goh BC, et al. Antitumor activity of nivolumab in recurrent and metastatic nasopharyngeal carcinoma: an international, multicenter study of the Mayo Clinic Phase 2 Consortium (NCI‐9742). J Clin Oncol. 2018;36:1412‐1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lin JC, Wang WY, Chen KY, et al. Quantification of plasma Epstein‐Barr virus DNA in patients with advanced nasopharyngeal carcinoma. N Engl J Med. 2004;350:2461‐2470. [DOI] [PubMed] [Google Scholar]
- 47. Stoker SD, Wildeman MA, Novalic Z, et al. Can Epstein‐Barr virus DNA load in nasopharyngeal brushings or whole blood predict recurrent nasopharyngeal carcinoma in a non‐endemic region? A prospective nationwide study of the Dutch head and neck oncology cooperative group. Eur Arch Otorhinolaryngol. 2016;273:1557‐1567. [DOI] [PubMed] [Google Scholar]
- 48. Lawrence MS, Sougnez C, Lichtenstein L, et al. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576‐582. [DOI] [PMC free article] [PubMed] [Google Scholar]


