Abstract
Background
BK polyomavirus (BKPyV) persistently infects the urinary tract and causes viremia and nephropathy in kidney transplantation (KTx), recipients. In a previous study, we observed an increased incidence and load of BKPyV viremia in KTx patients coinfected with human polyomavirus 9 (HPyV9). Here we sought confirmation of this observation and explored whether novel HPyVs that have been detected in urine (HPyV9 and trichodysplasia spinulosa polyomavirus [TSPyV]) potentially aggravate BKPyV infection.
Methods
A well‐characterized cohort of 209 KTx donor‐recipient pairs was serologically and molecularly analyzed for HPyV9 and TSPyV coinfection. These data were correlated with the occurrence of BKPyV viremia and BKPyVAN in the recipients within a year after KTx.
Results
Seropositivity for HPyV9 (19%) and TSPyV (89%) was comparable between donors and recipients and did not correlate with BKPyV viremia and BKPyVAN that developed in 25% and 3% of the recipients, respectively. Two recipients developed TSPyV viremia and none HPyV9 viremia. Modification of the predictive effect of donor BKPyV seroreactivity on recipient BKPyV viremia by HPyV9 and TSPyV was not observed.
Conclusions
Our data provide no evidence for a promoting effect of HPyV9 and TSPyV on BKPyV infection and BKPyVAN in renal allograft patients. Therefore, we do not recommend including HPyV9 and TSPyV screening in KTx patients.
Keywords: BK infection, BK polyomavirus, BKPyV‐associated nephropathy, human polyomavirus 9, kidney transplantation, trichodysplasia spinulosa polyomavirus
1. INTRODUCTION
Kidney transplant (KTx) recipients are at high risk of developing viral infections due to the immunosuppressive treatment required to prevent allograft rejection. BK polyomavirus (BKPyV) infection poses a threat to the transplanted kidney. Primary BKPyV infection occurs early in life and causes persistent (life‐long) asymptomatic infection, rendering ~90% of adults seropositive.1, 2 Thereafter, the virus resides in urothelium and renal tubular cells.3, 4 Reactivation of BKPyV occurs in immunocompromised patients indicated by the presence of viral DNA in urine (viruria) and blood (viremia), manifesting itself as hemorrhagic cystitis and BKPyV‐associated nephropathy (BKPyVAN). BKPyVAN of the renal allograft is seen in 1% to 10% of KTx recipients and ultimately causes allograft failure, unless immunosuppression is tapered.5, 6, 7 Recently we found BKPyV‐specific immunoglobulin G (IgG) levels, especially determined in donors before KTx, to be strongly predictive of BKPyV viremia and BKPyVAN in recipients after KTx.5
BKPyV is a member of the Polyomaviridae, which among others contains 13 species detected in humans,8 including human polyomavirus 9 (HPyV9) and trichodysplasia spinulosa polyomavirus (TSPyV). TSPyV primary infection causes trichodysplasia spinulosa in immunocompromised patients, in whom it can be found in large quantities in affected skin and blood, but also in kidney tissue and urine.9, 10, 11, 12, 13 TSPyV seroprevalence in healthy populations is high, approximately 75%,1, 14 which suggests that asymptomatic persistent TSPyV infection is common.
HPyV9 was discovered in serum of a KTx patient.15 A role for HPyV9 in hemorrhagic cystitis, bladder cancer, cutaneous T‐cell lymphoma, and cutaneous squamous cell carcinoma has been explored, but thus far no pathogenic effect of HPyV9 has been found. Seroprevalence for this virus is lower than most other HPyVs (~20%).1, 14, 16
We have previously observed an association between the HPyV9 and BKPyV infection in a small cohort of, mostly cadaveric, kidney(‐pancreas) transplant patients (n = 99). Approximately 20% of these patients became HPyV9 viremic after KTx, which preferentially occurred among BKPyV viremic recipients (P < 0.05), especially in those with high BKPyV DNA loads (>103 copies/mL).17 An association with Cytomegalovirus (CMV) viremia was not found, suggesting that the association between the HPyV9 and BKPyV was not merely the result of increased immunosuppression, but rather of a common risk factor shared between these polyomaviruses.
Since the detection of HPyV9 was associated with higher BKPyV loads, we wondered if HPyV9 somehow promotes BKPyV infection and could be involved in BKPyVAN development in KTx patients. In this study, we tested this hypothesis, although we are not aware of any documented complementarity/synergy between polyomaviruses, as for instance has been described between DNA viruses, such as hepatitis B and D virus.18 At the same time, this approach offered the possibility to confirm our previously reported HPyV9 viremia rates in KTx patients, which were not found in a number of other comparable studies.15, 19 Since TSPyV belongs to the same genus as HPyV9 (alpha polyomavirus), and is also detected in kidney tissue and urine, we included this polyomavirus in the analysis as well.
2. OBJECTIVE
The aim of this study was to investigate the association between the HPyV9 and TSPyV coinfection and the development of BKPyV viremia and BKPyVAN in KTx recipients.
3. MATERIALS AND METHODS
3.1. Study population and sample collection
The current study cohort was part of an established group of 407 KTx living adult (age > 18) donor‐recipient pairs transplanted at the Leiden University Medical Center between 2003 and 2012.5 Recipients for which no baseline or less than two plasma samples were available, or have been transplanted twice with different donors, were excluded from this cohort. All other recipients have been included. HPyV9 and TSPyV serology and polymerase chain reaction (PCR) was performed on the 209 donor‐recipient pairs with the oldest KTx dates, from 2003 to 2011. Blood samples for BKPyV DNA screening were collected as part of routine virus screening, before transplantation, and 1.5, 3, 6, 9, and 12 months after transplantation, with a median of 3.6‐time points analyzed per recipient. Wunderink et al5 reported a detailed description of study design and population, data collection, and medical ethical approval.
3.2. Seroresponsedetection of BKPyV, HPyV9, and TSPyV
An in‐house Luminex immunoassay (Austin, TX) detecting IgG reactivity against major viral protein 1 of BKPyV, HPyV9, and TSPyV expressed as median fluorescence intensity (MFI) was performed on serum and plasma samples of donors and recipients, as described.5, 20
3.3. DNA detection of BKPyV, TSPyV, and HPyV9
BKPyV, HPyV9, and TSPyV viremia was determined in serum and plasma with quantitative real‐time PCR, as described.5, 10, 17
3.4. Assessment of BKPyVAN development
BKPyVAN was determined by kidney biopsy, performed if clinically indicated according to the treating physician. The biopsy specimen was stained for BKPyV large T antigen with mouse monoclonal antibody PAb416 (Calbiochem; EMD Millipore, Billerica, MA) raised against large T antigen of SV40 polyomavirus.
3.5. Statistical analyses
Data were analyzed with IBM SPSS Statistics for Windows, Version 23.0 (IBM Corp, Armonk, NY). Descriptive analyses were used to report cohort characteristics, the comparison of the baseline characteristics and the association between the HPyV9 and TSPyV. The outcome variables were assessed using the χ2 test, the Fisher exact test, or the Student t test, as appropriate. Logistic regression was used to test whether donor and recipient HPyV9 or TSPyV seropositivity statistically modified the predictive effect of the BKPyV donor seroreactivity level on the development of BKPyV viremia. Tests were considered statistically significant with P < 0.05 in a two‐sided test.
4. RESULTS
4.1. Incidence of BKPyV viremia and BKPyVAN
In total, 209 KTx recipients were evaluated with regard to the incidence of BKPyV viremia and BKPyVAN within the first year after KTx. Fifty‐three recipients (25%) became BKPyV viremic, and four of them developed BKPyVAN (Table 1).
Table 1.
Basic and donor characteristics of KTx recipients sorted for the development of BKPyV viremia and BKPyVAN during the first year after transplantation
| All recipients (n = 209) | Viremic recipients(n = 53) | ||||||
|---|---|---|---|---|---|---|---|
| No BKPyV viremia (n = 156) | BKPyV viremia (n = 53) | P value | No BKPyVAN (n = 49) | BKPyVAN (n = 4) | P value | ||
| Recipient age, n (SD) | 49 (13.0) | 53 (14.1) | 0.113 | 52 (14.5) | 58 (5.8) | 0.131 | |
| Recipient male sex, n (%) | 96 (62) | 39 (74) | 0.113 | 36 (74) | 3 (75) | 1.000 | |
| Donor age, n (SD) | 51 (10.8) | 55 (11.3) | 0.028 | 55 (11.3) | 63 (8.6) | 0.160 | |
| Donor male sex, n (%) | 57 (37) | 21 (40) | 0.688 | 18 (37) | 3 (75) | 0.132 | |
Abbreviations: BKPyV, BK polyomavirus; BKPyVAN, BKPyV‐associated nephropathy; KTx, kidney transplantation; SD, standard deviation.
4.2. HPyV9 and TSPyV seropositivity and viremia among KTx donors and recipients
To investigate a possible effect on BKPyV infection and development of BKPyV viremia and BKPyVAN by coinfection with HPyV9 and TSPyV, pre‐KTx seropositivity for HPyV9 and TSPyV were determined in both donors and recipients. Forty‐five donors (22%) and 33 recipients (16%) were seropositive for HPyV9, and 184 donors (88%) and 186 recipients (89%) for TSPyV (Figure 1). HPyV9 and TSPyV seropositivity were distributed equally over age and sex of recipients (Table 2) and donors (not shown). When comparing HPyV9 and TSPyV seropositivity among donors and their corresponding recipients, associations were noticed between donor and recipient HPyV9 serostatus (P < 0.001), as well as TSPyV serostatus (P = 0.27) (Table 2). None of the recipients' developed HPyV9 viremia post‐KTx, while two developed TSPyV viremias.
Figure 1.
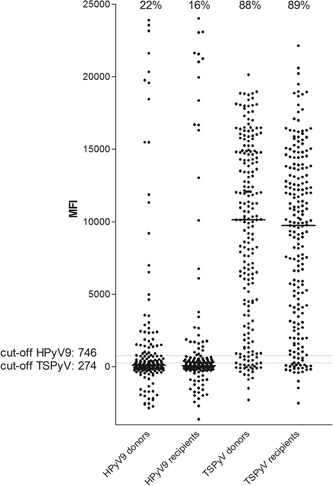
Pre‐KTx, HPyV9, and TSPyV seropositivity and seroreactivity of donors and recipients. HPyV9, human polyomavirus 9; KTx, kidney transplantation; MFI, median fluorescence intensity; TSPyV, trichodysplasia spinulosa polyomavirus
Table 2.
Distribution of TSPyV and HPyV9 seropositivity among 209 KTx recipients related to age, sex, and donor TSPyV and HPyV9 seropositivity
| Recipient HPyV9 serostatus | Recipient TSPyV serostatus | |||||
|---|---|---|---|---|---|---|
| Seronegative (n = 176) | Seropositive (n = 33) | P value | Seronegative (n = 23) | Seropositive (n = 186) | P value | |
| Age, n (SD) | 51 (13.5) | 48 (12.6) | 0.379 | 50 (12.0) | 50 (13.5) | 0.810 |
| Male sex, n (%) | 112 (64) | 23 (70) | 0.504 | 12 (52) | 123 (66) | 0.247 |
| Donor HPyV9 seropositivity, n (%) | 29 (17) | 16 (48) | <0.000 | 8 (35) | 37 (20) | 0.101 |
| Donor TSPyV seropositivity, n (%) | 154 (88) | 30 (91) | 0.580 | 17 (74) | 167 (90) | 0.027 |
Abbreviations: HPyV9, human polyomavirus 9; KTx, kidney transplantation; SD, standard deviation; TSPyV, trichodysplasia spinulosa polyomavirus.
4.3. Association between HPyV9 and TSPyV serostatus and development of BKPyV viremia and BKPyVAN
To investigate the association between TSPyV and HPyV9 serostatus and development of BKPyV infection and BKPyVAN in the first year after KTx, HPyV9 and TSPyV pre‐KTx seropositivity and seroreactivity (MFI value) of both donors and recipients were compared with the development of BKPyV viremia and BKPyVAN in recipients (Table 3). HPyV9 nor TSPyV serostatus was associated with the development of BKPyV viremia and BKPyVAN. For HPyV9, a positive association was found between pre‐KTx recipient seroreactivity and development of BKPyVAN within one year after transplantation (P = 0.007). This was not observed for BKPyV viremia. Donor nor recipient TSPyV seroreactivity was associated with the development of BKPyV viremia and BKPyVAN.
Table 3.
Pre‐KTx seropositivity and seroreactivity of donors and recipients sorted for BKPyV viremia and BKPyVAN during the first year after transplantation, shown for HPyV9 and TSPyV
| All recipients(n = 209) | Viremic recipients(n = 53) | |||||
|---|---|---|---|---|---|---|
| No BKPyV viremia (n = 156) | BKPyV viremia (n = 53) | P value | No BKPyVAN (n = 49) | BKPyVAN (n = 4) | P value | |
| HPyV9 | ||||||
| Donor seropositivity, n (%) | 36 (23) | 9 (17) | 0.351 | 40 (82) | 4 (100) | 1.000 |
| Donor seroreactivity, MFI value (SD) | 1458 (4769) | 1045 (4189) | 0.575 | 1123 (4350) | 90 (348) | 0.640 |
| Recipient seropositivity, n (%) | 28 (18) | 5 (9) | 0.142 | 4 (8) | 1 (25) | 0.336 |
| Recipient seroreactivity, MFI value (SD) | 1627 (5315) | 632 (3747) | 0.208 | 245 (2526) | 5371 (10432) | 0.007 |
| TSPyV | ||||||
| Donor seropositivity, n (%) | 138 (86) | 46 (87) | 0.746 | 42 (86) | 4 (100) | 1.000 |
| Donor seroreactivity, MFI value (SD) | 9384 (6396) | 8399 (5569) | 0.287 | 8601 (5676) | 5921 (3623) | 0.243 |
| Recipient seropositivity, n (%) | 141 (90) | 45 (85) | 0.271 | 41 (84) | 4 (100) | 1.000 |
| Recipient seroreactivity, MFI value (SD) | 8959 (5868) | 8321 (6083) | 0.507 | 8311 (6179) | 8446 (5523) | 0.965 |
Abbreviations: BKPyV, BK polyomavirus; BKPyVAN, BKPyV‐associated nephropathy; MFI, median fluorescence intensity; SD, standard deviation.
The strong association between pre‐KTx donor BKPyV seroreactivity and recipient BKPyV viremia and BKPyVAN, reported previously,5 was not influenced by the donor or recipient pre‐KTx HPyV9 or TSPyV serostatus or seroreactivity (data not shown).
5. DISCUSSION
Prompted by an increased BK viremia rate previously observed among HPyV9‐infected KTx recipients, we investigated whether HPyV coinfection, especially with HPyV9 and TSPyV is of influence to the development of BKPyV viremia and BKPyVAN in KTx recipients. Therefore, we determined the HPyV9 and TSPyV seroresponses in donor and recipient serum samples collected before KTx and detected the presence of HPyV9 and TSPyV DNA in blood plasma samples of KTx recipients collected within one year after KTx.
HPyV9 and TSPyV seroreactivity was comparable between donors and recipients, and showed the same distribution pattern as known from other immunocompetent and immunosuppressed populations: a high overall number of TSPyV seropositives, and approximately 20% seropositivity for HPyV9.2, 20 In comparing HPyV9 and TSPyV seroresponses among donors and recipients, we observed statistically significant associations between donor and recipient seropositivity for both viruses. Since the analyzed cohort consists of living donors, a considerable part of the donors and recipients is related as parent‐child or as siblings and might have had similar virus exposure at a younger age while sharing the same household. Indeed, Pedergnana et al21 found indication for TSPyV transmission from parents to children, and between siblings. Currently the mechanism for transmission of HPyV9 remains unidentified; however, Karachaliou et al22 identified younger age at day‐care entry as risk factor.23 Alternatively, household/family members might share genetic risk factors and glycan receptors relevant for these viruses, for example, the acquisition of the HPyV9 glycan receptor ligand, which Khan et al24 speculated to be acquired through the consumption of red meat and milk. Although we were informed about the (familial) relation between donor and recipient (sibling, parent, other families, partner, friends, or strangers), no trend towards a relationship with increased risk of HPyV9 infection was found (data not shown).
When we compared the incidence of BKPyV viremia and BKPyVAN in HPyV9 and TSPyV seropositive recipients, we did not observe specific associations to suggest that the BKPyV infection was related to previous or coincident HPyV9 or TSPyV infection. Only the MFI values for HPyV9 were significantly higher in BKPyV viremic recipients that developed BKPyVAN (P = 0.007). However, because this concerned only four recipients developing nephropathy, we are reluctant to conclude anything from this observation.
Previously we reported 21% HPyV9 DNA‐positive KTx recipients in a cohort of kidney‐pancreas transplant patients with deceased donors.17 This percentage is much higher than the prevalence found in this study where none of the recipients turned HPyV9 DNA‐positive. This discrepancy might be explained by a number of things. First, the cohort of the previous study contained primarily deceased donors, which conceivably could have a different health status than healthy living donors. Second, a substantial amount of the recipients in the previous cohort received kidney‐pancreas transplantation, which has been shown to have a higher chance of developing late‐onset BKPyVAN.25 Furthermore, the overrepresentation of diabetes mellitus type I patients in the kidney‐pancreas transplant cohort might have introduced additional, unknown risk factors for HPyV9 infection.
The 0.5% prevalence of TSPyV viremia that we observed in the KTx patients cohort reported here is lower than previously found by Urbano et al,26 who reported a prevalence of 27% in the KTx patients.27 We assume that different immunosuppressive regimens or geographical differences explain the discrepancy in the prevalence of TSPyV infection in the primarily Dutch and Argentinian cohorts.
Recent data from our lab published by Wunderink et al5 suggest that the BKPyV infection post‐KTx originates from the donor and that the level of donor BKPyV IgG seroreactivity can be used to predict the development of BKPyV viremia and BKPyVAN in the recipient. Our current data showed that the donor and recipient HPyV9 and TSPyV seroreactivity do not modify the predictive effect of donor BKPyV seroreactivity on the development of BKPyV infection in the recipient.
In summary, in this cohort of living KTx donor‐recipient pairs, we did not find evidence for relevant HPyV9 and TSPyV infections occurring in immunosuppressed recipients after KTx. Furthermore, we did not find an association between the HPyV9 or TSPyV infection and the development of BKPyV viremia and BKPyVAN, and also no modifying effect of HPyV9 or TSPyV serostatus or seroreactivity on the prediction of BKPyV viremia and BKPyVAN based on pre‐KTx donor BKPyV seroreactivity. Altogether, we did not find indication for complementarity or synergy between the HPyV9, TSPyV, and BKPyV infection. Therefore, further study into the role of HPyV9 and TSPyV in the development of BKPyVAN does not seem warranted.
ACKNOWLEDGMENT
Aline van Rijn was supported through an excellent student grant by the Leiden University Medical Center.
van Rijn AL, Wunderink HF, de Brouwer CS, van der Meijden E, Rotmans JI, Feltkamp MCW. Impact of HPyV9 and TSPyV coinfection on the development of BK polyomavirus viremia and associated nephropathy after kidney transplantation. J Med Virol. 2019;91:1142–1147. 10.1002/jmv.25397
References
REFERENCES
- 1. Gossai A, Waterboer T, Nelson HH, et al. Seroepidemiology of human polyomaviruses in a US population. Am J Epidemiol. 2016;183(1):61‐69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Van der Meijden E, Bialasiewicz S, Rockett RJ, Tozer SJ, Sloots TP, Feltkamp MCW. Different serologic behavior of MCPyV, TSPyV, HPyV6, HPyV7 and HPyV9 polyomaviruses found on the skin. PLOS One. 2013;8(11):e81078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chesters PM, Heritage J, McCance DJ. Persistence of DNA sequences of BK virus and JC virus in normal human tissues and in diseased tissues. J Infect Dis. 1983;147(4):676‐684. [DOI] [PubMed] [Google Scholar]
- 4. Boldorini R, Veggiani C, Barco D, Monga G. Kidney and urinary tract polyomavirus infection and distribution: molecular biology investigation of 10 consecutive autopsies. Arch Pathol Lab Med. 2005;129(1):69‐73. [DOI] [PubMed] [Google Scholar]
- 5. Wunderink HF, Van Der Meijden E, Van Der Blij‐de Brouwer CS, et al. Pretransplantation donor‐recipient pair seroreactivity against BK polyomavirus predicts viremia and nephropathy after kidney transplantation. Am J Transplant. 2017;17(1):161‐172. [DOI] [PubMed] [Google Scholar]
- 6. Sood P, Senanayake S, Sujeet K, et al. Management and outcome of BK viremia in renal transplant recipients: a prospective single‐center study. Transplantation. 2012;94(8):814‐821. [DOI] [PubMed] [Google Scholar]
- 7. Hirsch HH, Knowles W, Dickenmann M, et al. Prospective study of polyomavirus type BK replication and nephropathy in renal‐transplant recipients. N Engl J Med. 2002;347(7):488‐496. [DOI] [PubMed] [Google Scholar]
- 8. Moens U, Calvignac‐Spencer S, Lauber C, et al. ICTV virus taxonomy profile: polyomaviridae. J Gen Virol. 2017;98(6):1159‐1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Van Der Meijden E, Janssens RWA, Lauber C, Bouwes Bavinck JN, Gorbalenya AE, Feltkamp MCW. Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient. PLOS Pathog. 2010;6(7):e1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Van Der Meijden E, Horváth B, Nijland M, et al. Primary polyomavirus infection, not reactivation, as the cause of trichodysplasia spinulosa in immunocompromised patients. J Infect Dis. 2017;215(7):1080‐1084. [DOI] [PubMed] [Google Scholar]
- 11. Urbano PRP, Pannuti CS, Pierrotti LC, David‐Neto E, Romano CM. Rapid detection of trichodysplasia spinulosa‐associated polyomavirus in skin biopsy specimen. Genome Announc. 2014;2(4):e00694‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ho J, Jedrych JJ, Feng H, et al. Human polyomavirus 7‐associated pruritic rash and viremia in transplant recipients. J Infect Dis. 2015;211(10):1560‐1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tan BH, Busam KJ. Virus‐associated trichodysplasia spinulosa. Adv Anat Pathol. 2011;18(6):450‐453. [DOI] [PubMed] [Google Scholar]
- 14. Šroller V, Hamšíková E, Ludvíková V, Musil J, Němečková Š, Saláková M. Seroprevalence rates of HPyV6, HPyV7, TSPyV, HPyV9, MWPyV, and KIPyV polyomaviruses among the healthy blood donors. J Med Virol. 2016;88(7):1254‐1261. [DOI] [PubMed] [Google Scholar]
- 15. Scuda N, Hofmann J, Calvignac‐Spencer S, et al. A novel human polyomavirus closely related to the african green monkey‐derived lymphotropic polyomavirus. J Virol. 2011;85(9):4586‐4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kamminga S, Van Der Meijden E, Feltkamp MCW, Zaaijer HL. Seroprevalence of fourteen human polyomaviruses determined in blood donors. PLOS One. 2018;13:e0206273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Van Der Meijden E, Wunderink HF, Van Der Blij‐de Brouwer CS, et al. Human polyomavirus 9 infection in kidney transplant patients. Emerging Infect Dis. 2014;20(6):991‐999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Smedile A, Verme G, Cargnel A, et al. Influence of delta infection on severity of hepatitis B. Lancet. 1982;2(8305):945‐947. [DOI] [PubMed] [Google Scholar]
- 19. Csoma E, Bidiga L, Mehes G, Gergely L. No evidence of human polyomavirus 9, WU, and KI DNA in kidney and urinary bladder tumour tissue samples. Pathobiology: journal of immunopathology. Mol Cell Biol. 2016;83(5):252‐257. [DOI] [PubMed] [Google Scholar]
- 20. Kamminga S, Van Der Meijden E, Wunderink HF, Touzé A, Zaaijer HL, Feltkamp MCW. Development and evaluation of a broad bead‐based multiplex immunoassay to measure IgG seroreactivity against human polyomaviruses. J Clin Microbiol. 2018;56(4):e01566‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pedergnana V, Martel‐Jantin C, Nicol JTJ, et al. Trichodysplasia spinulosa polyomavirus infection occurs during early childhood with intrafamilial transmission, especially from mother to child. J Invest Dermatol. 2017;137(5):1181‐1183. [DOI] [PubMed] [Google Scholar]
- 22. Karachaliou M, Waterboer T, Casabonne D, et al. The Natural history of human polyomaviruses and herpesviruses in early life—the Rhea Birth Cohort in Greece. Am J Epidemiol. 2016;183(7):671‐679. [DOI] [PubMed] [Google Scholar]
- 23. Hampras SS, Giuliano AR, Lin HY, et al. Natural history of polyomaviruses in men: the HPV infection in men (HIM) study. J Infect Dis. 2015;211(9):1437‐1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Khan ZM, Liu Y, Neu U, et al. Crystallographic and glycan microarray analysis of human polyomavirus 9 VP1 identifies N‐glycolyl neuraminic acid as a receptor candidate. J Virol. 2014;88(11):6100‐6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Imlay H, Whitaker K, Fisher CE, Limaye AP. Clinical characteristics and outcomes of late‐onset BK virus nephropathy in kidney and kidney‐pancreas transplant recipients. Transpl Infect Dis. 2018;20:e12928. [DOI] [PubMed] [Google Scholar]
- 26. Urbano PR, Nali LHS, Bicalho CS, et al. New findings about trichodysplasia spinulosa‐associated polyomavirus (TSPyV)‐‐novel qPCR detects TSPyV‐DNA in blood samples. Diagn Microbiol Infect Dis. 2016;84(2):123‐124. [DOI] [PubMed] [Google Scholar]
- 27. Hirsch HH, Steiger J. Polyomavirus BK. Lancet Infect Dis. 2003;3(10):611‐623. [DOI] [PubMed] [Google Scholar]


