Abstract
Pulmonary fibrosis represents a leading cause of morbidity and mortality worldwide. Therapy induced lung fibrosis constitutes a pivotal dose‐limiting side effect of radiotherapy and other anticancer agents. We aimed to develop objective criteria for assessment of fibrosis and discover pathophysiological and molecular correlates of lung fibrosis as a function of fractionated whole thoracic irradiation. Dose–response series of fractionated irradiation was utilized to develop a non‐invasive and quantitative measure for the degree of fibrosis – the fibrosis index (FI). The correlation of FI with histopathology, blood‐gas, transcriptome and proteome responses of the lung tissue was analyzed. Macrophages infiltration and polarization was assessed by immunohistochemistry. Fibrosis development followed a slow kinetic with maximum lung fibrosis levels detected at 24‐week post radiation insult. FI favorably correlated with radiation dose and surrogates of lung fibrosis i.e., enhanced pro‐inflammatory response, tissue remodeling and extracellular matrix deposition. The loss of lung architecture correlated with decreased epithelial marker, loss of microvascular integrity with decreased endothelial and elevated mesenchymal markers. Lung fibrosis was further attributed to a switch of the inflammatory state toward a macrophage/T‐helper cell type 2‐like (M2/Th2) polarized phenotype. Together, the multiscale characterization of FI in radiation‐induced lung fibrosis (RILF) model identified pathophysiological, transcriptional and proteomic correlates of fibrosis. Pathological immune response and endothelial/epithelial to mesenchymal transition were discovered as critical events governing lung tissue remodeling. FI will be instrumental for deciphering the molecular mechanisms governing lung fibrosis and discovery of novel targets for treatment of this devastating disease with an unmet medical need.
Keywords: pulmonary fibrosis, M2 macrophages, Th2‐like response, fractionated radiotherapy, EMT/EndoMT, radiation‐induced lung fibrosis
Short abstract
What's new?
The development of fibrosis scar tissue in the lungs is a dose‐limiting effect of radiotherapy for thoracic malignancies. Molecular mechanisms driving radiation‐induced lung fibrosis (RILF), however, remain unclear. In this study, a fibrosis index (FI) was devised to quantitatively detect spatial and temporal kinetics of lung fibrosis development. Multi‐scale characterization of FI uncovered mechanisms governing lung fibrosis, including perturbation of immune balance and microvascular integrity. Radiation dose and FI were correlated with an inflammatory switch toward a macrophage/T‐helper cell type 2‐like polarized phenotype. The findings open the way for further mechanistic study and the discovery of therapeutic targets for RILF.
Introduction
Pulmonary fibrosis is as an undesired effect of radiotherapy, chemotherapy and some targeted anti‐cancer agents.1 Patient genetic predisposition, the volume of irradiated lung and combination regimens consisting of concurrent chemotherapy are correlated with increased risk of radiation‐induced toxicity in lung.2 The clinical sequelae of radiation injury consist of an acute/subacute onset of radiation induced inflammation (pneumonitis) that is often responsive to steroid therapy.3, 4 In contrast, activation of the fibrogenesis processes and emergence of pulmonary fibrosis is considered a therapy refractory late effect of irradiation with appearance of pathophysiologic events comparable with idiopathic pulmonary fibrosis (IPF).5 Therefore, a better understanding of molecular mechanisms governing radiation‐induced lung fibrosis (RILF) may lead to discovery of novel biomarkers and targets for a highly prevalent family of fibrosis related diseases of various etiologies.6, 7
Chemotherapy and radiation induced lung fibrosis models are frequently utilized for preclinical evaluation of novel biomarker and candidate modulators of the fibrosis process.2, 8, 9, 10 However, the settings are often artificial, e.g., fibrosis develops after a very short latency period of days post intratracheal installation, or mice are often irradiated with a single high lethal dose thoracic irradiation. Moreover, there is an urgent need of detailed dose–response studies allowing a thorough correlation of molecular‐, cellular‐ and tissue level effects with the severity of induced injury. Finally, no consensus exists on how to measure quantitatively the degree of fibrosis, limiting cross study comparisons. Recent advances of implementing computed tomography (CT) is error‐prone, particularly suffering from the subjectivity and experience of the examiner. For example, selection of the region of interest (ROI) by observers for determining the lung density (Hounsfield Units, HU) is highly dependent on the skills of the experimenter to identify representative regions for the entire lung and to distinguish between e.g., a vessel rich area, normal lung and true tissue remodeling.
The present study aimed to address these limitations by developing an objective and robust CT‐based criteria of pulmonary fibrosis – the fibrosis index (FI). This model considers both, the decrease in lung volume representing the restrictive lung function as well as enhanced lung density resulting from tissue remodeling. In contrast to selecting ROI, implementation of a 3D‐segmentation algorithm further allows lung histogram profiling providing a more representative overview of the entire lung including the distribution and variation of gains in tissue density in fibrotic lungs. Therefore, FI Model was used to systematically analyze the pathophysiological parameters with the degree of lung fibrosis development. This comparative dose–response analysis allowed us to gain insight into alterations of immune response, extracellular matrix (ECM) remodeling, and transition of epithelial and endothelial marker to mesenchymal phenotype that were attributed to the fibrotic switch. Development of FI will thus facilitate mechanistic studies in pulmonary fibrosis.
Methods
Whole thoracic irradiation
Whole thoracic irradiation was administrated to 8–10 weeks old female C57BL/6 mice as described previously.11 In brief, photons beam was delivered by a 6 MV Artiste Linac (Siemens) at a dose rate of 3 Gy/min. The prescribed doses were including a single dose of 20 Gy in the pilot study and a fractionated irradiation of 10–40 Gy total dose in 2, 4, 6, 8 Gy per fractions administered over 5 consecutive days. Prior to irradiation, mice were anesthetized by an intraperitoneal application of 0.36 ml/kg Rompun 2% (Bayer HealthCare) and 0.54 ml/kg ketamine 10% (Pfizer). Mice were randomized prior to irradiation to each dose group (n:12). Monitoring of lung fibrosis development via CT‐imaging as well as analysis of blood and tissue samples were performed in parallel to our previous observations after a follow‐up period of 24 weeks post irradiation. All animal work was approved and performed in compliance with rules outlined by the local and governmental animal care committees (Regierungspraesidium, Karlsruhe).
Assessment of lung fibrosis by computed tomography (CT)
A clinical PET/CT scanner (Biograph mCT, Siemens) was applied for quantitative CT imaging pre‐ and post‐irradiation. The standard protocol employed for the CT portion of PET/CT was as follow: 80 kV with 80 mAs, a pitch of 0.6 mm, slice thickness of 0.6 mm and acquisition time of 32 s. X‐ray exposure is approximately 4.14 mGy per scan. Images were reconstructed using the filter kernel H50s (Siemens) into a transaxial FOV of 138 × 138 mm2 as a 512 × 512 matrix.
Images acquired from the clinical CT scanner were viewed and analyzed with Medical Imaging Interaction Toolkit (MITK) software and OsiriX Imaging Software (OsiriX v.3.9.4, Switzerland). The lung tissue density was measured by average Hounsfield unit (HU) intensities. The lung, together with all the microstructures, was thereby segmented using a 3D regional growing algorithm with a lower threshold of −900 HU and an upper threshold of −100 HU. Trachea and primary bronchi were manually resected upon segmentation. Volume sizes and mean HU values within the segmented area were calculated for quantitative assessment of pulmonary toxicity. The fibrosis index was employed to assess the extent of fibrosis as the major endpoint. Briefly, the FI model is based on two critical parameters derived from CT segmented data: the relative increase in mean lung density (ΔHU) and decreased lung volume (ΔV) when compared to the mean of an age‐matched reference cohort. Biologically, the augmented ΔHU is an overall representation of collagen deposition and increased cellularity; whereas ΔV reflects the nature of fibrosis as a restrictive lung disease. The calculation of FI is based on the proposed equation as:
The derived density and volume parameters (ΔHU and ΔV) may be biased in cases of combined pulmonary fibrosis and emphysema syndrome (CPFE),12 due to the presence of trapped air in the lung. A histogram of the same lung region can be extracted and further smoothened using a negative exponential smoother with a sampling proportion of 0.2. Herein, the concept of the ‘peak position of smoothened histogram’ (PPSH) was introduced for differential diagnosis of CPFE independent of experimenter's expertise. To exclude emphysema regions and allow quantitative assessment of fibrosis alone, a lower threshold (−450 HU) was applied in the regional growing segmentation. PPSH is therefore considered as a supplementary parameter, integrated into the 3D segmentation algorithm.
The region‐of‐interest (ROI)‐based assessment of lung density as described in Plathow et al.13 was also performed for comparison. In this approach, eight circular ROIs on transaxial slices were selected in the after areas: the right upper anterior and posterior regions, the left upper anterior and posterior regions, the right lower anterior and posterior regions, and the left lower anterior and posterior regions. Mean HU values for each ROI and all the eight ROIs were calculated.
Vascular corrosion casting
To imaging the 3D pulmonary vasculature, mice were anesthetized and perfused with heparinized saline until all of the visceral blood volume was flushed out. Pre‐mixed MICROFIL (MV‐122, Flow Tech Inc.) solution was injected into the right atrium and perfused the pulmonary vessels according to the manufacturer's protocol. The cleared specimen was scanned ex vivo by Inveon SPECT/PET/CT (at DKFZ Imaging Core Facility). The 3D vasculature was further reconstructed into 40 μm.
Micro‐CT imaging
Micro‐CT imaging using both the micro‐CT component of a prototype SPECT–CT‐OT system14 and Inveon SPECT/PET/CT (at DKFZ Imaging Core Facility). The detailed CT acquisition protocol was described in Supporting Information Methods. For prototype SPECT–CT‐OT system, CT acquisitions were performed at 40 kV tube voltage, 0.4 mA anode current, 1 s acquisition time per projection, 240 projections per 360° rotation. Images were reconstructed into a matrix of 512 × 512 × 1,024 with an isotropic voxel size of 0.065 mm. For Inveon SPECT/PET/CT, CT acquisition were applied as 80 kV tube voltage, 0.5 mA anode current, 1 s acquisition time per projection, 720 projections per 360° rotation, with an effective pixel size of 19.29 μm.
Radiological diagnosis
Fibrosis (ROI) and emphysema were evaluated at least by two experimentators independently. The presence of pulmonary fibrosis was determined using several radiologic criteria: diffuse ground glass opacities, architectural distortion and traction bronchiectasis; emphysema was identified from well‐demarcated areas of decreased attenuation in comparison with contiguous normal lungs.6 Combined pulmonary fibrosis and emphysema syndrome (CPFE) in C57BL/6 mice typically manifested as apically condensed or fibrous tissue and, basally at both sides, rarefaction of lung parenchyma or emphysematous.12 Final decisions were reached by consensus.
Blood gas analysis
Blood was collected via retro‐orbital puncture before loading on a standard i‐STAT cartridge. Blood gas measurement series (VetScan i‐STAT EG7+) consisting of the clinical chemistry parameters pH, pCO2, base excess (BE), hematocrit (Hk), hemoglobin (Hb) was evaluated at the endpoint of 24 weeks post ionizing radiation (IR).
Histopathology and immunohistochemistry
The lung sections were stained with H&E for morphology and leukocytes detection and Trichrome for collagen deposition at the endpoint of 24 weeks. Mice lungs were inflated and fixed with 4% neutral‐buffered formalin via trachea. Formalin fixation and paraffin embedding (FFPE) was performed after fixation for 48 h at 4 °C, and blocks were sectioned into slices 5 μm thick. The sections were stained with hematoxylin and eosin (H&E), pico‐sirius red and Masson's trichrome stain (also known as Afog) to examine morphology and collagen deposition status. The pathological diagnosis and scoring was performed together with two trained pathologist (MK and WW). Whole slice sections were imaged and scanned using a NanoZoomer system (v2.0, Hamamatsu). Immunohistochemistry staining was used to identify different populations of macrophages. Lung sections were stained with different macrophage surface markers: anti‐mannose receptor antibody (CD206, MRC) (#ab64693, Abcam), CCL22 (#ab124768, Abcam), mouse Fc gamma RII/RIII (CD16/32, Fcgr2/3) (#AF1460, R&D), F4/80 (#ab6640, Abcam), Arg1 (#60176, Abcam), Nos2 (iNOS) (#ab3523, Abcam) and CD11b (#ab8878, Abcam). Stained tissues were examined using microscopy (Leica DM IL LED). Scoring of macrophage infiltration and fibrosis was performed semi‐quantitatively. Briefly, lung specimen was inspected for selection of representative areas. For semi‐quantitative analysis, these high‐power fields (HPF) were observed in 200x magnification and scored as follows: no increase in macrophages = 0, mild increase in macrophages = 1, moderate increase in macrophages = 2, severe increase in macrophages = 3. Fibrosis was scored accordingly: no fibrosis = 0, mild fibrosis = 1, moderate fibrosis = 2 and severe fibrosis = 3.
Microarray analysis
Microarray analysis was performed as described.15, 16, 17 Briefly, after total‐RNA isolation and DNase digestion probes were labeled and hybridized against Mouse Sentrix‐6 V2 Whole Genome BeadChip using illumina's protocol at DKFZ Genomics & Proteomics Core Facility. The expression level of 45,281 mouse transcripts, variants, and EST clusters was detected by an average 30x redundancy for each transcript using gene‐specific 50mer oligonucleotides. Generation of expression matrices, data annotation, filtering and processing were performed using TableButler software package.18 Microarray data are available online at ArrayExpress (http://www.ebi.ac.uk/arrayexpress), under the accession number: E‐MTAB‐5922. To account for intensity variations in whole data sets, mean normalization (condition) to the same total intensity was performed. Gene expression ratios were built vs. non‐irradiated control samples using the geometric mean and log2‐transformed to account for symmetric representation of both down‐regulation and up‐regulation in subsequent statistical tests. To analyze the expression signature of radiation dose response, supervised and non‐supervised clustering as well as statistical methods was performed using Statistical Utilities for Microarray and Omics data software package (SUMO v1.61 s, www.oncoexpress.org).
Cellular composition analysis via CIBERSORT
To characterize the cellular composition of leukocytes infiltrates in the lung tissue, gene expression data were analyzed via CIBERSORT according to guidelines by Newman et al.19 In brief, cell‐type identification was made by computationally estimating relative subsets of RNA transcripts (http://cibersort.stanford.edu/). The designed leukocyte gene signature matrix (LM22) contains 547 genes and is able to discriminate among a wide range of human hematopoietic cell phenotypes e.g., different T‐cells types, B cells, dendritic cells, mast cells, monocytes, macrophages, eosinophils, neutrophils and natural killer cells.
Proteome analysis by mass spectrometry
For proteome analysis protein lysates were separated by SDS‐PAGE. The resulting gel was sliced into 13 slices that were tryptically digested and analyzed by mass spectrometry as described previously.20 In brief, after extraction, the obtained peptide mixture was separated using a nanoAcquity UPLC (Water GmbH, Eschborn, Germany) coupled online to an LTQ Orbitrap (Thermo Fisher Scientific, Bremen, Germany) mass analyzer. LC–MS/MS raw data were analyzed using the MaxQuant (version 1.5.3.8) software with a mouse UniProt database (downloaded: 06/07/2015; 76.089 sequences). Quantification in MaxQuant was performed using the built in MaxLFQ algorithm. Data transformation and evaluation was performed with the freely available Perseus software (v1.5.2.4).
Statistics
Data were shown as mean ± SEM unless otherwise stated. Two tailed t‐test was to compare between two independent groups. Two‐way ANOVA with a subsequent Bonferroni correction was performed to determine the significance of multiple comparisons using the SUMO software package.15, 16, 17 To estimate the linear corrections, we used least‐squares regression as well as the Pearson R test (GraphPad Prism Software v5.0). p < 0.05 was considered statistically significant.
Results
Development of the fibrosis index model based quantitative computed tomography
The three‐dimensional (3D) segmentation using a regional growing algorithm represented a successful strategy to delineate lung parenchyma for quantitative assessment of lung volume and lung density. The success of this method could be attributed to the large amount of air contained in the lung allowing segmentation of a relatively homogenous density compared to the surrounding tissue (Fig. 1 a and c). By plotting the lung density distribution from the segmented matrix in 4 weeks intervals a gradual right shift was found in histogram and heatmap presentations correlating with the time dynamic of fibrosis development in 20 Gy irradiated lungs (Fig. 1 b and d). From 16 weeks post‐IR, a gradual reduction of the ventilated lung tissue and increased frequency of condensed lung parenchyma indicated the onset of the fibrosis process. A dramatic loss of air‐containing healthy tissue as a consequence of severe fibrosis and deteriorated pulmonary ventilation was established at 20 weeks onwards post‐IR. Increased mean lung density (ILD) represented by changes in CT Hounsfield Units (ΔHU), as well as decreased lung volume (DLV, represented as ΔV), both vs. non‐irradiated age matched control lung, were the two key parameters obtained from the 3D lung segmentation. ILD elevated as a function of weeks post radiotherapy, whereas the lung volumes gradually shrinked (DLV). Both parameters saturated around 24 weeks after IR (ILD = 124 HU, DLV = 0.327 cm3) (Fig. 1 e). In line with quantitative clinical CT data, morphological signs of lung fibrosis e.g., ground glass opacities and architectural distortion were incrementally observed in irradiated lungs after 16 weeks by high‐resolution micro‐CT analysis (Fig. 1 b). Based on the inverse relationship of the two central parameter, ILD and DLV (Fig. 1 e), we proposed a preclinical fibrosis index (FI) model for sophisticated characterization of fibrosis development. The FI developed with slow kinetics and raised sharply after 16 weeks (i.e., FI = 1.82, 5.78 for 16 and 20 weeks, respectively) until reaching the maximum at 24 weeks (FI = 6.34) post‐irradiation. As a result, 24‐week interval was revealed as a reliable and practical endpoint to quantifying pulmonary fibrosis after radiation exposure utilizing fibrosis index model (Fig. 1 f).
Figure 1.
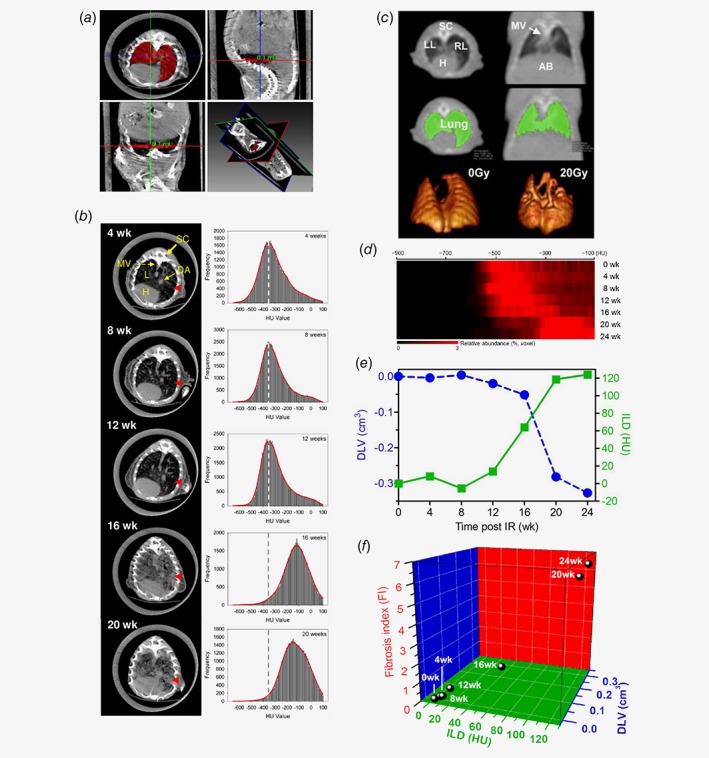
Development of the fibrosis index (FI) model based on longitudinal clinical CT assessment. (a) Manual segmentation of lung from micro‐CT. (b) Left: representative axial CT scan images of progressive lung fibrosis development after whole thoracic irradiation of 20 Gy. red arrow: massive pulmonary fibrosis in the lung parenchyma at the late stage. Right: lung tissue density (HU) histograms based on segmented lung micro‐CT showing a right‐shift over time indicating the temporal development kinetics of lung fibrosis after irradiation. Dash line: the baseline according to segmented CT matrix from mice prior to IR. (c) Representative three‐dimensional (3D) segmentation of the non‐irradiated and 20 Gy irradiated lungs based on clinical CT measurement. (d) Profiling of 20 Gy irradiated lung density histograms converted into a heatmap view, each raw representing averaged 3D segmented lung density of n:6 mice, indicates a robust right‐shift over a follow‐up period of 24 weeks, which is consistent with micro‐CT readouts. (e) Inverse pattern of mean differential lung density (ILD, ΔHU, dark green) and total lung volume (DLV, ΔV, blue) in irradiated vs. sham treated lungs. (f) Fibrosis index (FI) as a function of density (ILD) and volume (DLV) parameters over a follow‐up period of 24‐week post 20 Gy whole thoracic irradiation, i.e., 0, 4, 8, 12, 16, 20 and 24 weeks. The parameter volume is more weighted in the middle phase and the density in the end phase, both of which contributed to a significant increase of FI between 16 and 24 weeks. This 3D model is able to identify the best timepoint for establishment of the non‐invasive CT based fibrosis index at 24 weeks. Abbreviations: SC = spinal cord, LL = left lung, RL = right lung, L = lung, H = heart, MV = mediastinal vascular structures, AB = abdomen; DA = descending aorta, HU = Hounsfield unit. [Color figure can be viewed at wileyonlinelibrary.com]
Correlation of blood gas based clinical chemistry parameters with FI model
Lung is a radiation fractionation sensitive organ. We next sought to study FI in fractionated dose‐escalation model. At the endpoint of 24 weeks post‐IR severe pulmonary fibrosis was observed in mouse treated with high fractionated doses, i.e., X 6 Gy × 5 f. and X 8 Gy × 5 f. (Fig. 2 a). FI values for each dose group were determined (Fig. 2 b). Blood gas analysis was performed to evaluate respiratory function and acid/base balance. Direct measurements of hydrogen ion activity (pH), partial pressures of carbon dioxide (pCO2), base excess (BE), hematocrit (Hct) and total hemoglobin (Hb) with different doses were obtained (Fig. 2 c). Linear regression analysis between the clinical chemistry readouts and corresponding FI demonstrated an excellent correlation with R 2 = 0.96, 0.97, 0.81, 0.91, 0.97, respectively.
Figure 2.

Validation of the fibrosis index (FI) with blood gas analysis in a fractionated dose‐escalation model at the endpoint of 24‐week post IR. (a) A clinical CT transversal view of lungs irradiated with a range of dose schedules: 0 Gy, 2 Gy, 4 Gy, 6 Gy, 8 Gy per fraction for a total of 5 fractions. (b) The calculated FI with reference to each dose group. (c) FI correlates well with clinical chemistry readouts relevant to pulmonary fibrosis, i.e., potential of hydrogen (pH), partial pressure of CO 2 (pCO 2), base excess (BE), hematocrit (Hct) and hemoglobin (Hb), with correlation coefficient, R 2 = 0.96, 0.97, 0.81, 0.91 and 0.97, respectively.
Discovery of a dose‐dependent induction of a M2‐like polarized immune phenotype in fibrotic lungs
To determine the molecular and immunological fingerprint of FI, genome‐wide transcriptional analysis was applied to study the initiation and perpetuation of RILF. Lung inflammation‐associated signature genes (e.g., kynureninase (KYNU), integrin‐β2 (ITGB2), cathepsin D (CTSD), bone marrow stromal cell antigen 1 (BST1); correlation coefficient = 0.923, 0.914, 0.898, 0.861, respectively) were augmented as a function of radiation doses, particularly above 6 Gy × 5 fx. The activation of macrophages markers was found e.g., cathepsin K (CTSK), leukocyte immunoglobulin like receptor B4 (LILRB4), CD68, glycoprotein nmb (GPNMB), B‐cell leukemia/lymphoma 2 related protein A1D (BCL2A1D), B‐cell leukemia/lymphoma 2 related protein A1C (BCL2A1C), B‐cell leukemia/lymphoma 2 related protein A1B (BCL2A1B); correlation coefficient = 0.949, 0.935, 0.928, 0.880, 0.867, 0.861, 0.854, respectively, which correlated well with FIs (Pearson R = 0.75, p < 0.0001) (Fig. 3 a).
Figure 3.
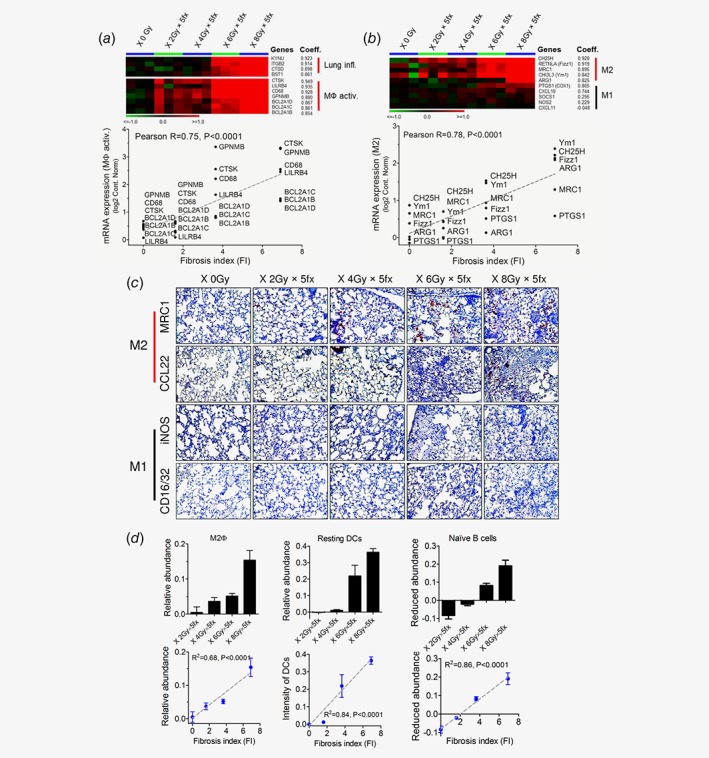
(a) Expression signatures for lung inflammation and pan‐macrophage activation correlated with the corresponding CT‐derived fibrosis index (Pearson R = 0.75, p < 0.0001). (b) A panel of markers for alternative activated macrophages (M2) was dose dependently up‐regulated in absence of M1 surrogates. Significant correlation between FI and regulation of the M2‐like gene‐signature was found (Peason R = 0.78, p < 0.0001). (c) Immunohistochemistry confirmed massive M2 macrophages infiltration in the fibrotic lung regions. MRC1/CD206 and CCL22 was employed as prototypic markers of M2 macrophages, whereas iNOS and CD16/32 were utilized as M1 macrophages surface markers (20× magnification). (d) Characterization of immunes cell compositions in irradiated lungs by CIBERSORT analysis algorithm. M2 macrophages (Left), resting dendritic cells (Middle) as well as naïve B cells (Right) levels well correlated with radiation dose/FI (R 2 = 0.68, 0.84 and 0.86, respectively, all p < 0.0001). [Color figure can be viewed at wileyonlinelibrary.com]
Induction of M2 macrophages was determined by a panel of M2‐like signature genes,21, 22 i.e., cholesterol 25‐hydroxylase (CH25H), resistin like β (Fizz1), mannose receptor C‐type 1 (MRC1), Chitinase‐3‐like protein 3 (Ym1), arginase 1 (ARG1), prostaglandin‐endoperoxide synthase 1 (COX1); correlation coefficient = 0.920, 0.919, 0.895, 0.842, 0.825, 0.805, respectively; in contrast, poor stimulation of classic macrophages M1 signature genes, i.e., C‐X‐C motif chemokine ligand 10 (CXCL10), suppressor of cytokine signaling 1 (SOCS1), nitric oxide synthase 2 (NOS2), C‐X‐C motif chemokine ligand 11 (CXCL11); correlation coefficient = 0.744, 0.295, 0.229, −0.048, respectively, was observed (Fig. 3 b). The activation of M2 macrophages was correlated well with FIs (Pearson R = 0.78, p < 0.0001, Fig. 3 b). This was further confirmed by massive infiltration of M2 macrophages (MRC1+, CCL22+) but not M1 macrophages (CD16/32+, iNOS+) into the fibrotic regions by immunohistochemistry (IHC) staining (Fig. 3 c). Collectively, M2‐like phenotype was demonstrated to be the dominant macrophage population in RILF.
Because our data indicated a prominent role of immune response in fibrogenesis, we next sought to characterize alterations in the diversity of leukocytes subsets using CIBERSORT analysis. The activation of M2 macrophages at higher doses was confirmed by a strong correlation with FIs (R 2 = 0.68, p < 0.0001, Fig. 3 d). Further, enhanced number of resting dendritic cells (DCs) was observed at higher doses, indicating a good linear correlation between resting DCs and FIs (R 2 = 0.84, p < 0.0001, Fig. 3 d). In contrast, a reduced level of naïve B cells was observed after increased radiation doses exposure. The infiltration status of naïve B cells correlated negatively with FIs (R 2 = 0.85, p < 0.0001, Fig. 3 d). The dose‐dependent modulations of these three populations serve as an immunological sign of a fibrotic switch after radiation exposure.
Correlation of a Th2‐biased immune response and extracellular matrix remodeling with FI
M2 macrophages are thought to promote T‐helper type 2 (Th2)‐like polarized immune response. In line, expression of a panel of genes attributed to the Th2‐like transcriptional signature was highly correlating with FI values i.e., coagulation factor VII (F7), C‐C motif chemokine ligand 6 (CCL6), thromboxane‐A synthase 1 (TBXAS1), suppressor of cytokine signaling 3 (SOCS3), solute carrier family 26 member 4 (SLC26A4), chitinase‐like protein 4 (CHI3L4), C‐type lectin domain containing 7A (CLEC7A); correlation coefficient = 0.925, 0.915, 0.912, 0.905, 0.890, 0.845, 0.803, respectively. Conversely, no regulation of Th1 related markers was found, i.e., signal transducer and activator of transcription 3 (STAT3), signal transducer and activator of transcription 4 (STAT4), interferon gamma (IFNG), signal transducer and activator of transcription 1 (STAT1), eomesodermin (EOMES), T‐Box 21 (TBX21); correlation coefficient = 0.249, 0.209, −0.075, −0.321, −0.408, −0.851, respectively (Fig. 4 a). This immune response was therefore Th2‐like polarized, and the extent of predominance correlated with FI (Pearson R = 0.83, p < 0.0001, Fig. 4 a). A number of Th2‐linked genes involved in inflammation and matrix remodeling were found to be dose‐dependently increased i.e., lysyl oxidase (LOX), matrix metalloproteinase‐2 (MMP2), tissue inhibitor of metalloproteinases 1 (TIMP1), heme oxygenase 1 (HMOX1), arginase 2 (ARG2), fibrillin 1 (FBN1); correlation coefficient = 0.934, 0.924, 0.916, 0.852, 0.837, 0.704, respectively. The expression of these Th2‐linked signature genes further well correlated with FI (Pearson R = 0.78, p < 0.0001, Fig. 4 b).
Figure 4.

Th2‐like polarized response and ECM remodeling correlated with FI. Expression of genes involved in Th2‐like response (a), linked to Th2‐like response (b) or tissue remodeling (c) showed high correlation with FI, Pearson R = 0.83, 0.78 and 0.78, respectively (p < 0.0001). (d) Dose dependent massive infiltration of inflammatory cells, collagen deposition and distorted lung parenchyma at 24‐week post IR assessed by H&E and Masson's trichrome staining (20× magnification). FI correlated well with the relative macrophage and fibrosis scores (Pearson R = 0.97 and 0.99, respectively, both p < 0.05). [Color figure can be viewed at wileyonlinelibrary.com]
A dose dependent shift in extracellular matrix (ECM) composition toward a fibrotic phenotype was observed. Genes encoding extracellular proteins and proteases were highly up‐regulated with escalated radiation doses i.e., tenascin C (TNC), cadherin related family member 1 (PCDH21), matrix metalloproteinase 14 (MMP14), collagen type I α2 chain (COL1A2), matrix metalloproteinase 12 (MMP12), fibulin 2 (FBLN2), elastin (ELN), fibrinogen α chain (FGA); correlation coefficient = 0.950, 0.949, 0.881, 0.834, 0.830, 0.824, 0.823, 0.738, respectively (Fig. 4 c). Further, a strong correlation between expression of these genes and FI was found (Pearson R = 0.78, p < 0.0001) (Fig. 4 c). Histopathological examinations of lung tissue revealed a dose‐dependent influx of leukocytes, collagen deposition (blue Afog/Trichrom stain) and distorted parenchymal structures (Fig. 4 d). This scenario was further confirmed by enhanced macrophage infiltration and fibrosis scores that strongly correlated with the degree of FI (Pearson R = 0.97 and 0.99, respectively; both p < 0.05) (Fig. 4 d).
Loss of microvascular integrity and gain of mesenchymal phenotype correlated with FI
Transcriptome analysis revealed a gradually dose‐dependent expression of mesenchymal markers in irradiated lungs i.e., fibronectin 1 (FN1), S100 calcium binding protein A14 (S100A14), S100 calcium binding protein A1 (S100A1), collagen type I α1 chain (COL1A1), collagen Type III α1 Chain (COL3A1), fibroblast‐specific protein‐1 (FSP‐1); correlation coefficient = 0.931, 0.920, 0.831, 0.789, 0.773, 0.627, respectively. In contrast, endothelial markers were clearly decreased with radiation doses (CD31, vascular endothelial cadherin (VE‐cadherin alias CDH5), TEK receptor tyrosine kinase (Tie2); correlation coefficient = −0.775, −0.948, −0.956, respectively). In addition, the epithelial tissue components were slightly reduced (desmoplakin (DSP), nidogen 1 (NID1), tight junction protein 1 (TJP1) and epithelial cadherin (E‐cadherin alias CDH1); correlation coefficient = 0.261, −0.157, −0.705, −0.735, respectively) (Fig. 5 a). In line, FI correlated well with the loss of endothelial (Pearson R = −0.93, p < 0.0001) and gain of mesenchymal markers (Pearson R = 0.61, p < 0.001) (Fig. 5 a ). Proteomics analysis confirmed elevated levels of mesenchymal proteins (COL1A1, vimentin and CD44) while epithelial (NID1, TJP1, E‐cadherin) and endothelial proteins (VE‐cadherin, CD31 and CD34) were significantly downregulated in fibrotic lungs (both p < 0.001) (Fig. 5 b ). Consistent with transcriptional and proteomic findings, a remarkable reduction of microvascular density and vascular 3D structures was found in irradiated (8Gy × 5fx) vs. non‐irradiated lungs by vascular corrosion casting and ex‐vivo ultra‐high‐resolution CT (Fig. 5 c ).
Figure 5.
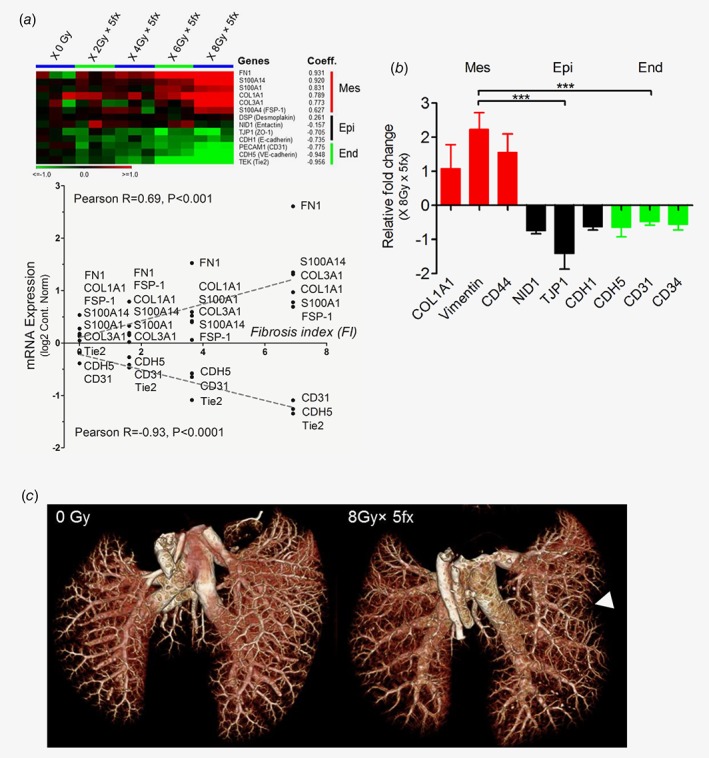
Epithelial and endothelial transition to mesenchymal phenotype correlated with a loss of vascular integrity. (a) Expression of epithelial and endothelial markers decreased as a function of radiation doses, in contrast to the significant increase of mesenchymal marker. Accordingly, FI correlated well with the gain of mesenchymal marker (Pearson R = 0.61, p < 0.001), while it was inversely correlated with expression of the endothelial markers (Pearson R = −0.93, p < 0.0001). (b) Proteomic analysis confirmed this EMT/EndoMT phenotype in fibrotic lungs post IR (***p < 0.001). (c) Vascular remodeling and reduced microvessel density (arrow) of the irradiated lung was observed by corrosion casting and high‐resolution micro‐CT ex vivo vascular imaging. [Color figure can be viewed at wileyonlinelibrary.com]
Discussion
In addition to the idiopathic pulmonary fibrosis (IPF) with high prevalence, therapy induced lung fibrosis constitutes a pivotal dose‐limiting side effect of radio‐(chemo)‐therapy and targeted anti‐cancer agents.1 To dissect the molecular mechanisms governing pulmonary fibrosis and develop novel biomarker and therapeutic targets robust models and reliable surrogates are needed. We show a time kinetics of fibrosis development (i.e., pixel histogram, CT heatmap, increased lung density, decreased lung volume), to identify the best time point for establishment of the novel non‐invasive CT based fibrosis index. The FI‐model circumvents current shortcomings of standard CT‐measures that employ subjective while operator dependent delineation of region of interests (ROI) to estimate lung density. our study was next to determine first the time point where the fibrosis was highest and then to determine the dose/FI vs. all other parameters. The time kinetics of FI was illustrated in Figure 1 (b, d‐f) based on a single dose exposure of 20 Gy. According to the Figure 1 f, the 3D model suggested that 24‐week is a reliable endpoint to quantifying pulmonary fibrosis after radiation exposure using FI model. Dose series were investigated by an arsenal of currently available surrogates of fibrosis, including investigation of tissue architecture, cellular composition and ECM deposition as well as clinical chemistry parameters evaluating respiratory function and acid/base balance. All parameters were studied in lungs irradiated with escalated fractionation doses (0Gy, 2Gy × 5‐fx, 4Gy × 5‐fx, 6Gy × 5‐fx, 8Gy × 5‐fx) at the endpoint of 24 weeks. The degree of fibrosis determined by FI correlated well with a panel of histological, clinical chemistry and molecular surrogates (Table. 1). In contrast to the standard invasive parameters like histology, FI‐model allows longitudinal investigation as e.g., it was utilized in 4‐week intervals in our study to determine the dynamic of fibrosis development over the period of 24 weeks (Fig. 1 d). Next, by interrogating the lung transcriptome, transcriptional signatures were discovered correlating with radiation dose and consequently with the degree of fibrosis determined by FI. These include, activation of Th2/M2‐like immune response, gain of mesenchymal marker, loss of microvascular integrity as well as excessive tissue remodeling in the lung parenchyma which was confirmed by immune staining, proteomics and vascular corrosion casting. A schematic picture is provided to visualizing the putative interactions of the multidimensional surrogates in RILF (Fig. 6).
Table 1.
A summary of multiscale parameters correlating well with fibrosis index (FI)
| Methods of examination | vs. FI Surrogates | Correlation coefficients | p Values |
|---|---|---|---|
| Blood gas analysis | pH ↓ | R 2 = 0.96 | < 0.05 |
| Blood gas analysis | pCO2 (mm Hg) ↑ | R 2 = 0.97 | < 0.05 |
| Blood gas analysis | Base excess (%) ↑ | R 2 = 0.81 | > 0.05 |
| Blood gas analysis | Hct (%PCV) ↑ | R 2 = 0.91 | < 0.05 |
| Blood gas analysis | Hb (g/dL) ↑ | R 2 = 0.97 | < 0.01 |
| Microarray | MΦ↑ | Pearson R = 0.75 | < 0.0001 |
| Microarray | M2Φ↑ | Pearson R = 0.78 | < 0.0001 |
| CIBERSORT | M2Φ↑ | R 2 = 0.68 | < 0.0001 |
| CIBERSORT | Resting DCs↑ | R 2 = 0.84 | < 0.0001 |
| CIBERSORT | Naïve B cells↓ | R 2 = 0.86 | < 0.0001 |
| Microarray | Th2↑ | Pearson R = 0.83 | < 0.0001 |
| Microarray | Th2‐related genes↑ | Pearson R = 0.78 | < 0.0001 |
| Microarray | ECM↑ | Pearson R = 0.78 | < 0.0001 |
| Histology | MΦ score↑ | Pearson R = 0.97 | < 0.05 |
| Histology | Fibrosis score ↑ | Pearson R = 0.99 | < 0.05 |
| Microarray | Mesenchymal sign. ↑ | Pearson R = 0.69 | < 0.001 |
| Microarray | Endothelial sign. ↓ | Pearson R = −0.93 | < 0.0001 |
All correlation coefficients for fibrosis index (FI) vs. a broad spectrum of physiological, transcriptional and histological surrogates were listed. The linear regression is determined by either least squares or Pearson's correlation analysis.
Figure 6.

A schematic diagram of fibrosis index development and multiscale characterizations. Diverse levels of investigations including radiological, clinical chemistry, pathophysiological, transcriptional and proteomic correlates of lung fibrosis were demonstrated. [Color figure can be viewed at wileyonlinelibrary.com]
The pathogenesis of fibrosis is a multistep process initiated by organ injury leading to an integrated response involving the recruitment of diverse inflammatory cells, activation of cascades of cytokines/chemokines/growth factors and ultimately ECM remodeling.23, 24, 25 Tasks executed by infiltrating immune cells are increasingly recognized to play a central role in development of fibrosis. Most recently it was shown that depletion of interstitial macrophages (IMs) by a neutralizing antibody abrogating the CSF‐CSF1R signaling inhibits RILF, whereas depletion of alveolar macrophages (AMs) by Clodrosome was not effective further underscoring the relevance of leukocyte subtypes in RILF development.26 A brief collection of recent studies investigating potential impact of different leukocyte subsets in RILF27, 28, 29, 30, 31, 32, 33, 34, 35, 36 is provided in supplemental Table S1, Supporting Information. Segregated‐nucleus‐containing atypical monocytes (SatM) with granulocyte characteristics regulated by CCAAT/enhancer binding protein β (C/EBPβ) as well as release of neutrophil extracellular traps (NETosis) consisting of extracellular chromatin orchestrated by peptidylarginine deiminase 4 (PAD4) in age related organ fibrosis.37, 38 However, we found no dose dependent regulation of surrogates for SatM and NETosis on transcriptional and proteome level in our RILF model (data not shown). In contrast, our data indicated a clear dose/FI dependent influx of macrophages with M2 polarization (Fig. 3). The impact of macrophages in development of fibrosis is controversially debated and pro‐ and anti‐fibrotic effects have been attributed to this immune cell population.39, 40 More recent data indicate the importance of different macrophage sub‐populations at different stages of lung fibrosis development.39 For example, depletion of macrophages at an early inflammatory stage does not affect fibrosis development; however, at the progressive fibrosis stage ameliorates fibrosis, while interference with ECM degradation and uptake at a resolution stage exacerbated fibrosis in Bleomycin and adenoviral transforming growth factor‐b (TGFβ) induced lung fibrosis models.41
It is hypothesized that alternative activated M2 macrophages may play a crucial role in development of fibrosis induced by different agents.41, 42 Alternatively activated M2 macrophages are characterized by expression of the mannose receptor 1 (MRC1 or CD206), arginase‐1 (ARG1),21, 22 found in inflammatory zone 1 (FIZZ1) and Chitinase‐3‐like protein 3 (Chi3l3 or Ym1). In line with these observations, we found a high correlation between the transcriptional expression of all these M2 markers and the degree of fibrosis determined by FI. The relevance of dose/FI dependent macrophage abundance and polarization was further validated by immunostaining of two M2 (MRC1/CD206 and C‐C motif chemokine 22 (CCL22)) and two M1 (CD16/32 and iNOS) markers. Functional studies aiming to modulate M2 specific signaling have produced inconclusive results. For example, targeting CCL22‐CCR4 axis activated in IPF and RILF43, 44 resulted in transient presence of M2 marker positive macrophages post injury but attenuated Bleomycin induced fibrosis in CCR4−/− mice.45 Likewise, knockout of FIZZ1 ameliorated Bleomycin induced fibrosis46 however IL4 knockout altered the composition of macrophages without attenuating RILF.47 Together, these data indicate that the context, i.e., time point of observation/intervention, disease stage, severity of injury, as well as heterogeneity and plasticity of macrophages in response to various stimuli are important parameters that need to be considered for proper interpretation of their impact in the fibrotic process.21, 48, 49 Moreover, a single marker alone may mislead interpretation of data, e.g., the universal macrophage marker CD68 that was also found to be upregulated on transcriptome level in our RILF study was recently shown to be also expressed on fibroblasts.50 Likewise, MRC1/CD206 is also expressed on immature dendritic cells. Of note, we found enhanced levels of resting dendritic cells as a function of FI by CIBERSORT analysis (Fig. 3 d). Interestingly, while aiming to deplete interstitial macrophages in CD11c‐DTR mice, also immature dendritic cells could have been efficiently depleted in this mouse model,51 contributing to the observed reduced lung remodeling in the Bleomycin induced lung fibrosis model.41 To this end, beyond utilizing dose series experiments, multilevel and systemic analysis of marker panels is another strength of our study allowing identification of distinct cellular populations and pathological states with relatively high confidence.
Our data equivocally indicate a correlation between the switch to M2/Th2‐like phenotype with radiation dose and the degree of fibrosis (FI). It is conceivable, that transient presence of M2 macrophages elicits an anti‐inflammatory effect by uptake of ECM and secretion of immunosuppressive factors such as TGFβ.39 In contrast, persistent activation of M2 population contributes to a pro‐fibrotic remodeling of the lung tissue as determined in our study in the late (24 weeks) fibrosis phase post irradiation. Dysregulated macrophage homeostasis toward a persistence M2‐like response is associated with the secretion of a variety of pro‐fibrotic factors orchestrating tissue remodeling via increase of extracellular matrix (ECM) proteins and proteases, loss of lung architecture (blood–air or alveolar–capillary barrier) with decreased epithelial and endothelial markers, and conversely gain of mesenchymal markers by e.g., epithelium/endothelium to mesenchymal transition (EMT/EndoMT).39, 52, 53, 54 Indeed, our data support this scenario (Fig. 4). The gradual FI/dose dependent switch to the M2/Th2‐like phenotype was associated with reduced level of epithelial and endothelial marker e.g., E‐Cadherin (CDH1), PECAM (CD31) and VE‐Cadherin (CDH5) and conversely gain of mesenchymal marker e.g., Vimentin, CD44, S100A4 (FSP‐1), and consequently increased ECM deposition e.g., fibronection (FN), collagen (COL1A1), tenascin C (TNC) as well as elevated levels of enzymes involved in matrix remodeling such as matrix metalloproteinases (MMPs) and lysyl oxidase (LOX). In line with these molecular observation, ex‐vivo ultra‐high‐resolution CT combined with vascular corrosion casting revealed significant loss of microvascular integrity with reduced density and complexity of vascular trees (Fig. 5 c). Intriguingly, it was recently shown that reversal of the mesenchymal phenotype via inhibition of CD44 signaling55 ameliorates Bleomycin induced lung fibrosis.56, 57 Although, radiation induced vascular effects are hypothesized to contribute to fibrosis development, little is known about the mechanism underlying vascular malfunction, the phenotype and functional consequences. Our data support previous findings reporting global vascular remodeling after low‐, intermediate or large lung volume after single high doses of proton irradiation,58 and sustained remodeling of the pulmonary vasculature after single whole thoracic high dose irradiation.59
Conclusion
Development of FI provides an objective non‐invasive criterion to assess the degree of lung fibrosis. Time kinetic studies identified 24 weeks post irradiation as an optimum time point for assessment of FI. Dose series of fractionated irradiation was then utilized to decipher cellular and molecular correlates of FI via a comprehensive multiscale study of candidate fibrosis surrogates. This analysis revealed a central role for a dose/FI dependent modulation of immune response. The relevance of M2‐polarized immune response was validated by immunostaining of four M1/M2 macrophage markers. These data confirmed the transcriptome derived hypothesis and supports previous reports on a potential role for immune polarization in development of RILF. Despite the vascular corrosion casting most parameters studied here are either part of the clinical routine e.g., blood gas or histological analysis or could be favorably implemented into the future diagnostics, e.g. molecular analysis. Therefore, the high correlation of FI with biological parameter evoke development of next‐generation multiscale integrative models embracing the full potential of mapping key principles such as immunological response with spatiotemporal imaging data and other meaningful radiomics parameters such as texture, shape etc.60 This work may be instrumental for deciphering the molecular mechanisms governing lung fibrosis and discovery of novel targets for treatment of this devastating disease with an unmet medical need.
Authors’ contributions
A.A. and C.Z. designed the experiments and wrote the study. C.Z. and M.M. performed experiments. L.C., C.S., M.W., T.B., M.S. and B.J. assisted in acquisition, analysis and interpretation of data for the work. S.W. and M.K. helped in animal surgery studies and pathological assessment. B.J., J.D., W.W., and P.Z. revised critically the study for important intellectual content. All authors have read and approved the final study.
Supporting information
Appendix S1: Supporting Information
Table S1 A collection of important investigations improving the understanding of radiation lung immune response.
Acknowledgements
We thank Claudia Rittmueller, Barbara Schwager, Ramona Galm, Christine Schmidt and Christiane Rutenberg for their excellent technical assistance. We further thank German Cancer Research Center (DKFZ) Genomics & Proteomics Core Facility (GPCF) and Imaging Core Facility (the Small Animal Imaging unit) for their excellent technical supports. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the study.
References
- 1. Abdollahi A, Li M, Ping G, et al. Inhibition of platelet‐derived growth factor signaling attenuates pulmonary fibrosis. J Exp Med 2005;201:925–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Graves PR, Siddiqui F, Anscher MS, et al. Radiation pulmonary toxicity: from mechanisms to management. Semin Radiat Oncol 2010;20:201–7. [DOI] [PubMed] [Google Scholar]
- 3. Benveniste MF, Welsh J, Godoy MC, et al. New era of radiotherapy: an update in radiation‐induced lung disease. Clin Radiol 2013;68:e275–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yarnold J, Brotons MC. Pathogenetic mechanisms in radiation fibrosis. Radiother Oncol 2010;97:149–61. [DOI] [PubMed] [Google Scholar]
- 5. Richeldi L, Collard HR, Jones MG. Idiopathic pulmonary fibrosis. Lancet 2017;389:1941–1952. [DOI] [PubMed] [Google Scholar]
- 6. Gross TJ, Hunninghake GW. Idiopathic pulmonary fibrosis. N Engl J Med 2001;345:517–25. [DOI] [PubMed] [Google Scholar]
- 7. Flechsig P, Dadrich M, Bickelhaupt S, et al. LY2109761 attenuates radiation‐induced pulmonary murine fibrosis via reversal of TGF‐beta and BMP‐associated proinflammatory and proangiogenic signals. Clin Cancer Res 2012;18:3616–27. [DOI] [PubMed] [Google Scholar]
- 8. du Bois RM. Strategies for treating idiopathic pulmonary fibrosis. Nat Rev Drug Discov 2010;9:129–40. [DOI] [PubMed] [Google Scholar]
- 9. Konigshoff M, Kramer M, Balsara N, et al. WNT1‐inducible signaling protein‐1 mediates pulmonary fibrosis in mice and is upregulated in humans with idiopathic pulmonary fibrosis. J Clin Invest 2009;119:772–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bickelhaupt S, Erbel C, Timke C, et al. Effects of CTGF blockade on attenuation and reversal of radiation‐induced pulmonary fibrosis. J Natl Cancer Inst 2017;109. [DOI] [PubMed] [Google Scholar]
- 11. Zhou C, Jones B, Moustafa M, et al. Quantitative assessment of radiation dose and fractionation effects on normal tissue by utilizing a novel lung fibrosis index model. Radiat Oncol 2017;12:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Oliva IB, Cortopassi F, Rochester CL, et al. Combined pulmonary fibrosis and emphysema syndrome: a radiologic perspective. Monaldi Arch Chest Dis 2011;75:220–34. [DOI] [PubMed] [Google Scholar]
- 13. Plathow C, Li M, Gong P, et al. Computed tomography monitoring of radiation‐induced lung fibrosis in mice. Invest Radiol 2004;39:600–9. [DOI] [PubMed] [Google Scholar]
- 14. Cao L, Breithaupt M, Peter J. Geometrical co‐calibration of a tomographic optical system with CT for intrinsically co‐registered imaging. Phys Med Biol 2010;55:1591–606. [DOI] [PubMed] [Google Scholar]
- 15. Almog N, Ma L, Raychowdhury R, et al. Transcriptional switch of dormant tumors to fast‐growing angiogenic phenotype. Cancer Res 2009;69:836–44. [DOI] [PubMed] [Google Scholar]
- 16. Domhan S, Schwager C, Wei Q, et al. Deciphering the systems biology of mTOR inhibition by integrative transcriptome analysis. Curr Pharm Des 2014;20:88–100. [DOI] [PubMed] [Google Scholar]
- 17. Abdollahi A, Schwager C, Kleeff J, et al. Transcriptional network governing the angiogenic switch in human pancreatic cancer. Proc Natl Acad Sci U S A 2007;104:12890–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schwager C, Wirkner U, Abdollahi A, et al. TableButler ‐ a windows based tool for processing large data tables generated with high‐throughput methods. BMC Bioinformatics 2009;10:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Winter M, Dokic I, Schlegel J, et al. Deciphering the acute cellular phosphoproteome response to irradiation with X‐rays, protons and carbon ions. Mol Cell Proteomics 2017;16:855–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep 2014;6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol 2011;11:723–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wick G, Backovic A, Rabensteiner E, et al. The immunology of fibrosis: innate and adaptive responses. Trends Immunol 2010;31:110–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meneghin A, Hogaboam CM. Infectious disease, the innate immune response, and fibrosis. J Clin Invest 2007;117:530–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hynes RO. The extracellular matrix: not just pretty fibrils. Science 2009;326:1216–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Meziani L, Mondini M, Petit B, et al. CSF1R inhibition prevents radiation pulmonary fibrosis by depletion of interstitial macrophages. Eur Respir J 2018;51:1702120. [DOI] [PubMed] [Google Scholar]
- 27. Cappuccini F, Eldh T, Bruder D, et al. New insights into the molecular pathology of radiation‐induced pneumopathy. Radiother Oncol 2011;101:86–92. [DOI] [PubMed] [Google Scholar]
- 28. Groves AM, Johnston CJ, Misra RS, et al. Whole‐lung irradiation results in pulmonary macrophage alterations that are subpopulation and strain specific. Radiat Res 2015;184:639–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jackson IL, Baye F, Goswami CP, et al. Gene expression profiles among murine strains segregate with distinct differences in the progression of radiation‐induced lung disease. Dis Model Mech 2017;10:425–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Paun A, Bergeron ME, Haston CK. The Th1/Th17 balance dictates the fibrosis response in murine radiation‐induced lung disease. Sci Rep 2017;7:11586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Paun A, Kunwar A, Haston CK. Acute adaptive immune response correlates with late radiation‐induced pulmonary fibrosis in mice. Radiat Oncol 2015;10:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paun A, Bergeron ME, Haston CK. NKT deficient mice are not spared lung disease after exposure to thoracic radiotherapy. Radiat Res 2014;181:369–75. [DOI] [PubMed] [Google Scholar]
- 33. Zhang H, Han G, Liu H, et al. The development of classically and alternatively activated macrophages has different effects on the varied stages of radiation‐induced pulmonary injury in mice. J Radiat Res 2011;52:717–26. [DOI] [PubMed] [Google Scholar]
- 34. Xiong S, Guo R, Yang Z, et al. Treg depletion attenuates irradiation‐induced pulmonary fibrosis by reducing fibrocyte accumulation, inducing Th17 response, and shifting IFN‐gamma, IL‐12/IL‐4, IL‐5 balance. Immunobiology 2015;220:1284–91. [DOI] [PubMed] [Google Scholar]
- 35. Chiang CS, Liu WC, Jung SM, et al. Compartmental responses after thoracic irradiation of mice: strain differences. Int J Radiat Oncol Biol Phys 2005;62:862–71. [DOI] [PubMed] [Google Scholar]
- 36. Wirsdorfer F, Jendrossek V. The role of lymphocytes in radiotherapy‐induced adverse late effects in the lung. Front Immunol 2016;7:591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Satoh T, Nakagawa K, Sugihara F, et al. Identification of an atypical monocyte and committed progenitor involved in fibrosis. Nature 2017;541:96–101. [DOI] [PubMed] [Google Scholar]
- 38. Martinod K, Witsch T, Erpenbeck L, et al. Peptidylarginine deiminase 4 promotes age‐related organ fibrosis. J Exp Med 2017;214:439–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Braga TT, Agudelo JS, Camara NO. Macrophages during the fibrotic process: M2 as friend and foe. Front Immunol 2015;6:602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Byrne AJ, Maher TM, Lloyd CM. Pulmonary macrophages: a new therapeutic pathway in Fibrosing lung disease? Trends Mol Med 2016;22:303–16. [DOI] [PubMed] [Google Scholar]
- 41. Gibbons MA, MacKinnon AC, Ramachandran P, et al. Ly6Chi monocytes direct alternatively activated profibrotic macrophage regulation of lung fibrosis. Am J Respir Crit Care Med 2011;184:569–81. [DOI] [PubMed] [Google Scholar]
- 42. de Leve S, Wirsdorfer F, Cappuccini F, et al. Loss of CD73 prevents accumulation of alternatively activated macrophages and the formation of prefibrotic macrophage clusters in irradiated lungs. FASEB J 2017;31:2869–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yogo Y, Fujishima S, Inoue T, et al. Macrophage derived chemokine (CCL22), thymus and activation‐regulated chemokine (CCL17), and CCR4 in idiopathic pulmonary fibrosis. Respir Res 2009;10:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Inoue T, Fujishima S, Ikeda E, et al. CCL22 and CCL17 in rat radiation pneumonitis and in human idiopathic pulmonary fibrosis. Eur Respir J 2004;24:49–56. [DOI] [PubMed] [Google Scholar]
- 45. Trujillo G, O'Connor EC, Kunkel SL, et al. A novel mechanism for CCR4 in the regulation of macrophage activation in bleomycin‐induced pulmonary fibrosis. Am J Pathol 2008;172:1209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu T, Yu H, Ullenbruch M, et al. The in vivo fibrotic role of FIZZ1 in pulmonary fibrosis. PLoS One 2014;9:e88362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Groves AM, Johnston CJ, Misra RS, et al. Effects of IL‐4 on pulmonary fibrosis and the accumulation and phenotype of macrophage subpopulations following thoracic irradiation. Int J Radiat Biol 2016;92:754–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wynn TA. Fibrotic disease and the T(H)1/T(H)2 paradigm. Nat Rev Immunol 2004;4:583–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature 2013;496:445–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kunz‐Schughart LA, Weber A, Rehli M, et al. The "classical" macrophage marker CD68 is strongly expressed in primary human fibroblasts. Verh Dtsch Ges Pathol 2003;87:215–23. [PubMed] [Google Scholar]
- 51. Fainaru O, Almog N, Yung CW, et al. Tumor growth and angiogenesis are dependent on the presence of immature dendritic cells. FASEB J 2010;24:1411–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim KK, Kugler MC, Wolters PJ, et al. Alveolar epithelial cell mesenchymal transition develops in vivo during pulmonary fibrosis and is regulated by the extracellular matrix. Proc Natl Acad Sci U S A 2006;103:13180–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Marmai C, Sutherland RE, Kim KK, et al. Alveolar epithelial cells express mesenchymal proteins in patients with idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol 2011;301:L71–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Willis BC, Dubois RM, Borok Z. Epithelial origin of myofibroblasts during fibrosis in the lung. Proc Am Thorac Soc 2006;3:377–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Matzke‐Ogi A, Jannasch K, Shatirishvili M, et al. Inhibition of tumor growth and metastasis in pancreatic cancer models by interference with CD44v6 signaling. Gastroenterology 2016;150:513–25. e10. [DOI] [PubMed] [Google Scholar]
- 56. Ghatak S, Hascall VC, Markwald RR, et al. Transforming growth factor beta1 (TGFbeta1)‐induced CD44V6‐NOX4 signaling in pathogenesis of idiopathic pulmonary fibrosis. J Biol Chem 2017;292:10490–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li Y, Jiang D, Liang J, et al. Severe lung fibrosis requires an invasive fibroblast phenotype regulated by hyaluronan and CD44. J Exp Med 2011;208:1459–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ghobadi G, Bartelds B, van der Veen SJ, et al. Lung irradiation induces pulmonary vascular remodelling resembling pulmonary arterial hypertension. Thorax 2012;67:334–41. [DOI] [PubMed] [Google Scholar]
- 59. Ghosh SN, Wu Q, Mader M, et al. Vascular injury after whole thoracic x‐ray irradiation in the rat. Int J Radiat Oncol Biol Phys 2009;74:192–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting Information
Table S1 A collection of important investigations improving the understanding of radiation lung immune response.


