Abstract
Background
Major depressive disorder (MDD) is a severe disease characterized by multiple pathological changes. However, there are no reliable diagnostic biomarkers for MDD. The aim of the current study was to investigate the gene network and biomarkers underlying the pathophysiology of MDD.
Methods
In this study, we conducted a comprehensive analysis of the mRNA expression profile of MDD using data from Gene Expression Omnibus (GEO). The MDD dataset (GSE98793) with 128 MDD and 64 control whole blood samples was divided randomly into two non-overlapping groups for cross-validated differential gene expression analysis. The gene ontology (GO) enrichment and gene set enrichment analysis (GSEA) were performed for annotation, visualization, and integrated discovery. Protein–protein interaction (PPI) network was constructed by STRING database and hub genes were identified by the CytoHubba plugin. The gene expression difference and the functional similarity of hub genes were investigated for further gene expression and function exploration. Moreover, the receiver operating characteristic curve was performed to verify the diagnostic value of the hub genes.
Results
We identified 761 differentially expressed genes closely related to MDD. The Venn diagram and GO analyses indicated that changes in MDD are mainly enriched in ribonucleoprotein complex biogenesis, antigen receptor-mediated signaling pathway, catalytic activity (acting on RNA), structural constituent of ribosome, mitochondrial matrix, and mitochondrial protein complex. The GSEA suggested that tumor necrosis factor signaling pathway, Toll-like receptor signaling pathway, apoptosis pathway, and NF-kappa B signaling pathway are all crucial in the development of MDD. A total of 20 hub genes were selected via the PPI network. Additionally, the identified hub genes were downregulated and show high functional similarity and diagnostic value in MDD.
Conclusions
Our findings may provide novel insight into the functional characteristics of MDD through integrative analysis of GEO data, and suggest potential biomarkers and therapeutic targets for MDD.
Keywords: Differentially expressed gene, Major depressive disorder, Inflammation, Correlation network analysis, Mitochondrial dysfunction, Diagnostic value
Introduction
The prevalence and incidence of major depressive disorder (MDD), which is ranked as the leading cause of the global disease burden and death by suicide (Ferrari et al., 2013; Hasin et al., 2018), are continuously increasing. MDD is a severe, recurrent, and debilitating disease characterized clinically by a multifactorial and multistage process (mild, moderate, or severe depression) associated with the interaction between genetic and environmental factors. Moreover, the duration, number, and pattern of episodes of MDD are variable, the term of “recovery” is used to describe patients that have regained their usual function and are no longer symptomatic after an episode of MDD in community settings. With timely and appropriate treatment, episodes last approximately 3–6 months, and most patients recover within 12 months (Lépine & Briley, 2011; Malhi & Mann, 2018). On the contrary, the probability of recurrence increases and the outcome is less favorable in longer-term episodes, and the recovery rate drops to approximately 60%, 40%, and 30% at 2, 4, and 6 years, respectively, with comorbid anxiety having an important role in limiting recovery (Malhi & Mann, 2018). It should be noted that early diagnosis and treatment would unquestionably decrease the morbidity and mortality associated with depression. A biomarkers is a measurable indicator of some biological condition or state. Identification of biomarkers would be a key step for MDD. C-reactive protein, an acute-phase protein, is widely used as biomarker in MDD and inflammation (Chamberlain et al., 2019). Recently, microRNAs and exosomes have been applied as diagnostic and therapeutic biomarkers in MDD patients (Tavakolizadeh et al., 2018). In addition, an interesting study identifies distinct “biotypes” of depression using fMRI, which could be diagnostic biomarkers and may predict treatment response (Wager & Woo, 2017). However, the accuracy of biomarkers for diagnosis and prognosis of MDD is still largely limited because the pathogenesis of depression is complex and heterogeneous. Thus, investigation of the molecular mechanisms underlying MDD is crucial, and may contribute to identification of the precise targets and essential biomarkers for MDD diagnosis.
A variety of differential diagnostic criteria are associated with MDD. The clinical standardized definitions, such as those provided by the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition, Text Revision; Hamilton Rating Scale for Depression (HAMD); and Montgomery–Asberg Depression Rating Scale (Nemeroff, 2007), are the most common classical methods for determining MDD, and can be applied either in the clinic or in treatment trials. For example, HAMD, a 17-item instrument, is used to examine the intensity and frequency of depression severity in individuals with MDD. Scores on HAMD represents the severity ranges of MDD: normal (0–7); mild depression (8–16); moderate depression (17–23); and severe depression (≥24). A potential problem of using threshold scores for identification and classification is low-accuracy in distinguishing the depression severity and prognoses across cases (Fitzgerald et al., 2018). Empirical methods such as positron emission tomography and functional magnetic resonance imaging have contributed to identification of the brain regions that are affected in MDD (Siegle, Carter & Thase, 2006). A challenge in these studies is to disentangle the different contribution of depression and other comorbidities to the overall clinical picture. Mostly, an accurate diagnosis can be achieved via detailed history-taking, mental status and physical examination, and laboratory tests. Notably, an emerging and powerful method for investigating the pathogenesis of this disorder is examining peripheral blood for verification of gene expression levels (Hepgul et al., 2013). Studies employing these peripheral blood examinations have analyzed biomarkers via relatively accessible and low-invasiveness procedures, and demonstrated that peripheral inflammation precedes the emergence of symptoms in patients with depression (Khandaker et al., 2014). Moreover, the differential expression of genes in venous blood samples can be measured using flow cytometry and polymerase chain reaction. These approaches have relatively low effectiveness and throughput, which can now be improved using microarray-based technologies for high-throughput functional genomic discovery. Microarray is a promising and popular method for large-scale gene expression profiling, greatly facilitating the analysis of thousands of mRNAs simultaneously in a single experiment.
Several studies have been performed to improve the understanding of the molecular mechanisms underlying microarray analysis. A meta-analysis of genome-wide expression studies on MDD has been conducted using different microarray platforms and tissues, such as blood, the amygdala, and the prefrontal cortex (Forero, Guio-Vega & González-Giraldo, 2017). However, multi-platform analyses, the use of various tissues, and a lack of important variables, such as postmortem intervals or severity of the disorder, in the raw data contribute to inevitable batch effects. Moreover, a previous study investigated differentially expressed genes (DEGs, a group of genes that differentially express in different experimental conditions) in peripheral blood samples from 38 patients with MDD and 14 healthy controls (Woo et al., 2018), but the small sample size diminished the reliability of the results. A more careful examination was performed in two case-control studies of MDD using microarray data from whole blood samples (GSE98793) to investigate changes in peripheral inflammation (Leday et al., 2018). In particular, they investigated DEGs focusing on the changes in innate and adaptive immune gene expression by comparing 113 patients with MDD (57 comorbid with anxiety disorder, 56 without anxiety) and 57 healthy controls. Moreover, although a series of bioinformatics analyses has thoroughly investigated the potential biomarkers of immunological stratification in patients with MDD, it remains to be examined how functional systems and molecules other than immunological biomarkers affect the pathophysiology of MDD. Thus, further analyses are warranted to identify more robust and reliable diagnostic biomarkers, with cross-validation, large samples to comprehensively consider the abnormalities in the molecular mechanisms involved in MDD.
Hence, the aim of this study was to identify potential diagnostic biomarkers and biological functions related to MDD from the Gene Expression Omnibus (GEO; Edgar, Domrachev & Lash, 2002). Further, DEGs were investigated to distinguish patients with MDD from healthy controls via cross-validation. Moreover, the biological processes (BPs) involved were analyzed using gene ontology (GO) enrichment and gene set enrichment analysis (GSEA) pathways for the DEGs. In addition, the top 20 hub genes screened via protein–protein interaction (PPI) network were selected for their functional similarity, and their diagnostic value was assessed. Our study may provide some insights into the molecular mechanisms underlying MDD based on its pathophysiology.
Materials and Methods
Data collection and preprocessing
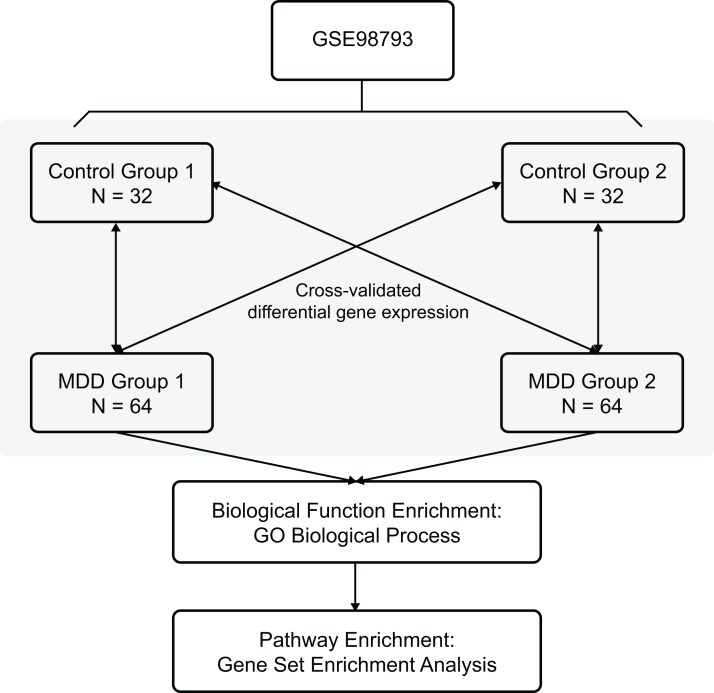
The Gene Expression Omnibus database is an international public repository which archives and distributes high-throughput gene expression and genomics data sets. The gene expression dataset GSE98793 (Leday et al., 2018) was downloaded from the GEO database (GPL570 (HG-U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array) and annotated in the R software (R Foundation for Statistical Computing, Vienna, Austria) using annotation files. The species selected was Homo sapiens, and the data type was microarray expression profiles. The whole blood samples included 128 MDD (diagnosed post hoc by the Mini-International Neuropsychiatric Interview) and 64 control healthy samples. Clinical and demographic characteristics of the MDD patients are shown in Table 1. The MDD and control blood samples were divided randomly into two non-overlapping groups each (MDD group 1, MDD group 2, control group 1, and control group 2) for cross-validation. The overview of the workflow is shown in Fig. 1.
Table 1. Clinical and demographic characteristics of the MDD patients.
| All subjects | MDD | Controls | |
|---|---|---|---|
| Patients | 192 | 128 | 64 |
| Gender | |||
| Male | 48 | 32 | 16 |
| Female | 144 | 96 | 48 |
| Comorbidities | |||
| Anxiety | 64 | 64 | 0 |
| Without anxiety | 64 | 64 | 0 |
| Age (years) | 52 ± 1 | 52 ± 1 | 52 ± 1 |
Figure 1. Flow chart of methodologies applied in the current study.
Screening of DEGs
The Affy (Gautier et al., 2004) and limma (Smyth, 2005) packages were applied to the microarray data to filter the DEGs by comparing both MDD groups to both healthy control groups using a multivariate linear model using moderated t-statistic. Data were corrected for multiple comparisons using false discovery rate adjustment, and genes with |logFC| (an absolute log2 value in the fold change of the expression of the genes) >0.6 and P-value < 0.05 (Dalman et al., 2012) in all four comparisons were identified as DEGs.
Functional and pathway enrichment analysis
The GO analysis serves as a bioinformatics tool that provides structured annotations, including BPs, molecular functions (MFs), and cellular components (CCs), for genes and gene products. Modules related to biological function were investigated using UpSetR (Conway, Lex & Gehlenborg, 2017) to determine the functional and pathway enrichment for BPs in GO. Functional and pathway enrichment were analyzed using hypergeometric test and Bonferroni correction. We also used GSEA which is a statistical approach for determining whether the genes from particular pathways or other predefined gene sets are differentially expressed in different phenotypes (Subramanian et al., 2005). Reactome pathways were analyzed with GSEA, using clusterProfiler (Yu et al., 2012) to define every functional cluster. C2.all.v6.2.symbols.gmt was selected as the reference gene set. False discovery rate <0.1, and P-value < 0.01 were set as the cut-off criteria.
PPI network construction
The PPI information available in the STRING network in the STRING database (http://string-db.org, version 10) (Szklarczyk et al., 2015) is useful for predicting physical and functional interactions. All DEGs were mapped to the STRING database, and the interactions with reliability scores more than 0.4 were selected to analyze the relationship of the DEGs. Cytoscape (Shannon et al., 2003) was used to select the key nodes with the strongest connectivity for visualizing molecular interaction networks. CytoHubba, a Cytoscape plugin, was used to identify the top 20 hub genes of the merged network (Chin et al., 2014). NetworkAnalyst (https://www.networkanalyst.ca/faces/home.xhtml) (Xia, Gill & Hancock, 2015) is a visual analytics platform for PPI networks. We inputted the 20 hub genes into NetworkAnalyst for visualization of PPI networks. The expression analysis of the top 20 hub genes with the highest ranking are shown in Table 2.
Table 2. The expression analysis of the top 20 hub genes with the highest ranking.
| Gene symbol | Entrez ID | logFC | P-value | Score |
|---|---|---|---|---|
| MRPS11 | 64963 | −0.698 | 6.49E-24 | 8.92E+13 |
| MRPS2 | 51116 | −0.502 | 6.07E-13 | 8.90E+13 |
| MRPL2 | 51069 | −1.105 | 4.37E-25 | 8.64E+13 |
| MRPL15 | 29088 | −0.823 | 3.60E-18 | 8.37E+13 |
| MRPL16 | 54948 | −0.828 | 8.73E-23 | 8.37E+13 |
| MRPS7 | 51081 | −0.905 | 5.15E-21 | 6.55E+13 |
| MRPS18 | 6222 | −0.835 | 1.15E-19 | 4.74E+13 |
| RPS3 | 6188 | −0.778 | 6.64E-21 | 4.74E+13 |
| RPL11 | 6135 | −0.927 | 1.14E-20 | 4.74E+13 |
| RPL26L1 | 51121 | −0.868 | 2.30E-21 | 4.73E+13 |
| RPL6 | 6128 | −0.689 | 1.1E-20 | 4.73E+13 |
| RPL19 | 6143 | −0.732 | 6.99E-24 | 4.73E+13 |
| RPS19 | 6223 | −0.714 | 1.01E-20 | 4.73E+13 |
| NAS2 | 10412 | −0.962 | 7.7E-23 | 4.73E+13 |
| NHP2 | 55651 | −1.344 | 3.1E-25 | 4.73E+13 |
| RPP38 | 10557 | −0.941 | 8.2E-23 | 4.73E+13 |
| RPL29 | 6159 | −1.065 | 7.18E-22 | 4.73E+13 |
| MRPL36 | 64979 | −0.757 | 4.01E-18 | 4.73E+13 |
| MRPL27 | 51264 | −1.257 | 1.84E-23 | 4.73E+13 |
| MRPL9 | 65005 | −0.752 | 9.09E-22 | 4.73E+13 |
Distributions of hub genes
The distributions of all DEGs in GSE98793 were identified. Moreover, the functional similarity among proteins was evaluated using the geometric mean of semantic similarities in CCs and MFs through the GOSemSim package (Yu et al., 2010).
Setting the cut-off score based on receiver operating characteristic curve analysis
Receiver operating characteristic (ROC) curve analysis, which yields indictors of accuracy such as the area under the curve (AUC), provides the basic principle and rationale for distinguishing between the specificity and sensitivity of diagnostic performance (Akobeng, 2007). The maximum value of the sum of specificity and sensitivity was used as the cut-off score for each hub gene. The “pROC” package of the R software was applied for ROC curve analysis (Robin et al., 2011).
Statistical analysis
All statistical analyses were performed as the means ± standard deviation. The R software (version 3.5.2) was utilized to measure the data. A P-value < 0.05 was considered statistically significant.
Results
Differentially expressed genes identification
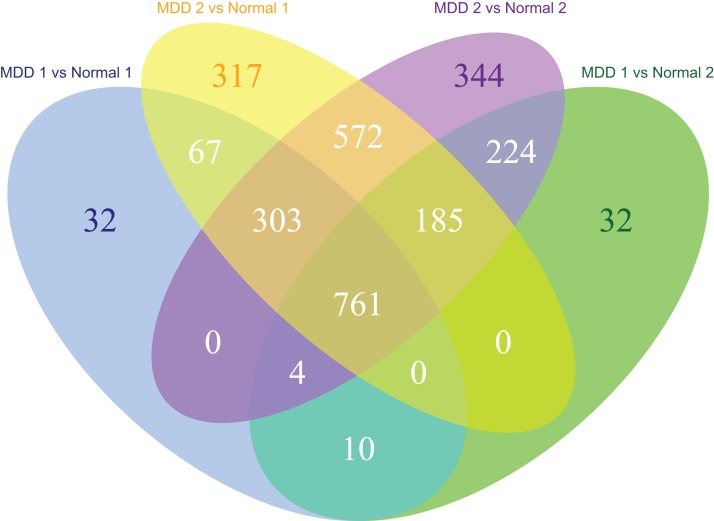
The MDD and control blood samples were divided into two groups for cross-validation. A total of 64 samples remained in both MDD groups and 32 samples remained in both control groups. Cross-validation of the data from MDD and control groups identified 761 DEGs in the MDD groups (Fig. 2).
Figure 2. Venn diagram of the differentially expressed genes significantly associated with major depression disorder which were short-listed for the cross-validation.
GO enrichment analysis of DEGs
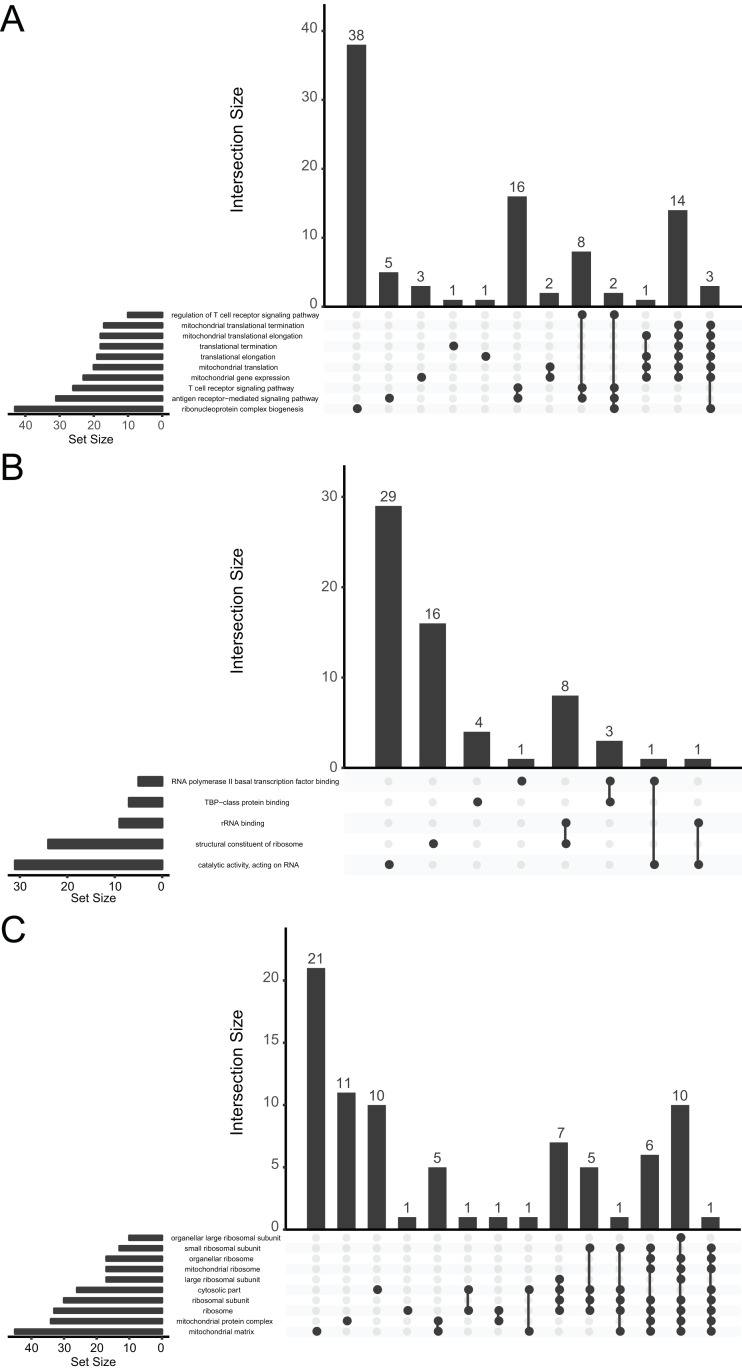
We performed a functional enrichment analysis for further investigation of DEGs. The data indicated that the DEGs were significantly enriched in GO terms. The GO analysis demonstrated that changes in BPs were mainly enriched in ribonucleoprotein complex biogenesis, antigen receptor-mediated signaling pathway, T-cell receptor signaling pathway, mitochondrial gene expression, mitochondrial translation, and translational elongation (Fig. 3A). Changes in MFs were significantly enriched in catalytic activity (acting on RNA), structural constituent of ribosomes, rRNA binding, TATA-binding protein-class protein binding, and RNA polymerase II basal transcription factor binding (Fig. 3B). Changes in CCs for the DEGs were enriched mainly in the mitochondrial matrix, mitochondrial protein complex, ribosome, ribosomal subunit, cytosolic part, and large ribosomal subunit (Fig. 3C).
Figure 3. UpSetR plot demonstrating distribution of the gene ontology annotations for major depressive disorder in (A) biological processes, (B) molecular functions, and (C) cellular components.
GSEA of MDD-related genes
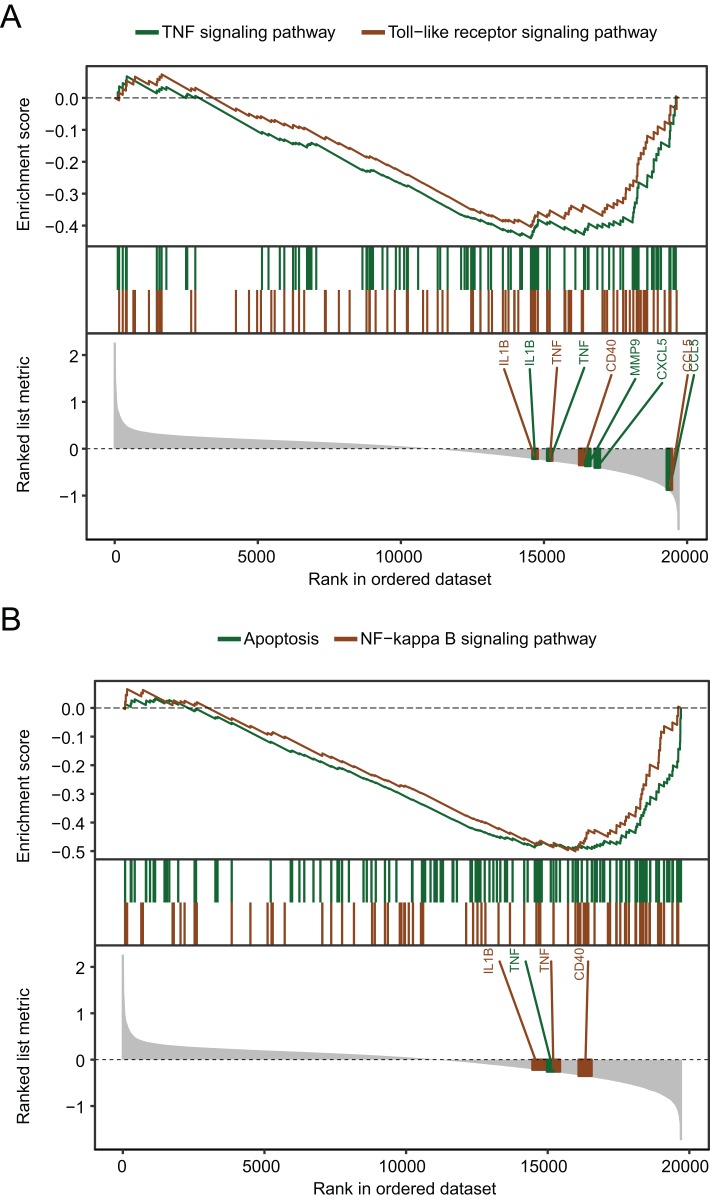
The biological pathways that were significantly altered in MDD blood samples compared with the control blood samples were determined using GSEA. The GSEA of GSE98793 gene expression profiles suggested that MDD is mainly related to the apoptosis pathway, and the tumor necrosis factor (TNF), Toll-like receptor (Fig. 4A), and nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) signaling pathways (Fig. 4B).
Figure 4. Gene set enrichment analysis of the gene expression profiles of the GSE98793 dataset.
(A) Gene set enrichment analysis demonstrated that the TNF signaling pathway and the Toll-like receptor signaling pathway were enriched in MDD. (B) Gene set enrichment analysis demonstrated that apoptosis and the NF-kappa B signaling pathway were enriched in MDD.
PPI network analysis of DEGs
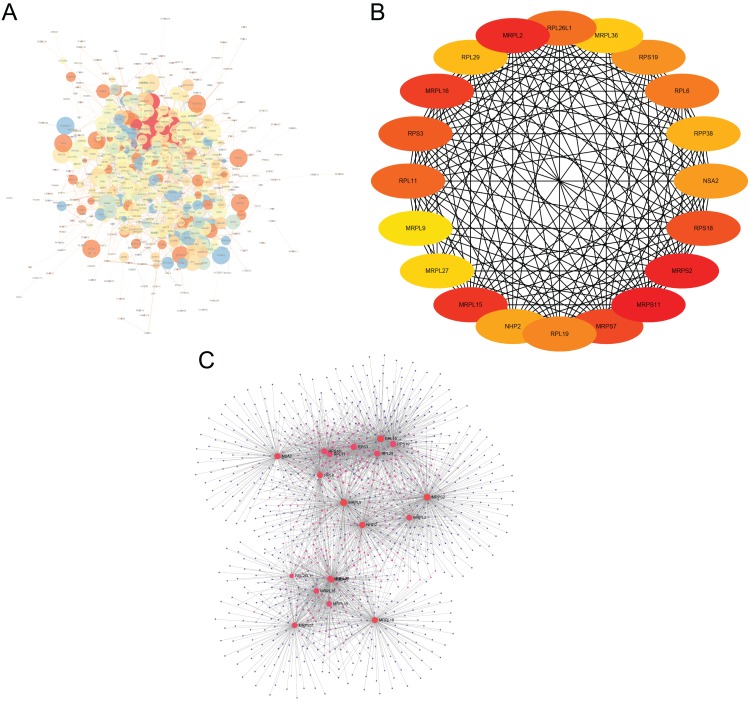
The interactions of 761 DEGs were analyzed using the STRING online database to investigate the PPI network underlying MDD. The obtained results were analyzed using the Cytoscape software (Fig. 5A). The cytoHubba plugin was then used to investigate the top 20 hub genes associated with MDD (Fig. 5B). Moreover, the visualized network of the hub genes is shown using the NetworkAnalyzer online tool (Fig. 5C).
Figure 5. Major depressive disorder-specific network.
(A) Protein–protein interaction network of differentially expressed genes using the STRING database. (B) The CytoHubba plugin was used to analyze the top 20 hub genes with maximum correlation criterion. (C) The hub genes with the top 20 scores were analyzed using the NetworkAnalyzer plugin.
Distributions of hub genes
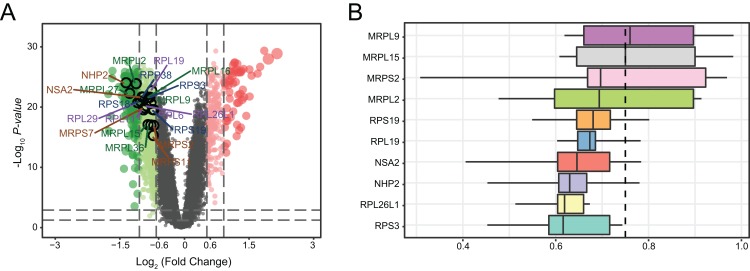
We determined the distributions of 761 DEGs from MDD and healthy control blood samples (Fig. 6A). Among all DEGs, the top 20 hub genes were identified as being downregulated in MDD. Notably, three hub genes with the highest ranking were found to be downregulated: mitochondrial ribosomal protein L2 (MRPL2), NHP2, and NOP-seven-associated 2 (NSA2). Moreover, we ranked top 10 genes among the 20 hub genes based on the average functional similarity (Fig. 6B). Mitochondrial ribosomal protein L9 (MRPL9), mitochondrial ribosomal protein L15 (MRPL15), and mitochondrial ribosomal protein S2 (MRPS2) were the top three proteins potentially playing key roles in MDD; MRPL9 was the only protein with a cut-off value >0.75.
Figure 6. Genetic screening for hub genes in patients with major depressive disorder.
(A) Volcano plot of fold changes in the expression of the hub genes. (B) Summary of functional similarities of the top 10 hub genes.
Using hub genes for MDD diagnosis
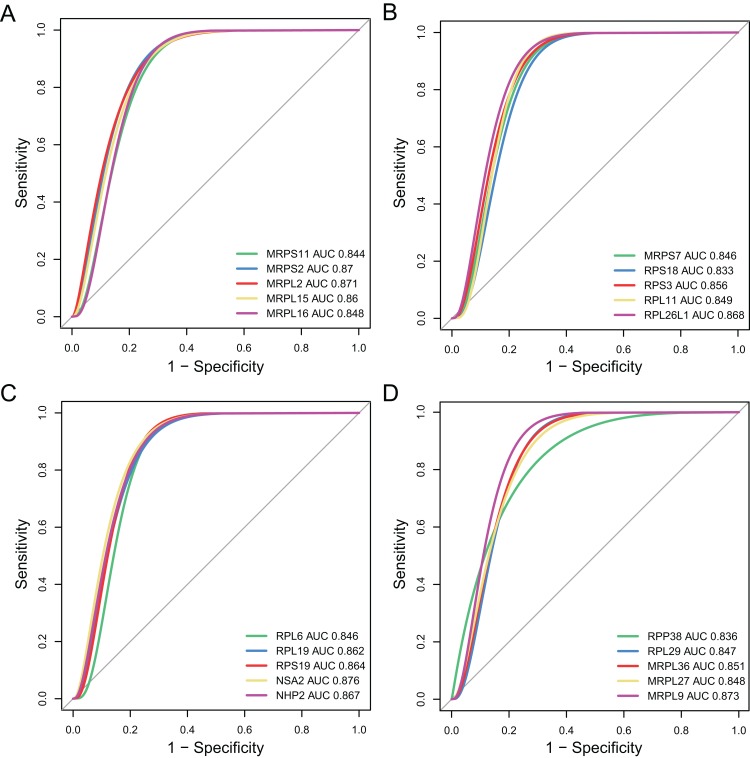
The diagnostic accuracy of the top 20 hub genes was assessed using ROC curve analysis (Fig. 7). The areas under the ROC curves were 0.844, 0.87, 0.871, 0.86, and 0.848 for MRPS11, MRPS2, MRPL2, MRPL15, and MRPL16, as shown in Fig. 7A. The areas under the ROC curves were 0.846, 0.833, 0.856, 0.849, and 0.868 for MRPS7, ribosomal protein S18 (RPS18), RPS3, ribosomal protein L11 (RPL11), and RPL26L1, as shown in Fig. 7B. The areas under the ROC curves were 0.846, 0.862, 0.864, 0.876, and 0.867 for RPL6, RPL19, RPS19, NSA2, and NHP2, as shown in Fig. 7C. The areas under the ROC curves were 0.836, 0.847, 0.851, 0.848, and 0.873 for RPP38, RPL29, MRPL36, MRPL27, and MRPL9, as shown in Fig. 7D.
Figure 7. Validation of diagnostic value of the hub genes for major depressive disorder (MDD).
(A–D) Receiver operating characteristic curve of the hub genes for diagnosis of MDD.
Discussion
Worldwide, MDD is a recurrent lifelong mental disorder of very high prevalence. The 12-month prevalence of MDD is approximately 6.6%, and the lifetime risk is 15–18% (Malhi & Mann, 2018). Increasing number of studies are being performed to develop a non-invasive and quantitative clinical test; however, no specific and sensitive biomarkers are available for the diagnosis and treatment of MDD yet. Therefore, in order to identify effective diagnostic biomarkers of MDD, we performed an integrated analysis on a large MDD cohort of 128 MDD patients and 64 healthy controls, using whole-genome microarray data for mRNA expression. A total of 761 DEGs were identified in the MDD group via cross-validation. Furthermore, GO enrichment analysis and GSEA showed that these enriched modules and pathways are closely related to the immune response and mitochondrial dysfunction observed in MDD. In addition, the top 20 hub genes associated with MDD, which were identified in the PPI network, showed high functional similarity and diagnostic values for MDD.
In the first part of the present study, we identified 761 DEGs in the GSE98793 dataset, collected from 128 MDD to 64 control whole blood samples, using cross-validation. To investigate the BPs of the DEGs involved in MDD, GO enrichment analyses were performed. Of the MF annotations, ribonucleoprotein complex biogenesis, antigen receptor-mediated signaling pathway, catalytic activity (acting on RNA), structural constituent of ribosomes, mitochondrial matrix, and mitochondrial protein complex were found to be significantly associated with the occurrence and development of MDD. Mitochondria play a critical role in the modulation of synaptic and neural plasticity required for the formation of novel neuronal synapses and pathways, as well as regulation of cellular Ca2+ homeostasis, oxidative stress, and apoptosis. Mitochondrial dysfunction has recently drawn considerable attention due to the postulation that impaired mitochondrial bioenergetics could be the basis for the pathophysiology of MDD through multiple potential pathways, including those related to genetics/genomics, oxidative stress, alterations in neuroplasticity, and inflammation (Klinedinst & Regenold, 2015). A previous study concluded that patients with mitochondrial disorders exhibit a higher rate of psychiatric illness than the general population; the authors reported that among 36 adults with mitochondrial cytopathies, the lifetime prevalence rate of psychiatric illnesses was up to 70% (Fattal et al., 2007). Taken together, these observations imply that mitochondrial dysfunction may be a major contributor to depression.
In the second part of the present study, in order to investigate the biological functions of the DEGs associated with MDD, GSEA was performed. The apoptosis pathway, and the TNF, Toll-like receptor, and NF-κB signaling pathways were the top four significantly enriched pathways. Interestingly, we noted that the most enriched pathway in our analysis was associated with immune response, inflammation, and apoptosis. Studies on rodent models and patients with depression show high levels of TNF-α, interleukin-1β, and interleukin-6, which are induced by infection, injury, and psychological stress (Haroon et al., 2018; Miller, Maletic & Raison, 2009; Wang et al., 2018). Consistent with our data, analysis of microarray results for MDD from other mRNA datasets also revealed that immune and inflammatory responses play a critical role in the regulation network of MDD (Woo et al., 2018). Additionally, it has been hypothesized that external stressors may induce depressive disorders via stimulation of inflammatory, oxidative, and apoptotic mechanisms, closely related to the pathways, such as TNF-α, NF-κB, Toll-like receptor, and apoptosis (Kubera et al., 2011). Our data-mining results further confirmed that inflammatory responses play a key role in the etiology of depression.
In the PPI network identified in the present study, 20 DEGs were highlighted as the most significant hub genes, with multiple interactions in the network. Further investigation of these genes may reveal the pathophysiology of MDD. All of these hub genes were identified as being significantly downregulated in MDD. Moreover, to identify the proteins involved in the pathophysiology of MDD, the top 10 genes were ranked among the 20 hub genes based on their average functional similarity. Moreover, MRPL9, MRPL15, and MRPS2 were ranked as the top three proteins potentially serving as central regulators in MDD. With regard to diagnostic value, the AUC of the 20 hub genes were analyzed. All the AUC values were in the range 0.830–0.900, suggesting that these genes possess moderate accuracy (Akobeng, 2007) in diagnostic examinations and may be promising targets for the diagnosis of MDD. Mitochondrial disorders may be induced by mutations in the mitochondrial and nuclear DNA contributing to impaired production of cellular energy (adenosine triphosphate) (Koene et al., 2009). NSA2, MRPL9, and MRPL2 showed the three highest prognostic values among the hub genes. These genes encode mitochondrial and cytosolic ribosomal proteins, including MRPL, MRPS, RPL, and RPS, which play critical roles in translation, transcription, proliferation, and neural plasticity. NSA2, also known as tumor growth factor-β inducible nuclear protein 1, is predicted to serve as a cell cycle repressor and plays a crucial role in cell proliferation (Zhang et al., 2010). A previous study has demonstrated that NSA2 is activated after permanent middle cerebral occlusion in an Alzheimer’s disease mouse model (Tseveleki et al., 2010); NSA2 was predicted to be related to brain defense and tissue repair in the pathological process of Alzheimer’s disease. Similar decreased NSA2 may also be found in MDD. Consistent with our results, another RNA-sequencing study reported that the expression of multiple ribosomal genes, including RPL6 and RPL29, downregulation in the hypothalamus of male mice under chronic social defeat stress, which contributes to the development of depression- and anxiety-like symptoms (Smagin et al., 2016). In addition, mutations in mitochondrial or nuclear DNA have been implicated in a variety of neurological diseases, such as depression or personality disorder (McFarland, Taylor & Turnbull, 2010). Especially, MRPS15 (chromosome 1p34.3) is a clinical candidate for depressive syndrome (O’Brien, O’Brien & Norman, 2005). Notably, depressive behavior is associated with mitochondrial disorder in children (Morava et al., 2010), which suggests that the genes encoding cytosolic and mitochondrial ribosomal proteins may be potential targets for early diagnosis of MDD.
Moreover, our current studies found that all of the top 20 hub genes were identified as being downregulated in MDD. We speculated that the reduced expression of NSA2 and the other ribosomal genes might play an important role in MDD, and these identified genes may be potential therapeutic targets for MDD. Further analyses were necessary to analyze the effect of these gene agonists on MDD and verify the mechanisms underlying the target gene agonist-induced improvements by evaluating the gene expression profile, the histological and biochemical parameters, and behavioral tests in MDD animal models and MDD patients.
Our study has a few limitations. First, to comprehensively identify the dysfunctions in MDD, integrated analysis of both venous blood samples and brain tissues is warranted; this was not performed in the present study. Second, in order to determine the diagnostic accuracy of the hub genes associated with MDD, it will be helpful to increase the sample size for further external validation. Third, single microarray analysis may contribute to high false-positive rate and one-sided results; thus, it is necessary to improve detection power by integrating multiple individual data in a future study. Fourth, due to the heterogeneity of depression and the lack of clinical data, we were unable to evaluate the associations between risk indicators and stratification of patients based on the severity of MDD. Fifth, not all depressed patients have mitochondrial dysfunction and inflammation. It is possible that these alterations are present only in specific subgroups of depressed patients, with specific clinical and pathophysiological features. For example, increased inflammation is identified in a subgroup of MDD patients who have a neurodevelopmental form of depression, deriving from exposure to stress early in childhood or in utero (Miller & Raison, 2016; Pariante, 2017). More clinical and demographic characteristics of MDD patients is needed to be included in the study for further subgroup analysis. Finally, further experimental evidence, such as real-time PCR, western blot, immunohistochemistry assays, is required to fully elucidate the role of hub genes and the underlying mechanisms of MDD.
These data suggest multiple pathways and biomarkers of MDD, consistent with our current knowledge of the pathophysiology of this disease. We believe that this hypothesis-generating study provides new insight into the molecular mechanisms underlying MDD, identifying several potential biomarkers for its diagnosis and treatment.
Conclusions
In conclusions, the aim of this study was to explore the molecular mechanisms underlying the progression of MDD via a comprehensive bioinformatics analysis that aimed to identify the associated biological functions and pathways involved in the development of MDD. Moreover, we also identified 20 candidate genes which could serve as potential diagnostic biomarkers through PPI network analysis, the functional similarity analysis, and ROC curve analysis. However, more molecular experiments are needed for further validation of the findings of current study.
Supplemental Information
Venn diagram of the number of DEGs identified as significant for the cross-validated comparisons.
The hub genes with the top 20 scores were analyzed using the NetworkAnalyzer plugin.
Acknowledgments
We would like to thank FigureYa (Xiao Ya Hua Tu) for the figure technology support.
Funding Statement
This work was supported by grants from the Natural Science Foundation of Fujian Province of China (Grant No. 2018J01169); the Scientific Research Personnel Training Project of Health and Family Planning Commission of Fujian Province (Grant No. 2017-02-03); the Science Foundation for Young Scientists of Fujian Health and Family Planning Commission (Grant No. 2018-2-17); and the Key R & D programs in Shandong (2016CYJS08A01-1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Yan Qi, Email: qiyankm@163.com.
Qiuyuan Yue, Email: circlessoo@sina.cn.
Additional Information and Declarations
Competing Interests
The authors declare that there are no conflicts of interest.
Author Contributions
Huimei Wang conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Mingwei Zhang conceived and designed the experiments, authored or reviewed drafts of the paper, approved the final draft.
Qiqi Xie conceived and designed the experiments, performed the experiments, analyzed the data, approved the final draft.
Jin Yu contributed reagents/materials/analysis tools, approved the final draft.
Yan Qi contributed reagents/materials/analysis tools, prepared figures and/or tables, approved the final draft.
Qiuyuan Yue contributed reagents/materials/analysis tools, authored or reviewed drafts of the paper, approved the final draft.
Microarray Data Deposition
The following information was supplied regarding the deposition of microarray data:
This work used a publicly available dataset, available at NCBI: GSE98793.
Data Availability
The following information was supplied regarding data availability:
The raw measurements are available in the Supplemental Files as Tables S1 and S2.
References
- Akobeng (2007).Akobeng AK. Understanding diagnostic tests 3: receiver operating characteristic curves. Acta Paediatrica. 2007;96(5):644–647. doi: 10.1111/j.1651-2227.2006.00178.x. [DOI] [PubMed] [Google Scholar]
- Chamberlain et al. (2019).Chamberlain SR, Cavanagh J, De Boer P, Mondelli V, Jones DNC, Drevets WC, Cowen PJ, Harrison NA, Pointon L, Pariante CM, Bullmore ET. Treatment-resistant depression and peripheral C-reactive protein. British Journal of Psychiatry. 2019;214(1):11–19. doi: 10.1192/bjp.2018.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin et al. (2014).Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Systems Biology. 2014;8(Suppl 4):S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway, Lex & Gehlenborg (2017).Conway JR, Lex A, Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33(18):2938–2940. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalman et al. (2012).Dalman MR, Deeter A, Nimishakavi G, Duan ZH. Fold change and P-value cutoffs significantly alter microarray interpretations. BMC Bioinformatics. 2012;13(Suppl 2):S11. doi: 10.1186/1471-2105-13-S2-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, Domrachev & Lash (2002).Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattal et al. (2007).Fattal O, Link J, Quinn K, Cohen BH, Franco K. Psychiatric comorbidity in 36 adults with mitochondrial cytopathies. CNS Spectrums. 2007;12(6):429–438. doi: 10.1017/S1092852900015303. [DOI] [PubMed] [Google Scholar]
- Ferrari et al. (2013).Ferrari AJ, Charlson FJ, Norman RE, Flaxman AD, Patten SB, Vos T, Whiteford HA. The epidemiological modelling of major depressive disorder: application for the Global Burden of Disease Study 2010. PLOS ONE. 2013;8(7):e69637. doi: 10.1371/journal.pone.0069637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald et al. (2018).Fitzgerald PB, Hoy KE, Elliot D, McQueen RS, Wambeek LE, Daskalakis ZJ. Accelerated repetitive transcranial magnetic stimulation in the treatment of depression. Neuropsychopharmacology. 2018;43(7):1565–1572. doi: 10.1038/s41386-018-0009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forero, Guio-Vega & González-Giraldo (2017).Forero DA, Guio-Vega GP, González-Giraldo Y. A comprehensive regional analysis of genome-wide expression profiles for major depressive disorder Journal of affective disorders. Journal of Affective Disorders. 2017;218:86–92. doi: 10.1016/j.jad.2017.04.061. [DOI] [PubMed] [Google Scholar]
- Gautier et al. (2004).Gautier L, Cope L, Bolstad BM, Irizarry RA. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Haroon et al. (2018).Haroon E, Daguanno AW, Woolwine BJ, Goldsmith DR, Baer WM, Wommack EC, Felger JC, Miller AH. Antidepressant treatment resistance is associated with increased inflammatory markers in patients with major depressive disorder. Psychoneuroendocrinology. 2018;95:43–49. doi: 10.1016/j.psyneuen.2018.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin et al. (2018).Hasin DS, Sarvet AL, Meyers JL, Saha TD, Ruan WJ, Stohl M, Grant BF. Epidemiology of adult DSM-5 major depressive disorder and its specifiers in the United States. JAMA Psychiatry. 2018;75(4):336–346. doi: 10.1001/jamapsychiatry.2017.4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepgul et al. (2013).Hepgul N, Cattaneo A, Zunszain PA, Pariante CM. Depression pathogenesis and treatment: what can we learn from blood mRNA expression? BMC Medicine. 2013;11(1):28. doi: 10.1186/1741-7015-11-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandaker et al. (2014).Khandaker GM, Pearson RM, Zammit S, Lewis G, Jones PB. Association of serum interleukin 6 and C-reactive protein in childhood with depression and psychosis in young adult life: a population-based longitudinal study. JAMA Psychiatry. 2014;71(10):1121–1128. doi: 10.1001/jamapsychiatry.2014.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinedinst & Regenold (2015).Klinedinst NJ, Regenold WT. A mitochondrial bioenergetic basis of depression. Journal of Bioenergetics and Biomembranes. 2015;47(1–2):155–171. doi: 10.1007/s10863-014-9584-6. [DOI] [PubMed] [Google Scholar]
- Koene et al. (2009).Koene S, Kozicz TL, Rodenburg RJT, Verhaak CM, De Vries MC, Wortmann S, Van De Heuvel L, Smeitink JA, Morava E. Major depression in adolescent children consecutively diagnosed with mitochondrial disorder. Journal of Affective Disorders. 2009;114(1–3):327–332. doi: 10.1016/j.jad.2008.06.023. [DOI] [PubMed] [Google Scholar]
- Kubera et al. (2011).Kubera M, Obuchowicz E, Goehler L, Brzeszcz J, Maes M. In animal models, psychosocial stress-induced (neuro) inflammation, apoptosis and reduced neurogenesis are associated to the onset of depression. Progress in Neuro-Psychopharmacology and Biological Psychiatry. 2011;35(3):744–759. doi: 10.1016/j.pnpbp.2010.08.026. [DOI] [PubMed] [Google Scholar]
- Leday et al. (2018).Leday GGR, Vertes PE, Richardson S, Greene JR, Regan T, Khan S, Henderson R, Freeman TC, Pariante CM, Harrison NA, Perry VH, Drevets WC, Wittenberg GM, Bullmore ET. Replicable and coupled changes in innate and adaptive immune gene expression in two case-control studies of blood microarrays in major depressive disorder. Biological Psychiatry. 2018;83(1):70–80. doi: 10.1016/j.biopsych.2017.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lépine & Briley (2011).Lépine JP, Briley M. The increasing burden of depression. Neuropsychiatric Disease and Treatment. 2011;7(Suppl 1):3–7. doi: 10.2147/NDT.S19617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhi & Mann (2018).Malhi GS, Mann JJ. Depression. Lancet. 2018;392(10161):2299–2312. doi: 10.1016/S0140-6736(18)31948-2. [DOI] [PubMed] [Google Scholar]
- McFarland, Taylor & Turnbull (2010).McFarland R, Taylor RW, Turnbull DM. A neurological perspective on mitochondrial disease. Lancet Neurology. 2010;9(8):829–840. doi: 10.1016/S1474-4422(10)70116-2. [DOI] [PubMed] [Google Scholar]
- Miller, Maletic & Raison (2009).Miller AH, Maletic V, Raison CL. Inflammation and its discontents: the role of cytokines in the pathophysiology of major depression. Biological Psychiatry. 2009;65(9):732–741. doi: 10.1016/j.biopsych.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller & Raison (2016).Miller AH, Raison CL. The role of inflammation in depression: from evolutionary imperative to modern treatment target. Nature Reviews Immunology. 2016;16(1):22–34. doi: 10.1038/nri.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morava et al. (2010).Morava E, Gardeitchik T, Kozicz T, De Boer L, Koene S, De Vries MC, McFarland R, Roobol T, Rodenburg RJT, Verhaak CM. Depressive behaviour in children diagnosed with a mitochondrial disorder. Mitochondrion. 2010;10(5):528–533. doi: 10.1016/j.mito.2010.05.011. [DOI] [PubMed] [Google Scholar]
- Nemeroff (2007).Nemeroff CB. The burden of severe depression: a review of diagnostic challenges and treatment alternatives. Journal of Psychiatric Research. 2007;41(3–4):189–206. doi: 10.1016/j.jpsychires.2006.05.008. [DOI] [PubMed] [Google Scholar]
- O’Brien, O’Brien & Norman (2005).O’Brien TW, O’Brien BJ, Norman RA. Nuclear MRP genes and mitochondrial disease. Gene. 2005;354:147–151. doi: 10.1016/j.gene.2005.03.026. [DOI] [PubMed] [Google Scholar]
- Pariante (2017).Pariante CM. Why are depressed patients inflamed? A reflection on 20 years of research on depression, glucocorticoid resistance and inflammation. European Neuropsychopharmacology. 2017;27(6):554–559. doi: 10.1016/j.euroneuro.2017.04.001. [DOI] [PubMed] [Google Scholar]
- Robin et al. (2011).Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C, Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12(1):77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon et al. (2003).Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegle, Carter & Thase (2006).Siegle GJ, Carter CS, Thase ME. Use of FMRI to predict recovery from unipolar depression with cognitive behavior therapy. American Journal of Psychiatry. 2006;163(4):735–738. doi: 10.1176/ajp.2006.163.4.735. [DOI] [PubMed] [Google Scholar]
- Smagin et al. (2016).Smagin DA, Kovalenko IL, Galyamina AG, Bragin AO, Orlov YL, Kudryavtseva NN. Dysfunction in ribosomal gene expression in the hypothalamus and hippocampus following chronic social defeat stress in male mice as revealed by RNA-seq. Neural Plasticity. 2016;2016:3289187. doi: 10.1155/2016/3289187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth (2005).Smyth GK. Bioinformatics & Computational Biology Solutions Using R & Bioconductor. New York: Springer; 2005. Llimma: linear models for microarray data; pp. 397–420. [Google Scholar]
- Subramanian et al. (2005).Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk et al. (2015).Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, Von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Research. 2015;43(D1):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavakolizadeh et al. (2018).Tavakolizadeh J, Roshanaei K, Salmaninejad A, Yari R, Nahand JS, Sarkarizi HK, Mousavi SM, Salarinia R, Rahmati M, Mousavi SF, Mokhtari R, Mirzaei H. MicroRNAs and exosomes in depression: Potential diagnostic biomarkers. Journal of Cellular Biochemistry. 2018;119(5):3783–3797. doi: 10.1002/jcb.26599. [DOI] [PubMed] [Google Scholar]
- Tseveleki et al. (2010).Tseveleki V, Rubio R, Vamvakas S-S, White J, Taoufik E, Petit E, Quackenbush J, Probert L. Comparative gene expression analysis in mouse models for multiple sclerosis, Alzheimer’s disease and stroke for identifying commonly regulated and disease-specific gene changes. Genomics. 2010;96(2):82–91. doi: 10.1016/j.ygeno.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wager & Woo (2017).Wager TD, Woo C-W. Imaging biomarkers and biotypes for depression. Nature Medicine. 2017;23(1):16–17. doi: 10.1038/nm.4264. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2018).Wang YL, Han QQ, Gong WQ, Pan DH, Wang LZ, Hu W, Yang M, Li B, Yu J, Liu Q. Microglial activation mediates chronic mild stress-induced depressive- and anxiety-like behavior in adult rats. Journal of Neuroinflammation. 2018;15(1):21. doi: 10.1186/s12974-018-1054-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo et al. (2018).Woo HI, Lim S-W, Myung W, Kim DK, Lee S-Y. Differentially expressed genes related to major depressive disorder and antidepressant response: genome-wide gene expression analysis. Experimental and Molecular Medicine. 2018;50(8):92. doi: 10.1038/s12276-018-0123-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, Gill & Hancock (2015).Xia J, Gill EE, Hancock RE. Network Analyst for statistical, visual and network-based meta-analysis of gene expression data. Nature Protocols. 2015;10(6):823–844. doi: 10.1038/nprot.2015.052. [DOI] [PubMed] [Google Scholar]
- Yu et al. (2010).Yu G, Li F, Qin Y, Bo X, Wu Y, Wang S. GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics. 2010;26(7):976–978. doi: 10.1093/bioinformatics/btq064. [DOI] [PubMed] [Google Scholar]
- Yu et al. (2012).Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: A Journal of Integrative Biology. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2010).Zhang H, Ma X, Shi T, Song Q, Zhao H, Ma D. NSA2, a novel nucleolus protein regulates cell proliferation and cell cycle. Biochemical and Biophysical Research Communications. 2010;391(1):651–658. doi: 10.1016/j.bbrc.2009.11.114. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Venn diagram of the number of DEGs identified as significant for the cross-validated comparisons.
The hub genes with the top 20 scores were analyzed using the NetworkAnalyzer plugin.
Data Availability Statement
The following information was supplied regarding data availability:
The raw measurements are available in the Supplemental Files as Tables S1 and S2.