Abstract
Deinococcus radiodurans R1 is extremely resistant to ionizing radiation and oxidative stress. In this study, we characterized DR0846, a candidate peroxiredoxin in D. radiodurans. DR0846 is a peroxiredoxin Q containing two conserved cysteine residues. DR0846 exists mainly in monomeric form with an intramolecular disulfide bond between the two cysteine residues. We found that DR0846 functions as a molecular chaperone as well as a peroxidase. A mutational analysis indicates that the two cysteine residues are essential for enzymatic activity. A double‐deletion mutant lacking DR0846 and catalase DR1998 exhibits decreased oxidative and heat shock stress tolerance with respect to the single mutants or the wild‐type cells. These results suggest that DR0846 contributes to resistance against oxidative and heat stresses in D. radiodurans.
Keywords: Deinococcus radiodurans R1, DR0846, molecular chaperone, peroxidase, peroxiredoxin Q
Abbreviations
AMS, 4‐acetamido‐4′‐maleimidylstilbene‐2,2′‐disulfonic acid
DSB, DNA double‐strand break
GR, glutathione reductase
GSH, reduced glutathione
H 2 O 2, hydrogen peroxide
HMW, high molecular weight
IR, ionizing radiation
MDH, malate dehydrogenase
NADH, nicotinamide adenine dinucleotide
PrxQ, peroxiredoxin Q
ROS, reactive oxygen species
SEC, size exclusion chromatography
SOD, superoxide dismutases
TCA, trichloroacetic acid
TF, trigger factor
TR, thioredoxin reductase
Trx, thioredoxin
WT, wild‐type
Deinococcus radiodurans R1 is well‐known for its extreme resistance to ionizing radiation (IR) 1, 2, 3, 4. IR induces DNA double‐strand breaks and reactive oxygen species (ROS) in bacteria 3, 5, 6, 7. Although the IR resistance of D. radiodurans is due to its highly condensed nucleoid structure and enhanced DNA repair systems 3, some evidence suggests that it requires ROS scavenging systems 8, 9. D. radiodurans can remove ROS by nonenzymatic antioxidants, such as manganese complexes or carotenoids, and enzymatic antioxidants, such as catalases, superoxide dismutases (SOD), and peroxidases 3, 10. Catalases and peroxidases decompose hydrogen peroxide to water, whereas SOD converts superoxide radicals to hydrogen peroxide from the cells. D. radiodurans encodes two catalases (DR1998 and DRA0259), three SODs (DR1279, DR1546, and DRA0202), a cytochrome c peroxidase (DRA0301), an iron‐dependent peroxidase (DRA0145), and four peroxiredoxins (Prxs) (DR0846, DR1208, DR1209, and DR2242) 2, 3, 11.
Peroxiredoxins are a family of antioxidant enzymes that detoxifying hydrogen peroxide, alkylhydroperoxide, and peroxynitrite using thiols as electron donors 12, 13, 14. Prxs have highly conserved peroxidatic (C P) and resolving (C R) cysteine residues, which are essential for peroxidase activity 15, 16. Based on the absence or presence of conserved catalytic cysteine residues, Prxs are divided into three groups, typical 2‐Cys Prx, atypical 2‐Cys Prx, which are subdivided into type II Prx and PrxQ, and 1‐Cys Prx 17, 18. Typical and atypical 2‐Cys Prx have two Cys residues and 1‐Cys Prx lacks the resolving cysteine residue 17. PrxQ is a homolog of bacterioferritin co‐migratory protein (BCP) in Escherichia coli 19. PrxQ possesses two conserved cysteine residues (CXXXXC), with an intramolecular disulfide bond between the two cysteine residues, which is reduced by Trx 19. PrxQ usually functions as a monomeric peroxidase having high reactivity to H2O2 and butyl hydroperoxide 13. However, PrxQ4 (SsBcp4) in Sulfolobus solfataricus forms a non‐covalent dimeric structure and adopts an atypical 2‐Cys catalytic mechanism 20. Some PrxQ proteins function as peroxidases in a Grx‐dependent manner and only have a functional catalytic cysteine residue 21, 22. PrxQ plays an important role in E. coli and Helicobacter pylori under oxidative stress 21, 23. Despite many recent studies, the physiological functions, electron donors, and substrate specificity of PrxQs are largely unknown.
In this study, we characterized the expression and functions of DR0846 of D. radiodurans. We found that DR0846 is a PrxQ with two cysteine residues at positions 60 and 65. Additionally, we demonstrated that DR0846 exhibits a dual function as a peroxidase and a molecular chaperone in vivo and in vitro.
Materials and methods
Cell strains, growth conditions, and medium
Deinococcus radiodurans R1 (ATCC13939) was cultured in TGY broth (0.5% tryptone, 0.1% glucose, and 0.3% yeast extract) or on TGY plates at 30 °C. A stationary‐phase culture grown overnight with shaking was used as the seed culture. The seed culture was inoculated in TGY broth at a 1 : 100 dilution. E. coli DH5α (Promega, Madison, WI, USA) and BL21‐DE3 (Invitrogen, Carlsbad, CA, USA) strains were grown in Luria–Bertani (LB) broth (DB, Franklin Lakes, NJ, USA) or on LB plate at 37 °C. Antibiotics were used when necessary at the following concentrations: kanamycin (50 μg·mL−1) for E. coli and kanamycin (8 μg·mL−1) or chloramphenicol (3 μg·mL−1) for D. radiodurans.
Bioinformatics analysis of peroxiredoxins
Analyses of the amino acid sequences, the isoelectric point, and molecular mass of proteins were performed using National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/nucleotide/) and UCSC Archaea Genome Browser (http://archaea.ucsc.edu/lists/deinRadi/refSeq-list.html) databases. mega 7.0 was used to determine sequence identities and to build a phylogenetic tree using the neighbor‐joining method 24, 25, 26. Evolutionary distances were computed using the number of differences method 27 and are expressed as the number of amino acid differences between sequences. The analysis involved 34 amino acid sequences. All ambiguous positions were removed for each sequence pair.
qRT‐PCR analysis of peroxiredoxin genes
For the qRT‐PCR analysis, cells were grown on TGY medium for 16 h at 30 °C. The seed culture was inoculated in TGY broth at a 1 : 100 dilution and grown at 30 °C until OD600 reached ~ 0.5 in TGY broth. For the oxidative stress treatment, cells were incubated for 5, 10, 20, or 30 min after treatment with 20 mm H2O2 at 30 °C. For the gamma ray treatment, cells were irradiated at 1, 3, or 5 kGy for 1 h. Total RNA was isolated from treated or untreated cells using TRI Reagent® (Molecular Research Center, Inc., Cincinnati, OH, USA) according to the manufacturer's instructions.
Cloning of DR0846 and DR0846 cysteine mutants
DR0846 and its mutants were cloned in the pET‐28a (+) expression vector (Novagen, Madison, WI, USA). To construct pET‐28a:DR0846, the coding region was isolated from the genomic DNA of D. radiodurans R1 by PCR with primers harboring BamHI (N terminus) and HindIII (C terminus) sites (Table S1) using Pfu DNA polymerase (Solgent, Gyeonggi‐do, Korea). The PCR products were inserted into the pGEM‐T Easy vector (Promega). The DNA fragments were cut with their corresponding restriction enzymes and cloned into the pET‐28a vector. The pET‐28a:DR0846 plasmid was used as a template to generate the cysteine mutants C60S, C65S, and C60S/C65S by substituting Cys for Ser60, Ser65, and Ser60, 65 by PCR‐mediated site‐directed mutagenesis. All constructs were confirmed by DNA sequencing.
Expression and purification of recombinant proteins
The pET‐28a:DR0846, pET‐28a:DR0846C60S, pET‐28a:DR0846C65S, and pET‐28a:DR0846C60S/C65S vectors were transformed into BL21‐DE3 and plated on LB plates containing 50 μg·mL−1 kanamycin. A single colony was inoculated in 5 mL of LB medium and grown overnight at 37 °C to obtain the seed culture. Then, the seed culture was inoculated in LB broth at a 1 : 100 dilution. The expression of His‐tagged DR0846 was induced with 0.1 mm isopropyl‐β‐d‐thiogalactopyranoside (IPTG) for 4 h at 30 °C. The recombinant proteins were purified using a nickel‐nitrilotriacetate‐agarose (Ni‐NTA) column (Peptron, Daejeon, Korea) following the manufacturer's instructions. The proteins were eluted by thrombin at 4 °C overnight. Purified proteins were dialyzed using 50 mm Tris/HCl (pH 7.5) for biochemical analyses.
Size exclusion chromatography
Size exclusion chromatography (SEC) was performed at 25 °C to determine the size of DR0846 by fast protein liquid chromatography (AKTA, Amersham Biosciences, Uppsala, Sweden) using a Superdex 200 10/300 GL gel‐filtration column (Amersham Biosciences) following previously described methods, with minor modifications 28. The column was equilibrated and run with 50 mm Tris/HCl (pH 7.5) buffer at a flow rate of 0.5 mL·min−1 at 4 °C. Absorbance was monitored at 280 nm.
Peroxidase activity assay
The peroxidase activity of purified DR0846 was measured by nicotinamide adenine dinucleotide phosphate (NADH) oxidation at 340 nm as described previously, with minor modifications 29. For thioredoxin‐dependent peroxidase activity, various concentrations of DR0846 were incubated with 0.3 mm NADH, 5 μm yeast thioredoxin reductase (TR), and 1 μm yeast thioredoxin (Trx) in 50 mm HEPES buffer (pH 8.0). For glutaredoxin‐dependent peroxidase activity, various concentrations of DR0846 were incubated with 0.3 mm NADH, 5 μm glutathione reductase (GR), and 1 mm reduced glutathione (GSH) in 50 mm Tris/HCl (pH 8.0), followed by the addition of 1 mm H2O2. NADH oxidation was monitored by measuring the change in absorbance at 340 nm for 10 min using a UV‐Visible spectrophotometer (Evolution 300 UV‐Vis Spectrophotometer; Thermo Scientific, Worcester, MA, USA).
Molecular chaperone activity assay
Holdase chaperone activity was determined as described previously 29 by assessing the ability of recombinant DR0846 to inhibit the thermal aggregation of substrate proteins 30, 31, 32, 33. Briefly, malate dehydrogenase (MDH) was incubated in 50 mm HEPES buffer (pH 8.0) with various concentrations of recombinant DR0846. The reaction mixture was incubated at 42 °C for 15 min, and thermal aggregation of the substrate was estimated by monitoring the degree of turbidity at 340 nm using an Evolution 300 Spectrophotometer (Thermo Scientific) equipped with a thermostatic cell holder. The thermal aggregation of MDH was used as the control. The holdase chaperone activity of DR0846 was determined at 1 : 1, 1 : 2, and 1 : 3 molar ratios between substrate (MDH) and DR0846.
AMS modification of DR0846 and DR0846 Cys mutant proteins
4‐Acetamido‐4′‐maleimidylstilbene‐2,2′‐disulfonic acid (AMS) modification was performed as described previously 34 with minor modifications. The proteins were precipitated by the addition of one volume of trichloroacetic acid (TCA), followed by incubation for 1 h at −20 °C. After 1 h, precipitates were collected by centrifugation at 18 400 g for 5 min and washed with ice‐cold acetone three times. Final pellets were dissolved in 20 μL of AMS working solution (50 mm Tris/HCl, pH 7.5, 0.1% SDS, 10 mm EDTA, 20 mm AMS) and incubated for 1 h at 25 °C in the dark. Samples were separated by reducing SDS/PAGE and analyzed by western blotting. For western blot, we generate antibody using purified DR0846 recombinant protein. DR0846 protein was immunize to mice and the antiserum was used for immunoblotting.
Construction of deletion mutants in D. radiodurans
The ∆dr0846 and ∆dr1998 disruption mutants were constructed by targeted mutagenesis using the double cross‐over recombination method described previously 35, 36. Two amplified 1 kb fragments from upstream and downstream of the targeted genes were digested with appropriate restriction enzymes (Table S1), and ligated into the corresponding sites of kanamycin resistance cassette in pKatAPH3. The recombinant plasmids were transformed into D. radiodurans cells. The mutant strains were selected on TGY agar plates supplemented with 8 μg·mL−1 kanamycin. To construct the ∆dr0846/∆dr1998 double‐deletion mutant, digested upstream and downstream fragments of dr1998 gene were ligated into the corresponding sites of chloramphenicol resistance cassette in pKatCAT5. The recombinant plasmids were transformed into ∆dr0846 mutant strain, and the transformant was screened on TGY agar plate containing kanamycin (8 μg·mL−1) and chloramphenicol (4 μg·mL−1). The deletions of genes were verified by diagnostic PCR and nucleotide sequencing.
Hydrogen peroxide (H2O2) and heat stress tolerance assay
The sensitivity of D. radiodurans cells to hydrogen peroxide was assayed as described previously 37 with minor modifications. Cells were harvested in early stationary phase, washed twice and resuspended in phosphate buffer (20 mm, pH 7.4). The cells were serially diluted from 10−1 to 10−4. Cells were spotted on TGY plates containing 0 or 0.2 mm H2O2 and incubated at 30 °C for 16 h.
To study the heat resistance of ∆dr0846, early stationary‐phase cells were used for the seed culture. The seed culture was inoculated in TGY broth at a 1 : 100 dilution. Cells were grown at 30 °C until OD600 of ~ 0.5 in TGY broth with or without 8 μg·mL−1 kanamycin. For thermal stress, cells were incubated at 30 °C or 42 °C for 30 min. The cells were serially diluted from 10−1 to 10−4. Diluted cells were spotted on TGY plates and incubated at 30 °C for 16 h.
Results
Phylogenetic analysis of peroxiredoxins from D. radiodurans R1
The peroxiredoxin (Prx) family is classified into four groups based on sequence properties: 1‐Cys Prx, 2‐Cys Prx, type II Prx, and PrxQ 38, 39. 1‐Cys Prxs contain only one conserved cysteine residue. 2‐Cys Prxs contain two conserved cysteine residues and both residues are essential for enzyme activity. Type II Prxs, also named atypical 2‐Cys Prxs, have two cysteine residues but the position of one of these cysteines is not conserved 38, 39. In addition, PrxQs contain two cysteine residues 38, 39.
The D. radiodurans R1 genome encodes four putative peroxiredoxins (DR0846, DR1208, DR1209, and DR2242) 3. To investigate the relationship between D. radiodurans Prxs and those of diverse organisms, a phylogenetic analysis was conducted using Prx family members from Arabidopsis, humans, yeast, Synechocystis sp., and Chlamydomonas (Fig. S1). The evolutionary history was inferred using the neighbor‐joining method 24, 26. All peroxiredoxins in D. radiodurans were assigned to the PrxQ, suggesting that PrxQs may have a vital role for the viability of D. radiodurans under oxidative stress.
Expression analysis of peroxiredoxins in response to oxidative stress or gamma rays
Peroxiredoxins are a family of antioxidant enzymes involved in sensing and detoxifying hydrogen peroxide (H2O2) and other ROS 40. To investigate the expression of Deinococcus peroxiredoxins in response to oxidative stress, we evaluated cells by qRT‐PCR after oxidative stress treatment. After 20 mm H2O2 treatment, DR0846 and DR1209 expression was induced, whereas DR1208 and DR2242 expression levels were not different from those in the control group (Fig. 1A). To test the expression of peroxiredoxin genes in response to gamma rays, we irradiated cells with 1, 3, and 5 kGy. The transcript expression levels of DR0846 and DR1208 increased gradually with increasing gamma irradiation, whereas DR1209 and DR2242 expression levels were unchanged until 3 kGy and decreased at higher doses (Fig. 1B). Based on these results, we selected PrxQ (DR0846), which is simultaneously induced by both H2O2 and gamma rays and showed a higher expression level than that of DR1208, for further analyses (Fig. 1B).
Figure 1.
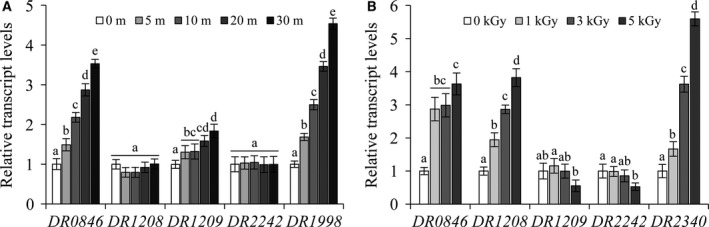
Expression analysis of Prx genes in response to oxidative stress or gamma rays. Cells were grown on TGY medium for 16 h at 30 °C. mRNA levels of peroxiredoxin genes were determined by qRT‐PCR after treatment with 20 mm H2O2 for 5–30 min (A) or treatment with gamma rays at 1–5 kGy (B). Data are means ± SE from three replications for each treatment. Different letters indicate significant differences at P < 0.05 between the groups by one‐way ANOVA with Tukey's test. DR1343 (gap) was used as a loading control. DR1998 (KatE1) and DR2340 (recA) were used as positive controls for oxidative stress and gamma rays, respectively.
Protein sequence and oligomeric state of DR0846
The sequence of DR0846 was compared with those of homologous PrxQ proteins from diverse organisms by a multiple sequence alignment. The conserved peroxidatic cysteine (C p) of DR0846 was located around position 60 in a PxxxTxxC‐motif (Fig. S2) 41. The sequence alignment also showed that there is an additional cysteine at position 65 in DR0846.
For biochemical studies of DR0846 encoding a protein of 175 amino acids with a theoretical molecular mass of 19.1 kDa, the recombinant protein was overexpressed in E. coli BL21 (DE3) and purified. The purified proteins were analyzed by SDS/PAGE (Fig. 2A) or native‐PAGE (Fig. 2B). As shown in Fig. 2A, the purified recombinant DR0846 protein showed a single band with a molecular mass of approximately 19 kDa by 12% SDS/PAGE in the presence (reducing) or absence (nonreducing) of DTT. To further determine the oligomeric status of the native DR0846 protein, we performed native‐PAGE and SEC (Fig. 2B,C). As shown in Fig. 2B,C, DR0846 existed primarily in monomeric form, with a small quantity of oligomeric structures without the formation of intermolecular disulfide bonds.
Figure 2.
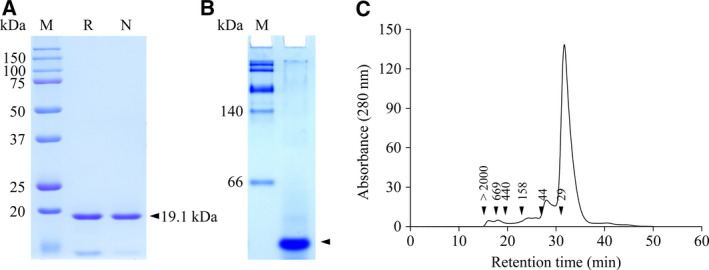
Purity and oligomeric state of DR0846 based on SDS/PAGE (A), native‐PAGE (B), and SEC (C). The proteins were denatured by heating in the presence (R) or absence (N) of 5 mm DTT. DR0846 was separated by 12% SDS/PAGE (A) or 10% native‐PAGE (B) and the gel was stained with Coomassie Blue. M, Marker; R, Reducing; N, Nonreducing. SEC was performed using a Superdex 200 10/300 column. The numbers in the chromatogram represent the molecular weights of the standard proteins; blue dextran (> 2000 kDa), thyroglobulin (669 kDa), ferritin (440 kDa), aldolase (158 kDa), ovalbumin (44 kDa), and carbonic anhydrase (29 kDa).
Effect of cysteine residues on enzymatic activity of DR0846
PrxQ proteins are thiol‐based peroxidase that catalyzes the reduction of hydrogen peroxide 19, 22. To investigate whether DR0846 possesses peroxidase activity, we conducted peroxidase activity assays (Fig. 3A). We measured the peroxidase enzymatic activity of DR0846 by monitoring the reduction of H2O2 by coupled NADH oxidation at 340 nm using the Trx (Trx, TR, and NADH) system. As shown in Fig. 3A, DR0846 showed peroxidase activity in a concentration‐dependent manner in the presence of the Trx system (Trx, TR, and NADH). Typical 2‐Cys Prx and some atypical 2‐Cys Prx‐type peroxidases use thioredoxin as a reductant. However, the donor substrate specificity of PrxQ remains unclear. It has been reported that Burkholderia cenocepacia BCP (BcBCP) uses thioredoxin as a reductant for the sulfenic acid intermediate 22. However, greater peroxidase activity is observed when using glutathione as an electron donor 22. Therefore, we assayed the efficiency of the GSH (GSH, GR, and NADH) systems in providing reducing power for DR0846 in the reduction of H2O2 (Fig. S3). No peroxidase activity was detected for DR0846 in the presence of the GSH system (Fig. S3). In contrast, erythrocyte GPx showed significant catalytic activity in the same system. These results indicated that DR0846 is a thioredoxin‐dependent PrxQ that uses thioredoxin as an electron donor and consistent with the fact that D. radiodurans has no GSH.
Figure 3.
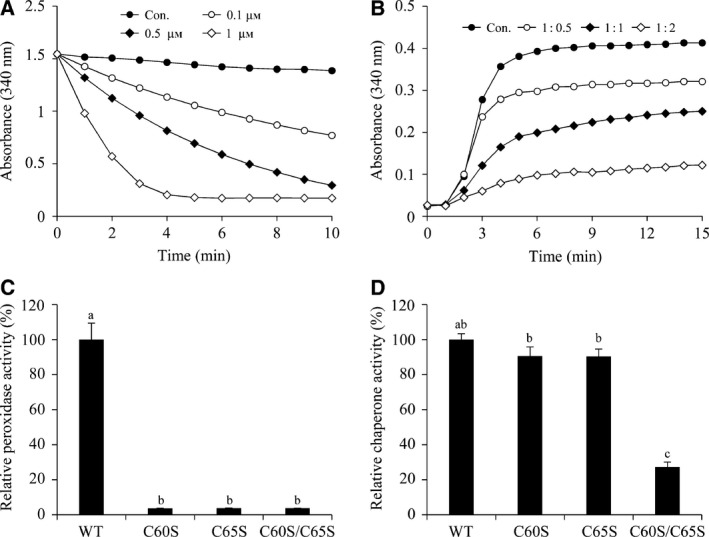
Peroxidase and chaperone activities of DR0846 and DR0846 Cys mutant proteins. (A) Peroxidase enzyme assay of DR0846. Peroxidase enzyme activity was measured using the yeast Trx system at various concentrations. (B) Molecular chaperone assay of DR0846. Chaperone activity was measured by the aggregation of MDH at 42 °C at different molar ratios of MDH/DR0846. Con., 1 : 0 (●); 1 : 0.5 DR0846 (○); 1 : 1 DR0846 (♦); 1 : 2 DR0846 (♢). Peroxidase enzyme assay (C) and chaperone enzyme assay (D) of DR0846 Cys mutant. C60S, C65S, and C60S/C65S are cysteine substitution mutants. Data are means ± SD of three independent experiments. Different letters indicate significant differences at P < 0.05 between the WT and mutant proteins by one‐way ANOVA with Tukey's test.
Recent studies have revealed that some peroxiredoxins have dual functions as a peroxidase and a chaperone 31, 42, 43. To investigate whether DR0846 act as a molecular chaperone, we performed a holdase chaperone activity assay using MDH as a heat‐sensitive substrate. The incubation MDH with increasing amounts of DR0846 prevented thermal aggregation of MDH at 42 °C, and aggregation was completely inhibited at a 1 : 2 molar ratio of MDH to DR0846 (Fig. 3B). These results indicate that DR0846 act as a peroxidase and a molecular chaperone.
Thiol peroxidase of E. coli has a functional cysteine residue that is a crucial for enzymatic activity 44. To investigate the physiological function of the cysteine residue on DR0846 enzymatic activity, we generated various substitution mutants of DR0846 by replacing cysteine residues with serine at the C60 and C65 positions. All of the mutants, i.e., C60S, C65S, and C60S/C65S, exhibited complete loss of peroxidase activity (Fig. 3C). The holdase chaperone activity of C60S and C65S single mutants was not altered, whereas the holdase chaperone activity of C60S/C65S was almost blocked (Fig. 3D). These results suggest that each Cys residues are important for peroxidase activity, whereas both Cys residues are required for molecular chaperone activity.
Analysis of disulfide bond formation in DR0846
PrxQ possesses the PXXXXC motif, forms an intramolecular disulfide bond between two cysteine residues, of the catalytic site and adopts an atypical 2‐Cys catalytic mechanism 19. The amino acid sequence of DR0846 has only two cysteine residues (Fig. S2) and did not form an intermolecular disulfide bond (Fig. 2A). To test whether the cysteine in DR0846 forms an intramolecular disulfide bond, trapping experiments of free thiol (‐SH) groups were performed using wild‐type (WT) DR0846 and its Cys‐to‐Ser mutants (C60S, C65S, and C60S/C65S). The thiol alkylation agent AMS reacts with a free thiol (‐SH) group in the protein resulting from increasing the molecular mass by approximately 540 Da per AMS molecule. AMS‐modified and unmodified DR0846 were separated by reducing SDS/PAGE and detected by immunoblotting using DR0846 antibody (Fig. 4). In the presence of AMS, two bands were detected for WT DR0846 corresponding to the fully reduced form, showing a weight consistent with the binding of two AMS molecules (upper band) and oxidized form (lower band) with an intramolecular disulfide bond. Two forms of the protein were purified in atmospheric conditions, i.e., oxidized and reduced forms, but the oxidized form was slightly more highly represented. The C60S and C65S proteins exhibited only reduced forms showing a weight consistent with the binding of single AMS molecules (upper band). However, only one band, which was not modified by AMS, was observed for the C60S/C65S double mutant (Fig. 4). These results indicate that the two cysteines (C60 and C65) of DR0846 exhibit the free thiol groups or intramolecular disulfide bonds under redox status.
Figure 4.
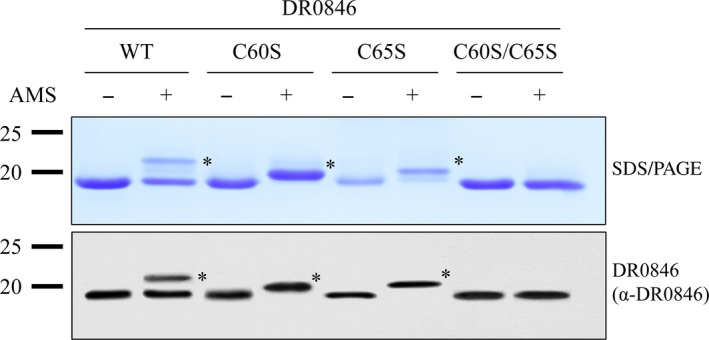
Redox properties of DR0846 and Cys mutant proteins. AMS shift assays were performed using purified WT DR0846 and cysteine mutant proteins. Purified proteins were precipitated with TCA, and treated without (−) or with (+) AMS. The proteins were resolved by reducing SDS/PAGE and subjected to a western blot analysis. Asterisks indicate the oxidized forms of each protein.
Phenotypic characterization of the dr0846 disruption mutant
DR0846 showed peroxidase and molecular chaperone activities in vitro (Fig. 3A,B). To assess the role of DR0846 in vivo, we generated a dr0846 disruption mutant and evaluated it by oxidative and heat stress tolerance assays (Fig. 5). However, as shown in Fig. 5, the sensitivity to H2O2 or heat stress of the ∆dr0846 mutant was similar to that of the WT. Catalases and peroxidases remove H2O2 and catalase activity is correlated with the lethal effects of heat shock stress 45. Catalase activities of exponential and stationary‐phase D. radiodurans were greater than those of E. coli 46. D. radiodurans possesses two catalases 3; among them, the typical monofunctional heme‐containing DR1998 is a major catalase 11. To rule out the possibility of DR1998 catalase function, we constructed catalase‐ and peroxidase‐deficient double‐mutant strains and conducted H2O2 and heat stress tolerance assays using log phase cells were subjected oxidative and heat stress tolerance assay. The ∆dr1998 catalase single‐deletion mutants showed reduced growth compared with that of the WT after treatment with 0.2 mm H2O2 (Fig. 5A) or heat treatment (Fig. 5B). Furthermore, the ∆dr0846 ∆dr1998 double mutants showed severe growth retardation in response to H2O2 or heat stresses (Fig. 5). These results suggest that DR0846 has dual functions as a peroxidase and a molecular chaperone in vivo.
Figure 5.
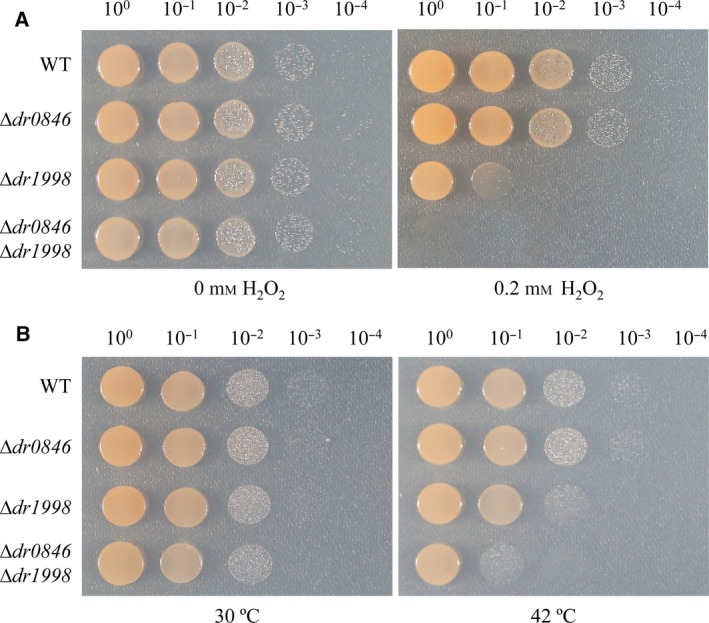
Hydrogen peroxide (H2O2) and heat stress tolerance assays of the ∆dr0846 mutant. Exponential‐phase WT Deinococcus radiodurans and the deletion mutant (Δdr0846) were grown on TGY plates containing different concentrations of H2O2 for 16 h (A) or incubated at 30 °C or 42 °C for 30 min and grown on TGY plates for 16 h, followed by serial dilution (B).
Discussion
Peroxiredoxins catalyze the reduction of hydrogen peroxide and alkyl hydroperoxides 40. Although Prxs of various organisms have been studied, little is known about PrxQ D. radiodurans R1. These studies on D. radiodurans PrxQ, a member of peroxiredoxin, provide a new insight into the structure and function of peroxiredoxin. Here, we reports the substrate and donor specificity and in vivo function of DR0846 from D. radiodurans. As expected, D. radiodurans PrxQ acts as a thioredoxin‐dependent monomeric peroxidase and molecular chaperone, which has two cysteine residues important for its activity.
The typical 2‐Cys Prx have been reported to have physiological functions as peroxidase and molecular chaperone 28, 29, 33, 42, 47, 48. Ionic interactions play an important role in the oligomerization and function of Prx proteins 49, 50. The typical 2‐Cys Prx dynamically regulates the structure between low molecular weight and high molecular weight (HMW) in response to cellular redox states and this structural change is important for the switch between peroxidase and chaperone function 28, 29, 31, 42, 48. 1‐Cys Prx, PrxQ, and Type II Prx act as monomers, whereas 2‐Cys Prxs act as dimers. However, the oligomeric complex has been detected for 1‐Cys Prx and 2‐Cys Prx, whereas PrxQ and Type II Prx have been observed as monomers or dimers 47, 50, 51. The Corynebacterium glutamicum PrxQ (CgPrxQ) and E. coli BCP are predominantly present as monomers in the native or functional states 19, 52. PrxQ4 (ScBCP4) forms a dimer with a non‐covalent dimeric structure 20.
Molecular chaperones prevent the aggregation and assist the covalent folding of proteins and oligomeric architectures are important for chaperone functions 53, 54. While most typical 2‐Cys Prx, some 1‐Cys Prx and CgPrxQ have been reported to have peroxidase activity and oligomeric or dimeric chaperone activity 28, 29, 31, 33, 42, 55, 56, this is the first report for the dual activity of a monomeric atypical 2‐Cys Prx. Although it is not known in Prx, this observation is similar to the results of other proteins reported previously. The FanE is a monomeric chaperone that is present in the periplasm of E. coli 57. Trigger Factor (TF) from Psychrobacter frigidicola (TFPf) displays no dimerization and it can promote refold of RNase T1 58. The monomeric 14‐3‐3ζ protein has a chaperone‐like activity and the 14‐3‐3ζ more effectively prevents aggregation of myosin subfragment 1 compared with its dimeric form. In addition, HSP27, most abundant small heat shock protein in humans, the reduced form was more effective than its oxidized form in preventing protein aggregation 59. However, the molecular mechanism underlying the chaperone activity of monomeric proteins needs to be further studied.
Recent studies revealed that the resistance of D. radiodurans to variety of stresses conditions can be explained by high antioxidant activity to protect cells 1, 3, 5, 10, 11. Heat stress causes the production of ROS to induce oxidative stress and affects antioxidant enzyme activities such as catalase, SOD, and peroxidase in plant species 60, 61. In plants, catalase activities and intensities of catalase isozymes may be important determinants of antioxidant resistance to heat stress 62. The peroxidase enzyme has been associated with the emergence of physiological injuries and its activity was enhanced by high‐temperature stress in mulberry and strawberry 63, 64. Despite previous studies of antioxidant enzymes in relation to heat tolerance, specific antioxidant enzymes have not been well‐characterized. In this study, DR0846 showed strong peroxidase and chaperone activity and exhibited a sensitive phenotype for heat stress based on a mutation analysis (Figs 2 and 4). These results suggest that PrxQ (DR0846) may be an important antioxidant enzyme involved in thermal stress resistance.
Author contributions
CC and SSL conceived and designed the study; CC, GWL, SHH, SK, and JJ performed the experiments; CC, KJ, JJ, SL, BYC, and SSL analyzed the data; CC, SL, and SSL wrote the manuscript.
Supporting information
Fig. S1. Phylogenetic analysis of peroxiredoxins in D. radiodurans R1 and diverse organisms.
Fig. S2. Alignment and sequence comparison of DR0846 with PrxQ proteins from diverse organisms.
Fig. S3. GSH‐dependent peroxidase activity of DR0846 under GSH system.
Table S1. List of primers used in this study.
Acknowledgement
This work was supported by the Nuclear R&D Program of the Ministry of Science and ICT (MSIT), Republic of Korea.
Edited by Miguel De la Rosa
References
- 1. Cox MM and Battista JR (2005) Deinococcus radiodurans – the consummate survivor. Nat Rev Microbiol 3, 882–892. [DOI] [PubMed] [Google Scholar]
- 2. Makarova KS, Aravind L, Wolf YI, Tatusov RL, Minton KW, Koonin EV and Daly MJ (2001) Genome of the extremely radiation‐resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol Mol Biol Rev 65, 44–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Slade D and Radman M (2011) Oxidative stress resistance in Deinococcus radiodurans . Microbiol Mol Biol Rev 75, 133–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tanaka M, Earl AM, Howell HA, Park MJ, Eisen JA, Peterson SN and Battista JR (2004) Analysis of Deinococcus radiodurans's transcriptional response to ionizing radiation and desiccation reveals novel proteins that contribute to extreme radioresistance. Genetics 168, 21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ghosal D, Omelchenko MV, Gaidamakova EK, Matrosova VY, Vasilenko A, Venkateswaran A, Zhai M, Kostandarithes HM, Brim H, Makarova KS et al (2005) How radiation kills cells: survival of Deinococcus radiodurans and Shewanella oneidensis under oxidative stress. FEMS Microbiol Rev 29, 361–375. [DOI] [PubMed] [Google Scholar]
- 6. Imlay JA (2008) Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem 77, 755–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Luan H, Meng N, Fu J, Chen X, Xu X, Feng Q, Jiang H, Dai J, Yuan X, Lu Y et al (2014) Genome‐wide transcriptome and antioxidant analyses on gamma‐irradiated phases of Deinococcus radiodurans R1. PLoS One 9, e85649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Daly MJ (2009) A new perspective on radiation resistance based on Deinococcus radiodurans . Nat Rev Microbiol 7, 237–245. [DOI] [PubMed] [Google Scholar]
- 9. Li M, Sun H, Feng Q, Lu H, Zhao Y, Zhang H, Xu X, Jiao J, Wang L and Hua Y (2013) Extracellular dGMP enhances Deinococcus radiodurans tolerance to oxidative stress. PLoS One 8, e54420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Abreu IA, Hearn A, An H, Nick HS, Silverman DN and Cabelli DE (2008) The kinetic mechanism of manganese‐containing superoxide dismutase from Deinococcus radiodurans: a specialized enzyme for the elimination of high superoxide concentrations. Biochemistry 47, 2350–2356. [DOI] [PubMed] [Google Scholar]
- 11. Jeong SW, Jung JH, Kim MK, Seo HS, Lim HM and Lim S (2016) The three catalases in Deinococcus radiodurans: only two show catalase activity. Biochem Biophys Res Commun 469, 443–448. [DOI] [PubMed] [Google Scholar]
- 12. Kim K, Kim IH, Lee KY, Rhee SG and Stadtman ER (1988) The isolation and purification of a specific “protector” protein which inhibits enzyme inactivation by a thiol/Fe(III)/O2 mixed‐function oxidation system. J Biol Chem 263, 4704–4711. [PubMed] [Google Scholar]
- 13. Dietz KJ (2011) Peroxiredoxins in plants and cyanobacteria. Antioxid Redox Signal 15, 1129–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cui H, Wang Y, Wang Y and Qin S (2012) Genome‐wide analysis of putative peroxiredoxin in unicellular and filamentous cyanobacteria. BMC Evol Biol 12, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chae HZ, Robison K, Poole LB, Church G, Storz G and Rhee SG (1994) Cloning and sequencing of thiol‐specific antioxidant from mammalian brain: alkyl hydroperoxide reductase and thiol‐specific antioxidant define a large family of antioxidant enzymes. Proc Natl Acad Sci U S A 91, 7017–7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kang SW, Baines IC and Rhee SG (1998) Characterization of a mammalian peroxiredoxin that contains one conserved cysteine. J Biol Chem 273, 6303–6311. [DOI] [PubMed] [Google Scholar]
- 17. Dietz KJ (2003) Plant peroxiredoxins. Annu Rev Plant Biol 54, 93–107. [DOI] [PubMed] [Google Scholar]
- 18. Wood ZA, Schroder E, Robin Harris J and Poole LB (2003) Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci 28, 32–40. [DOI] [PubMed] [Google Scholar]
- 19. Kong W, Shiota S, Shi Y, Nakayama H and Nakayama K (2000) A novel peroxiredoxin of the plant Sedum lineare is a homologue of Escherichia coli bacterioferritin co‐migratory protein (Bcp). Biochem J 351, 107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Limauro D, D'Ambrosio K, Langella E, De Simone G, Galdi I, Pedone C, Pedone E and Bartolucci S (2010) Exploring the catalytic mechanism of the first dimeric Bcp: functional, structural and docking analyses of Bcp4 from Sulfolobus solfataricus . Biochimie 92, 1435–1444. [DOI] [PubMed] [Google Scholar]
- 21. Jeong W, Cha MK and Kim IH (2000) Thioredoxin‐dependent hydroperoxide peroxidase activity of bacterioferritin comigratory protein (BCP) as a new member of the thiol‐specific antioxidant protein (TSA)/Alkyl hydroperoxide peroxidase C (AhpC) family. J Biol Chem 275, 2924–2930. [DOI] [PubMed] [Google Scholar]
- 22. Clarke DJ, Ortega XP, Mackay CL, Valvano MA, Govan JR, Campopiano DJ, Langridge‐Smith P and Brown AR (2010) Subdivision of the bacterioferritin comigratory protein family of bacterial peroxiredoxins based on catalytic activity. Biochemistry 49, 1319–1330. [DOI] [PubMed] [Google Scholar]
- 23. Wang G, Olczak AA, Walton JP and Maier RJ (2005) Contribution of the Helicobacter pylori thiol peroxidase bacterioferritin comigratory protein to oxidative stress resistance and host colonization. Infect Immun 73, 378–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. [DOI] [PubMed] [Google Scholar]
- 25. Kumar S, Stecher G and Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saitou N and Nei M (1987) The neighbor‐joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4, 406–425. [DOI] [PubMed] [Google Scholar]
- 27. Nei M and Kumar S (2000) Molecular Evolution and Phylogenetics ISBN: 9780195135855. Oxford University Press, Oxford, UK. [Google Scholar]
- 28. An BC, Lee SS, Lee EM, Lee JT, Wi SG, Jung HS, Park W, Lee SY and Chung BY (2011) Functional switching of a novel prokaryotic 2‐Cys peroxiredoxin (PpPrx) under oxidative stress. Cell Stress Chaperones 16, 317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee EM, Lee SS, Tripathi BN, Jung HS, Cao GP, Lee Y, Singh S, Hong SH, Lee KW, Lee SY et al (2015) Site‐directed mutagenesis substituting cysteine for serine in 2‐Cys peroxiredoxin (2‐Cys Prx A) of Arabidopsis thaliana effectively improves its peroxidase and chaperone functions. Ann Bot 116, 713–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cheong NE, Choi YO, Lee KO, Kim WY, Jung BG, Chi YH, Jeong JS, Kim K, Cho MJ et al (1999) Molecular cloning, expression, and functional characterization of a 2Cys‐peroxiredoxin in Chinese cabbage . Plant Mol Biol 40, 825–834. [DOI] [PubMed] [Google Scholar]
- 31. Chuang MH, Wu MS, Lo WL, Lin JT, Wong CH and Chiou SH (2006) The antioxidant protein alkylhydroperoxide reductase of Helicobacter pylori switches from a peroxide reductase to a molecular chaperone function. Proc Natl Acad Sci U S A 103, 2552–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee GJ, Roseman AM, Saibil HR and Vierling E (1997) A small heat shock protein stably binds heat‐denatured model substrates and can maintain a substrate in a folding‐competent state. EMBO J 16, 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moon JC, Hah YS, Kim WY, Jung BG, Jang HH, Lee JR, Kim SY, Lee YM, Jeon MG et al (2005) Oxidative stress‐dependent structural and functional switching of a human 2‐Cys peroxiredoxin isotype II that enhances HeLa cell resistance to H2O2‐induced cell death. J Biol Chem 280, 28775–28784. [DOI] [PubMed] [Google Scholar]
- 34. Appenzeller‐Herzog C and Ellgaard L (2008) In vivo reduction‐oxidation state of protein disulfide isomerase: the two active sites independently occur in the reduced and oxidized forms. Antioxid Redox Signal 10, 55–64. [DOI] [PubMed] [Google Scholar]
- 35. Funayama T, Narumi I, Kikuchi M, Kitayama S, Watanabe H and Yamamoto K (1999) Identification and disruption analysis of the recN gene in the extremely radioresistant bacterium Deinococcus radiodurans . Mutat Res 435, 151–161. [DOI] [PubMed] [Google Scholar]
- 36. Joe MH, Lee KH, Lim SY, Im SH, Song HP, Lee IS and Kim DH (2012) Pigment‐based whole‐cell biosensor system for cadmium detection using genetically engineered Deinococcus radiodurans . Bioprocess Biosyst Eng 35, 265–272. [DOI] [PubMed] [Google Scholar]
- 37. Brenot A, King KY and Caparon MG (2005) The PerR regulon in peroxide resistance and virulence of Streptococcus pyogenes . Mol Microbiol 55, 221–234. [DOI] [PubMed] [Google Scholar]
- 38. Bréhélin C, Meyer EH, de Souris JP, Bonnard G and Meyer Y (2003) Resemblance and dissemblance of Arabidopsis type II peroxiredoxins: similar sequences for divergent gene expression, protein localization, and activity. Plant Physiol 132, 2045–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rouhier N and Jacquot JP (2005) The plant multigenic family of thiol peroxidases. Free Radic Biol Med 38, 1413–1421. [DOI] [PubMed] [Google Scholar]
- 40. Rhee SG, Yang KS, Kang SW, Woo HA and Chang TS (2005) Controlled elimination of intracellular H(2)O(2): regulation of peroxiredoxin, catalase, and glutathione peroxidase via post‐translational modification. Antioxid Redox Signal 7, 619–626. [DOI] [PubMed] [Google Scholar]
- 41. Hofmann B, Hecht HJ and Flohe L (2002) Peroxiredoxins. Biol Chem 383, 347–364. [DOI] [PubMed] [Google Scholar]
- 42. Jang HH, Lee KO, Chi YH, Jung BG, Park SK, Park JH, Lee JR, Lee SS, Moon JC et al (2004) Two enzymes in one; two yeast peroxiredoxins display oxidative stress‐dependent switching from a peroxidase to a molecular chaperone function. Cell 117, 625–635. [DOI] [PubMed] [Google Scholar]
- 43. Jang HH, Chi YH, Park SK, Lee SS, Lee JR, Park JH, Moon JC, Lee YM, Kim SY et al (2006) Structural and functional regulation of eukaryotic 2‐Cys peroxiredoxins including the plant ones in cellular defense‐signaling mechanisms against oxidative stress. Physiol Plant 126, 549–559. [Google Scholar]
- 44. Cha MK, Kim HK and Kim IH (1996) Mutation and mutagenesis of thiol peroxidase of Escherichia coli and a new type of thiol peroxidase family. J Bacteriol 178, 5610–5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Noventa‐Jordão MA, Couto RM, Goldman MH, Aguirre J, Iyer S, Caplan A, Terenzi HF and Goldman GH (1999) Catalase activity is necessary for heat‐shock recovery in Aspergillus nidulans germlings. Microbiology 145, 3229–3234. [DOI] [PubMed] [Google Scholar]
- 46. Wang P and Schellhorn HE (1995) Induction of resistance to hydrogen peroxide and radiation in Deinococcus radiodurans . Can J Microbiol 41, 170–176. [DOI] [PubMed] [Google Scholar]
- 47. König J, Baier M, Horling F, Kahmann U, Harris G, Schürmann P and Dietz KJ (2002) The plant‐specific function of 2‐Cys peroxiredoxin‐mediated detoxification of peroxides in the redox‐hierarchy of photosynthetic electron flux. Proc Natl Acad Sci U S A 99, 5738–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Banerjee M, Chakravarty D and Ballal A (2015) Redox‐dependent chaperone/peroxidase function of 2‐Cys‐Prx from the cyanobacterium Anabaena PCC7120: role in oxidative stress tolerance. BMC Plant Biol 15, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chauhan R and Mande SC (2001) Characterization of the Mycobacterium tuberculosis H37Rv alkyl hydroperoxidase AhpC points to the importance of ionic interactions in oligomerization and activity. Biochem J 354, 209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Barranco‐Medina S, Lazaro JJ and Dietz KJ (2009) The oligomeric conformation of peroxiredoxins links redox state to function. FEBS Lett 583, 1809–1816. [DOI] [PubMed] [Google Scholar]
- 51. Stork T, Laxa M, Dietz MS and Dietz KJ (2009) Functional characterisation of the peroxiredoxin gene family members of Synechococcus elongatus PCC 7942. Arch Microbiol 191, 141–151. [DOI] [PubMed] [Google Scholar]
- 52. Su T, Si M, Zhao Y, Liu Y, Yao S, Che C and Chen C (2018) A thioredoxin‐dependent peroxiredoxin Q from Corynebacterium glutamicum plays an important role in defense against oxidative stress. PLoS One 13, e0192674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bukau B, Weissman J and Horwich A (2006) Molecular chaperones and protein quality control. Cell 125, 443–451. [DOI] [PubMed] [Google Scholar]
- 54. Kim YE, Hipp MS, Bracher A, Hayer‐Hartl M and Hartl FU (2013) Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem 82, 323–355. [DOI] [PubMed] [Google Scholar]
- 55. Si M, Wang T, Pan J, Lin J, Chen C, Wei Y, Lu Z, Wei G and Shen X (2016) Graded response of the multifunctional 2‐cysteine peroxiredoxin, CgPrx, to increasing levels of hydrogen peroxide in Corynebacterium glutamicum . Antioxid Redox Signal 26, 1–14. [DOI] [PubMed] [Google Scholar]
- 56. Tailor V and Ballal A (2017) Novel molecular insights into the function and the antioxidative stress response of a Peroxiredoxin Q protein from cyanobacteria. Free Radic Biol Med 106, 278–287. [DOI] [PubMed] [Google Scholar]
- 57. Mol O, Fokkema H and Oudega B (1996) The Escherichia coli K99 periplasmic chaperone FanE is a monomeric protein. FEMS Microbiol Lett 138, 185–189. [DOI] [PubMed] [Google Scholar]
- 58. Robin S, Togashi DM, Ryder AG and Wall JG (2009) Trigger factor from the psychrophilic bacterium Psychrobacter frigidicola is a monomeric chaperone. J Bacteriol 191, 1162–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sluchanko NN, Artemova NV, Sudnitsyna MV, Safenkova IV, Antson AA, Levitsky DI and Gusev NB (2012) Monomeric 14‐3‐3ζ has a chaperone‐like activity and is stabilized by phosphorylated HspB6. Biochemistry 51, 6127–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Foyer CH, Descourviéres P and Kunert KJ (1994) Protection against oxygen radicals: an important defense mechanism studied in transgenic plants. Plant Cell Environ 17, 507–523. [Google Scholar]
- 61. Gong M, Chen S, Song Y and Li Z (1997) Effect of calcium and calmodulin on intrinsic heat tolerance in relation to antioxidant systems in maize seedlings. Funct Plant Biol 24 (3), 371–379. [Google Scholar]
- 62. He Y and Huang B (2010) Differential responses to heat stress in activities and isozymes of four antioxidant enzymes for two cultivars of Kentucky Bluegrass contrasting in heat tolerance. J Am Soc Hort Sci 135, 116–124. [Google Scholar]
- 63. Chaitanya KV, Sundar D, Masilamani S and Reddy AR (2002) Variation in heat stress‐induced antioxidant enzyme activities among three mulberry cultivars. Plant Growth Regul 36, 175–180. [Google Scholar]
- 64. Gulen H and Eris A (2004) Effect of heat stress on peroxidase activity and total protein content in strawberry plants. Plant Sci 166, 739–744. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Phylogenetic analysis of peroxiredoxins in D. radiodurans R1 and diverse organisms.
Fig. S2. Alignment and sequence comparison of DR0846 with PrxQ proteins from diverse organisms.
Fig. S3. GSH‐dependent peroxidase activity of DR0846 under GSH system.
Table S1. List of primers used in this study.


