Abstract
Human gene GLIS family zinc finger 2 (GLIS2) is a member of GLI-similar zinc finger protein family. Previous studies indicated GLIS2 gene involved in tumorigenesis mechanisms. However, the association between GLIS2 expression and radiosensitivity of gastric cancer has not been well understood. In this study, we used the gastric cancer database in TCGA, and significant association was observed between the low expression of GLIS2 and radiosensitivity of patients with gastric cancer. The adjusted HR values for radiotherapy were 0.162(0.035-0.756) and 0.089(0.014-0.564), with p values 0.021 and 0.010, respectively, in training and testing data, for these patients with low expression of GLIS2, while for patients with high expression of GLIS2, there was no significant survival difference between radiotherapy and nonradiotherapy groups. The adjusted HR were 0.676(0.288-1.586) and 0.508(0.178-1.450), with p values 0.368 and 0.206 in training and testing data, respectively. Further study showed that, for low expression patients, radiotherapy did not significantly increase new tumor event rate and disease progression rate, which partially supported our assumption. These results suggested that low expression of GLIS2 might significantly associate with the radiosensitivity of patients with gastric cancer. The GLIS2 gene might be a potential effective molecular marker of gastric cancer for precise radiotherapy.
1. Introduction
Gastric cancer accounts for a large proportion of cancer death worldwide. GLOBOCAN2018 data showed that over 1000,000 people were newly diagnosed with gastric cancer in 2018 [1], nearly 783,000 people died. According to Chinese cancer statistics in 2015 [2], gastric cancer is one of the top four common malignant tumors in China, with the second highest morbidity and mortality. Clinically, the treatment of gastric cancer mainly focuses on surgery, radiotherapy, chemotherapy, immunotherapy, and combined treatment of traditional Chinese medicine as adjuvant treatment methods. Based on extensive application of radiotherapy in clinical practice worldwide, many researchers have paid great attention to how to utilize radiotherapy to improve the life quality of patients with gastric cancer in different directions. Chung et al. did retrospective analysis on the efficacy of treatment of gastric cancer by volumetric-modulated arc therapy (VMAT) and found that VMAT could effectively reduce the treatment time and the irradiation dose at the liver and kidney to alleviate the toxicity of radiotherapy [3]. Some clinical trials had focused on providing optimal radiotherapy strategies for patients with different stages of gastric cancer [4–7]. However, they did not reach consistent results.
Due to diverse sensitivity to radiotherapy in individuals, clinical doctors would like to screen radiosensitive patients who can obtain higher survival benefits. Based on the genome sequence technology, researchers could find potential biomarkers to predict radiosensitive patients, then oncologists and surgeons could adjust their treatment strategy to reduce adverse reactions and improve radiotherapy efficacy [8]. Therefore, it is particularly imperative to find gene biomarkers that can accurately and sensitively predict whether patients with gastric cancer are sensitive to radiotherapy, so as to provide evidence to formulate targeted radiotherapy regimens.
GLIS2 gene is a dual-function transcriptional regulator; its regulation would play an important role during embryonic development [9]. GLIS2 gene could regulate self-renewal capacity in hematopoietic progenitor cells and promote differentiation of megakaryocytes [10, 11], which was identified as one of several genes required for optimal repopulation [12]. Nevertheless, overexpression of human GLIS2 had a negative effect on reprogramming [13, 14], leading to a decreased number of ESC-like colonies, denoting that GLIS2 gene might be associated with cancers. Moreover, expression levels of GLIS2 gene make crucial contribution to maintaining normal renal structure and function [15]; previous studies reported that loss of GLIS2 could lead to increased renal cell apoptosis and fibrosis in human and mice [16]. Mutant mice lacking GLIS2 function showed anterior bowel defects, including esophageal and tracheal stenosis, as well as pulmonary hypoplasia and pulmonary function defects [17].
Recent study had shown that overexpression of GLIS2 had significant association with chemoresistance of gastric cancer [18]. Relationship between the expression of GLIS2 and the sensitivity of radiotherapy for patients suffering from gastric cancer has not been well studied. We assumed the expression level of GLIS2 associated with radiosensitivity of patients. Sensitive patients could obtain survival benefits after radiotherapy. To verify this hypothesis, we analyzed the relationship between GLIS2 expression and radiotherapy sensitivity based on gastric cancer data from TCGA, to provide references for clinical treatment of gastric cancer.
2. Data Sources and Methods
2.1. Data Sources
In the present study, we analyzed normalized mRNA sequencing data of GLIS2 of the patients with gastric cancer. The data was downloaded in December 2016 from TCGA (The Cancer Genome Atlas, http://cancergenome.nih.gov/) [19, 20], by using TCGA assembler [21]. To clean data, we combined the clinical survival information from several raw data files and eliminated the data of patients with no survival time or survival outcome to obtain effective patient survival information. Then we screened the clinical factors needed among the available data and combined them to obtain a complete clinical data file. In addition, we kept the patients with clear information on radiotherapy. Furthermore, we deleted repeated expression information of normal tissue and combined the mRNA sequencing data of GLIS2 with the clinical data obtained in previous steps to obtain the data used for the present study, which contained 371 patients.
2.2. Analysis Method
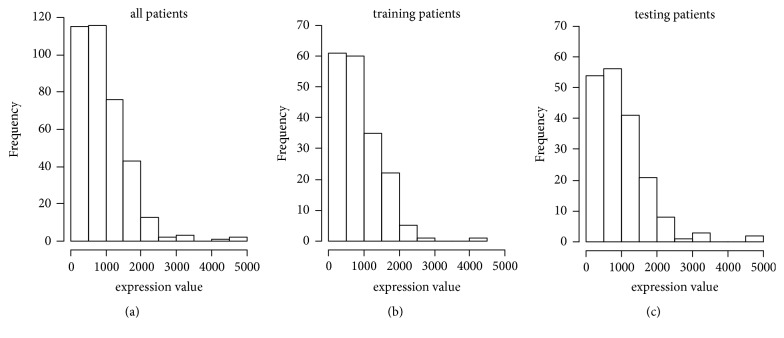
In the present study, radiotherapy sensitivity was defined as the improved survival benefits of the patients receiving radiotherapy, compared with the patients who did not receive radiotherapy [22, 23]. The gene that can predict individual radiosensitivity could be a potential biomarker for radiosensitivity prediction. In order to validate the hypothesis of this study, we randomly split the data into training data and testing data. Firstly, we generated random number between 0 and 1 for all patients, by using the R function runif(). Then, we picked up a half of patients with small random number, as training data. The rest patients were treated as testing data. The same analysis was performed for both training and testing data. Since the expression level of GLIS2 gene showed skewness distribution in the training data (Figure 1), the median values in the training data were defined as the critical threshold of high and low expression. Then, univariate and multivariate Cox regression analysis were performed for patients with high and low expression. Supplemental Table S1 showed the basic patient characteristics.
Figure 1.
Expression distribution of GLIS2 gene of patients with gastric cancer. (a) Expression distribution of GLIS2 gene in all data. (b) Expression distribution of GLIS2 gene in training data. (c) Expression distribution of GLIS2 gene in testing data.
In the present study, survival analysis model of R packages survival was adopted for analysis, and survival curves were plotted by R packages rms. P value of 0.05 was taken as the criterion of significance. Missing values were multiple imputed by R package mice [24].
3. Results
3.1. Correlation Analysis of GLIS2 Expression Level and Clinical Indicators with Survival
In this study, the overall survival was the main observation outcome. The Cox proportional hazard model was used to evaluate the association between GLIS2 expression level and clinical factors with survival. Tables 1 and 2 illustrated the analysis results of training and testing data, respectively. The analysis results showed that radiotherapy could improve the overall survival of patients. However, in both datasets, there were no significant associations between expression level of GLIS2 and overall survival.
Table 1.
Associations of clinical indicators and GLIS2 expression level with total survival in training data.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| HR | P values | HR | P values | |
| Radiotherapy | ||||
| Yes | 0.458(0.247-0.849) | 0.013 | 0.453(0.226-0.909) | 0.026 |
| No | 1.000 | 1.000 | ||
| Gender | ||||
| Male | 1.422(0.886-2.283) | 0.145 | 1.645(0.994-2.724) | 0.053 |
| Female | 1.000 | 1.000 | ||
| Age | ||||
| >60 | 1.498(0.894-2.509) | 0.125 | 1.569(0.895-2.751) | 0.116 |
| ≤60 | 1.000 | 1.000 | ||
| Histologic type | ||||
| NOS | 0.956(0.556-1.641) | 0.868 | 0.969(0.550-1.706) | 0.912 |
| DT/MT/SRT | 0.774(0.405-1.480) | 0.438 | 0.724(0.356-1.473) | 0.373 |
| PT/TT | 1.000 | 1.000 | ||
| T Stage | ||||
| T3/T4 | 1.891(1.088-3.285) | 0.024 | 1.642(0.870-3.098) | 0.126 |
| T1/T2 | 1.000 | 1.000 | ||
| N Stage | ||||
| N1/N2/N3 | 1.837(1.086-3.105) | 0.023 | 1.433(0.666-3.080) | 0.357 |
| N0 | 1.000 | 1.000 | ||
| M Stage | ||||
| M1 | 3.178(1.616-6.250) | 0.001 | 3.305(1.435-7.614) | 0.005 |
| M0 | 1.000 | 1.000 | ||
| Pathological stage | ||||
| III/IV | 1.810(1.129-2.902) | 0.014 | 1.268(0.609-2.639) | 0.526 |
| I/II | 1.000 | 1.000 | ||
| Targeted therapy | ||||
| Yes | 0.753(0.481-1.176) | 0.212 | 1.010(0.352-2.898) | 0.986 |
| No | 1.000 | 1.000 | ||
| Chemotherapy | ||||
| Yes | 0.771(0.497-1.196) | 0.246 | 0.792(0.291-2.160) | 0.649 |
| No | 1.000 | 1.000 | ||
| GLIS2 expression | ||||
| High | 1.232(0.794-1.910) | 0.352 | 1.353(0.854-2.144) | 0.197 |
| Low | 1.000 | 1.000 | ||
Note: HR: hazard ratio; NOS: not otherwise specified; DT: diffuse type; MT: mucinous type; SRT: signet ring type; PT: papillary type; TT: tubular type.
Table 2.
Associations of clinical indicators and GLIS2 expression level with total survival in testing data.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| HR | P values | HR | P values | |
| Radiotherapy | ||||
| Yes | 0.334(0.161-0.690) | 0.003 | 0.309(0.128-0.744) | 0.009 |
| No | 1.000 | 1.000 | ||
| Gender | ||||
| Male | 1.100(0.668-1.813) | 0.708 | 1.193(0.711-2.003) | 0.504 |
| Female | 1.000 | 1.000 | ||
| Age | ||||
| >60 | 1.309(0.793-2.160) | 0.293 | 1.346(0.794-2.282) | 0.270 |
| ≤60 | 1.000 | 1.000 | ||
| Histologic type | ||||
| NOS | 1.521(0.811-2.853) | 0.191 | 1.843(0.962-3.534) | 0.065 |
| DT/MT/SRT | 1.065(0.520-2.180) | 0.863 | 1.570(0.739-3.333) | 0.240 |
| PT/TT | 1.000 | 1.000 | ||
| T Stage | ||||
| T3/T4 | 1.700(0.909-3.183) | 0.097 | 0.994(0.472-2.091) | 0.986 |
| T1/T2 | 1.000 | 1.000 | ||
| N Stage | ||||
| N1/N2/N3 | 2.227(1.169-4.240) | 0.015 | 2.280(0.995-5.228) | 0.051 |
| N0 | 1.000 | 1.000 | ||
| M Stage | ||||
| M1 | 1.167(0.559-2.439) | 0.681 | 1.162(0.536-2.520) | 0.704 |
| M0 | 1.000 | 1.000 | ||
| Pathological stage | ||||
| III/IV | 1.952(1.162-3.279) | 0.012 | 1.703(0.831-3.490) | 0.146 |
| I/II | 1.000 | 1.000 | ||
| Targeted therapy | ||||
| Yes | 0.580(0.356-0.946) | 0.029 | 0.997(0.340-2.924) | 0.996 |
| No | 1.000 | 1.000 | ||
| Chemotherapy | ||||
| Yes | 0.608(0.376-0.982) | 0.042 | 0.679(0.249-1.856) | 0.451 |
| No | 1.000 | 1.000 | ||
| GLIS2 expression | ||||
| High | 1.401(0.869-2.260) | 0.166 | 1.184(0.725-1.936) | 0.500 |
| Low | 1.000 | 1.000 | ||
Note: abbreviations were the same with Table 1.
These results showed radiation therapy was significantly associated with overall survival. However, we argued that not all patients benefitted from radiation therapy. More accurate radiotherapy can be achieved if sensitive patients are effectively screened. In other words, poorer sensitive patients could be protected from noneffective radiotherapy and the adverse reactions.
3.2. Relationship between GLIS2 Expression Levels and Clinical Indicators
We analyzed the relationship between expression levels of GLIS2 and clinical factors by using the chi-square test. The analysis results in Table 3 showed that there were no significant associations between the expressions of GLIS2 and any clinical indicator.
Table 3.
Relationship between expression levels of GLIS2 and clinical indicators.
| Training data(n=185) | Testing data(n=186) | |||||||
|---|---|---|---|---|---|---|---|---|
| High | Low | χ 2 | P values | High | Low | χ 2 | P values | |
| Gender | 0.264 | 0.608 | 0.978 | 0.323 | ||||
| Male | 56 | 61 | 71 | 53 | ||||
| Female | 36 | 32 | 30 | 32 | ||||
| Age | 0.000 | 1.000 | 2.287 | 0.131 | ||||
| >60 | 64 | 63 | 59 | 59 | ||||
| ≤60 | 28 | 28 | 42 | 25 | ||||
| Histologic type | 5.681 | 0.058 | 7.173 | 0.028 | ||||
| NOS | 44 | 54 | 50 | 42 | ||||
| MT/DT/SRT | 30 | 16 | 33 | 17 | ||||
| PT/TT | 18 | 22 | 16 | 26 | ||||
| T Stage | 1.518 | 0.218 | 0.080 | 0.777 | ||||
| T3/T4 | 69 | 59 | 77 | 63 | ||||
| T1/T2 | 23 | 31 | 23 | 22 | ||||
| N Stage | 0.000 | 1.000 | 0.243 | 0.622 | ||||
| N1/N2/N3 | 64 | 63 | 73 | 57 | ||||
| N0 | 28 | 29 | 28 | 27 | ||||
| M Stage | 1.278 | 0.258 | 0.330 | 0.565 | ||||
| M1 | 4 | 9 | 12 | 7 | ||||
| M0 | 88 | 84 | 89 | 78 | ||||
| Pathological Stage | 0.019 | 0.891 | 0.013 | 0.909 | ||||
| III/IV | 48 | 44 | 51 | 42 | ||||
| I/II | 42 | 42 | 46 | 41 | ||||
Note: abbreviations were the same with Table 1.
3.3. Relationship between Radiotherapy and Survival in High and Low Expression Groups
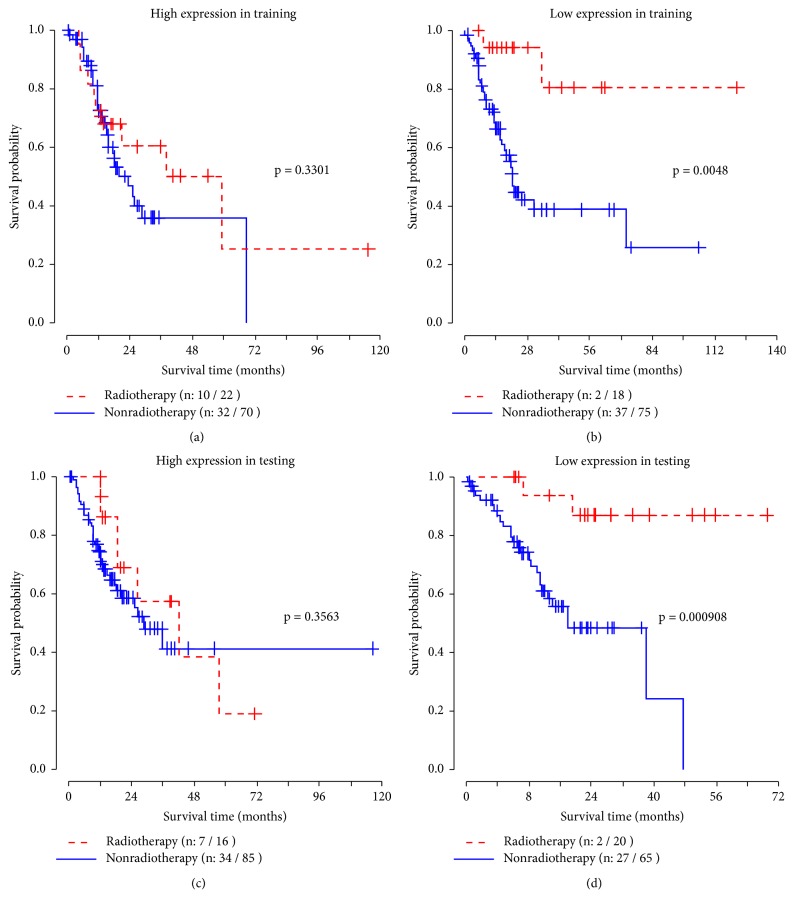
The main idea of this study was to discuss whether the patients with low expression of GLIS2 were sensitive to radiotherapy. In order to obtain reliable results, we split the data to two part, training data and testing data, and performed survival analysis respectively. Table 4 demonstrated that, in the training and testing data, for low expression subgroup, there were significant associations between radiotherapy and overall survival. The similar results could be found between univariate and multivariate analysis. The adjusted HR for radiotherapy vs nonradiotherapy were 0.162(0.035-0.756) and 0.089(0.014-0.564) in training and testing data, respectively. For the patients with high expression in training and testing data, radiotherapy could not significantly improve the overall survival, with the adjusted HR 0.676(0.288-1.586) and 0.508(0.178-1.450), respectively.
Table 4.
Association analysis between radiotherapy and survival under different expressions of GLIS2.
| Data | GLIS2 expression | Unadjusted(RT vs NRT) | Adjusted(RT vs NRT) | ||
|---|---|---|---|---|---|
| HR | P values | HR | P values | ||
| Training | High (n=92) | 0.694(0.332-1.452) | 0.332 | 0.676(0.288-1.586) | 0.368 |
| Low (n=93) | 0.165(0.040-0.686) | 0.013 | 0.162(0.035-0.756) | 0.021 | |
|
| |||||
| Testing | High (n=101) | 0.677(0.294-1.558) | 0.359 | 0.508(0.178-1.450) | 0.206 |
| Low (n=85) | 0.116(0.027-0.509) | 0.004 | 0.089(0.014-0.564) | 0.010 | |
|
| |||||
| All Data | High (n=193) | 0.694(0.401-1.200) | 0.191 | 0.673(0.360-1.257) | 0.214 |
| Low (n=178) | 0.145(0.053-0.401) | <0.001 | 0.170(0.055-0.521) | 0.002 | |
Note: adjusted factors: gender, age, histologic type, TNM stage, pathological stage, chemotherapy, and targeted therapy.
Figure 2 illustrated the survival curves of radiotherapy and nonradiotherapy groups based on different expression levels of GLIS2 gene in the training data and testing data. In the low expression group, the survival time of the patients receiving radiotherapy was significantly prolonged, shown in Figures 2(b) and 2(d). In the high expression group, Figures 2(a) and 2(c), radiotherapy had no significant effect on the overall survival.
Figure 2.
Survival curves under different expression levels of GLIS2 in training and testing data. Log-rank test was used to estimate the P values.
We further performed survival analysis by combined training and testing data (Table 4). The same conclusion could be reached. The unadjusted and adjusted HR were 0.694(0.401-1.200) and 0.673(0.360-1.257) for high expression subgroup, respectively. However, for low expression group, the unadjusted and adjusted HR were 0.145(0.053-0.401) and 0.170(0.055-0.521), respectively. For low expression group, radiotherapy exhibited significant clinical efficacy.
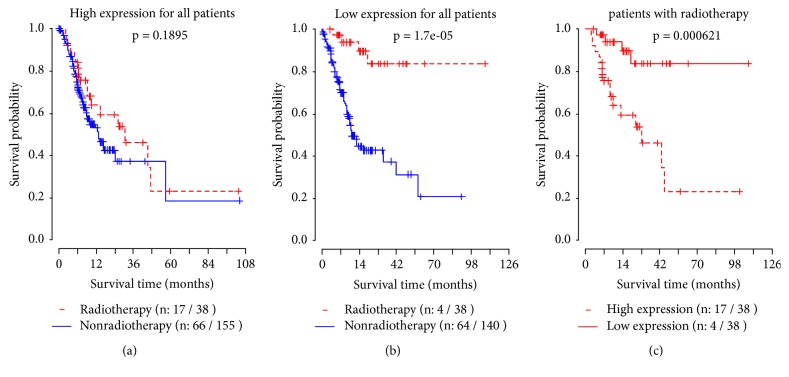
Survival curves for all high and low expression patients were shown in Figures 3(a) and 3(b), respectively. Figure 3(c) contained the survival curves for patients who received radiotherapy. It can be seen that the survival rate of patients with low expression was significantly prolonged after receiving radiotherapy.
Figure 3.
Survival curves under different expression levels of GLIS2 for all patients.
In summary, in the high expression group, patients who received radiotherapy achieved no significant survival benefits than those who were not treated by radiotherapy, while in the low expression group, the survival rate of patients was significantly improved if they received radiotherapy. The results suggested that the low expression of GLIS2 gene might effectively indicate the radiosensitivity of patients. These patients would obtain significant survival benefits from radiotherapy.
3.4. Associations among GLIS2 Expressions and Clinical Assessment Factors after Adjuvant Therapy
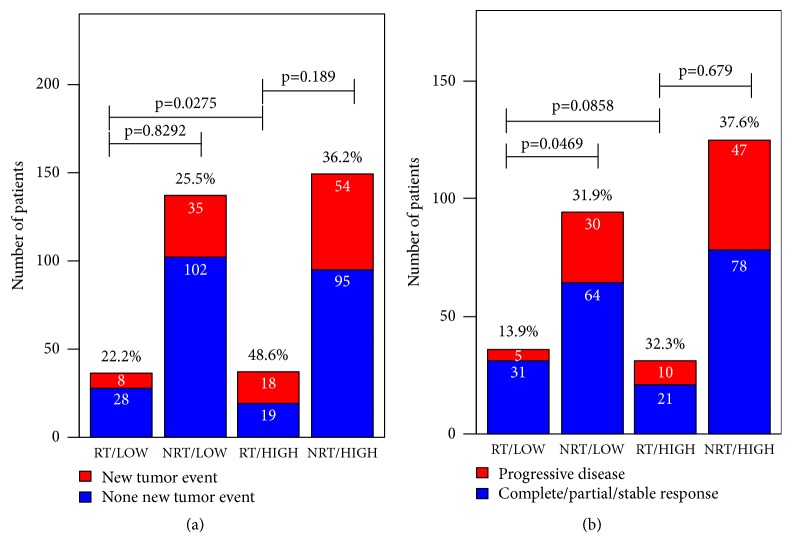
Figure 4 showed the associations among the expression levels of GLIS2 and two clinical assessment indexes: the new tumor event and progressive disease. There was a significant difference in new tumor event rate between high and low expression group patients under radiotherapy. The low expression group hold lower new tumor event rate 22.2%, which was a half of high expression group (48.6%). Disease progression rate was also approximately significant lower (13.9%) in low expression group, compared with high expression group (32.3%), for patients who received radiotherapy. The results in Figure 4 suggested that, for low expression patients, radiotherapy did not increase new tumor event rate and disease progression rate and even decreased the two rates. These results partially supported that patients with low expression of GLIS2 might be sensitive to radiotherapy.
Figure 4.
Associations among GLIS2 expressions and clinical assessment factors. Chi-square test was used for comparisons of rates of different groups. RT: radiotherapy; NRT: nonradiotherapy; HIGH: high expression of GLIS2 gene; LOW: low expression of GLIS2 gene.
4. Discussion
Radiotherapy is an essential part of adjuvant treatment to cancers, whereas it is also a double-edged sword. It not only kills tumor cells, but also promotes radioimmunity, induces distant metastasis, and damages normal tissues [25]. Studies had reported that increased irradiation had toxic effects on the skin and other normal tissues [26]. Therefore, improving the efficacy of radiotherapy and reducing toxicity had attracted worldwide attention. According to variations of radiosensitivity in individuals, dividing patients with gastric cancer and giving radiation treatment to patients with significant radiotherapy sensitivity would make radiotherapy more accurate, while eliminating the adverse reactions of patients who are not sensitive to radiotherapy after radiotherapy [27]. It can be seen that finding appropriate biomarkers to distinguish sensitive populations is of great importance for clinical treatment. However, effective molecular markers had not been found in gastric cancer so far.
We made use of public data from TCGA in the present study. Due to the difficulties of collecting clinical samples and the large number of potential genes available for external validation, the development of a reliable diagnostic classifier using early nonrandomized phase II data is often not feasible. To overcome these difficulties, we performed an internal validation procedure that randomly divided the data into two parts and analyzed them separately. We found significant associations between expression levels of GLIS2 gene and survival outcome. Low expression of GLIS2 gene might indicate radiosensitivity of patients. Table S2 illustrated the research results of data. Radiotherapy did have significant effect on improving total survival time of patients suffering from gastric cancer. However, we argued that, in clinical practice, not all patients, some patients with gastric cancer will benefit from radiotherapy. The need for radiation depends on what type of surgery, whether the cancer has spread to somewhere else of body, and in some cases, the age or other clinical factors. If clinical doctors can predict radiosensitivity of patients, they could evaluate sensitive patients more effectively and perform accurate radiotherapy.
In our analysis, we chose median as cutoff to divide high and low expression group. We also performed analysis on other cutoffs, as shown in Figure S1. The results suggested that, when the cutoff larger than median was selected to divide high and low expression group, for patients with high expression, there was no significant survival difference between radiotherapy and nonradiotherapy, while for patients with low expression, patients who received radiotherapy had better survival than who did not receive radiotherapy. We also found that if we selected other cutoffs lower than median, like 10% to 40% quantiles, for these patients with high expression, the HR of radiotherapy were statistically significant, which may be caused by including too many relative lower expression patients. These results were consistent with our conclusion, that low expression patients could be predicted as radiosensitive patients.
It is known that radiotherapy type in gastric cancer includes preoperative, postoperative, and palliative therapy. Preoperative radiotherapy is mainly used in patients with locally advanced gastric cancer to reduce tumor burden and control tumor progression for surgery. TCGA did not provide clear information about radiotherapy type. But, if preoperative therapy was used, the expression data of cancer tissue would be not useful.
Postoperative radiotherapy was the main direction of our research, which is usually combined with chemotherapy to treat patients with resectable gastric cancer as adjuvant therapy. Macdonald et al. conducted Gastrointestinal Cancer Intergroup phase III Trial (INT 0116) and found that postoperative chemoradiotherapy (CRT) could significantly improve the survival rate of patients after radical gastrectomy, though lack of strict trial control [28]. Furthermore, the result of phase III trial led by Lee et al. suggested that additional postoperative radiotherapy to surgery and postoperative chemotherapy could effectively reduce the local recurrence rate and improve the progression-free survival time of patients with positive pathologic lymph nodes [29, 30].
Palliative radiotherapy is mainly applied to the treatment of patients with advanced gastric cancer, focusing on reducing bleeding, pain and other symptoms to improve the quality of life of patients. For M1 stage patients, palliative radiotherapy might be used to improve survival of these patients. We further performed analysis on M0 stage patients and removed M1 stage patients. The results (Table S3) were consistent with our previous results in Table 4. We treated the M stage of patients as a covariate in our analysis.
The mechanisms of the association between GLIS2 and radiotherapy are still not clear. According to the report published by Masetti et al. [31], CBFA2T3-GLIS2 is the most frequent chimeric oncogene identified in non-Down's syndrome acute megakaryocytic leukemia (non-DS-AMKL). It regulated molecules involved in the Hedgehog pathway and Wingless/Integrated (WNT)/β-catenin pathways, such as GATA3, HHIP and β-catenin. GATA3 was demonstrated to interact with HIF-1α to enhance cancer cell invasiveness [32], and inhibition of HHIP promoter methylation suppressed human gastric cancer cell proliferation and migration [33], which would affect the treatment and prognosis of patients with gastric cancer. β-catenin regulated cell adhesion to impair DNA repair [34], leading to increased DNA damage and sensitivity of treatment for cancers [35, 36]. GLIS2 could also regulate the interaction between β-catenin and T-cell factor/Lymphoid enhancer factor (TCF/LEF) to affect the activation of cyclin D1, which may have association with poor tumor differentiation and prognosis in gastric cancer [37]. These findings suggested that GLIS2 might be an important gene associated with tumor DNA repair and tumor cell cycle. The changes of tumor DNA repair and tumor cell cycle also associated with another important clinical treatment, the radiotherapy. Therefore, GLIS2 may also involve in molecular response under adjuvant radiotherapy. Mechanisms of GLIS2 and radiosensitivity of gastric cancer require further study.
In the present study, expression levels of GLIS2 gene did not associate with overall survival. However, in the subgroup analysis, we came to conclude that gastric cancer patients with low expression of GLIS2 were supposed to possess high radiosensitivity, while patients with high expression of GLIS2 gene were not sensitive to radiotherapy. Population selectivity of radiotherapy has certain guiding value for treatment of gastric cancer.
In clinical work, it was found that the degree of tumor retraction was significantly different after radiotherapy, mainly because of the large individual differences in radiosensitivity. DNA is the main target of ionizing radiation. Cancer risk is usually associated with changes in DNA repair, cell cycle, or apoptotic pathways [38], which plays important roles in radiosensitivity. Gene mutations, polymorphisms, and epigenetic modifications related to DNA repair function can make radiosensitivity variant [39]. In radiotherapy, the survival time varies greatly due to sensitivity of radiotherapy. If cancer patients can be predicted to exhibit radiotherapy sensitivity or resistance, oncologists and surgeons can then alter the treatment to reduce adverse reactions or improve the efficacy of radiotherapy. CBFA2T3-GLIS2 is an important prognostic factor for patients with non-DS-AMKL [10, 40, 41]. Studies had linked the function of GLIS2 to autosomal recessive kidney disease and found that GLIS2 was the most common genetic cause of end-stage renal failure [42]. These studies had demonstrated that the GLIS2 gene had important links to cancers. In our research, we concluded that GLIS2 gene might be an effective molecular marker which was independent of tumor clinical indicators and an indicator of prognostic assessment indexes, which was consistent with the conclusion of previous study [18].
Studies on the association between radiosensitivity of gastric cancer and GLIS2 expression had not been reported before. In this study, internal validation strategy was used to make up for the small sample size and the defects of data only from TCGA. The relationship between GLIS2 and radiosensitivity of gastric cancer was explored, which provided a new reference for clinical improvement of the therapeutic effect on gastric cancer, and an important clue for basic research on radiosensitivity of gastric cancer. Furthermore, we should mention that there were some limitations of this study. We used the data only from TCGA. Sample size was small. In addition, there was no external validation study on our results, like real data from clinical study. Although the limitations existed, the present study still provided a potential helpful clue for further study.
Acknowledgments
This work was supported in part by the National Natural Science Foundation of China (81773541 and 81573253), Project of State Key Laboratory of Radiation Medicine and Protection at Soochow University to Zaixiang Tang, Key Scientific Development Program of China (2016YFC0904702) to Jianping Cao, and Jiangsu Provincial Key Project in Research and Development of Advanced Clinical Technique (BL2018657) to Ye Tian. We acknowledge the contributions of the TCGA Research Network.
Data Availability
The datasets used in the present study are available from The Cancer Genome Atlas database (http://cancergenome.nih.gov/).
Disclosure
The funding body did not play any roles in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Authors' Contributions
Haitong Sun and Jincheng Gu contributed equally to this work.
Supplementary Materials
Table S1: basic patient characteristics. Table S2: associations of clinical indicators and GLIS2 expression level with total survival in all data. Table S3: association analysis between radiotherapy and survival under different expressions of GLIS2 for M0 stage patients. Figure S1: the HR values of radiotherapy along with different cutoffs. We chose different quantiles as cutoff to divide high and low expression group for further survival analysis.
References
- 1.Bray F., Ferlay J., Soerjomataram I. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Chen W., Zheng R., Baade P. D., et al. Cancer statistics in China, 2015. CA: A Cancer Journal for Clinicians. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Chung J., Na K., Kim I. H. Benefit of volumetric-modulated arc therapy over three-dimensional conformal radiotherapy for stage I-II extranodal marginal zone B-cell lymphoma of mucosa-associated lymphoid tissue in the stomach: a dosimetric comparison. Radiation Oncology Journal. 2018;36(4):332–340. doi: 10.3857/roj.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van den Ende T., ter Veer E., Machiels M., et al. The Efficacy and safety of (Neo) Adjuvant therapy for gastric cancer: a network meta-analysis. Cancers. 2019;11(1):p. 80. doi: 10.3390/cancers11010080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kondoh C., Shitara K., Nomura M., et al. Efficacy of palliative radiotherapy for gastric bleeding in patients with unresectable advanced gastric cancer: A retrospective cohort study. BMC Palliative Care. 2015;14(1):p. 37. doi: 10.1186/s12904-015-0034-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu J. I., Lim D. H., Ahn Y. C., et al. Effects of adjuvant radiotherapy on completely resected gastric cancer: A radiation oncologist's view of the ARTIST randomized phase III trial. Radiotherapy & Oncology. 2015;117(1):171–177. doi: 10.1016/j.radonc.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 7.Zhu W.-G., Xua D.-F., Pu J., et al. A randomized, controlled, multicenter study comparing intensity-modulated radiotherapy plus concurrent chemotherapy with chemotherapy alone in gastric cancer patients with D2 resection. Radiotherapy & Oncology. 2012;104(3):361–366. doi: 10.1016/j.radonc.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 8.Rattay T., Talbot C. J. Finding the genetic determinants of adverse reactions to radiotherapy. Clinical Oncology. 2014;26(5):301–308. doi: 10.1016/j.clon.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Zhang F., Nakanishi G., Kurebayashi S., et al. Characterization of Glis2, a novel gene encoding a Gli-related, Krüppel-like transcription factor with transactivation and repressor functions. Roles in kidney development and neurogenesis. The Journal of Biological Chemistry. 2002;277(12):10139–10149. doi: 10.1074/jbc.M108062200. [DOI] [PubMed] [Google Scholar]
- 10.Gruber T. A., Larson Gedman A., Zhang J., et al. An Inv(16)(p13.3q24.3)-Encoded CBFA2T3-GLIS2 fusion protein defines an aggressive subtype of pediatric acute megakaryoblastic leukemia. Cancer Cell. 2012;22(5):683–697. doi: 10.1016/j.ccr.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scoville D. W., Kang H. S., Jetten A. M. GLIS1-3: Emerging roles in reprogramming, stem and progenitor cell differentiation and maintenance. Stem Cell Investigation. 2017;4(9) doi: 10.21037/sci.2017.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmfeldt P., Ganuza M., Marathe H., et al. Functional screen identifies regulators of murine hematopoietic stem cell repopulation. The Journal of Experimental Medicine. 2016;213(3):433–449. doi: 10.1084/jem.20150806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pells S., Koutsouraki E., Morfopoulou S., et al. Novel Human embryonic stem cell regulators identified by conserved and distinct CpG island methylation state. PLoS ONE. 2015;10(7):p. e0131102. doi: 10.1371/journal.pone.0131102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee S.-Y., Noh H. B., Kim H.-T., Lee K.-I., Hwang D.-Y. Glis family proteins are differentially implicated in the cellular reprogramming of human somatic cells. Oncotarget . 2017;8(44):77041–77049. doi: 10.18632/oncotarget.20334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim Y., Kang H. S., Herbert R., et al. Kruppel-Like zinc finger protein Glis2 is essential for the maintenance of normal renal functions. Molecular and Cellular Biology. 2008;28(7):2358–2367. doi: 10.1128/MCB.01722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Attanasio M., Uhlenhaut N. H., Sousa V. H., et al. Loss of GLIS2 causes nephronophthisis in humans and mice by increased apoptosis and fibrosis. Nature Genetics. 2007;39(8):1018–1024. doi: 10.1038/ng2072. [DOI] [PubMed] [Google Scholar]
- 17.Motoyama J., Liu J., Mo R., Ding Q., Post M., Hui C.-C. Essential function of Gli2 and Gli3 in the formation of lung, trachea and oesophagus. Nature Genetics. 1998;20(1):54–57. doi: 10.1038/1711. [DOI] [PubMed] [Google Scholar]
- 18.Yuan J., Tan L., Yin Z., et al. GLIS2 redundancy causes chemoresistance and poor prognosis of gastric cancer based on co-expression network analysis. Oncology Reports. 2019;41(1):191–201. doi: 10.3892/or.2018.6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chia N.-Y., Tan P. Molecular classification of gastric cancer. Annals of Oncology. 2016;27(5):763–769. doi: 10.1093/annonc/mdw040. [DOI] [PubMed] [Google Scholar]
- 21.Zhu Y., Qiu P., Ji Y. TCGA-assembler: Open-source software for retrieving and processing TCGA data. Nature Methods. 2014;11(6):599–600. doi: 10.1038/nmeth.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou J., Wu X., Li G., et al. Prediction of radiosensitive patients with gastric cancer by developing gene signature. International Journal of Oncology. 2017;51(4):1067–1076. doi: 10.3892/ijo.2017.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji Y., Jiang Q., Jian G., et al. Developing a radiosensitivity gene signature for Caucasian patients with breast cancer. Oncology Reports. 2018;40(3):1695–1705. doi: 10.3892/or.2018.6567. [DOI] [PubMed] [Google Scholar]
- 24.van Buuren S., Groothuis-Oudshoorn K. Mice: multivariate imputation by chained equations in R. Journal of Statistical Software . 2011;45(3):1–67. [Google Scholar]
- 25.Liu W., Huang Y., Liu C., et al. Inhibition of TBK1 attenuates radiation-induced epithelial–mesenchymal transition of A549 human lung cancer cells via activation of GSK-3β and repression of ZEB1. Laboratory Investigation. 2014;94(4):362–370. doi: 10.1038/labinvest.2013.153. [DOI] [PubMed] [Google Scholar]
- 26.Lin J., Liu C., Gao F., et al. MiR-200c enhances radiosensitivity of human breast cancer cells. Journal of Cellular Biochemistry. 2013;114(3):606–615. doi: 10.1002/jcb.24398. [DOI] [PubMed] [Google Scholar]
- 27.Bentzen S. M., Overgaard J. Patient-to-patient variability in the expression of radiation-induced normal tissue injury. Seminars in Radiation Oncology. 1994;4(2):68–80. doi: 10.1016/S1053-4296(05)80034-7. [DOI] [PubMed] [Google Scholar]
- 28.Macdonald J. S., Smalley S. R., Benedetti J., et al. Chemoradiotherapy after surgery compared with surgery alone for adenocarcinoma of the stomach or gastroesophageal junction. The New England Journal of Medicine. 2001;345(10):725–730. doi: 10.1056/NEJMoa010187. [DOI] [PubMed] [Google Scholar]
- 29.Lee J., Lim D. H., Kim S., et al. Phase III trial comparing capecitabine plus cisplatin versus capecitabine plus cisplatin with concurrent capecitabine radiotherapy in completely resected gastric cancer with D2 lymph node dissection: the ARTIST trial. Journal of Clinical Oncology. 2012;30(3):268–273. doi: 10.1200/jco.2011.39.1953. [DOI] [PubMed] [Google Scholar]
- 30.Park S. H., Sohn T. S., Lee J., et al. Phase III trial to compare adjuvant chemotherapy with capecitabine and cisplatin versus concurrent chemoradiotherapy in gastric cancer: final report of the adjuvant chemoradiotherapy in stomach tumors trial, including survival and subset analyses. Journal of Clinical Oncology. 2015;33(28):3130–3136. doi: 10.1200/jco.2014.58.3930. [DOI] [PubMed] [Google Scholar]
- 31.Masetti R., Bertuccio S. N., Pession A., Locatelli F. CBFA2T3-GLIS2-positive acute myeloid leukaemia. A peculiar paediatric entity. British Journal of Haematology. 2019;184(3):337–347. doi: 10.1111/bjh.15725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin M. C., Lin J. J., Hsu C. L., et al. GATA3 interacts with and stabilizes HIF-1alpha to enhance cancer cell invasiveness. Oncogene. 2017;36(30):p. 4380. doi: 10.1038/onc.2017.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuo Y., Lv Y., Qian X., et al. Inhibition of HHIP promoter methylation suppresses human gastric cancer cell proliferation and migration. Cellular Physiology and Biochemistry. 2018;45(5):1840–1850. doi: 10.1159/000487875. [DOI] [PubMed] [Google Scholar]
- 34.Gavert N., Ben-Ze'ev A. β-Catenin signaling in biological control and cancer. Journal of Cellular Biochemistry. 2007;102(4):820–828. doi: 10.1002/jcb.21505. [DOI] [PubMed] [Google Scholar]
- 35.Eke I., Cordes N. Focal adhesion signaling and therapy resistance in cancer. Seminars in Cancer Biology. 2015;31:65–75. doi: 10.1016/j.semcancer.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 36.Cortesio C. L., Boateng L. R., Piazza T. M., Bennin D. A., Huttenlocher A. Calpain-mediated proteolysis of paxillin negatively regulates focal adhesion dynamics and cell migration. The Journal of Biological Chemistry. 2011;286(12):9998–10006. doi: 10.1074/jbc.m110.187294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shan Y.-S., Hsu H.-P., Lai M.-D., et al. Cyclin D1 overexpression correlates with poor tumor differentiation and prognosis in gastric cancer. Oncology Letters. 2017;14(4):4517–4526. doi: 10.3892/ol.2017.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hornhardt S., Rößler U., Sauter W., et al. Genetic factors in individual radiation sensitivity. DNA Repair. 2014;16(1):54–65. doi: 10.1016/j.dnarep.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 39.Parliament M. B., Murray D. Single nucleotide polymorphisms of DNA repair genes as predictors of radioresponse. Seminars in Radiation Oncology. 2010;20(4):232–240. doi: 10.1016/j.semradonc.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 40.De Rooij J. D. E., Branstetter C., Ma J., et al. Pediatric non-Down syndrome acute megakaryoblastic leukemia is characterized by distinct genomic subsets with varying outcomes. Nature Genetics. 2017;49(3):451–456. doi: 10.1038/ng.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hara Y., Shiba N., Ohki K., et al. Prognostic impact of specific molecular profiles in pediatric acute megakaryoblastic leukemia in non-Down syndrome. Genes, Chromosomes and Cancer. 2017;56(5):394–404. doi: 10.1002/gcc.22444. [DOI] [PubMed] [Google Scholar]
- 42.Lu D., Rauhauser A., Li B., et al. Loss of Glis2/NPHP7 causes kidney epithelial cell senescence and suppresses cyst growth in the Kif3a mouse model of cystic kidney disease. Kidney International. 2016;89(6):1307–1323. doi: 10.1016/j.kint.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: basic patient characteristics. Table S2: associations of clinical indicators and GLIS2 expression level with total survival in all data. Table S3: association analysis between radiotherapy and survival under different expressions of GLIS2 for M0 stage patients. Figure S1: the HR values of radiotherapy along with different cutoffs. We chose different quantiles as cutoff to divide high and low expression group for further survival analysis.
Data Availability Statement
The datasets used in the present study are available from The Cancer Genome Atlas database (http://cancergenome.nih.gov/).