Summary
Human endogenous retroviruses (HERVs) are widely believed to be remnants of ancestral germ line infections by exogenous retroviruses. Although HERVs are deemed as “nonfunctional DNAs” due to loss of most of their viral protein coding capacity during evolution as part of the human genome, cumulative evidences are showing the expressional activation and potential roles of HERVs in diseases especially cancers. Work by other researchers and us has observed the dysregulation of HERVs in cancers, identified new HERV‐related genes, and revealed their potential importance in cancer development. Here, we summarized the current knowledge on the mechanisms of the expressional activation and functional roles of HERVs, with a focus on the H family HERV (HERV‐H), in carcinogenesis. HERV expression is regulated by external chemical or physical substances and exogenous virus infection, as well as host factors such as epigenetic DNA methylation, transcription factors, cytokines, and small RNAs. Diverse roles of HERVs have been proposed by acting in the forms of noncoding RNAs, proteins, and transcriptional regulators during carcinogenesis. However, much remains to be learnt about the contributions of HERVs to human cancers. More investigation is warranted to elucidate the functions of these “fossil remnants” yet important viral DNAs in the human genome.
Keywords: aberrant expression, cancer, human endogenous retrovirus (HERV), long terminal repeat (LTR), noncoding RNA
Abbreviations
- CSF1R
colony‐stimulating factor 1 receptor
- dsRNA
double‐stranded RNA
- GSDML
gasdermin‐like protein
- HERV‐H
H family HERV
- HERV‐K
K family HERV
- HERVs
human endogenous retroviruses
- HSV‐1
herpes simplex virus‐1
- IFN
interferon
- IL
interleukin
- LTR
long terminal repeat
- MLV
murine leukemia virus
- MMTV
mouse mammary tumor virus
- MSRV
multiple sclerosis‐associated retrovirus
- ncRNA
noncoding RNA
- ORF
open reading frame
- PHA
phytohemagglutinin
- PMA
phorbol‐12‐myristate‐13‐acetate
- PSF
polypyrimidine tract‐binding protein‐associated splicing factor
- SNP
single nucleotide polymorphism
- TSERV
tumor‐specific ERV
1. INTRODUCTION
1.1. Human endogenous retroviruses and their aberrant expressional activation
Human endogenous retroviruses (HERVs) originate from retroviral infections into the germ cell more than a million years ago.1 They are estimated to comprise more than 8% of the human genome.2 HERVs are classified into different groups or families based on sequence homology and similarity of their primer binding sites to host tRNAs. For example, H family HERV (HERV‐H) is primed by tRNAHis, while HERV‐F is primed by tRNAPhe. HERVs share the same genomic structure as exogenous retroviruses: 5′LTR‐gag‐pro‐pol‐env‐3′LTR, in which the four genes encode structural/functional proteins essential to a replication‐competent retrovirus, and long terminal repeats (LTRs) contain transcriptional regulation sequences. The gag gene encodes the major structural polyprotein Gag, which functions in the assembly of noninfectious and immature viral‐like particles. The pro gene encodes the viral protease that is responsible for facilitating the maturation of viral particles. Multiple products are encoded by the pol gene, including reverse transcriptase, RNase H, and integrase. The envelope protein encoded by env, consisting of the viral surface glycoprotein and transmembrane subunits, is responsible for mediating cellular receptor binding and membrane fusion. Evolution has adapted several thousand copies of HERV elements and even more LTRs to their present locations. However, most open reading frames (ORFs) of the coding genes in HERVs have been degraded by deletions or mutations during evolution as part of the human genome,3 and only very few of them retain all structural features necessary for viral replication.4 In contrast to the coding genes, many HERV LTRs retain their potential to regulate gene transcription with the viral promoter, enhancer, and polyadenylation signals.5
Physiological expression of HERV‐H elements has been observed in embryonic stem cells.5, 6 The placenta has some peculiarities in the expression of all kinds of HERVs,1, 2 for which lower methylation content in the genome and some unique properties in epigenetic gene regulation may contribute to this phenomenon.4 However, in more circumstances, aberrant expression of HERVs is associated with a wide range of diseases, including cancers, fetal malformations, and autoimmune diseases.7, 8, 9, 10 HERVs have been investigated in a large number of autoimmune disorders due to their role in innate and adaptive immunity based on molecular mimicry.11 Multiple sclerosis‐associated retrovirus (MSRV) might have originated from the recombination of different HERV‐W transcripts,12 which was associated with poor prognosis in patients with multiple sclerosis and a higher rate of clinical re‐exacerbations.13, 14, 15 Syncytin‐1, a HERV‐W env protein, has been found to regulate neuroinflammation in multiple sclerosis.16 HERV‐W env gene products have also been detected in plasma of schizophrenia and bipolar disorders patients.17, 18 Recently, Levet et al demonstrated that HERV‐W env expression was significantly upregulated in patients suffering from type 1 diabetes,19 suggesting new therapies for type 1 diabetes.20 Besides HERV‐W, single‐nucleotide polymorphisms (SNPs) around HERV‐Fc1 showed a highly significant association with multiple sclerosis.21 HERV‐H/env59 protein was found to be upregulated in patients with systemic lupus erythematosus and negatively correlated with pathogenetic factors of human autoimmune rheumatic diseases.22
Aberrant expression of various HERVs has been demonstrated in human cancer cell lines and primary tumor tissues, such as HERV‐H,23 HERV‐K,24 HERV‐F,25 HERV‐R,26 and HERV‐S.27 Rooney et al systematically mapped the RNA sequencing data from 18 different types of solid tumors to 66 HERV family members and identified numerous HERVs to show enhanced transcription in tumor tissues as compared with normal tissues.28, 29 Among them, a conservative set of three tumor‐specific endogenous retroviruses (HERVH‐5, HERVH48‐1, and HERVE‐4) all showed minimal to undetectable expression in normal tissues and elevated expression in tumor tissues. Another array‐based study demonstrated that numerous HERV family members exhibited lower levels of DNA methylation in head and neck cancers as compared with adjacent nontumor tissues, with the most pronounced changes in methylation levels observed in HERV‐H, HERV‐W, and HERV‐K families.30 Due to the hypomethylation status of CpG dinucleotides, high expression of HERV‐K gag gene, LINE‐1, and HERV‐W was detected in ovarian cancer cell lines and tumor tissues.31, 32, 33 Moreover, the methylation levels of HERV‐K, HERV‐E, and LINE‐1 were found to be decreased during ovarian cancer progression.34 However, the pathogenic roles and mechanisms of HERVs in cancers remain elusive. Further investigations on individual families of HERVs are also warranted.
1.2. H family HERV
H family HERV (HERV‐H) is one of the most abundant HERV families, with about 1000 members. Previous studies on the function of HERVs have been focused on the envelope protein encoded by the env gene as it might possess immunosuppressive properties and is suspected to be the most important gene of HERVs in disease.31, 35, 36 However, very few HERV‐H family members have been shown to encode intact envelope proteins currently. Only three HERV‐H integrations retain env ORFs, including HERV‐H/env62 (or HERV‐H19), HERV‐H/env60, and HERV‐H/env59.36, 37 HERV‐H19, reported to be a coding competent member, contains a 1752‐bp env‐ORF that could produce a 77‐kDa envelope protein during in vitro translation reactions.38
The transcriptional analysis of specific HERV‐H copies is challenging due to the high similarity among the 1000 members of the HERV‐H family distributing throughout the human genome. However, there are still a variety of researches focusing on the expressional activation, especially the noncoding spliced transcripts, of HERV‐H elements.
1.3. Expression of HERV‐H in cancers, especially colorectal cancer
HERV‐H transcripts have been detected in cell lines of different cancer types, including teratocarcinoma, bladder carcinoma, testicular tumors, lung cancer, and colorectal cancer.39 Expression of the HERV‐H env gene was not detected in normal tissues of the brain, prostate, kidney, liver, and lung but was detected in cancer cells.40, 41 Using massively parallel signature sequencing, Stauffer et al revealed preferential expression of HERV‐H elements in cancerous tissues of the colon, small intestine, and stomach among various cancer types tested.41 Alves et al reported that a specific HERV‐H element on X chromosome was selectively transcribed in 60% of colon cancers23 and in a higher proportion of metastatic colon cancers. Wentzensen et al also reported a HERV‐H RNA sequence overexpressing in a series of gastrointestinal cancers,40 which was correlated to demethylation of the HERV‐H LTR. Pérot et al detected colorectal cancer‐specific HERV‐H transcripts throughout the disease progression from adenoma to liver metastasis, with a significantly higher level in tumors with lymph node invasion.42 Therefore, according to literature, the aberrant expression of HERV‐H element is associated with cancers, with a preference in colorectal cancer.
Our team has been investigating HERV‐Hs in cancers. In one of our studies searching for genes differentially expressed in colorectal tumors compared with adjacent nontumor tissues using microarray expressional profiling, we identified and characterized a HERV‐H element located at Xp22, designated as HERV‐HX, which is specifically upregulated in colon tumor tissues. Further analysis and promoter activity assays on the 5′ LTR of HERV‐HX demonstrated that a 17‐bp unique sequence within the 5′ U3 region contributed pivotally to the transcriptional activity of HERV‐HX.43 Although no exact chromosomal locations could be obtained for the X‐linked HERV‐H elements reported by Alves et al19 and Wentzensen et al40 we infer that these are the same one as HERV‐HX according to the rough chromosomal location and expression pattern. The functional importance of HERV‐HX, transcribing in a nonspliced format, needs further investigation.
In another study, we characterized the expression of noncoding spliced transcripts of HERV‐H elements in colon cancer using a targeted PCR and cloning strategy.44 We observed that many env‐deleted HERV‐Hs were transcriptionally active, with spliced noncoding RNAs actively transcribed in both tumorous and normal colon tissues, as well as in colon cancer cell lines. Nevertheless, the expression patterns of HERV‐H spliced transcripts differ among colon cancer cell lines and could be affected by genomic DNA methylation levels. More importantly, besides the commonly accepted view of upregulation of HERV‐H expression in colon tumor tissues, we found more active HERV‐H loci in colon tumors as compared with adjacent normal tissues.
Besides the investigation of HERV‐Hs in colon cancer, we have identified two other new HERV‐H‐related genes active in other cancer types, psiTPTE22‐HERV 45 and HERV‐H4p15.2.46 psiTPTE22‐HERV is located near the centromere of chromosome 22, adjacent to the gene psiTPTE22, and contains a HERV‐H element. The expression of psiTPTE22‐HERV is regulated by methylation of the CpG island promoter, while its transcription termination involves the transcription termination signal contained in the 3′ LTR of the HERV‐H element. Similarly, transcription of HERV‐H4p15.2 also terminates using the transcription termination signal contained in the 3′ LTR of the HERV‐H element involved, while its promoter region is an Alu element. Interestingly, the two HERV‐H–related genes involving adjacent DNA sequences are differentially downregulated in tumor tissues compared with adjacent nontumorous tissues of multiple cancer types, including stomach and kidney. Interestingly, the noncoding RNA produced by HERV‐H4p15.2 is almost silenced in colon tumors but actively transcribed in nontumorous colon tissues, while the gene psiTPTE22‐HERV shows no differential expression between colon tumor and adjacent non‐tumor tissues; these are opposed to HERV‐HX and the general view that HERV‐Hs are upregulated in colon cancers. Further investigations are needed to elucidate how the expression of HERV‐H family is regulated.
1.4. Other HERV families in cancers
Besides HERV‐H, expression of various HERVs has been demonstrated in human cancer cell lines and primary tumor tissues, such as HERV‐K,24, 47, 48 HERV‐W,47 HERV‐F,25 HERV‐R,26 and HERV‐S.27 Among these, the HERV‐K family is remarkable since many of them still retain full‐length coding genes, with their viral particles and env protein detected in various cancers. The two representative proteins of HERV‐K, Np9 and Rec, which are encoded by env gene with the presence or absence of a 292‐bp deletion, have been intensively studied for their possible role in human cancers.49, 50 Both Np9 and Rec interact with the promyelocytic leukemia zinc finger protein (PLZF) tumor suppressor, abrogating the transcriptional repression of the c‐myc proto‐oncogene by PLZF and ultimately promoting cell proliferation.51 Zhao et al observed that the HERV‐K env protein was expressed in the majority of primary breast tumors.52 Furthermore, HERV‐K antibodies and mRNA are elevated in blood of patients at an early stage of breast cancer and further increased in patients who are at risk of developing metastatis.53 An in vitro experiment showed that HERV‐K env protein effectively activated the ERK1/2 pathway and induced epithelial‐mesenchymal transition in a nontumorigenic human breast epithelial cell line, suggesting that this HERV‐K env protein has oncogenic properties.54 Moreover, HERV‐K expression was significantly associated with poor outcome in hepatocellular carcinoma patients,55 as well as cancer progression in soft tissue sarcoma.56 Elevated HERV‐K gag expression in PBMC was associated with prostate cancer diagnosis particularly in older men and smokers who tend to develop a more aggressive disease.57 Recently, HERV‐K env expression was detected in pancreatic cancer cell lines and in 80% of pancreatic cancer patient biopsies, but not in normal pancreatic cell lines or uninvolved normal tissues.58 The significantly elevated expression of numerous HERV elements in various cancers has accelerated their use as biomarkers for cancer aggressiveness and malignancy.
2. REGULATORY MECHANISMS OF HERV EXPRESSION
The human host has developed a long‐standing relationship with HERVs since their insertion into the human genome over a million years ago. Activation of HERVs occurs under certain circumstances and plays pivotal roles in certain cellular processes and disease development including cancer. Mechanisms have been proposed to explain the activation of HERVs, including regulation by external factors and host factors (Figure 1).
Figure 1.
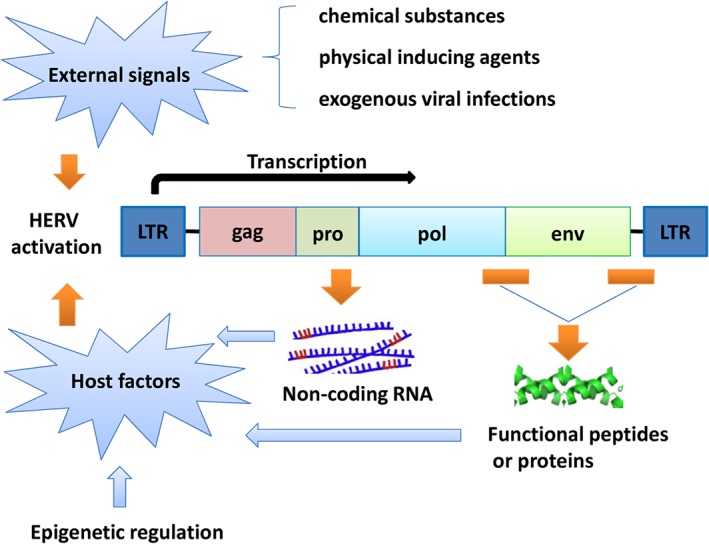
Proposed model for HERV activation. HERV expression can be induced by environmental factors and host factors
2.1. Activation of HERVs by external factors
There is substantial evidence showing that HERV expression can be upregulated by environmental factors, such as external chemical substances, physical agents, and viral infections. Treatment of normal peripheral T‐cells with phytohemagglutinin (PHA) alone or in combination with phorbol‐12‐myristate‐13‐acetate (PMA) induced HERV‐H transcription.59 Increased expressions of HERV‐W, HERV‐K, and HERV‐H in primary macrophages and monocytoid cells were found to be induced by PMA treatment.60 Moreover, physical agents, such as UVB irradiation and X‐rays, have also been shown to act as transducers for HERV expression.61, 62
Besides chemical and physical agents, HERVs can also be reactivated by exogenous viral infections. Brudek et al demonstrated that herpes virus antigens activated HERVs, as measured by reverse transcriptase activity, which subsequently contributed to the synergistic immune responses in multiple sclerosis patients.63 Expression of HERVs has also been shown to be induced by exogenous retroviruses such as HIV‐1 in vitro, and higher levels of HERV expression were also observed in HIV‐1 patients.64, 65 HERV LTRs have been observed to be activated by the Tax protein of human T‐lymphotropic virus 1.64 Herpes simplex virus‐1 (HSV‐1) infection induced transcription of HERV‐K, which was transactivated by the HSV‐1 immediate‐early protein ICP0.66 Remarkably, Frank et al observed an increased transcriptional activity of HERVs in neuroepithelial cells infected with the protozoan parasite Toxoplasma gondii.67 Therefore, HERV expression may be activated not only by viruses but also by other infectious agents. However, the specific microbial species affecting HERV expression and the relevant mechanism need to be elucidated in further studies.
2.2. Activation of HERVs by host factors
Besides the activation by external factors, HERV expression is also regulated by host factors during cellular processes. Epigenetic mechanisms including DNA methylation and histone modifications,68 regulatory proteins such as transcription factors,69 cytokines,70 and small RNAs71, 72, 73 are involved in the activation of HERVs.
DNA methylation and histone modifications are the most common epigenetic mechanisms that influence HERVs activity. A landmark genome‐wide methylation study reported that some HERVs were demethylated in human embryonic stem cells as compared with fetal lung fibroblasts, and this was associated with transcriptional activity.74 Recent studies have demonstrated the general trend of HERV demethylation in cancers. Some HERVs can be affected by global DNA demethylation and such DNA demethylation‐mediated HERV expression has functional effects on tumor progression; this may mainly contribute to the increased expression of some full‐length HERVs in cancers as compared with normal tissues.75, 76 By analyzing the DNA methylation profiles of tissue samples obtained from patients with head and neck squamous carcinoma, it was found that all HERV families appeared to be heavily methylated in normal tissues, while HERV‐H and HERV‐17 families exhibited apparent loss of DNA methylation in tumors than adjacent nontumor tissues and normal tissues.30 Similarly, other DNA methylation profiling analyses also showed aberrant demethylation of certain HERV families in testicular cancer, ovarian carcinoma, and urothelial and renal cell carcinomas.77, 78 These findings support a general trend of aberrant demethylation, which may result in aberrant expressional activation, of HERVs during carcinogenesis.
The epigenetic signatures associated with changes to histone tails are far more complex than DNA methylation. Current knowledge on how histone modifications specifically regulate HERVs in the human genome is limited. Nevertheless, studies on mouse embryonic stem cells demonstrated an important role for histone methylation (including H3K9me3 and H4K20me3) in silencing active ERVs during embryogenesis.79, 80, 81 On the other hand, treatment with histone deacetylase inhibitors suggests that histone acetylation is not crucial for controlling HERV expression in experimental cell lines and human T cells.82 Therefore, the regulatory effects of different types of histone modifications on HERV expression need further studies to elucidate.
HERV expression can be regulated in a tissue‐specific manner by transcription factors. Transcription of HERV is usually initiated by a TATA box motif and other key promoter elements. Fuchs et al found the HERV‐K LTR acted as a TATA‐less promoter, and their transcription activation was mediated by the transcription factors Sp1 and Sp3.69 HERV‐K promoter activity was significantly reduced after knockdown Sp1 or Sp3 by RNA interference in teratocarcinoma cells and melanoma cells.
Some cytokines and small RNAs also affect the expression of HERVs. It has been reported that the expression of HERV‐R was significantly upregulated by treatment with tumor necrosis factor‐α, interleukin (IL)‐1α, and IL‐1β and downregulated by treatment with interferon (IFN)‐γ.70 The important role for small RNAs (such as Piwi‐interacting small RNA) in silencing ERVs via homology‐dependent mechanisms has been confirmed in mouse.73, 83 However, the role of small RNAs in regulating the expression of HERVs in human needs further investigation.
3. FUNCTIONAL ROLES OF HERVs IN CARCINOGENESIS
The aberrant expression of HERVs has inferred potential roles in the development of cancers. Although the functional roles of HERVs in carcinogenesis remain not comprehensively conclusive, diverse roles of HERVs have been reported by acting in the forms of noncoding RNAs, proteins, transcriptional regulators, and so on (Figure 2).
Figure 2.
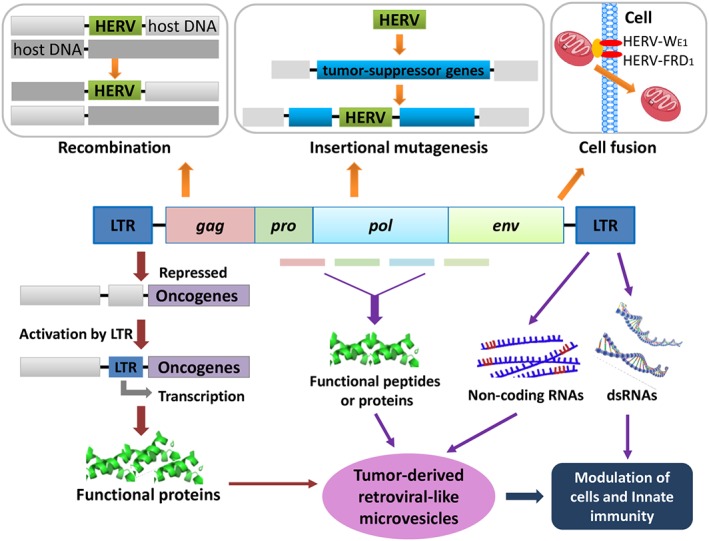
Potential mechanisms of HERV‐mediated oncogenesis. HERV‐mediated homologous recombination leads to chromosomal re‐arrangements. HERV integrations may disrupt the tumor‐suppressor gene. HERV‐LTR can function as the alternative promoters for the downstream oncogenes. Noncoding RNAs and functional peptides produced by HERV may have effect on tumorigenesis. Certain tumor‐derived retroviral‐like microvesicles may also be involved in tumor growth and metastasis. dsRNA transcripts from HERV‐LTRs, which could be induced by DNA methylation inhibitors or specific genes, may activate innate immunity such as interferon response in tumor cells
3.1. Noncoding RNA transcripts
Due to the accumulated deletions and mutations in the viral protein ORFs during evolution as parts of the human genome, most of the HERVs are transcriptionally silenced or may only produce noncoding RNA (ncRNA) transcripts. Kelley and Rinn found a close association between HERVs and ncRNAs and identified 127 ncRNAs originating from HERVs.84 They believed that HERV‐Hs were responsible for the birth of new ncRNAs by inserting active promoters into previously inactive genomic regions. The long ncRNA ROR, with emerging roles being found in human cancers,85, 86 was one of the ncRNAs promoted by a HERV‐H element.87
The ncRNAs from HERVs have various potential to affect genome functions; some of the effects may benefit the host, while others may be pathogenic.88, 89 The protein PSF (polypyrimidine tract‐binding protein‐associated splicing factor) functions to repress proto‐oncogene transcription.90 The ncRNA produced by a HERV‐K11 provirus binds to PSF directly, reversing the PSF‐mediated repression of proto‐oncogene transcription and subsequently driving transformation and tumorigenesis.90, 91 Lv and Zhao also observed that LINE‐1 RNA specifically bound to PSF and released its target gene, proto‐oncogene G antigen 6 (GAGE6), from the DNA‐binding domain of PSF, consequently promoting cell proliferation and colony formation of hepatocellular carcinoma cells.92 In this regard, HERV transcription and proto‐oncogene activation can be connected. Besides such HERV‐related ncRNAs with oncogenic function, others with tumor‐suppressive potential have also been identified. The intronic RNAs arising from LTRs of ERV‐9 are expressed as both sense and antisense transcripts. It has been reported that a lower level of the antisense transcript of ERV‐9 LTR was expressed in malignant cells than normal cells.93 This antisense RNA was found to physically bind to key transcription factors for cellular proliferation, including nuclear factor Y (NF‐Y), p53, and sp1, which may ultimately inhibit cancer cell growth via functioning as decoy targets or traps for NF‐Y.94
3.2. Transcriptional regulation of adjacent genes by LTR
The majority (90%) of LTR elements exists as solitary LTRs due to recombination between the 5′ and 3′ LTRs.24 HERV‐LTRs regulate the transcriptional activity of HERVs, with the 5′ LTR containing enhancer and promoter sequences to provide signals that can be recognized by the cellular machinery for transcription initiation and the 3′ LTR providing a polyadenylation signal for transcription termination. Solitary LTRs, as well as some HERV‐LTRs, may function in transcriptional regulation of adjacent cellular genes. There are a number of examples for LTRs as transcriptional control elements for cellular genes, such as ZNF80,95 PLA2L,96 PTN,97 and two of the HERV‐related genes (psiTPTE22‐HERV and HERV‐H4p15.2) identified by us.45, 46
There are two ways for LTRs to regulate gene expression. The most widely studied way is to function as alternative promoters, which may result in different functional effects. Different protein isoforms or transcription patterns produced by alternative LTR promoters have been reported.87, 98, 99 In some cases, transcriptional activation of HERV‐LTRs leads to activation of cancer‐related genes. The myeloid‐specific proto‐oncogene CSF1R (colony‐stimulating factor 1 receptor) was observed to be driven by an aberrantly activated LTR promoter to express in Hodgkin lymphoma,100 suggesting that activation of the usually silenced repeat elements may play a role in the pathogenesis of human malignancies. An LTR element of HERV‐H with reverse orientation has been identified as an alternative promoter of the GSDML (gasdermin‐like protein) gene,101 which is located near an oncogenomic recombination hotspot.102 The GSDML transcripts derived from this LTR promoter were widely distributed in various human tissues and cancer cells.
The other way for LTRs to regulate cellular gene expression is to facilitate gene transcription by donating binding sites for transcription factors or other regulatory proteins, especially for activation of oncogenes. p53 binding sites within HERV‐LTRs have been found to lead to the transcriptional activation of p53 downstream genes in human cancer cells.103 Importantly, all the p53‐containing LTRs are primate‐specific.104 Therefore, LTR elements are involved in regulating the primate‐specific transcriptional network of p53‐related genes. A genome‐wide analysis demonstrated that the binding sites for the transcription factors CTCF, TP53, Pou5F1, Sox2 and ESR1 are enriched within individual LTRs and are also present in LTR consensus sequences.105 The binding sites for Myb protein and Sp1 family proteins have been identified in the LTRs of the HERV‐H.106, 107 HERV‐H LTRs also appeared to function as enhancers for pluripotency‐related genes.6 Therefore, by serving as promoters or providing transcriptional regulatory sites, LTRs participate in regulating cellular gene expression and may affect carcinogenesis when cancer‐related genes are regulated.
3.3. HERV‐mediated nonallelic homologous recombination
HERV families contain a large number of long and highly homogeneous copies, providing an ideal substrate for nonallelic homologous recombination between members that are located on the same or different chromosomes. A systematic phylogenetic sequence analysis on the most recently active HERV members showed evidence for HERV‐mediated genomic rearrangements during primate evolution; at least 16% of these elements had undergone apparent rearrangements.108 HERV elements throughout the genome have been shown to serve as substrates for genomic instability and mediate human copy‐number variation.109 Nonallelic homologous recombination may result in chromosomal anomalies that are ubiquitous in cancers, including translocations, duplications, deletions, inversions, and rearrangements. Tomlins et al110 found a fusion between a HERV‐K provirus and a dormant oncogene of the E26 transformation‐specific family in human prostate cancer. A recurrent translocation was identified to be mediated by homologous recombination between HERV‐H elements on chromosomes 4 and 18 in children with intellectual disability.111 In one of the most serious radiological accidents occurred at a radiation therapy unit in Brazil, nonallelic recombination between two HERVs, caused by exposure to ionizing radiations, contributed to chromosomal rearrangements in the AZF region in patients.112 More systematic analyses are needed to elucidate the importance of HERV‐mediated rearrangements during carcinogenesis.
3.4. HERV‐mediated insertional mutagenesis
Exogenous infectious retroviruses‐mediated insertional mutagenesis can led to the activation of cellular oncogenes or disruption of tumor‐suppressor genes.113 Some ERVs retaining infectivity may share the transforming potential. The role of infectious copies of mouse mammary tumor virus (MMTV) or murine leukemia virus (MLV) has been well studied in the insertional activation of oncogenes (Wnt1 and Notch4) and disruption of tumor suppressor genes in mice.114 The strong link between MMTV infection and mammary tumorigenesis in mice leads to a hypothesis for retroviruses in causing human cancers. Some researchers have succeeded in reconstituting the human‐specific active HERV‐K provirus,115, 116 indicating that human cells still have the potential to produce infectious retroviruses. Although these reconstituted viral particles are not highly infectious and no infectious HERV‐derived retrovirus has been discovered in vivo until now, these studies support the hypothesis regarding the potential oncogenic mechanism of insertional mutagenesis by an infectious HERV.
3.5. HERV‐mediated cell fusion
The most well‐known physiological role of HERV is that of the Syncytin‐1 protein, encoded by HERV‐W, in mediating trophoblast fusion.117 Syncytin‐1 expression on the surface of cytotrophoblasts and syncytiotrophoblasts mediates fusion; this cell‐cell fusion is essential for normal placental development.118 Recently, another HERV‐FRD–derived protein, syncytin‐2, was identified and shown to be sufficient for mediating cell fusion.119, 120
Besides mediating trophoblast fusion, the HERV protein syncytins have recently been found to mediate cell‐to‐cell transfer of mitochondria. Mitochondria are crucial organelles that regulate not only the energy metabolism but also the survival and fate of eukaryotic cells. Mitochondria were recently discovered to be able to translocate from one cell to another. This phenomenon was observed in vitro and in vivo, both in physiological and pathophysiological conditions including tissue injury and cancer. Mitochondrial transfer from donor cells to cells of injured tissues is a promising cell‐based therapy that effectively brings about the recovery of tissue bioenergetics. Syncytin‐1/syncytin‐2 also function to mediate the cell‐to‐cell transfer of mitochondria, and this could be blocked by anti‐syncytin‐1 and anti‐syncytin‐2 antibodies.121
Huang et al demonstrated that Env proteins encoded by HERV‐K could mediated intercellular fusion of melanoma cells,122 which may lead to syncytia and drive genetic changes such as loss of heterozygosity. This may ultimately contribute to the malignant growth of tumor cells, thus promoting tumor progression and treatment resistance. Function of other HERV proteins in mediating cell fusion is rarely observed and is worthy of attention.
3.6. Tumor‐derived retroviral‐like microvesicles
Living cells can release a variety of vesicles into the extracellular fluids, such as blood, urine, and breast milk, acting as important intercellular communicators. Microvesicles are one subtype of extracellular vesicles, deriving from the plasma membrane. Microvesicles carry RNA (mRNA, miRNA, and ncRNA), genomic sequences, and a large component of proteins and lipids.123 Their contents vary according to the cellular composition and physiologic state. Retroviral‐like microvesicles have been found in patients with cancers, including lymphomas,124 breast cancer,125 melanoma,126 and teratomas.127 The levels of reverse transcriptase and retroviral‐like microvesicles drop dramatically with cancer treatment, suggesting that these retroviral‐like microvesicles are tumor‐derived. More interestingly, HERV RNA transcripts were found to be particularly enriched in tumor‐derived microvesicles, and these tumor‐derived HERV RNAs could be transferred to other cells via microvesicles.128 Therefore, tumor cells may modulate neighboring or distant normal cells and microenvironment via microvesicles containing HERV RNAs to stimulate tumor growth and metastasis.
3.7. Regulation of innate immunity by HERVs
The ability of HERVs in regulating the immune system may have opposite effects on cancer development, relevant to both oncogenic processes and anticancer defenses. Some HERV‐env proteins have been shown to exert immunosuppressive properties, such as the human syncytin‐2.129 Cianciolo et al also reported that some ERV‐env proteins have an immunosuppressive domain, which may contribute to cancer progression by promoting the escape of tumor cells from immune surveillance.130 Infection of murine ERV‐env expression vector presenting the envelope immunosuppressive domain into cancer cells promoted tumor growth in vivo by preventing the activity of innate immune system that clears tumors.131 The immunosuppressive peptide H17, deriving from HERV‐H, was recently identified to induce epithelial‐mesenchymal transition and stimulate expression of the chemokine CCL19 in tumor cells.132
In recent years, epigenetic therapies are under active investigation in diverse solid tumors, inducing immune responses and reactivating silenced tumor suppressors.133, 134 It has been shown that low doses of DNA methylation inhibitors 5‐aza‐2′‐deoxycytidine (decitabine) or CDK4/6 inhibitors can induce the transcription of HERV‐derived double‐stranded RNAs (dsRNAs) and IFN‐response genes in tumor cells.135, 136, 137, 138 Moreover, the upregulation of IFN‐response genes was observed in breast cancer and colorectal cancer patients treated with low‐dose DNA methylation inhibitors. Recent findings suggest that HERV‐derived dsRNAs exhibit antitumor effects by leading to a “viral mimicry” state in human tumors, which subsequently activate the responses including TLR3, MAVS, MDA5, IRF7, and IFNs.139, 140 A major trigger of the DNA methylation inhibitor–induced “viral mimicry” appears to be bidirectional transcription of HERVs that are known to fold into dsRNA secondary structures.140 Chuong et al also found that HERVs played a crucial role in the transcriptional network underlying IFN response and induced innate immunity, and the expression of IFN‐induced genes was significantly decreased after the CRISPR‐Cas9 deletion of these HERV elements in human genome.141
Besides the expression of HERVs induced by DNA methylation inhibitors, some specific genes were identified to be associated with the induction of HERV‐derived dsRNAs. Cañadas et al found that expression of a novel subclass of ERVs was triggered in mesenchymal tumor subpopulations with high AXL/MET expression and low EZH2 levels when exposed to IFN‐γ.142 Expression of such ERVs promoted MHC class 1 upregulation, T‐cell infiltration, activation of immune checkpoints, and myeloid infiltration, thus promoting tumor immune suppression. On the other hand, Sheng et al demonstrated that expression of ERVs was increased following ablation of the histone demethylase LSD1 in cancer cells, which led to dsRNA stress and activation of type 1 IFNs, stimulated antitumor T‐cell immunity, and restrained tumor growth in return.143 Further studies are still warranted to elucidate the detailed role of HERVs in regulating immunity.
4. SUMMARY AND PROSPECTS
Increasing evidences demonstrate that HERVs are upregulated in a variety of cancers, such as HERV‐Hs in colorectal cancer. However, their potential effects on the stability of host genomes and cancer progression are still poorly understood. Here, we summarize the potential consequences of HERV deregulation in cancers by mechanisms such as noncoding transcripts, transcriptional regulation, and HERV‐mediated genomic rearrangements, which are shared by HERV‐H and other HERV groups. Firstly, we discuss the activation of HERVs induced by host factors and external signals. Then we focus on the tumorigenic mechanisms of HERVs. Upregulation of HERVs may result in oncogenic effects through transcribing noncoding RNAs or acting as regulators of oncogenes. HERV‐LTRs can serve as alternative promoters or binding sites of oncogenic transcription factors. HERV‐mediated recombination and insertional mutagenesis (activation of oncogenes or disruption of tumor suppressor genes) may also contribute to tumorigenesis. Tumor‐derived HERV RNAs/proteins can be transferred to normal cells through retroviral‐like microvesicles, the effects of which on tumor growth and metastasis still need to be further studied. Moreover, HERVs may also regulate immunity, such as by activating IFN response. However, much remains to be learnt about the connections between HERVs and human cancers. Nevertheless, the effects of HERV‐produced RNAs/proteins and HERV‐induced transcriptional enhancement of oncogenes or interference of tumor suppressor genes on tumor initiation and progression may provide us new insights into the management of cancers.
Zhang M, Liang JQ, Zheng S. Expressional activation and functional roles of human endogenous retroviruses in cancers. Rev Med Virol. 2019;29:e2025 10.1002/rmv.2025
Contributor Information
Jessie Qiaoyi Liang, Email: jessieqy@cuhk.edu.hk.
Shu Zheng, Email: zhengshu@zju.edu.cn.
REFERENCES
- 1. Lower R, Lower J, Kurth R. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci U S A. 1996;93:5177‐5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860‐921. [DOI] [PubMed] [Google Scholar]
- 3. Larsson E, Kato N, Cohen M. Human endogenous proviruses. Curr Top Microbiol Immunol. 1989;148:115‐132. [DOI] [PubMed] [Google Scholar]
- 4. Ono M, Yasunaga T, Miyata T, Ushikubo H. Nucleotide sequence of human endogenous retrovirus genome related to the mouse mammary tumor virus genome. J Virol. 1986;60:589‐598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Theunissen TW, Friedli M, He Y, et al. Molecular criteria for defining the naive human pluripotent state. Cell Stem Cell. 2016;19:502‐515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lu X, Sachs F, Ramsay L, et al. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat Struct Mol Biol. 2014;21:423‐425. [DOI] [PubMed] [Google Scholar]
- 7. Antony JM, Deslauriers AM, Bhat RK, Ellestad KK, Power C. Human endogenous retroviruses and multiple sclerosis: innocent bystanders or disease determinants? Biochim Biophys Acta. 1812;2011:162‐176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Balada E, Ordi‐Ros J, Vilardell‐Tarres M. Molecular mechanisms mediated by human endogenous retroviruses (HERVs) in autoimmunity. Rev Med Virol. 2009;19:273‐286. [DOI] [PubMed] [Google Scholar]
- 9. Moyes D, Griffiths DJ, Venables PJ. Insertional polymorphisms: a new lease of life for endogenous retroviruses in human disease. Trends Genet. 2007;23:326‐333. [DOI] [PubMed] [Google Scholar]
- 10. Lower R. The pathogenic potential of endogenous retroviruses: facts and fantasies. Trends Microbiol. 1999;7:350‐356. [DOI] [PubMed] [Google Scholar]
- 11. Trela M, Nelson PN, Rylance PB. The role of molecular mimicry and other factors in the association of human endogenous retroviruses and autoimmunity. Apmis. 2016;124:88‐104. [DOI] [PubMed] [Google Scholar]
- 12. Komurian‐Pradel F, Paranhos‐Baccala G, Bedin F, et al. Molecular cloning and characterization of MSRV‐related sequences associated with retrovirus‐like particles. Virology. 1999;260:1‐9. [DOI] [PubMed] [Google Scholar]
- 13. Schmitt K, Richter C, Backes C, et al. Comprehensive analysis of human endogenous retrovirus group HERV‐W locus transcription in multiple sclerosis brain lesions by high‐throughput amplicon sequencing. J Virol. 2013;87:13837‐13852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Perron H, Garson JA, Bedin F, et al. Molecular identification of a novel retrovirus repeatedly isolated from patients with multiple sclerosis. The Collaborative Research Group on Multiple Sclerosis. Proc Natl Acad Sci U S A. 1997;94:7583‐7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Laufer G, Mayer J, Mueller BF, Mueller‐Lantzsch N, Ruprecht K. Analysis of transcribed human endogenous retrovirus W env loci clarifies the origin of multiple sclerosis‐associated retrovirus env sequences. Retrovirology. 2009;6:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Antony JM, Ellestad KK, Hammond R, et al. The human endogenous retrovirus envelope glycoprotein, syncytin‐1, regulates neuroinflammation and its receptor expression in multiple sclerosis: a role for endoplasmic reticulum chaperones in astrocytes. J Immunol. 2007;179:1210‐1224. [DOI] [PubMed] [Google Scholar]
- 17. Frank O, Giehl M, Zheng C, et al. Human endogenous retrovirus expression profiles in samples from brains of patients with schizophrenia and bipolar disorders. J Virol. 2005;79:10890‐10901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Karlsson H, Bachmann S, Schroder J, et al. Retroviral RNA identified in the cerebrospinal fluids and brains of individuals with schizophrenia. Proc Natl Acad Sci U S A. 2001;98:4634‐4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Levet S, Medina J, Joanou J, et al. An ancestral retroviral protein identified as a therapeutic target in type‐1 diabetes. JCI Insight. 2017;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Curtin F, Bernard C, Levet S, et al. A new therapeutic approach for type 1 diabetes: rationale for GNbAC1, an anti‐HERV‐W‐Env monoclonal antibody. Diabetes Obes Metab. 2018;20:2075‐2084. [DOI] [PubMed] [Google Scholar]
- 21. Nexo BA, Christensen T, Frederiksen J, et al. The etiology of multiple sclerosis: genetic evidence for the involvement of the human endogenous retrovirus HERV‐Fc1. Plos One. 2011;6:e16652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Laska MJ, Troldborg A, Hauge EM, Bahrami S, Stengaard‐Pedersen K. Human endogenous retroviral genetic element with immunosuppressive activity in both human autoimmune diseases and experimental arthritis. Arthritis Rheumatol. 2017;69:398‐409. [DOI] [PubMed] [Google Scholar]
- 23. Alves PM, Levy N, Stevenson BJ, et al. Identification of tumor‐associated antigens by large‐scale analysis of genes expressed in human colorectal cancer. Cancer Immun. 2008;8:11. [PMC free article] [PubMed] [Google Scholar]
- 24. Buscher K, Trefzer U, Hofmann M, et al. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res. 2005;65:4172‐4180. [DOI] [PubMed] [Google Scholar]
- 25. Yi JM, Kim HS. Expression analysis of endogenous retroviral elements belonging to the HERV‐F family from human tissues and cancer cells. Cancer Lett. 2004;211:89‐96. [DOI] [PubMed] [Google Scholar]
- 26. Kim HS, Yi JM, Hirai H, et al. Human endogenous retrovirus (HERV)‐R family in primates: chromosomal location, gene expression, and evolution. Gene. 2006;370:34‐42. [DOI] [PubMed] [Google Scholar]
- 27. Yi JM, Kim TH, Huh JW, et al. Human endogenous retroviral elements belonging to the HERV‐S family from human tissues, cancer cells, and primates: expression, structure, phylogeny and evolution. Gene. 2004;342:283‐292. [DOI] [PubMed] [Google Scholar]
- 28. Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48‐61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mayer J, Blomberg J, Seal RL. A revised nomenclature for transcribed human endogenous retroviral loci. Mob DNA. 2011;2:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Szpakowski S, Sun X, Lage JM, et al. Loss of epigenetic silencing in tumors preferentially affects primate‐specific retroelements. Gene. 2009;448:151‐167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang‐Johanning F, Liu J, Rycaj K, et al. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int J Cancer. 2007;120:81‐90. [DOI] [PubMed] [Google Scholar]
- 32. Menendez L, Benigno BB, McDonald JF. L1 and HERV‐W retrotransposons are hypomethylated in human ovarian carcinomas. Mol Cancer. 2004;3:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gotzinger N, Sauter M, Roemer K, Mueller‐Lantzsch N. Regulation of human endogenous retrovirus‐K Gag expression in teratocarcinoma cell lines and human tumours. J Gen Virol. 1996;77(Pt 12):2983‐2990. [DOI] [PubMed] [Google Scholar]
- 34. Iramaneerat K, Rattanatunyong P, Khemapech N, Triratanachat S, Mutirangura A. HERV‐K hypomethylation in ovarian clear cell carcinoma is associated with a poor prognosis and platinum resistance. Int J Gynecol Cancer. 2011;21:51‐57. [DOI] [PubMed] [Google Scholar]
- 35. Mangeney M, de Parseval N, Thomas G, Heidmann T. The full‐length envelope of an HERV‐H human endogenous retrovirus has immunosuppressive properties. J Gen Virol. 2001;82:2515‐2518. [DOI] [PubMed] [Google Scholar]
- 36. de Parseval N, Casella J, Gressin L, Heidmann T. Characterization of the three HERV‐H proviruses with an open envelope reading frame encompassing the immunosuppressive domain and evolutionary history in primates. Virology. 2001;279:558‐569. [DOI] [PubMed] [Google Scholar]
- 37. Lindeskog M, Mager DL, Blomberg J. Isolation of a human endogenous retroviral HERV‐H element with an open env reading frame. Virology. 1999;258:441‐450. [DOI] [PubMed] [Google Scholar]
- 38. Hirose Y, Takamatsu M, Harada F. Presence of env genes in members of the RTVL‐H family of human endogenous retrovirus‐like elements. Virology. 1993;192:52‐61. [DOI] [PubMed] [Google Scholar]
- 39. Yi JM, Kim HM, Kim HS. Human endogenous retrovirus HERV‐H family in human tissues and cancer cells: expression, identification and phylogeny. Cancer Lett. 2006;231:228‐239. [DOI] [PubMed] [Google Scholar]
- 40. Wentzensen N, Coy JF, Knaebel HP, et al. Expression of an endogenous retroviral sequence from the HERV‐H group in gastrointestinal cancers. Int J Cancer. 2007;121:1417‐1423. [DOI] [PubMed] [Google Scholar]
- 41. Stauffer Y, Theiler G, Sperisen P, Lebedev Y, Jongeneel CV. Digital expression profiles of human endogenous retroviral families in normal and cancerous tissues. Cancer Immun. 2004;4:2. [PubMed] [Google Scholar]
- 42. Perot P, Mullins CS, Naville M, et al. Expression of young HERV‐H loci in the course of colorectal carcinoma and correlation with molecular subtypes. Oncotarget. 2015;6:40095‐40111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liang Q, Ding J, Xu R, Xu Z, Zheng S. Identification of a novel human endogenous retrovirus and promoter activity of its 5′ U3. Biochem Biophys Res Commun. 2009;382:468‐472. [DOI] [PubMed] [Google Scholar]
- 44. Liang Q, Xu Z, Xu R, Wu L, Zheng S. Expression patterns of non‐coding spliced transcripts from human endogenous retrovirus HERV‐H elements in colon cancer. Plos One. 2012;7:e29950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liang Q, Ding J, Xu R, Xu Z, Zheng S. The novel human endogenous retrovirus‐related gene, psiTPTE22‐HERV, is silenced by DNA methylation in cancers. Int J Cancer. 2010;127:1833‐1843. [DOI] [PubMed] [Google Scholar]
- 46. Liang Q, Ding J, Zheng S. Identification and detection of a novel human endogenous retrovirus‐related gene, and structural characterization of its related elements. Genet Mol Biol. 2009;32:704‐708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bergallo M, Montanari P, Mareschi K, et al. Expression of the pol gene of human endogenous retroviruses HERV‐K and ‐W in leukemia patients. Arch Virol. 2017;162:3639‐3644. [DOI] [PubMed] [Google Scholar]
- 48. Johanning GL, Malouf GG, Zheng X, et al. Expression of human endogenous retrovirus‐K is strongly associated with the basal‐like breast cancer phenotype. Sci Rep. 2017;7:41960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Armbruester V, Sauter M, Krautkraemer E, et al. A novel gene from the human endogenous retrovirus K expressed in transformed cells. Clin Cancer Res. 2002;8:1800‐1807. [PubMed] [Google Scholar]
- 50. Lower R, Tonjes RR, Korbmacher C, Kurth R, Lower J. Identification of a Rev‐related protein by analysis of spliced transcripts of the human endogenous retroviruses HTDV/HERV‐K. J Virol. 1995;69:141‐149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Denne M, Sauter M, Armbruester V, et al. Physical and functional interactions of human endogenous retrovirus proteins Np9 and rec with the promyelocytic leukemia zinc finger protein. J Virol. 2007;81:5607‐5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhao J, Rycaj K, Geng S, et al. Expression of human endogenous retrovirus type K envelope protein is a novel candidate prognostic marker for human breast cancer. Genes Cancer. 2011;2:914‐922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang‐Johanning F, Li M, Esteva FJ, et al. Human endogenous retrovirus type K antibodies and mRNA as serum biomarkers of early‐stage breast cancer. Int J Cancer. 2014;134:587‐595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lemaitre C, Tsang J, Bireau C, Heidmann T, Dewannieux M. A human endogenous retrovirus‐derived gene that can contribute to oncogenesis by activating the ERK pathway and inducing migration and invasion. PLoS Pathog. 2017;13:e1006451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ma W, Hong Z, Liu H, et al. Human endogenous retroviruses‐K (HML‐2) expression is correlated with prognosis and progress of hepatocellular carcinoma. Biomed Res Int. 2016;2016, 8201642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Giebler M, Staege MS, Blauschmidt S, et al. Elevated HERV‐K expression in soft tissue sarcoma is associated with worsened relapse‐free survival. Front Microbiol. 2018;9:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wallace TA, Downey RF, Seufert CJ, et al. Elevated HERV‐K mRNA expression in PBMC is associated with a prostate cancer diagnosis particularly in older men and smokers. Carcinogenesis. 2014;35:2074‐2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li M, Radvanyi L, Yin B, et al. Downregulation of human endogenous retrovirus type K (HERV‐K) viral env RNA in pancreatic cancer cells decreases cell proliferation and tumor growth. Clin Cancer Res. 2017;23:5892‐5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kelleher CA, Wilkinson DA, Freeman JD, Mager DL, Gelfand EW. Expression of novel‐transposon‐containing mRNAs in human T cells. J Gen Virol. 1996;77(Pt 5):1101‐1110. [DOI] [PubMed] [Google Scholar]
- 60. Johnston JB, Silva C, Holden J, et al. Monocyte activation and differentiation augment human endogenous retrovirus expression: implications for inflammatory brain diseases. Ann Neurol. 2001;50:434‐442. [DOI] [PubMed] [Google Scholar]
- 61. Lee JR, Ahn K, Kim YJ, Jung YD, Kim HS. Radiation‐induced human endogenous retrovirus (HERV)‐R env gene expression by epigenetic control. Radiat Res. 2012;178:379‐384. [DOI] [PubMed] [Google Scholar]
- 62. Reiche J, Pauli G, Ellerbrok H. Differential expression of human endogenous retrovirus K transcripts in primary human melanocytes and melanoma cell lines after UV irradiation. Melanoma Res. 2010;20:435‐440. [DOI] [PubMed] [Google Scholar]
- 63. Brudek T, Luhdorf P, Christensen T, Hansen HJ, Moller‐Larsen A. Activation of endogenous retrovirus reverse transcriptase in multiple sclerosis patient lymphocytes by inactivated HSV‐1, HHV‐6 and VZV. J Neuroimmunol. 2007;187:147‐155. [DOI] [PubMed] [Google Scholar]
- 64. Toufaily C, Landry S, Leib‐Mosch C, Rassart E, Barbeau B. Activation of LTRs from different human endogenous retrovirus (HERV) families by the HTLV‐1 tax protein and T‐cell activators. Viruses. 2011;3:2146‐2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Contreras‐Galindo R, Lopez P, Velez R, Yamamura Y. HIV‐1 infection increases the expression of human endogenous retroviruses type K (HERV‐K) in vitro. AIDS Res Hum Retroviruses. 2007;23:116‐122. [DOI] [PubMed] [Google Scholar]
- 66. Kwun HJ, Han HJ, Lee WJ, Kim HS, Jang KL. Transactivation of the human endogenous retrovirus K long terminal repeat by herpes simplex virus type 1 immediate early protein 0. Virus Res. 2002;86:93‐100. [DOI] [PubMed] [Google Scholar]
- 67. Frank O, Jones‐Brando L, Leib‐Mosch C, Yolken R, Seifarth W. Altered transcriptional activity of human endogenous retroviruses in neuroepithelial cells after infection with Toxoplasma gondii . J Infect Dis. 2006;194:1447‐1449. [DOI] [PubMed] [Google Scholar]
- 68. Romanish MT, Cohen CJ, Mager DL. Potential mechanisms of endogenous retroviral‐mediated genomic instability in human cancer. Semin Cancer Biol. 2010;20:246‐253. [DOI] [PubMed] [Google Scholar]
- 69. Fuchs NV, Kraft M, Tondera C, et al. Expression of the human endogenous retrovirus (HERV) group HML‐2/HERV‐K does not depend on canonical promoter elements but is regulated by transcription factors Sp1 and Sp3. J Virol. 2011;85:3436‐3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Katsumata K, Ikeda H, Sato M, et al. Cytokine regulation of env gene expression of human endogenous retrovirus‐R in human vascular endothelial cells. Clin Immunol. 1999;93:75‐80. [DOI] [PubMed] [Google Scholar]
- 71. Kuramochi‐Miyagawa S, Watanabe T, Gotoh K, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908‐917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Aravin AA, Hannon GJ. Small RNA silencing pathways in germ and stem cells. Cold Spring Harb Symp Quant Biol. 2008;73:283‐290. [DOI] [PubMed] [Google Scholar]
- 73. Aravin AA, Sachidanandam R, Girard A, Fejes‐Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744‐747. [DOI] [PubMed] [Google Scholar]
- 74. Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315‐322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Oricchio E, Sciamanna I, Beraldi R, et al. Distinct roles for LINE‐1 and HERV‐K retroelements in cell proliferation, differentiation and tumor progression. Oncogene. 2007;26:4226‐4233. [DOI] [PubMed] [Google Scholar]
- 76. Taruscio D, Mantovani A. Factors regulating endogenous retroviral sequences in human and mouse. Cytogenet Genome Res. 2004;105:351‐362. [DOI] [PubMed] [Google Scholar]
- 77. Gimenez J, Montgiraud C, Pichon JP, et al. Custom human endogenous retroviruses dedicated microarray identifies self‐induced HERV‐W family elements reactivated in testicular cancer upon methylation control. Nucleic Acids Res. 2010;38:2229‐2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Florl AR, Lower R, Schmitz‐Drager BJ, Schulz WA. DNA methylation and expression of LINE‐1 and HERV‐K provirus sequences in urothelial and renal cell carcinomas. Br J Cancer. 1999;80:1312‐1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Maksakova IA, Mager DL, Reiss D. Keeping active endogenous retroviral‐like elements in check: the epigenetic perspective. Cell Mol Life Sci. 2008;65:3329‐3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Mikkelsen TS, Ku M, Jaffe DB, et al. Genome‐wide maps of chromatin state in pluripotent and lineage‐committed cells. Nature. 2007;448:553‐560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Martens JH, O'Sullivan RJ, Braunschweig U, et al. The profile of repeat‐associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800‐812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hurst T, Pace M, Katzourakis A, et al. Human endogenous retrovirus (HERV) expression is not induced by treatment with the histone deacetylase (HDAC) inhibitors in cellular models of HIV‐1 latency. Retrovirology. 2016;13:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Peterlin BM, Liu P, Wang X, et al. Hili Inhibits HIV replication in activated T cells. J Virol. 2017;91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kelley D, Rinn J. Transposable elements reveal a stem cell‐specific class of long noncoding RNAs. Genome Biol. 2012;13:R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Pan Y, Li C, Chen J, et al. The emerging roles of long noncoding RNA ROR (lincRNA‐ROR) and its possible mechanisms in human cancers. Cell Physiol Biochem. 2016;40:219‐229. [DOI] [PubMed] [Google Scholar]
- 86. Gao S, Wang P, Hua Y, et al. ROR functions as a ceRNA to regulate Nanog expression by sponging miR‐145 and predicts poor prognosis in pancreatic cancer. Oncotarget. 2016;7:1608‐1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Shephard EA, Chandan P, Stevanovic‐Walker M, Edwards M, Phillips IR. Alternative promoters and repetitive DNA elements define the species‐dependent tissue‐specific expression of the FMO1 genes of human and mouse. Biochem J. 2007;406:491‐499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Jern P, Coffin JM. Effects of retroviruses on host genome function. Annu Rev Genet. 2008;42:709‐732. [DOI] [PubMed] [Google Scholar]
- 89. Garen A, Song X. Regulatory roles of tumor‐suppressor proteins and noncoding RNA in cancer and normal cell functions. Int J Cancer. 2008;122:1687‐1689. [DOI] [PubMed] [Google Scholar]
- 90. Li L, Feng T, Lian Y, et al. Role of human noncoding RNAs in the control of tumorigenesis. Proc Natl Acad Sci U S A. 2009;106:12956‐12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wang G, Cui Y, Zhang G, Garen A, Song X. Regulation of proto‐oncogene transcription, cell proliferation, and tumorigenesis in mice by PSF protein and a VL30 noncoding RNA. Proc Natl Acad Sci U S A. 2009;106:16794‐16798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lv J, Zhao Z. Binding of LINE‐1 RNA to PSF transcriptionally promotes GAGE6 and regulates cell proliferation and tumor formation in vitro. Exp Ther Med. 2017;14:1685‐1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Xu L, Elkahloun AG, Candotti F, et al. A novel function of RNAs arising from the long terminal repeat of human endogenous retrovirus 9 in cell cycle arrest. J Virol. 2013;87:25‐36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Benatti P, Basile V, Merico D, et al. A balance between NF‐Y and p53 governs the pro‐ and anti‐apoptotic transcriptional response. Nucleic Acids Res. 2008;36:1415‐1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Di Cristofano A, Strazzullo M, Longo L, La Mantia G. Characterization and genomic mapping of the ZNF80 locus: expression of this zinc‐finger gene is driven by a solitary LTR of ERV9 endogenous retroviral family. Nucleic Acids Res. 1995;23:2823‐2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Feuchter‐Murthy AE, Freeman JD, Mager DL. Splicing of a human endogenous retrovirus to a novel phospholipase A2 related gene. Nucleic Acids Res. 1993;21:135‐143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Schulte AM, Lai S, Kurtz A, et al. Human trophoblast and choriocarcinoma expression of the growth factor pleiotrophin attributable to germ‐line insertion of an endogenous retrovirus. Proc Natl Acad Sci U S A. 1996;93:14759‐14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Singer GA, Wu J, Yan P, et al. Genome‐wide analysis of alternative promoters of human genes using a custom promoter tiling array. BMC Genomics. 2008;9:349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Puomila K, Simell O, Huoponen K, Mykkanen J. Two alternative promoters regulate the expression of lysinuric protein intolerance gene SLC7A7. Mol Genet Metab. 2007;90:298‐306. [DOI] [PubMed] [Google Scholar]
- 100. Lamprecht B, Walter K, Kreher S, et al. Derepression of an endogenous long terminal repeat activates the CSF1R proto‐oncogene in human lymphoma. Nat Med. 2010;16:571‐579. 1p‐579p [DOI] [PubMed] [Google Scholar]
- 101. Sin HS, Huh JW, Kim DS, et al. Transcriptional control of the HERV‐H LTR element of the GSDML gene in human tissues and cancer cells. Arch Virol. 2006;151:1985‐1994. [DOI] [PubMed] [Google Scholar]
- 102. Saeki N, Kuwahara Y, Sasaki H, Satoh H, Shiroishi T. Gasdermin (Gsdm) localizing to mouse Chromosome 11 is predominantly expressed in upper gastrointestinal tract but significantly suppressed in human gastric cancer cells. Mamm Genome. 2000;11:718‐724. [DOI] [PubMed] [Google Scholar]
- 103. Wang T, Zeng J, Lowe CB, et al. Species‐specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci U S A. 2007;104:18613‐18618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Sundaram V, Cheng Y, Ma Z, et al. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 2014;24:1963‐1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Bourque G, Leong B, Vega VB, et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18:1752‐1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. de Parseval N, Alkabbani H, Heidmann T. The long terminal repeats of the HERV‐H human endogenous retrovirus contain binding sites for transcriptional regulation by the Myb protein. J Gen Virol. 1999;80(Pt 4):841‐845. [DOI] [PubMed] [Google Scholar]
- 107. Sjottem E, Anderssen S, Johansen T. The promoter activity of long terminal repeats of the HERV‐H family of human retrovirus‐like elements is critically dependent on Sp1 family proteins interacting with a GC/GT box located immediately 3′ to the TATA box. J Virol. 1996;70:188‐198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Hughes JF, Coffin JM. Evidence for genomic rearrangements mediated by human endogenous retroviruses during primate evolution. Nat Genet. 2001;29:487‐489. [DOI] [PubMed] [Google Scholar]
- 109. Campbell IM, Gambin T, Dittwald P, et al. Human endogenous retroviral elements promote genome instability via non‐allelic homologous recombination. BMC Biol. 2014;12:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Tomlins SA, Laxman B, Dhanasekaran SM, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595‐599. [DOI] [PubMed] [Google Scholar]
- 111. Hermetz KE, Surti U, Cody JD. Rudd MK. A recurrent translocation is mediated by homologous recombination between HERV‐H elements. Mol Cytogenet. 2012;5:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Arruda JT, Silva DM, Silva CC, Moura KK, Da CA. Homologous recombination between HERVs causes duplications in the AZFa region of men accidentally exposed to cesium‐137 in Goiania. Genet Mol Res. 2008;7:1063‐1069. [DOI] [PubMed] [Google Scholar]
- 113. Moore PS, Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat Rev Cancer. 2010;10:878‐889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Fan H, Johnson C. Insertional oncogenesis by non‐acute retroviruses: implications for gene therapy. Viruses. 2011;3:398‐422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Lee YN, Bieniasz PD. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 2007;3:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Dewannieux M, Harper F, Richaud A, et al. Identification of an infectious progenitor for the multiple‐copy HERV‐K human endogenous retroelements. Genome Res. 2006;16:1548‐1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Blond JL, Lavillette D, Cheynet V, et al. An envelope glycoprotein of the human endogenous retrovirus HERV‐W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol. 2000;74:3321‐3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Lu X, Wang R, Zhu C, et al. Fine‐tuned and cell‐cycle‐restricted expression of fusogenic protein syncytin‐2 maintains functional placental syncytia. Cell Rep. 2017;21:1150‐1159. [DOI] [PubMed] [Google Scholar]
- 119. Blaise S, de Parseval N, Benit L, Heidmann T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc Natl Acad Sci U S A. 2003;100:13013‐13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Esnault C, Priet S, Ribet D, et al. A placenta‐specific receptor for the fusogenic, endogenous retrovirus‐derived, human syncytin‐2. Proc Natl Acad Sci U S A. 2008;105:17532‐17537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Diaz‐Carballo D, Klein J, Acikelli AH, et al. Cytotoxic stress induces transfer of mitochondria‐associated human endogenous retroviral RNA and proteins between cancer cells. Oncotarget. 2017;8:95945‐95964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Huang G, Li Z, Wan X, Wang Y, Dong J. Human endogenous retroviral K element encodes fusogenic activity in melanoma cells. J Carcinog. 2013;12:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43‐51. [DOI] [PubMed] [Google Scholar]
- 124. Contreras‐Galindo R, Kaplan MH, Leissner P, et al. Human endogenous retrovirus K (HML‐2) elements in the plasma of people with lymphoma and breast cancer. J Virol. 2008;82:9329‐9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Seifarth W, Skladny H, Krieg‐Schneider F, et al. Retrovirus‐like particles released from the human breast cancer cell line T47‐D display type B‐ and C‐related endogenous retroviral sequences. J Virol. 1995;69:6408‐6416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Serafino A, Balestrieri E, Pierimarchi P, et al. The activation of human endogenous retrovirus K (HERV‐K) is implicated in melanoma cell malignant transformation. Exp Cell Res. 2009;315:849‐862. [DOI] [PubMed] [Google Scholar]
- 127. Bronson DL, Fraley EE, Fogh J, Kalter SS. Induction of retrovirus particles in human testicular tumor (Tera‐1) cell cultures: an electron microscopic study. J Natl Cancer Inst. 1979;63:337‐339. [PubMed] [Google Scholar]
- 128. Balaj L, Lessard R, Dai L, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Mangeney M, Renard M, Schlecht‐Louf G, et al. Placental syncytins: genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proc Natl Acad Sci U S A. 2007;104:20534‐20539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Cianciolo GJ, Copeland TD, Oroszlan S, Snyderman R. Inhibition of lymphocyte proliferation by a synthetic peptide homologous to retroviral envelope proteins. Science. 1985;230:453‐455. [DOI] [PubMed] [Google Scholar]
- 131. Mangeney M, Heidmann T. Tumor cells expressing a retroviral envelope escape immune rejection in vivo. Proc Natl Acad Sci U S A. 1998;95:14920‐14925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Kudo‐Saito C, Yura M, Yamamoto R, Kawakami Y. Induction of immunoregulatory CD271+ cells by metastatic tumor cells that express human endogenous retrovirus H. Cancer Res. 2014;74:1361‐1370. [DOI] [PubMed] [Google Scholar]
- 133. Li H, Chiappinelli KB, Guzzetta AA, et al. Immune regulation by low doses of the DNA methyltransferase inhibitor 5‐azacitidine in common human epithelial cancers. Oncotarget. 2014;5:587‐598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Tsai HC, Li H, Van Neste L, et al. Transient low doses of DNA‐demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell. 2012;21:430‐446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Goel S, DeCristo MJ, Watt AC, et al. CDK4/6 inhibition triggers anti‐tumour immunity. Nature. 2017;548:471‐475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Leonova KI, Brodsky L, Lipchick B, et al. p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc Natl Acad Sci U S A. 2013;110:E89‐E98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Karpf AR, Lasek AW, Ririe TO , et al. Limited gene activation in tumor and normal epithelial cells treated with the DNA methyltransferase inhibitor 5‐aza‐2′‐deoxycytidine. Mol Pharmacol. 2004;65:18‐27. [DOI] [PubMed] [Google Scholar]
- 138. Karpf AR, Peterson PW, Rawlins JT, et al. Inhibition of DNA methyltransferase stimulates the expression of signal transducer and activator of transcription 1, 2, and 3 genes in colon tumor cells. Proc Natl Acad Sci U S A. 1999;96:14007‐14012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Roulois D, Loo YH, Singhania R, et al. DNA‐demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 2015;162:961‐973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Chiappinelli KB, Strissel PL, Desrichard A, et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2015;162:974‐986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Chuong EB, Elde NC, Feschotte C. Regulatory evolution of innate immunity through co‐option of endogenous retroviruses. Science. 2016;351:1083‐1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Canadas I, Thummalapalli R, Kim JW, et al. Tumor innate immunity primed by specific interferon‐stimulated endogenous retroviruses. Nat Med. 2018;24:1143‐1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Sheng W, LaFleur MW, Nguyen TH, et al. LSD1 ablation stimulates anti‐tumor immunity and enables checkpoint blockade. Cell. 2018;174:549. [DOI] [PMC free article] [PubMed] [Google Scholar]


