Abstract
To achieve guide RNA (gRNA) multiplexing and an efficient delivery of tens of distinct gRNAs into single cells, we developed a molecular assembly strategy termed chimeric array of gRNA oligonucleotides (CARGO). We coupled CARGO with dCas9 (catalytically dead Cas9) imaging to quantitatively measure the movement of enhancers and promoters that undergo differentiation-associated activity changes in live embryonic stem cells. Whereas all examined functional elements exhibited subdiffusive behavior, their relative mobility increased concurrently with transcriptional activation. Furthermore, acute perturbation of RNA polymerase II activity can reverse these activity-linked increases in loci mobility. Through quantitative CARGO-dCas9 imaging, we provide direct measurements of cis-regulatory element dynamics in living cells and distinct cellular and activity states and uncover an intrinsic connection between cis-regulatory element mobility and transcription.
C is-regulatory DNA elements such as promoters and long-range enhancers mediate precise spatiotemporal control of gene expression (1–4). Understanding regulatory elements dynamics in living cells and their changes in relation to transcriptional status and cellular states is important for comprehending gene expression control. To explore these questions, we first had to address technical limitations associated with the labeling of small, nonrepetitive genomic elements. Previous strategies typically relied on inserting heterologous arrays of bacterial operator sequences that are much larger than a typical individual enhancer or promoter and, owing to their highly repetitive nature, may be subject to regulation specific for repetitive sequences (5–10). Alternatively, dCas9 (catalytically dead Cas9) imaging can facilitate RNA-guided labeling of native genomic regions (11), but similarly to other methods, it requires a large number of fluorescent molecules bound to the target locus at any given time to enable microscopic visualization. Consequently, in the absence of approaches allowing for highly multiplexed and uniform delivery of guide RNAs (gRNAs) to target cells, dCas9 imaging has been practically limited to the labeling of repetitive sequences (12–15).
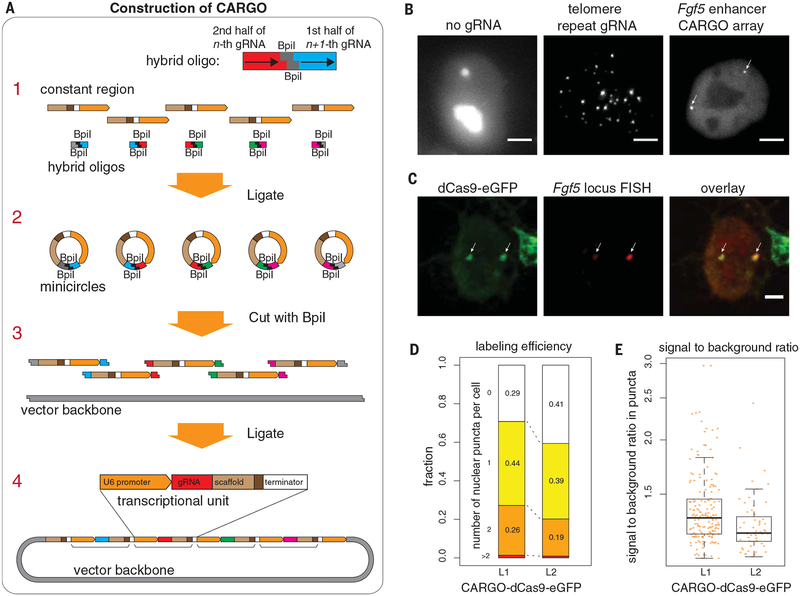
To overcome this bottleneck, we developed a molecular assembly strategy, termed chimeric array of gRNA oligonucleotides (CARGO), that can achieve highly multiplexed gRNA delivery into single cells (diagram in Fig. 1A). With this strategy, gRNA 12- and 18-nucleotide monomers can be readily assembled in a single step with 70% and 60% efficiencies, respectively, and an even higher degree of multiplexing can be readily accomplished (fig. S1; see materials and methods and other supplementary materials for details and protocol). We then asked whether robust labeling of nonrepetitive cis-regulatory elements can be achieved in mammalian cells by combining CARGO with dCas9 imaging. To this end, we chose mouse embryonic stem cells (mESCs) and their in vitro differentiation to epiblast-like cells (mEpiLCs) as a cell fate transition model (16–18) (fig. S2A). We have previously shown that during this transition a cluster of enhancers located ~30 to 54 kb downstream from the Fgf5 promoter is activated de novo (17) (fig. S2B) and that these changes are accompanied by the transcriptional induction of the Fgf5 gene (17). To label this developmentally regulated enhancer cluster, we designed three CARGO arrays, each harboring 12 different gRNAs, spanning a 2-kb window immediately upstream of the first enhancer (E1) within the cluster (fig. S2B). We generated clonal mESC lines with a stably integrated, inducible dCas9-eGFP transgene expressed at relatively low (albeit clone-to-clone–variable) level upon induction with doxycycline (Dox) (fig. S3, A and B). As expected (11), dCas9-eGFP displayed previously observed nucleolar retention in the absence of gRNA and robustly labeled telomeric repeats upon transfection with telomere gRNA (Fig. 1B). Upon transfection of the three Fgf5 enhancer CARGO arrays, one or two puncta were clearly visible in live cells to which arrays were successfully introduced, as measured by a fluorescent marker encoded by the CARGO plasmid (Fig. 1B). The labeling was specific, as confirmed by the colocalization of the dCas9-eGFP immunofluorescence and Fgf5 DNA fluorescence in situ hybridization (FISH) signals (Fig. 1C), and efficient, as 60% to 70% of cells across different lines were consistently labeled with one or two puncta with a sufficiently high signal to background ratio (Fig. 1, D and E).
Fig. 1. CARGO-dCas9 imaging enables robust and noninvasive labeling of cis-regulatory elements in living cells.
(A) CARGO assembly of a multiplexed gRNA array. Hybrid DNA oligonucleotides are synthesized with the first half of nth gRNA sequence, followed by the second half of the (n – 1)th gRNA separated by two BpiI restriction sites, with distinct sticky ends for each gRNA. Step 1: Synthetic DNA oligonucleotides are mixed and ligated with a permuted expression unit constant region (gRNA scaffold, Pol III termination signal, and human U6 promoter). Step 2: Resulting mini circles are cut with BpiI, exposing complementary sticky ends from different circles. Step 3: Digested products and destination vector are ligated to produce an array of gRNA expression units (shown in step 4) in a single-pot reaction. (B) Representative examples of dCas9 imaging of genomic loci in mESCs. (Left) No gRNA control. (Middle) Single-gRNA–targeting telomere repeats. (Right) CARGO-array–targeting Fgf5 enhancer. Scale bars, 5 mm. (C) Representative images showing colocalization of the Fgf5 enhancer CARGO dCas9-eGFP signal [as visualized by anti-eGFP (anti–enhanced GFP) immunofluorescence] with the DNA FISH signal (position of a BAC FISH probe is shown in fig. S3C). Scale bar, 2 mm. Colocalization was confirmed by Fisher’s exact test: P < 3 × 10−28; odds ratio = 1.49 × 104 of nonrandom association between sparsely sampled dCas9 and DNA FISH image pixels. (D and E) CARGO-dCas9 locus labeling efficiency (D) and signal-to-background ratio (E) in two clonal mESC lines (L1 and L2) bearing dCas9-GFP fusion and transfected with Fgf5 enhancer CARGO arrays. In (E), the bold line at the center of each box denotes the median value; top and bottom edges of the box denote the 25th and 75th percentiles, respectively.
We next investigated both the precision of the dCas9 targeting and its potential interference with enhancer activation during differentiation. We performed anti-GFP (green fluorescent protein) and anti-H3K27ac (acetylation of histone H3 at lysine 27) chromatin immunoprecipitation (ChIP)–quantitative polymerase chain reaction (qPCR) analyses from dCas9-eGFP mESCs with or without a CARGO array, and from mEpiLCs derived from them through 48 hours of differentiation. Using qPCR amplicons spanning either the CARGO-array targeted region (a to c) or enhancer E1 itself (d and e) (fig. S2B), we observed that: (i) dCas9 recruitment is dependent on the array and localized to the targeted region, and (ii) enhancer activation, as indirectly measured by H3K27ac, retains its developmental dynamics and is not considerably affected by dCas9-eGFP recruitment in the vicinity (fig. S2, C and D). In agreement, Fgf5 mRNA single-molecule FISH (smFISH) analysis of mEpiLC dCas9-eGFP cells with or without a CARGO array showed no significant differences in transcript numbers between the two populations (fig. S4A). Taken together, these results demonstrate that CARGO-dCas9 imaging provides a specific and noninvasive strategy to label functional cis-regulatory sequences in their native chromosomal context.
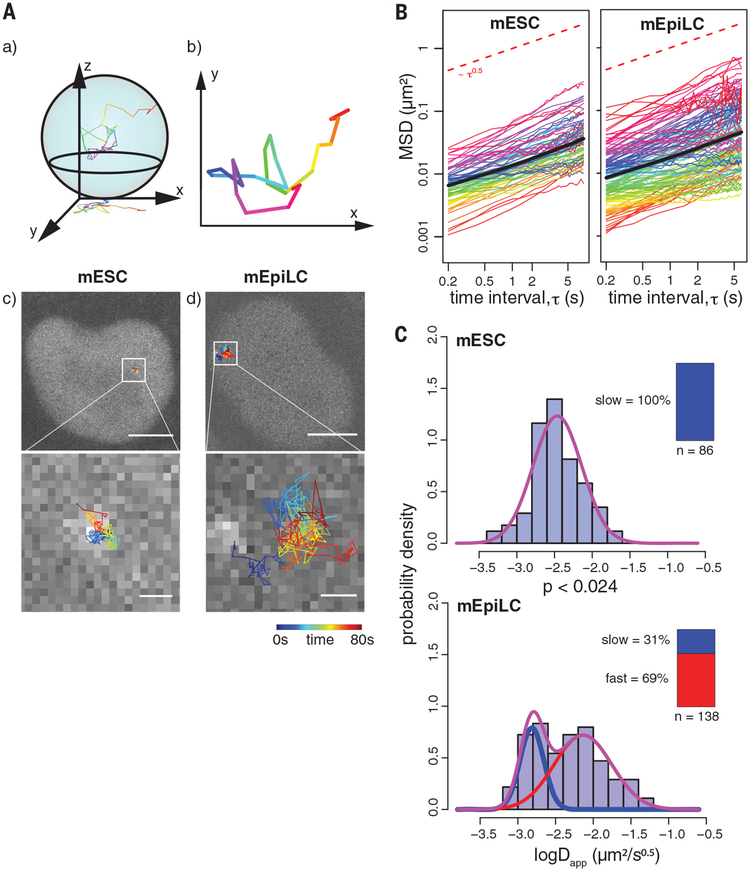
To follow the movement of the Fgf5 enhancer in its active and inactive state, we performed live CARGO-dCas9 imaging in mESCs or in mEpiLCs after 48 hours of differentiation and tracked centers of labeled loci with high temporal resolution (Fig. 2A and movie S1). Visual inspection of the recorded time-lapse images revealed that a substantial fraction of Fgf5 enhancers displayed increased mobility in mEpiLCs compared with mESCs (compare movies S2 and S3), and this was also evident when we examined individual trajectories over a fixed time interval (Fig. 2A, compare c and d). To characterize the Fgf5 enhancer movement quantitatively, we computed the mean square displacement (MSD) for these trajectories (Fig. 2B) and extracted two parameters: the scaling exponent a and the apparent diffusion coefficient Dapp. Time-averaged MSD (tMSD) plots show an anomalous scaling exponent of α= 0.53 ± 0.21 in mESCs and α = 0.51 ± 0.26 in mEpiLCs (Fig. 2B), suggesting subdiffusive behavior of the enhancer, similar to chromosome movement in bacteria, yeast, and B cells (8, 19, 20). Additionally, a comparable scaling exponent α (0.54 in mESCs and 0.51 in mEpiLCs) was obtained from the time- and ensemble-averaged MSD (eMSD) plots (Fig. 2B, shaded area denotes SEM), indicating that the loci movements are ergodic.
Fig. 2. Live-cell CARGO-dCas9 imaging and tracking of the Fgf5 enhancer during differentiation of mESCs to mEpiLCs.
(A) Live-cell two-dimensional (2D) tracking of the CARGO-dCas9–labeled Fgf5 enhancer in mESCs and mEpiLCs. (a and b) Movies of the cell nuclei are recorded as 2D projections of the 3D movement of the labeled loci. (c and d) Representative images of a single mESC (c) or mEpiLC (d) nucleus, overlaid with recorded trajectories color-coded by time (0 to 80 s). The bottom panels show zoomed-in views of the inset areas in the top images. Scale bars, 5 mm (top); 500 nm (bottom). (B) Subdiffusive motion of the Fgf5 enhancer locus. tMSD for each tracked enhancer allele (colored curves) and eMSD (bold black curve, shaded area indicates ± SEM) as a function of the time interval (t) between observations. Ninety-one and 130 observed alleles are plotted for the mESC and mEpiLC state, respectively. The red dashed reference line has a slope of 0.5. (C) Appearance of a fast-moving Fgf5 enhancer population in the mEpiLC state. Histograms of fitted apparent anomalous diffusion coefficients calculated from tMSD trajectories in mESCs or mEpiLCs, as indicated, are overlaid with fitted Gaussian mixture distribution curves in purple. Individual slow and fast components are plotted as blue and red curves, respectively. The inset bar plots indicate the number of recorded trajectories, together with the mixing proportion of slow and fast population components for each individual Gaussian. Bayesian information criterion (BIC) values for different component fittings are listed in table S1. The difference in distributions is supported by a two-sample Kolmogorov-Smirnov test.
Whereas the scaling exponent extracted from our MSD measurements was similar for the two examined cell states, the apparent anomalous diffusion coefficient of the Fgf5 enhancer exhibited a significant increase in mEpiLCs compared with ESCs (Fig. 2C and table S2). Moreover, distribution of Dapp was unimodal in the mESC state, whereas in the mEpiLC state the observed data can be better explained by the appearance of an additional “fast” population with a higher apparent anomalous diffusion coefficient Dapp (8.6 ×10−3 ± 0.38 μm2/s0.5) representing ~69% of the tracked enhancer alleles (with the remaining alleles showing “slow” behavior with Dapp of 1.6 × 10−3 ± 0.16 μm2/s0.5) (Fig. 2C). These measurements are consistent with the increased mobility of the enhancer in mEpiLCs in our time-lapse images.
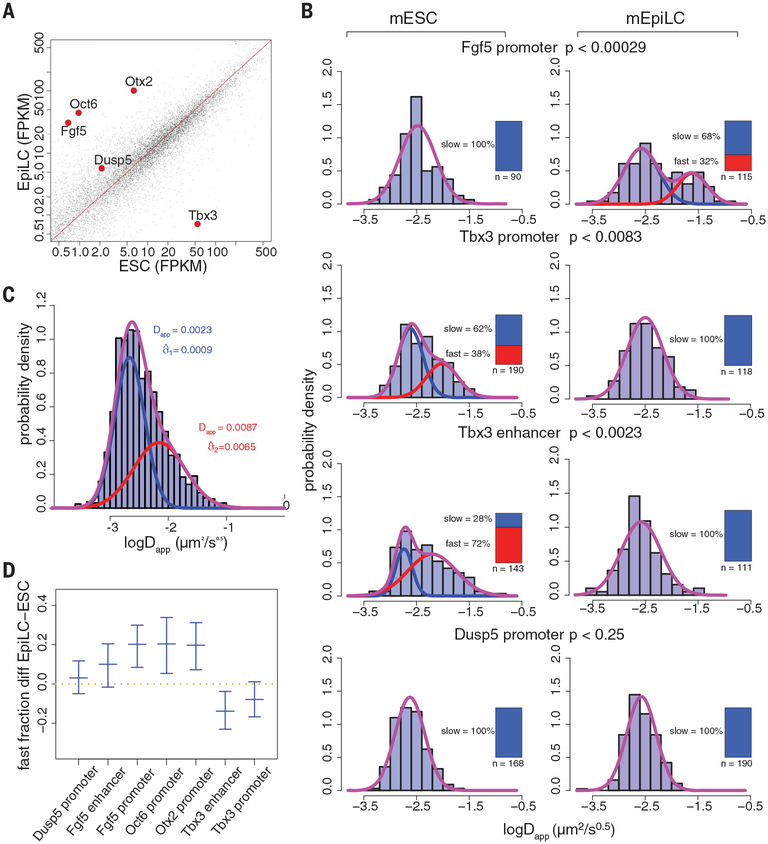
We next used CARGO-dCas9 to label the Fgf5 promoter, which is transcriptionally induced during the mESC-to-mEpiLC transition [Fig. 3A, fig. S4 (for smFISH showing that labeling does not interfere with Fgf5 expression), and fig. S5, top panel (for the CARGO array position in relation to the transcription start site, TSS)]. Tracking and MSD measurements of Fgf5 promoter movement in mESCs showed a and Dapp values consistent with the slow subdiffusive behavior observed for the Fgf5 enhancer in mESCs (table S2). Fur thermore, an overall increase in mobility was observed for the Fgf5 promoter in the mEpiLC state, which can also be attributed to the appearance of a fast subpopulation, with Dapp of 26 × 10−3 ± 0.28 mm2/s0.5 (Fig. 3B, first row). Two other promoters that become transcriptionally induced during the mESC-to-mEpiLC transition, Otx2 and Oct6, also show elevated mobility in the active state, whereas the Dusp5 promoter, which maintains a consistently low transcriptional activity, shows unimodal distribution of Dapp with slow diffusivity in both cellular states [Fig. 3, A and B; fig. S5, bottom panel; fig. S6 (for tMSD and eMSD plots); and fig. S7D]. These observations suggest that mobility of cis-regulatory elements may be linked to their activity status and that the underlying heterogeneity of this dynamics could reflect the heterogeneity of the transcriptional state. Alternatively, these mobility shifts could be explained by the global differences in chromatin properties and/or compaction associated with mESC and mEpiLC states.
Fig. 3. Mobility of cis-regulatory elements changes with the transcriptional status of their associated genes.
(A) Expression changes of genes whose cis-regulatory elements were analyzed by live-cell tracking. Ordinate: mean expression in mESC state; abscissa: mean expression in mEpiLC state, as measured by RNA sequencing. All genes are shown, with investigated genes highlighted in red. FPKM, fragments per kilobase of transcript per million mapped reads. (B) Histograms of fitted apparent anomalous diffusion coefficients from tMSD of the indicated regulatory regions in mESCs (left panels) or mEpiLCs (right panels), overlaid with fitted Gaussian mixture distributions (purple) along with slow (blue) and fast (red) components. The inset bar plots indicate the number of recorded trajectories and the individual Gaussian mixing proportion of slow and fast population components. BIC values for different component fittings are listed in table S1. Differences in the distribution between the mESC and mEpiLC states are supported by a two-sample Kolmogorov-Smirnov test. (C) Histogram of combined fitted apparent anomalous diffusion coefficients from tMSD of all loci in both mESC and mEpiLC states (n = 1271) overlaid with the fitted Gaussian mixture distribution (purple) along with slow (blue) and fast (red) components. Log-normal means and standard deviations of the slow and fast components (in μm2 s−0.5) are denoted on the plot. (D) Difference in fractions of fast populations between the mESC and mEpiLC states. Center bars indicate the median value; upper and lower bars indicate the 95% confidence interval of the estimates.
To distinguish between these two possibilities, we labeled the promoter and distal superenhancer of Tbx3, a gene that becomes down-regulated during the mESC-to-mEpiLC transition (Fig. 3A and fig. S5, middle panel). Notably, CARGO-dCas9 imaging of the Tbx3 promoter or superenhancer located ~90 kb away from the TSS revealed changes in mobility that were opposite to those observed at the Fgf5 locus [Fig. 3B, fig. S6 (for tMSD and eMSD plots), and movies S4 and S5].
Analysis of the combined live-cell tracking data of all seven regulatory elements in both cellular states suggests that as a first-order approximation, the observed heterogeneity of cis-regulatory element dynamics can be better explained by a two-population model [ΔBIC = −143.2; BIC, Bayesian information criterion] versus a single-population model, where the apparent diffusivity of the fast population is three to four times higher than that of the slow population (Fig. 3C). We compared the extracted fast versus slow population fractions for all seven tested loci in either mESCs or mEpiLCs and observed a statistically significant increase of the fast population in the transcriptionally active state (Fig. 3D and fig. S7A). However, because the fitted standard deviations of Dapp distribution in the fast state are broad, it remains an open question whether a single Dapp value for the fast state exists or whether there is a continuum that is dependent on the level of transcription and locus-specific effects.
The scaling exponent a was comparable (~0.5) for all examined elements in both cellular states (fig. S7B and table S2) and close to the a values obtained through measurements of chromatin movement in mammalian cells, yeast, and bacteria. These observations suggest that the subdiffusive behavior and scaling laws governing the movement of cis-regulatory elements in the interphase nuclei are similar to those of other classes of chromatin and are conserved across the tree of life. Furthermore, the comparable shape and negative dip of the time-lag–rescaled velocity autocorrelation function indicate that the viscoelastic nature of nucleoplasm and chromatin fibers can explain the subdiffusive scaling behavior of cis-regulatory elements (8) in both the active and inactive states (fig. S7C).
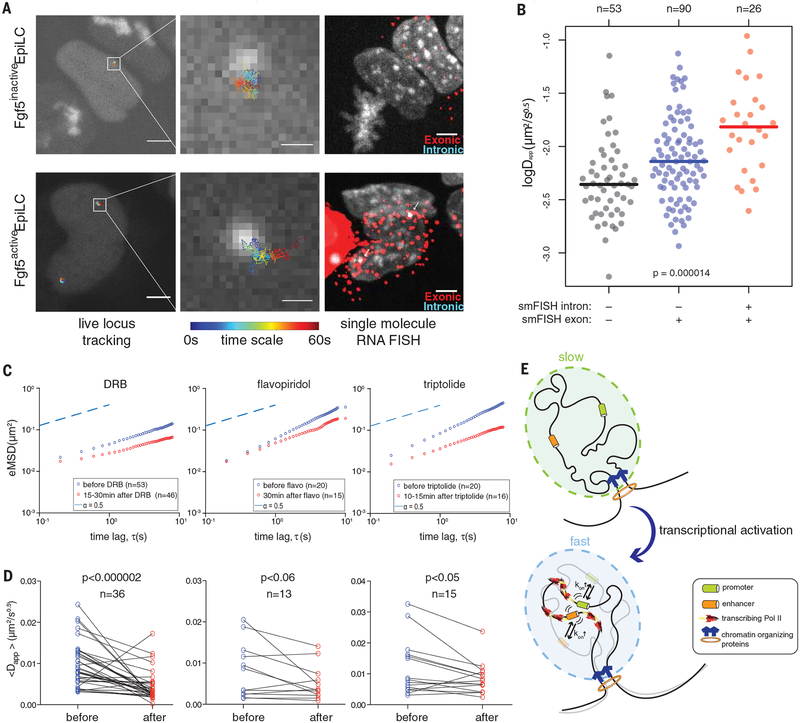
To gain more insight into the relationship between the cis-regulatory element mobility and the transcriptional status of its cognate gene at the single-cell level, we collected matched live–fixed cell data by tracking Fgf5 enhancer movement in live mEpiLCs and measuring transcriptional status in the same cells by smFISH with the intronic and exonic Fgf5 mRNA probes (see fig. S8 for specificity validation). We took advantage of the expression heterogeneity during differentiation and classified cells into three categories: actively transcribing (Fgf5 intronic +, Fgf5 exonic +), recently active (Fgf5 intronic −, Fgf5 exonic +), or inactive (Fgf5 intronic −, Fgf5 exonic −). By analyzing the smFISH signals in relation to the Fgf5 enhancer mobility, we observed that in cells with Fgf5 mRNA transcripts, the enhancer explores a larger nuclear space over a given time compared with in the inactive cells, with the highest mobility observed for enhancer alleles that are actively transcribing at the time of measurement (Fig. 4, A and B). This strong association suggests a direct connection between transcriptional activity and cis-regulatory element mobility.
Fig. 4. Inhibition of RNA polymerase II reverses activity-associated changes in enhancer mobility.
(A) Live-cell locus tracking correlation with multiplexed single-molecule RNA FISH. Representative images of a cell with an inactive (top) or active (bottom) Fgf5 locus are shown. (Left) Snapshot of live-cell imaging in grayscale overlaid with fitted Fgf5 enhancer trajectories color-coded by frame number from 1 to 300 (corresponding to time 200 ms to 60 s). Scale bars, 5 mm. (Middle) Magnified view of the highlighted regions in the left panel. Scale bars, 500 nm. (Right) Multiplexed smFISH of Fgf5 mRNA from the same cells. Gray channel: DAPI (4′,6-diamidino-2-phenylindole) staining for cell nucleus; red channel: smFISH probe targeting the first exon of Fgf5 mRNA (the mCherry marker in the CARGO array is also visible); blue channel: smFISH probe targeting the first intron of Fgf5 mRNA. Colocalized intronic and exonic smFISH signals are highlighted by arrows. Scale bars, 5 mm. (B) Mobility of the Fgf5 enhancer correlates with nascent transcription of the Fgf5 locus at the single-cell level. Cells were binned into three groups according to the transcription status of the Fgf5 locus, as measured by multiplexed smFISH and indicated at the bottom. Individual dots represent apparent anomalous diffusion coefficients extracted from the corresponding live-imaging tMSD data. Statistical significance is supported by a Kruskal-Wallis test. (C) Increased mobility of the Fgf5 enhancer in mEpiLCs is reversed by Pol II inhibition. eMSD of the Fgf5 enhancer trajectories in mEpiLCs is shown before (blue circles) and after (red circles) treatment with DRB, flavopiridol, and triptolide, as indicated. (D) Increased mobility of the Fgf5 enhancer in mEpiLCs is reversed by Pol II inhibition at the single-cell level. Anomalous diffusion coefficient of the Fgf5 enhancer is shown for the same cells before and after the corresponding drug treatment. Differences are supported by a paired Wilcoxon test, as indicated by P values in the plots. (E) The stirring model provides an explanation for observed transcription-coupled changes in the mobility of cis-regulatory elements. The ground state (slow) is characterized by subdiffusive behavior with low apparent diffusivity governed by thermal forces. The activated (fast) state is characterized by an increased apparent diffusivity, which may be due to nonthermal agitation by transcribing RNA Pol II and/or its associated ATPases. Under the assumption that the radius of a local 3D chromosomal domain remains relatively invariant in the slow and fast states, elevated mobility of cis-regulatory elements would lead to decreased time to the first encounter between distally located enhancer and promoter regions, resulting in an increased enhancer-promoter contact frequency.
To directly test the relationship between the status of RNA polymerase II (Pol II) and the anomalous diffusive behavior of the Fgf5 enhancer, we acutely (for 10 to 30 min) perturbed Pol II activity by using chemical inhibitors targeting either transcriptional elongation (DRB and flavopiridol) (21, 22) or transcriptional initiation (triptolide) (23, 24). Tracking Fgf5 enhancer mobility revealed that inhibition of either initiation or elongation results in a significant reduction of Dapp, both in bulk measurements of eMSD across all cells (Fig. 4C) and at the single-cell level, as measured by tMSD in matched single cells before and after the treatment (Fig. 4D). Notably, alleles showing higher Dapp values in untreated cells (and thus representing the fast subpopulation) were the most affected by the Pol II inhibition. Similar decreases in Dapp upon Pol II inhibition were also observed for the Tbx3 promoter in mESCs (fig.S9). Together, our observations indicate that the increased nuclear mobility of cis-regulatory elements is directly coupled to the Pol II activity and transcriptional status of their cognate genes.
The CARGO assembly method described here has a wide application potential beyond imaging, in techniques such as CRISPR interference, CRISPR activation, and in simultaneous editing of multiple genomic regions, where multiplexed recruitment of dCas9 or Cas9 to genomic loci of interest is required or desired. Through CARGO-dCas9 visualization and quantitative measurement of cis-regulatory element dynamics in distinct activity states, we uncovered an unexpected relationship between the mobility of cis-regulatory elements and local transcriptional activity. What may be the underlying causes of the observed transcription-coupled increases in mobility? Although the exact formulations vary with different polymer models (25), Dapp generally rises with increased molecular agitation (e.g., absolute temperature in the simplest form but also nonthermal sources of agitation) and with decreased friction and/or viscosity and polymer size. Several lines of evidence suggest that the increase in Dapp may be due to a nonthermal molecular motion generated by an energy-dissipating biochemical process (26). The main candidate for such a process in our studies is the activity of RNA Pol II—a major molecular motor that trans-locates through the chromatin and dissipates energy via pyrophosphate release—although transcription-coupled adenosine triphosphate (ATP)–dependent chromatin remodelers may also contribute to the local movement. Collectively, we propose that ongoing transcription may provide a source of nonthermal molecular agitation that can “stir” the chromatin within the local chromosomal domain, leading to an increase in anomalous Dapp (Fig. 4E). We will hereafter refer to this hypothesis as the “stirring model.”
The stirring model may have implications for transcription regulation: Under the assumption that the radius of the local chromosomal domain [such as a topologically associated domain (TAD)] does not substantially change in the examined cell state(s), the time to the first encounter between distally located enhancer and promoter regions should decrease along with the increased mobility within the domain. In other words, enhancer-promoter contact frequencies may increase upon transcriptional activation due to the increased probability of the stochastic encounters within the TAD, rather than due to the formation of stable enhancer-promoter loops (Fig. 4E). This type of mechanism could provide a positive-feedback loop facilitating gene expression robustness once transcription is initiated, as increased mobility would boost enhancer-promoter contact frequencies, in turn leading to more transcription.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Theriot, J. Ferrell, and E. Calo for comments on the manuscript; Y. Fan for assistance with single-particle tracking (IDL tracking package) matlab script modification; R. Greenberg for coining the CARGO acronym; R. Srinivasan for contributing to the initial phases of CARGO assembly optimization; and members of the Wysocka and Meyer laboratories for discussions. Funding: This work was supported in part by the Howard Hughes Medical Institute, NIH R01 grant GM112720–01 and Ludwig Institute Funds (to J.W.), R35GM127026 and S10OD018073 (to T.M.), and a Henry Fan Stanford Graduate Fellowship (to B.G.).
Footnotes
Competing interests: B.G., T.S., and J.W. have filed a U.S. provisional patent application relating to CARGO methodology. All other authors declare no competing interests.
Data and materials availability: All live-cell time-lapse images will be provided upon request.
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/359/6379/1050/suppl/DC1
REFERENCES AND NOTES
- 1.Long HK, Prescott SL, Wysocka J, Cell 167, 1170–1187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine M, Curr. Biol 20, R754–R763 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dekker J, Mirny L, Cell 164, 1110–1121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Remeseiro S, Hörnblad A, Spitz F, Dev. Biol 5, 169–185 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Nielsen HJ, Li Y, Youngren B, Hansen FG, Austin S, Mol. Microbiol 61, 383–393 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Robinett CC et al. , J. Cell Biol 135, 1685–1700 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marshall WF et al. , Curr. Biol 7, 930–939 (1997). [DOI] [PubMed] [Google Scholar]
- 8.Lucas JS, Zhang Y, Dudko OK, Murre C, Cell 158, 339–352 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vazquez J, Belmont AS, Sedat JW, Curr. Biol 11, 1227–1239 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Mitsuda SH, Shimizu N, PLOS ONE 11, e0161288 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen B et al. , Cell 155, 1479–1491 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen B et al. , Nucleic Acids Res 44, e75 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma H et al. , Proc. Natl. Acad. Sci. U.S.A 112, 3002–3007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma H et al. , Nat. Biotechnol 34, 528–530 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu Y et al. , Nat. Commun 7, 11707 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M, Cell 146, 519–532 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Buecker C et al. , Cell Stem Cell 14, 838–853 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalkan T, Smith A, Philos. Trans. R. Soc. Lond. B Biol. Sci 369, 20130540 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber SC, Spakowitz AJ, Theriot JA, Phys. Rev. Lett 104, 238102 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cabal GG et al. , Nature 441, 770–773 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Chao S-H et al. , J. Biol. Chem 275, 28345–28348 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Yankulov K, Yamashita K, Roy R, Egly J-M, Bentley DL, J. Biol. Chem 270, 23922–23925 (1995). [DOI] [PubMed] [Google Scholar]
- 23.Titov DV et al. , Nat. Chem. Biol 7, 182–188 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vispé S et al. , Mol. Cancer Ther 8, 2780–2790 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Dudko OK, Annu. Rev. Biophys 45, 117–134 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Weber SC, Spakowitz AJ, Theriot JA, Proc. Natl. Acad. Sci. U.S.A 109, 7338–7343 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.