Abstract
Inhibitors of the 20S proteasome such as bortezomib are cytotoxic to tumor cells and have been proven to be valuable for the clinical management of multiple myeloma. The therapeutic efficacy of bortezomib is, however, hampered by the emergence of acquired resistance. Available data suggest that blocking proteasome activity at the level of proteasome-associated deubiquitinases (DUBs) provides a mechanism to overcome resistance to bortezomib and also to other cancer therapies. The small molecule b-AP15 is an inhibitor of proteasome-associated DUB activity that induces both proteotoxic stress and increases in the levels of reactive oxygen species (ROS) in tumor cells. Antioxidants have been shown to decrease apoptosis induction by b-AP15 and we here addressed the question of the mechanism of redox perturbation by this compound. We show that oxidative stress induction by b-AP15 is abrogated in cells deprived of mitochondrial DNA (ρ 0 cells). We also show associations between the level of proteotoxic stress, the degree of mitochondrial dysfunction, and the extent of induction of hemeoxygenase-1 (HO-1), a target of the redox-regulated Nrf-2 transcription factor. Decreased expression of COX5b (cytochrome c oxidase subunit 5b) and TOMM34 (translocase of outer mitochondrial membrane 34) was observed in b-AP15-treated cells. These findings suggest a mitochondrial origin of the increased levels of ROS observed in cells exposed to the DUB inhibitor b-AP15.
1. Introduction
Aberrant accumulation of misfolded or damaged proteins is associated with reduced cell survival [1]. Protein quality control is primarily mediated by the ubiquitin-proteasome system (UPS), the major eukaryotic proteolytic pathway, and is essential for cell viability [2, 3]. The proteasome degrades the bulk of cellular protein and is instrumental to the regulation of essential cellular processes such as cell cycle progression [4–7]. Misfolded, damaged, or temporally regulated proteins are marked for removal by the destruction tag ubiquitin that signals traffic to the proteasome for degradation. Once at the proteasome, ubiquitin is removed from polyubiquitinated proteins by deubiquitinases (DUBs) localized in the 19S regulatory particle, to facilitate translocation into the 20S core particle where degradation takes place [8, 9]. Cancer cells, characterized by rapid protein synthesis and unlimited proliferation, face an extreme load of misfolded proteins and therefore have an increased requirement for UPS-mediated protein turnover [10, 11]. Under conditions of proteasome inhibition, misfolded proteins accumulate in tumor cells, resulting in pleiotropic effects such as induction of cytosolic chaperones, endoplasmic reticulum (ER) stress, and oxidative stress [12–14].
Inhibitors of the 20S proteasome such as bortezomib and carfilzomib are used for treatment of multiple myeloma and have changed the clinical course of this disease [15, 16]. However, both intrinsic and acquired resistance to bortezomib limit its therapeutic efficacy [17]. A number of different mechanisms have been described to result in bortezomib resistance, including mutations in the PSMB5 subunit and overexpression of this subunit [17].
The redox state is important for cell survival, proliferation, and apoptosis [18]. Reactive oxygen species (ROS) may be harmful to cells leading to oxidative damage such as lipid peroxidation but may also be second messengers controlling signaling pathways [19]. The three major sources of ROS in the cell are mitochondria, peroxisomes, and the endoplasmic reticulum (ER) [20, 21]. Oxidative stress has been described to be induced by proteasome inhibitors [13, 14, 22], and antioxidants have been shown to decrease the apoptotic effects of these drugs [12, 13]. Oxidative stress resulting from proteasome inhibition has been attributed to ER stress [23] and to mitochondrial dysfunction [13] by various investigators.
We and the others have shown that the dienone compounds b-AP15, VLX1570, and RA-9 inhibit the activities of proteasome-associated DUBs [24–27], in particular USP14 [27]. This class of compounds induce apoptosis in tumor cells defective in TP53 [28] and overexpressing BCL2 [24, 29]. The ability of these compounds to selectively kill tumor cells while being largely insensitive to TP53 mutational status and defects in apoptotic machinery is interesting both from a mechanistic and therapeutic point of view, in particular considering their anticancer activities in vitro and in animal models [24, 25, 27, 29–41]. Interestingly, b-AP15 shows antiproliferative activity on myeloma cells resistant to bortezomib [33] and melanoma cells resistant to MAPK-targeting therapies [27]. We recently showed that the strong proteotoxicity induced by b-AP15 resulted in mitochondrial toxicity [42]. We and the others have shown that b-AP15 induces reactive oxygen species in tumor cells and that antioxidants decrease the apoptotic response [22, 27, 43]. These findings prompted us to examine whether oxidative stress induction by b-AP15 is mechanistically linked to mitochondrial dysfunction. We here provide experimental evidence in support of this notion.
2. Material and Methods
2.1. Chemicals and Antibodies
b-AP15 was obtained from OnTarget Chemistry (Uppsala, Sweden), Velcade (bortezomib, Selleck Chem) and CpdA [44] from Novartis. Antibodies used were anti-actin (Sigma-Aldrich catalogue number A5441), anti-Ub-K48 (Merck Millipore catalogue number 05-1307), anti-HMOX (BD Biosciences catalogue number 610713), anti-Hsp60 (Cell Signaling catalogue number 12165), anti-HSP40 (Cell Signaling catalogue number 4868), anti-Nrf-2 (Cell Signaling catalogue number 12721), anti-CHOP (Cell Signaling, catalogue number 5554), anti-HSP70B′ (Abcam catalogue number ab69408), and anti-MTCOXII2 (Abcam catalogue number ab110258).
2.2. Cell Culture and Drug Treatment
HCT116 colon carcinoma cells were maintained in McCoy's 5A modified medium with 10% FBS and 1% penicillin. HeLa cells were cultured in DMEM medium with supplemented 10% FBS and 1% penicillin. Cell lines were used at low passage numbers and checked for absence of mycoplasma. Drugs were dissolved in DMSO for final concentrations of DMSO 0.5%. CpdA was used at a concentration of 10 μM as described previously [45].
2.3. Western Blot Analysis
Cell extract proteins were resolved by 3-8% Tris-Acetate protein gels (Invitrogen, Carlsbad, CA) to detect polyubiquitinated proteins and 4-12% Bis-Tris protein gels to detect other proteins mentioned in the text, then transferred onto a PVDF membrane for western blotting [46]. Blots were developed by enhanced chemiluminescence (Amersham, Arlington Heights, IL).
2.4. Electron Microscopy
Cells were treated with b-AP15 for different times and fixed with 2.5% glutaraldehyde. Cells were postfixed in 1% osmium tetroxide, dehydrated, and embedded in epoxy resin. Ultrathin sections were prepared for analysis in a transmission electron microscope. Electron microscopy was performed by Kjell Hultenby at the Department of Laboratory Medicine, Clinical Research Center, Karolinska Universitetssjukhuset Huddinge, Sweden.
2.5. Glutathione Assays
For measurement of glutathione, cells were treated with 1.0 μM b-AP15 for 6 h. Cells were collected and concentrations of GSSG and total glutathione (GSH + GSSG) were analyzed using the quantification kit for oxidized and reduced glutathione (#38185, Sigma) as described. The final concentration of GSH was determined by equation of GSH = total glutathione (GSH + GSSG)–GSSG × 2. The data was analyzed using GraphPad Prism 7.
2.6. Measurements of G6PD, Glutathione Peroxidase, and Malondialdehyde
Drug-treated cells were washed with PBS and frozen at -80°C as cell pellets. Pellets were shipped to Biochemikon SAS, 94000 Créteil, France (study director Marc Conti). Cell pellets were sonicated, and enzymatic activities and substrate concentration measurements were performed. Glutathione peroxidase activity was measured according to Paglia and Valentine [47]. G6PD activity was adapted/optimized from Beutler [48]. MDA measurements were determined according to Conti et al. [49].
2.7. Measurements of Oxygen Consumption
OCR (oxygen consumption rates) were measured using a Seahorse XF24 extracellular flux analyzer in real time as recommended by the manufacturer (Seahorse Bioscience, North Billerica, MA, USA). Cells (60,000 cells/well) were plated in 100 μL culture medium in XF24-well cell plates with blank control wells. Prior to the measurements, the medium was replaced with 500 μL Seahorse assay media (1 mM pyruvate, 25 mM glucose, and 2 mM glutamine) at 37°C without CO2 for 1 h.
2.8. Generation of HeLa Rho0 (ρ 0) Cells
Hela cells were grown in DMEM medium supplemented with 100 ng/mL EtBr and 50 μg/mL uridine [50]. DNA was isolated using PureLink® Genomic DNA Mini Kit (Thermo Fisher Scientific), and mtDNA and nDNA were amplified by Human Mitochondrial DNA (mtDNA) Monitoring Primer Set (Takara). Copy number was measured using a 7500/7500 Fast Real-Time PCR System (Applied Biosystems), and mitochondria DNA to nuclear DNA ratios were calculated by the program supported by Takara. The absence of mtDNA-encoded protein MTCOXII2 in Rho0 cells was confirmed by immune blotting.
2.9. Isolation of Mitochondria
Mitochondrial isolation process was performed as [42].
2.10. Proteomics
Proteomic analysis was performed as described [42]. The raw data from LC-MS were analyzed by MaxQuant, version 1.5.6.5 [51]. The Andromeda search engine [52] searched MS/MS data against the International Protein Index (human, version UP000005640_9606, 92957 entries). Protein abundances were normalized by the total protein abundance in each sample. Mitochondrial proteins were further selected from total detected protein pool using MitoCarta (http://www.broad.mit.edu/publications/MitoCarta) supplied by [53].
2.11. Statistical Analysis
Statistical significance was evaluated by Student's two-tailed paired t-test (parametric) or Mann-Whitney U test (nonparametric). Protein expression data were compared using Spearman correlation coefficients.
3. Results
3.1. The Deubiquitinase Inhibitor b-AP15 Affects Mitochondrial Structure and Function
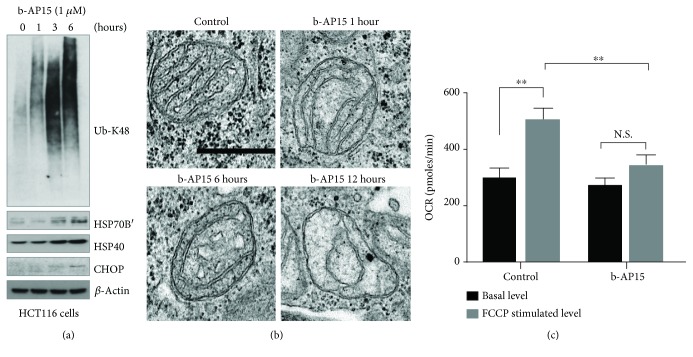
We have previously reported that b-AP15, an inhibitor of proteasome-associated deubiquitinases, generates both proteotoxic stress and oxidative stress [22, 24, 26, 27] and also induces mitochondrial dysfunction [42]. As shown in Figure 1(a), treatment of HCT116 cells with 1 μM b-AP15 resulted in increased levels of K48-linked polyubiquitin conjugates and induction of the chaperones HSP70B′ and HSP40 as well as the ER marker CCAAT-enhancer-binding protein homologous protein (CHOP) [54, 55]. Consistent with previous results, mitochondria became increasingly deformed during exposure to b-AP15 (Figure 1(b)). Mitochondrial function was examined by monitoring oxygen consumption rates using a Seahorse XF24 analyzer. Confirming previous results [42], the stimulation of oxygen consumption by carbonyl cyanide p-trifluoromethoxy-phenylhydrazone (FCCP) was reduced in b-AP15-exposed cells (Figure 1(c)), showing a decrease in maximal respiration capacity.
Figure 1.
Induction of mitochondrial dysfunction in HCT116 cells by the deubiquitinase inhibitor b-AP15. (a) HCT116 cells were exposed to 0.5% DMSO or 1 μM b-AP15 for 1, 3, and 6 hours, and extracts were prepared and subjected to immunoblotting using the indicated antibodies. (b) Electron micrographs of HCT116 cells treated with b-AP15 for 1, 6, and 12 h. Scale bar = 0.5 μm. (c) Basal and maximal oxygen consumption rates (OCR) were measured after a 5-hour exposure of HCT116 cells to 1 μM b-AP15 using a Seahorse XF analyzer. Uncoupled respiration was measured after exposure to carbonyl cyanide-4-(trifluoromethoxy)-phenylhydrazone (FCCP) (mean ± S.D.; ∗∗p < 0.01).
3.2. b-AP15 Induces Oxidative Stress but Not Lipid Peroxidation
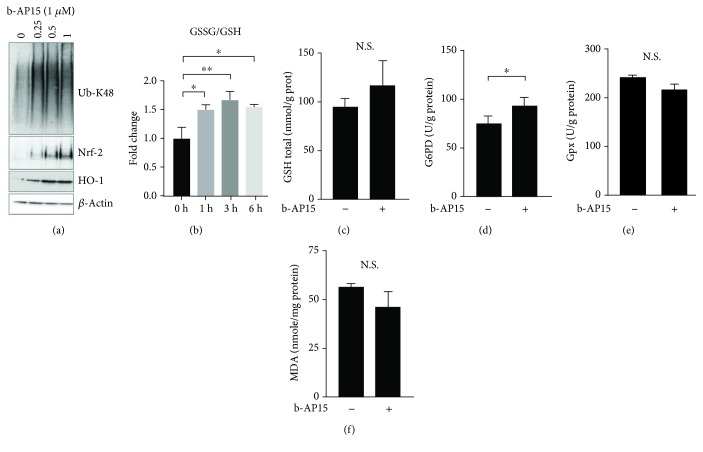
Previous studies demonstrated increased levels of intracellular ROS in b-AP15-treated HCT116 cells [22] and in melanoma cells [27]. Exposure of HCT116 cells to b-AP15 resulted in increased levels of the redox-regulated transcription factor Nrf-2 (nuclear factor erythroid 2-related factor 2) and its downstream target HO-1. This increases occurred at doses that induced the accumulation of high molecular weight K48-linked polyubiquitin conjugates (Figure 2(a)). Furthermore, a significant increase in the GSSG/GSH ratio was observed in b-AP15-exposed HCT116 cells, whereas no significant increases in total GSH levels were observed (Figures 2(b) and 2(c)). Increased glucose 6-phosphate dehydrogenase (G6PD) enzyme activity, the rate-limiting enzyme of the pentose phosphate pathway, was observed in b-AP15-treated HCT116 cells (p = 0.015) (Figure 2(d)). In contrast, glutathione peroxidase (GPx) activity levels were not significantly altered by b-AP15 treatment (Figure 2(e)). Malondialdehyde is a product of lipid peroxidation and a marker of oxidative damage [56]. Increased levels of malondialdehyde were not observed in HCT116 cells exposed to b-AP15 for 6 h (Figure 2(f)).
Figure 2.
Evidence of oxidative stress in b-AP15-exposed HCT116 cells. (a) HCT116 cells were exposed to 0.5% DMSO or b-AP15 (0.25, 0.5, and 1.0 μM in 0.5% DMSO) for 6 h, and extracts were prepared and subjected to immunoblotting using the indicated antibodies. (b) Increases in the ratio of GSSG/GSH in HCT116 cells exposed to 1 μM b-AP15 for the indicated times (mean ± S.D.; ∗p < 0.05, ∗∗p < 0.01; n = 3). (c) Total levels of GSH were determined in vehicle-treated cells and in cells exposed to 1 μM b-AP15 for 6 h (mean ± S.D.; n = 3; N.S.: not significant at p < 0.05). (d) Glucose-6-phosphate dehydrogenase activity in HCT116 cells exposed to 1 μM b-AP15 for 6 h compared to vehicle-treated cells (mean ± S.D.; ∗p < 0.05; n = 3). (e) Glutathione peroxidase activity in HCT116 cells exposed to 1 μM b-AP15 for 6 h compared to vehicle-treated cells (mean ± S.D.; n = 3). (f) Malondialdehyde levels in HCT116 cells exposed to 1 μM b-AP15 or vehicle for 6 h. Statistical significance was calculated using Student's t-test in (b)–(f).
3.3. Induction of Oxidative Stress Is Dependent on Functional Mitochondria
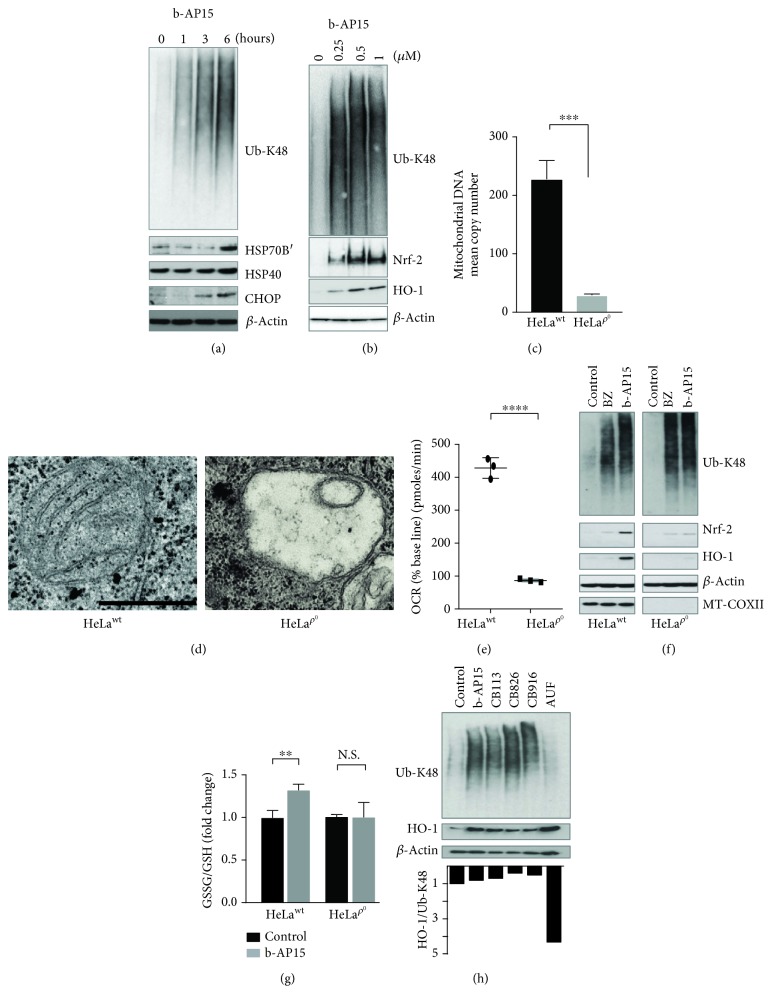
Attempts to generate HCT116 cells deficient in mitochondrial DNA (ρ 0 cells) by exposure to ethidium bromide were unsuccessful (not shown), possibly due to HCT116 cells being dependent on oxidative phosphorylation [57]. We therefore used HeLa cells, for which ρ 0 derivatives have been described [58]. Similar to the response in HCT116 cells, increases in polyubiquitinated proteins, chaperones, and CHOP were observed in HeLa cells exposed to b-AP15 (Figure 3(a)) and b-AP15 induces an apoptotic response in HeLa cells (Supplementary Fig. 1). Furthermore, Nrf-2 and HO-1 induction was observed also in b-AP15-treated HeLa cells (Figure 3(b)). Continuous exposure of HeLa cells to low doses of ethidium bromide resulted in cells with a reduced copy number of mitochondrial DNA (Figure 3(c)), aberrant mitochondrial morphology (Figure 3(d)), low oxygen consumption rates (Figure 3(e)), and no detectable expression of the mitochondria genome-encoded protein MTCOXII (Figure 3(f)). Exposure of HeLa ρ 0 cells to b-AP15 resulted in a dramatic abrogation of Nrf-2 and HO-1 induction (Figure 3(f)). Furthermore, and in contrast to the response observed in HeLa parental cells, exposure of HeLa ρ 0 cells to b-AP15 did not result in an increased GSSG/GSH ratio (Figure 3(g)). These findings are consistent with the notion that oxidative stress induction by b-AP15 is dependent on functional mitochondria. We considered the possibility of decreased levels of protein synthesis in ρ 0 cells, resulting in reduced proteotoxic stress and, as a consequence, lower oxidative stress. However, the levels of polyubiquitinated proteins induced by b-AP15 or bortezomib were comparable in HeLa parental and ρ 0 cells (Figure 3(f)).
Figure 3.
HeLa Rho0 (ρ 0) cells show a decreased oxidative stress response to b-AP15. (a) HeLa cells were exposed to 0.5% DMSO or 1 μM b-AP15 for 1, 3, and 6 hours, and extracts were prepared and subjected to immunoblotting using the indicated antibodies. All cultures received 0.5% DMSO. (b) HeLa cells were exposed to 0.5% DMSO or b-AP15 (0.25, 0.5, and 1.0 μM in 0.5% DMSO) for 6 h, and extracts were prepared and subjected to immunoblotting using the indicated antibodies. (c) HeLa cells were exposed to EtBr and uridine to generate mitochondrial DNA depleted cells (HeLa ρ 0). The ratio of mtDNA to nDNA was compared in HeLa parental and ρ 0 cells using RT-PCR (∗∗∗p < 0.001). (d) Electron micrographs of mitochondria in HeLa parental and ρ 0 cells. Scale bar = 0.5 μm. (e) Basal oxygen consumption rates (OCR) of HeLa parental and ρ 0 cells (n = 3; mean ± S.D.; ∗∗∗∗p < 0.0001). (f) HeLa ρ 0 cells were treated with 100 nM bortezomib (BZ) or 1μM b-AP15 for 5 h followed by western blot analysis for K48-linked polyubiquitin chains, Nrf-2, HO-1, MT-COXII, and β-actin. Note the impaired induction of Nrf-2 and HO-1 by UPS inhibitors in ρ 0 cells. (g) The ratio of GSSG/GSH was determined in parental HeLa and ρ 0 cells exposed to 1 μM b-AP15 or vehicle for 6 h (n = 3; mean ± S.D.; ∗∗p < 0.01). (h) HCT116 cells were exposed to 0.5% DMSO, 1 μM b-AP15, 5 μM CB113, 5 μM CB826, 5 μM CB916, and 1.5 μM auranofin (AUF) for 6 h, and extracts were prepared and subjected to immunoblotting using the indicated antibodies.
Our observations suggest an association between oxidative stress and mitochondria perturbation as a result of proteotoxic stress generated by b-AP15. One alternative mechanism of oxidative stress induction is inhibition of thioredoxin reductase (TrxR) activity, previously shown for b-AP15 [43]. To examine this possibility, we used a number of recently identified inhibitors of proteasome-associated DUBs that do not inhibit TrxR (Supplementary Fig. 2). We found that three different and chemically unrelated molecules that do not inhibit TrxR all induced the expression of the Nrf-2 target HO-1 (Figure 3(h)). Auranofin, a well-documented inhibitor of TrxR [59], induced HO-1 expression but did not induce accumulation of polyubiquitinated proteins (Figure 3(h)). These findings show that induction of the Nrf-2 target protein HO-1 by inhibitors of proteasome-associated DUBs does not require inhibition of TrxR.
3.4. Increased Levels of Proteotoxic Stress Result in Decreased Oxygen Consumption and Increased Expression of HO-1
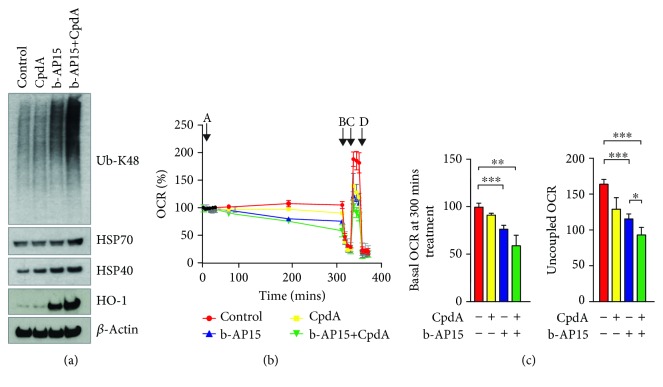
Further increases in the levels of proteotoxic stress are expected to result in increased mitochondrial damage and elevated oxidative stress. We used the CpdA, an inhibitor of Sec61-mediated anterograde protein translocation over the ER membrane [44], to test this prediction. Consistent with previous results [45], cotreatment of b-AP15 with CpdA induced strong accumulation of polyubiquitinated proteins and overexpression of HSP70 and HSP40 chaperones in HCT116 cells (Figure 4(a)). Cotreatment resulted in stronger reductions in oxygen consumption rates compared to treatments with b-AP15 or CpdA alone (Figure 4(b)). Cotreatment with b-AP15 and CpdA also resulted in stronger increases in HO-1 (Figure 4(a)), consistent with the notion of an association between proteotoxic stress and oxidative stress.
Figure 4.
Increased levels of proteotoxic stress are associated with decreased mitochondrial function and increased induction of HO-1. (a) HCT116 cells were exposed to 0.5% DMSO, 1 μM b-AP15, and 10 μM CpdA for 6 h, as indicated. Extracts were prepared and subjected to immunoblotting using the indicated antibodies. Note the increased levels of polyubiquitinated proteins, Hsp70, and HO-1 in cells exposed to b-AP15 and the ER translocation inhibitor CpdA. (b, c) HCT116 cells were treated with b-AP15 (1 μM) and/or CpdA (10 μM) for 5 hours and oxygen consumption rates were measured using a Seahorse XF analyzer (n = 3 in each group). A: DMSO or compounds; B: oligomycin; C: FCCP; D: antimycin and rotenone. (b) Measurement of OCR in real time after exposure to different compounds; (c) left: basal OCR after 300 min of treatment with compounds (mean ± S.D.; ∗∗∗p < 0.0001; n = 3); right: uncoupled OCR after addition of FCCP (mean ± S.D.; ∗∗∗p < 0.0001; ∗p < 0.05; n = 3).
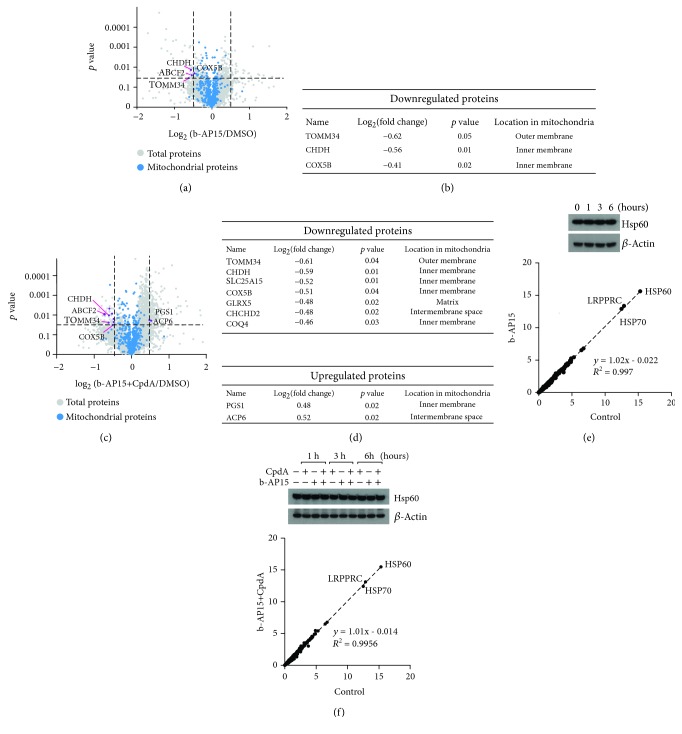
3.5. Alterations of the Mitochondrial Proteome Reveal Decreased Expression of COX5b
Damaged mitochondria in b-AP15-treated cells are not cleared by mitophagy and can be purified and analyzed by proteomics and other methods [42]. Three mitochondrial proteins were found to be significantly downregulated in mitochondrial preparations from b-AP15-treated cells: TOMM34 (translocase of outer mitochondrial membrane 34), CHDH (choline dehydrogenase), and COX5b (cytochrome c oxidase subunit 5B) (Figures 5(a) and 5(b)). Cotreatment with b-AP15 and CpdA resulted in a similar or larger decrease in the levels of these proteins and significant downregulation of some additional proteins (Figures 5(c) and 5(d)). COX5b is a component of the electron transport chain and the decrease of this protein may explain the decreases in mitochondrial oxidative phosphorylation observed in b-AP15-treated cells.
Figure 5.
Proteomic analysis of mitochondrial proteins. (a) Volcano plot showing log2(fold change) versus p values for proteins from isolated mitochondria prepared from HCT116 cells treated with DMSO or 1 μM b-AP15 for 6 h. (b) Top candidates with significant changes from (a) (p ≤ 0.05, log2 ≥ 0.4 or log2 < −0.4). (c) Volcano plot showing log2(fold change) versus p values for proteins from isolated mitochondria prepared from HCT116 cells treated with DMSO or 1 μM b-AP15 and 10 μM CpdA for 6 h. (d) Top candidates with significant changes from (c) (p ≤ 0.05, log2 ≥ 0.4 or log2 < −0.4). (e, f) Upper part: HCT116 cells were exposed to 0.5% DMSO, 1 μM b-AP15 in the presence or absence of 10 μM CpdA for 1, 3, and 6 h, as indicated. Extracts were prepared and subjected to immunoblotting using antibodies to Hsp60 and β-actin. (e, f) Lower part: mitochondria were purified from cells treated with 1 μM b-AP15 in the presence or absence of 10 μM CpdA and analyzed by shotgun proteomics. Data was normalized to control samples (treated with 0.5% DMSO).
Mitochondria possess a protein folding machinery (HSP60, HSP10, TRAP1, and mtHSP70) to respond to the misfolding stress inside of mitochondria (UPRmt) [60]. We addressed the question of whether b-AP15 affects mitochondrial protein homeostasis, leading to induction of HSP60. However, HSP60 expression was not affected by treatment with b-AP15 in the absence or presence of CpdA (Figures 5(e) and 5(f)).
4. Discussion
b-AP15 and similar compounds have been shown to induce apoptotic responses in tumor cells overexpressing BCL2 family proteins and cells defective in TP53 [24, 29]. Previous reports have demonstrated induction of both strong oxidative stress and proteotoxicity by b-AP15 [22, 27, 61] and also showed evidence of mitochondrial toxicity [42]. These findings led to the hypothesis that oxidative stress induction by this class of compounds is mechanistically linked to mitochondrial dysfunction. We here found weaker induction of the Nrf-2 target HO-1 and decreased elevation of GSSG/GSH ratios in ρ 0 cells exposed to b-AP15, consistent with a mitochondrial involvement in b-AP15-induced oxidative stress. We also found that increasing the level of proteotoxic stress by inhibiting anterograde ER translocation resulted in increased induction of expression of HO-1. These findings are consistent with the hypothesis of a mitochondrial origin of the increased levels of ROS observed in cells exposed to the DUB inhibitor b-AP15.
We and the others have shown that b-AP15 induces phosphorylation of JNK and that inhibition of JNK signaling decreases the apoptotic response [22, 27]. Both JNK signaling and apoptosis are decreased by antioxidant treatment [22]. These findings point to a perturbation of the intracellular redox state being involved in induction of apoptosis. The levels of malondialdehyde, a product of lipid peroxidation of polyunsaturated fatty acids [56], did not increase during b-AP15 treatment and available data suggest that activation of antioxidant systems prevents direct oxidative damage. We here found significant increases in glucose 6-phosphate dehydrogenase (G6PD) activity, leading to a larger potential of NADPH generation [62]. The lack of detectable lipid peroxidation can be argued to mean that b-AP15 does not induce “oxidative stress” by a more stringent definition but merely induces redox imbalances. Although these imbalances are sufficient to induce Nrf-2 and phosphorylation of JNK, they appear to be contained by antioxidant defenses. It should be pointed out, however, that the lack of detectable increases in lipid peroxidation does not necessarily mean that oxidative damage to macromolecules does not occur in specific cellular compartments. For a discussion of redox perturbations, oxidative stress, and oxidative damage, see [63].
We previously presented evidence favoring that the mitochondrial damage that occurs during exposure to b-AP15 is due to the accumulation of misfolded proteins on the outer mitochondrial membrane [42]. This observation did not explain the decrease in oxidative phosphorylation that occurs during drug treatment. We here show downregulation of the COX5b protein, an essential component of cytochrome c oxidase [64]. Cytochrome c oxidase is a key enzyme in the overall regulation of cellular energy production in eukaryotes [65]. Decreases in COX5b have been associated with mitochondrial dysfunction in various conditions [66], and upregulation of COX5b has been observed in energy-demanding cell types and healthy tissues. It has also been demonstrated that downregulation of COX5b by siRNA increases mitochondrial ROS generation [67]. The levels of the yeast homologue of COX5b, COX IV-1, have been shown to be posttranscriptionally regulated by the cardiolipin content of the mitochondria [68], and COX5b has also been reported to be regulated by carbon source and oxygen [64, 69].
TOMM34 (34 kDa translocase of the outer mitochondrial membrane) was originally identified as a component of the mitochondrial import machinery for nucleus-encoded mitochondrial proteins and has been reported to form a complex with both Hsp70 and Hsp90 as a cytosolic scaffolding cochaperone [70–72]. We observed decreased levels of TOMM34 in mitochondrial preparations in parallel with elevated levels of HSP70B′. It is possible that an elevated demand of TOMM34 in assisting Hsp70/Hsp90 in different folding processes in the cytosol limits the availability of TOMM34 proteins on the outer mitochondrial membrane.
Despite its strong cytotoxicity to tumor cells, b-AP15 and similar compounds show limited activity against normal cells [3, 6] and its cytotoxicity is likely to be dependent on the elevated rate of protein turnover in tumor cells. The results presented here suggest that oxidative stress is coupled to proteotoxic stress, leading to an enhancement of the effects on proteasome inhibition. b-AP15 has shown activity in a number of tumor models, including multiple myeloma [12, 13], Ewing's carcinoma [14], Waldenström's macroglobulinaemia [15], melanoma [9], and colon cancer [3]. The in vivo efficacy of b-AP15 is limited by the poor solubility of the compound, and efforts are ongoing to improve the pharmacological properties of this class of molecules. If these efforts are crowned by success, inhibitors of proteasome-associated DUBs could be important drugs in an increasing arsenal of therapeutic options for cancer.
Acknowledgments
We are grateful to Kjell Hultenby for electron microscopy analysis. The study was supported by the Swedish Cancer Society, Radiumhemmets Forskningsfonder, Vetenskapsrådet, Barncancerfonden, and Knut och Alice Wallenbergs Stiftelse.
Data Availability
The data used to support the findings of this study are included within the article.
Conflicts of Interest
SL is a consultant of Vivolux AB. No other potential conflicts of interest were disclosed.
Authors' Contributions
X.Z. and S.L. were responsible for the conception and design. X.Z., B.E., and A.S. were responsible for the acquisition of data. X.Z., B.E., and A.S. were responsible for the analysis and interpretation of data. The manuscript was written by X.Z. and S.L. and reviewed/revised by X.Z., B.E., A.S., P.D., R.Z., and S.L.
Supplementary Materials
Supplementary Fig. 1: induction of caspase cleavage activity by b-AP15 in HeLa cells. HeLa cells were exposed to 1 μM b-AP15 for 24 hours in the presence of absence of BIK siRNA or scrambled control siRNA as indicated. Exponentially growing HeLa cells were seeded in 100 mm dishes at 106 cells per plate and grown for 24 h and then transfected with BIK siRNA (Qiagen) at a final concentration of 10 nM using Lipofectamine RNAi MAX (Invitrogen). Cells were incubated for another 72 h and then treated with DMSO or b-AP15 for another 18 h. Apoptosis was determined by the measurement of the accumulation of caspase-cleaved apoptosis product in cells and culture medium using the M30 Apoptosense ELISA assay (VLVbio, Stockholm). The assay measures caspase-cleaved keratin 18 (K18-Asp396) (Bivén et al., Apoptosis 8 (2003) 263). BIK is a BH3-only protein that has been described to be associated with oxidative stress-induced apoptosis (Bodet et al., Br J Cancer 12 (2010) 1808). Means + S.D. are shown (n = 3). Statistical significance was determined using Student's t-test. Supplementary Fig. 2: oxidative stress induced by b-AP15 is not due to the inhibition of thioredoxin reductase (TrxR) activity. CB113 (ChemBridge ID 6943113), CB826 (ChemBridge ID 6556826), or CB916 (ChemBridge ID 6237916) was added to thioredoxin reductase enzyme assay mixes at 20 μM in Tris-EDTA and incubated for 5 hours. Reactions contained 50 nM rTrxR (21 U/mg), 200 μM NADPH, and 1 mg/mL BSA. Reactions were started by the addition of 2.5 mM of DTNB and absorbance was read at 412 nms. The TrxR inhibitor auranofin and Juglone were used as positive controls. Note, none of these three proteasome inhibitors targeted the TrxR activity at the concentration of 20 μM.
References
- 1.Chiti F., Dobson C. M. Protein misfolding, functional amyloid, and human disease. Annual Review of Biochemistry. 2006;75(1):333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 2.Rock K. L., Gramm C., Rothstein L., et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78(5):761–771. doi: 10.1016/S0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 3.Ciechanover A. The ubiquitin–proteasome pathway: on protein death and cell life. The EMBO Journal. 1998;17(24):7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suraweera A., Münch C., Hanssum A., Bertolotti A. Failure of amino acid homeostasis causes cell death following proteasome inhibition. Molecular Cell. 2012;48(2):242–253. doi: 10.1016/j.molcel.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanaka K. The proteasome: overview of structure and functions. Proceedings of the Japan Academy, Series B. 2009;85(1):12–36. doi: 10.2183/pjab.85.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciechanover A. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Best Practice & Research Clinical Haematology. 2017;30(4):341–355. doi: 10.1016/j.beha.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Varshavsky A. Regulated protein degradation. Trends in Biochemical Sciences. 2005;30(6):283–286. doi: 10.1016/j.tibs.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Rechsteiner M., Hoffman L., Dubiel W. The multicatalytic and 26 S proteases. Journal of Biological Chemistry. 1993;268(9):6065–6068. [PubMed] [Google Scholar]
- 9.Hershko A., Ciechanover A. The ubiquitin system. Annual Review of Biochemistry. 1998;67(1):425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 10.DeBerardinis R. J., Lum J. J., Hatzivassiliou G., Thompson C. B. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metabolism. 2008;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Hsu P. P., Sabatini D. M. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134(5):703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 12.Fribley A., Zeng Q., Wang C. Y. Proteasome inhibitor PS-341 induces apoptosis through induction of endoplasmic reticulum stress-reactive oxygen species in head and neck squamous cell carcinoma cells. Molecular and Cellular Biology. 2004;24(22):9695–9704. doi: 10.1128/MCB.24.22.9695-9704.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ling Y. H., Liebes L., Zou Y., Perez-Soler R. Reactive oxygen species generation and mitochondrial dysfunction in the apoptotic response to bortezomib, a novel proteasome inhibitor, in human H460 non-small cell lung cancer cells. Journal of Biological Chemistry. 2003;278(36):33714–33723. doi: 10.1074/jbc.M302559200. [DOI] [PubMed] [Google Scholar]
- 14.Paniagua Soriano G., de Bruin G., Overkleeft H. S., Florea B. I. Toward understanding induction of oxidative stress and apoptosis by proteasome inhibitors. Antioxidants & Redox Signaling. 2014;21(17):2419–2443. doi: 10.1089/ars.2013.5794. [DOI] [PubMed] [Google Scholar]
- 15.Attar E. C., Amrein P. C., Fraser J. W., et al. Phase I dose escalation study of bortezomib in combination with lenalidomide in patients with myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML) Leukemia Research. 2013;37(9):1016–1020. doi: 10.1016/j.leukres.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Varga C., Laubach J., Hideshima T., Chauhan D., Anderson K. C., Richardson P. G. Novel targeted agents in the treatment of multiple myeloma. Hematology/Oncology Clinics of North America. 2014;28(5):903–925. doi: 10.1016/j.hoc.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Niewerth D., Jansen G., Assaraf Y. G., Zweegman S., Kaspers G. J. L., Cloos J. Molecular basis of resistance to proteasome inhibitors in hematological malignancies. Drug Resistance Updates. 2015;18:18–35. doi: 10.1016/j.drup.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Gorrini C., Harris I. S., Mak T. W. Modulation of oxidative stress as an anticancer strategy. Nature Reviews Drug Discovery. 2013;12(12):931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- 19.Matsuzawa A., Ichijo H. Redox control of cell fate by MAP kinase: physiological roles of ASK1-MAP kinase pathway in stress signaling. Biochimica et Biophysica Acta (BBA) - General Subjects. 2008;1780(11):1325–1336. doi: 10.1016/j.bbagen.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 20.Holmstrom K. M., Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nature Reviews Molecular Cell Biology. 2014;15(6):411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 21.Zeeshan H., Lee G., Kim H. R., Chae H. J. Endoplasmic reticulum stress and associated ROS. International Journal of Molecular Sciences. 2016;17(3):p. 327. doi: 10.3390/ijms17030327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brnjic S., Mazurkiewicz M., Fryknäs M., et al. Induction of tumor cell apoptosis by a proteasome deubiquitinase inhibitor is associated with oxidative stress. Antioxidants & Redox Signaling. 2014;21(17):2271–2285. doi: 10.1089/ars.2013.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung T., Hohn A., Grune T. The proteasome and the degradation of oxidized proteins: part II – protein oxidation and proteasomal degradation. Redox Biology. 2014;2:99–104. doi: 10.1016/j.redox.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D'Arcy P., Brnjic S., Olofsson M. H., et al. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nature Medicine. 2011;17(12):1636–1640. doi: 10.1038/nm.2536. [DOI] [PubMed] [Google Scholar]
- 25.Coughlin K., Anchoori R., Iizuka Y., et al. Small-molecule RA-9 inhibits proteasome-associated DUBs and ovarian cancer in vitro and in vivo via exacerbating unfolded protein responses. Clinical Cancer Research. 2014;20(12):3174–3186. doi: 10.1158/1078-0432.CCR-13-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X., Mazurkiewicz M., Hillert E. K., et al. The proteasome deubiquitinase inhibitor VLX1570 shows selectivity for ubiquitin-specific protease-14 and induces apoptosis of multiple myeloma cells. Scientific Reports. 2016;6(1, article 26979) doi: 10.1038/srep26979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Didier R., Mallavialle A., Ben Jouira R., et al. Targeting the proteasome-associated deubiquitinating enzyme USP14 impairs melanoma cell survival and overcomes resistance to MAPK-targeting therapies. Molecular Cancer Therapeutics. 2018;17(7):1416–1429. doi: 10.1158/1535-7163.MCT-17-0919. [DOI] [PubMed] [Google Scholar]
- 28.Erdal H., Berndtsson M., Castro J., Brunk U., Shoshan M. C., Linder S. Induction of lysosomal membrane permeabilization by compounds that activate p53-independent apoptosis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(1):192–197. doi: 10.1073/pnas.0408592102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aleo E., Henderson C. J., Fontanini A., Solazzo B., Brancolini C. Identification of new compounds that trigger apoptosome-independent caspase activation and apoptosis. Cancer Research. 2006;66(18):9235–9244. doi: 10.1158/0008-5472.CAN-06-0702. [DOI] [PubMed] [Google Scholar]
- 30.Adams B. K., Cai J., Armstrong J., et al. EF24, a novel synthetic curcumin analog, induces apoptosis in cancer cells via a redox-dependent mechanism. Anti-Cancer Drugs. 2005;16(3):263–275. doi: 10.1097/00001813-200503000-00005. [DOI] [PubMed] [Google Scholar]
- 31.Bazzaro M., Anchoori R. K., Mudiam M. K. R., et al. α,β-Unsaturated carbonyl system of chalcone-based derivatives is responsible for broad inhibition of proteasomal activity and preferential killing of human papilloma virus (HPV) positive cervical cancer cells. Journal of Medicinal Chemistry. 2011;54(2):449–456. doi: 10.1021/jm100589p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anchoori R. K., Karanam B., Peng S., et al. A bis-benzylidine piperidone targeting proteasome ubiquitin receptor RPN13/ADRM1 as a therapy for cancer. Cancer Cell. 2013;24(6):791–805. doi: 10.1016/j.ccr.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian Z., D'Arcy P., Wang X., et al. A novel small molecule inhibitor of deubiquitylating enzyme USP14 and UCHL5 induces apoptosis in multiple myeloma and overcomes bortezomib resistance. Blood. 2014;123(5):706–716. doi: 10.1182/blood-2013-05-500033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shukla N., Somwar R., Smith R. S., et al. Proteasome addiction defined in Ewing sarcoma is effectively targeted by a novel class of 19S proteasome inhibitors. Cancer Research. 2016;76(15):4525–4534. doi: 10.1158/0008-5472.CAN-16-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chitta K., Paulus A., Akhtar S., et al. Targeted inhibition of the deubiquitinating enzymes, USP14 and UCHL5, induces proteotoxic stress and apoptosis in Waldenström macroglobulinaemia tumour cells. British Journal of Haematology. 2015;169(3):377–390. doi: 10.1111/bjh.13304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kropp K. N., Maurer S., Rothfelder K., et al. The novel deubiquitinase inhibitor b-AP15 induces direct and NK cell-mediated antitumor effects in human mantle cell lymphoma. Cancer Immunology, Immunotherapy. 2018;67(6):935–947. doi: 10.1007/s00262-018-2151-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding Y., Chen X., Wang B., Yu B., Ge J. Deubiquitinase inhibitor b-AP15 activates endoplasmic reticulum (ER) stress and inhibits Wnt/Notch1 signaling pathway leading to the reduction of cell survival in hepatocellular carcinoma cells. European Journal of Pharmacology. 2018;825:10–18. doi: 10.1016/j.ejphar.2018.02.020. [DOI] [PubMed] [Google Scholar]
- 38.Liu H., Liang Y., Wang L., et al. In vivo and in vitro suppression of hepatocellular carcinoma by EF24, a curcumin analog. PLoS One. 2012;7(10, article e48075) doi: 10.1371/journal.pone.0048075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai J., Xia X., Liao Y., et al. A novel deubiquitinase inhibitor b-AP15 triggers apoptosis in both androgen receptor-dependent and -independent prostate cancers. Oncotarget. 2017;8(38):63232–63246. doi: 10.18632/oncotarget.18774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vogel R. I., Coughlin K., Scotti A., et al. Simultaneous inhibition of deubiquitinating enzymes (DUBs) and autophagy synergistically kills breast cancer cells. Oncotarget. 2015;6(6):4159–4170. doi: 10.18632/oncotarget.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cersosimo U., Sgorbissa A., Foti C., et al. Synthesis, characterization, and optimization for in vivo delivery of a nonselective isopeptidase inhibitor as new antineoplastic agent. Journal of Medicinal Chemistry. 2015;58(4):1691–1704. doi: 10.1021/jm501336h. [DOI] [PubMed] [Google Scholar]
- 42.Zhang X., Pellegrini P., Saei A. A., et al. The deubiquitinase inhibitor b-AP15 induces strong proteotoxic stress and mitochondrial damage. Biochemical Pharmacology. 2018;156:291–301. doi: 10.1016/j.bcp.2018.08.039. [DOI] [PubMed] [Google Scholar]
- 43.Wang X., Stafford W., Mazurkiewicz M., et al. The 19S deubiquitinase inhibitor b-AP15 is enriched in cells and elicits rapid commitment to cell death. Molecular Pharmacology. 2014;85(6):932–945. doi: 10.1124/mol.113.091322. [DOI] [PubMed] [Google Scholar]
- 44.Besemer J., Harant H., Wang S., et al. Selective inhibition of cotranslational translocation of vascular cell adhesion molecule 1. Nature. 2005;436(7048):290–293. doi: 10.1038/nature03670. [DOI] [PubMed] [Google Scholar]
- 45.Sun C., Roboti P., Puumalainen M. R., et al. Elevation of proteasomal substrate levels sensitizes cells to apoptosis induced by inhibition of proteasomal deubiquitinases. PLoS One. 2014;9(10, article e108839) doi: 10.1371/journal.pone.0108839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Penna A., Cahalan M. Western blotting using the Invitrogen NuPage Novex Bis Tris minigels. Journal of Visualized Experiments. 2007;(7, article e264) doi: 10.3791/264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paglia D. E., Valentine W. N. Studies on the quantitative and qualitative characterization of erythrocyte glutathione peroxidase. The Journal of Laboratory and Clinical Medicine. 1967;70(1):158–169. [PubMed] [Google Scholar]
- 48.Beutler E. Red Cell Metabolism, a Manual of Biochemical Methods. New York, NY, USA: Grune and Stratton; 1984. [Google Scholar]
- 49.Conti M., Morand P. C., Levillain P., Lemonnier A. Improved fluorometric determination of malonaldehyde. Clinical Chemistry. 1991;37(7):1273–1275. [PubMed] [Google Scholar]
- 50.Hashiguchi K., Zhang-Akiyama Q. M. Mitochondrial DNA. Vol. 554. Humana Press; 2009. Establishment of human cell lines lacking mitochondrial DNA; pp. 383–391. (Methods in Molecular Biology). [DOI] [PubMed] [Google Scholar]
- 51.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnology. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 52.Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. Journal of Proteome Research. 2011;10(4):1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 53.Pagliarini D. J., Calvo S. E., Chang B., et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134(1):112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nature Reviews Molecular Cell Biology. 2013;14(10):630–642. doi: 10.1038/nrm3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han J., Back S. H., Hur J., et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nature Cell Biology. 2013;15(5):481–490. doi: 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pryor W. A., Stanley J. P. Suggested mechanism for the production of malonaldehyde during the autoxidation of polyunsaturated fatty acids. Nonenzymic production of prostaglandin endoperoxides during autoxidation. The Journal of Organic Chemistry. 1975;40(24):3615–3617. doi: 10.1021/jo00912a038. [DOI] [PubMed] [Google Scholar]
- 57.Zhang X., Fryknäs M., Hernlund E., et al. Induction of mitochondrial dysfunction as a strategy for targeting tumour cells in metabolically compromised microenvironments. Nature Communications. 2014;5(1):p. 3295. doi: 10.1038/ncomms4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schauen M., Spitkovsky D., Schubert J., Fischer J. H., Hayashi J. I., Wiesner R. J. Respiratory chain deficiency slows down cell-cycle progression via reduced ROS generation and is associated with a reduction of p21CIP1/WAF1 . Journal of Cellular Physiology. 2006;209(1):103–112. doi: 10.1002/jcp.20711. [DOI] [PubMed] [Google Scholar]
- 59.Gandin V., Fernandes A. P., Rigobello M. P., et al. Cancer cell death induced by phosphine gold(I) compounds targeting thioredoxin reductase. Biochemical Pharmacology. 2010;79(2):90–101. doi: 10.1016/j.bcp.2009.07.023. [DOI] [PubMed] [Google Scholar]
- 60.Bulteau A. L., Mena N. P., Auchère F., et al. Dysfunction of mitochondrial Lon protease and identification of oxidized protein in mouse brain following exposure to MPTP: implications for Parkinson disease. Free Radical Biology & Medicine. 2017;108:236–246. doi: 10.1016/j.freeradbiomed.2017.03.036. [DOI] [PubMed] [Google Scholar]
- 61.Hillert E. K., Brnjic S., Zhang X., et al. Proteasome inhibitor b-AP15 induces enhanced proteotoxicity by inhibiting cytoprotective aggresome formation. Cancer Letters. 2019;448:70–83. doi: 10.1016/j.canlet.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 62.Stanton R. C. Glucose-6-phosphate dehydrogenase, NADPH, and cell survival. IUBMB Life. 2012;64(5):362–369. doi: 10.1002/iub.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Burton G. J., Jauniaux E. Oxidative stress. Best Practice & Research Clinical Obstetrics & Gynaecology. 2011;25(3):287–299. doi: 10.1016/j.bpobgyn.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trueblood C. E., Wright R. M., Poyton R. O. Differential regulation of the two genes encoding Saccharomyces cerevisiae cytochrome c oxidase subunit V by heme and the HAP2 and REO1 genes. Molecular and Cellular Biology. 1988;8(10):4537–4540. doi: 10.1128/MCB.8.10.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Poyton R. O., Trueblood C. E., Wright R. M., Farrell L. E. Expression and function of cytochrome c oxidase subunit isologues. Modulators of cellular energy production? Annals of the New York Academy of Sciences. 1988;550:289–307. doi: 10.1111/j.1749-6632.1988.tb35344.x. [DOI] [PubMed] [Google Scholar]
- 66.Hinkelbein J., Böhm L., Braunecker S., Adler C., de Robertis E., Cirillo F. Decreased tissue COX5B expression and mitochondrial dysfunction during sepsis-induced kidney injury in rats. Oxidative Medicine and Cellular Longevity. 2017;2017:24. doi: 10.1155/2017/8498510.8498510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Campian J., Gao X., Qian M., Eaton J. W. Cytochrome c oxidase activity and oxygen tolerance. Journal of Biological Chemistry. 2007;282(17):12430–12438. doi: 10.1074/jbc.M604547200. [DOI] [PubMed] [Google Scholar]
- 68.Su X., Dowhan W. Translational regulation of nuclear gene COX4 expression by mitochondrial content of phosphatidylglycerol and cardiolipin in Saccharomyces cerevisiae . Molecular and Cellular Biology. 2006;26(3):743–753. doi: 10.1128/MCB.26.3.743-753.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hodge M. R., Kim G., Singh K., Cumsky M. G. Inverse regulation of the yeast COX5 genes by oxygen and heme. Molecular and Cellular Biology. 1989;9(5):1958–1964. doi: 10.1128/MCB.9.5.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Faou P., Hoogenraad N. J. Tom34: a cytosolic cochaperone of the Hsp90/Hsp70 protein complex involved in mitochondrial protein import. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2012;1823(2):348–357. doi: 10.1016/j.bbamcr.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 71.Chewawiwat N., Yano M., Terada K., Hoogenraad N. J., Mori M. Characterization of the novel mitochondrial protein import component, Tom34, in mammalian cells. The Journal of Biochemistry. 1999;125(4):721–727. doi: 10.1093/oxfordjournals.jbchem.a022342. [DOI] [PubMed] [Google Scholar]
- 72.Trcka F., Durech M., Man P., Hernychova L., Muller P., Vojtesek B. The assembly and intermolecular properties of the Hsp70-Tomm34-Hsp90 molecular chaperone complex. Journal of Biological Chemistry. 2014;289(14):9887–9901. doi: 10.1074/jbc.M113.526046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1: induction of caspase cleavage activity by b-AP15 in HeLa cells. HeLa cells were exposed to 1 μM b-AP15 for 24 hours in the presence of absence of BIK siRNA or scrambled control siRNA as indicated. Exponentially growing HeLa cells were seeded in 100 mm dishes at 106 cells per plate and grown for 24 h and then transfected with BIK siRNA (Qiagen) at a final concentration of 10 nM using Lipofectamine RNAi MAX (Invitrogen). Cells were incubated for another 72 h and then treated with DMSO or b-AP15 for another 18 h. Apoptosis was determined by the measurement of the accumulation of caspase-cleaved apoptosis product in cells and culture medium using the M30 Apoptosense ELISA assay (VLVbio, Stockholm). The assay measures caspase-cleaved keratin 18 (K18-Asp396) (Bivén et al., Apoptosis 8 (2003) 263). BIK is a BH3-only protein that has been described to be associated with oxidative stress-induced apoptosis (Bodet et al., Br J Cancer 12 (2010) 1808). Means + S.D. are shown (n = 3). Statistical significance was determined using Student's t-test. Supplementary Fig. 2: oxidative stress induced by b-AP15 is not due to the inhibition of thioredoxin reductase (TrxR) activity. CB113 (ChemBridge ID 6943113), CB826 (ChemBridge ID 6556826), or CB916 (ChemBridge ID 6237916) was added to thioredoxin reductase enzyme assay mixes at 20 μM in Tris-EDTA and incubated for 5 hours. Reactions contained 50 nM rTrxR (21 U/mg), 200 μM NADPH, and 1 mg/mL BSA. Reactions were started by the addition of 2.5 mM of DTNB and absorbance was read at 412 nms. The TrxR inhibitor auranofin and Juglone were used as positive controls. Note, none of these three proteasome inhibitors targeted the TrxR activity at the concentration of 20 μM.
Data Availability Statement
The data used to support the findings of this study are included within the article.