Abstract
Daratumumab is a CD38‐targeted human monoclonal antibody with direct anti‐myeloma cell mechanisms of action. Flow cytometry in relapsed and/or refractory multiple myeloma (RRMM) patients treated with daratumumab revealed cytotoxic T‐cell expansion and reduction of immune‐suppressive populations, suggesting immune modulation as an additional mechanism of action. Here, we performed an in‐depth analysis of the effects of daratumumab on immune‐cell subpopulations using high‐dimensional mass cytometry. Whole‐blood and bone‐marrow baseline and on‐treatment samples from RRMM patients who participated in daratumumab monotherapy studies (SIRIUS and GEN501) were evaluated with high‐throughput immunophenotyping. In daratumumab‐treated patients, the intensity of CD38 marker expression decreased on many immune cells in SIRIUS whole‐blood samples. Natural killer (NK) cells were depleted with daratumumab, with remaining NK cells showing increased CD69 and CD127, decreased CD45RA, and trends for increased CD25, CD27, and CD137 and decreased granzyme B. Immune‐suppressive population depletion paralleled previous findings, and a newly observed reduction in CD38+ basophils was seen in patients who received monotherapy. After 2 months of daratumumab, the T‐cell population in whole‐blood samples from responders shifted to a CD8 prevalence with higher granzyme B positivity (P = 0.017), suggesting increased killing capacity and supporting monotherapy‐induced CD8+ T‐cell activation. High‐throughput cytometry immune profiling confirms and builds upon previous flow cytometry data, including comparable CD38 marker intensity on plasma cells, NK cells, monocytes, and B/T cells. Interestingly, a shift toward cytolytic granzyme B+ T cells was also observed and supports adaptive responses in patients that may contribute to depth of response. © 2018 The Authors. Cytometry Part A published by Wiley Periodicals, Inc. on behalf of International Society for Advancement of Cytometry.
Keywords: CyTOF, immune profiling, cytometry by time of flight
Short abstract

Introduction
Daratumumab is a human monoclonal antibody that targets CD38, a type II transmembrane glycoprotein that is highly and ubiquitously expressed on myeloma cells 1, 2, 3, 4. In clinical studies, daratumumab has shown impressive anti‐myeloma activity in patients with relapsed and/or refractory multiple myeloma (RRMM), both when administered as monotherapy and when given in combination with standard‐of‐care agents 5, 6, 7, 8, 9, 10. In Part 2 of the GEN501 monotherapy study, at a median follow‐up of 10.2 months, patients in the 16 mg/kg daratumumab cohort had an overall clinical response rate of 36%. The median duration of response was not reached, and progression‐free survival was 5.6 months 5. Additionally, in the phase 2 SIRIUS monotherapy study, patients who received 16 mg/kg daratumumab had an overall response rate of 29.2% and median progression‐free survival of 3.7 months at a median follow‐up of 9.3 months 6. These studies led to the first approval of daratumumab by the US Food and Drug Administration for patients with multiple myeloma who have received at least three prior lines of therapy, including a proteasome inhibitor and an immunomodulatory agent, or who are double refractory to a proteasome inhibitor and an immunomodulatory agent 11.
Daratumumab induces deep and durable clinical responses in patients with multiple myeloma through a multifaceted mechanism of action. Direct on‐tumor actions include complement‐dependent cytotoxicity, antibody‐dependent cellular cytotoxicity, antibody‐dependent cellular phagocytosis, and induction of apoptosis via crosslinking 1, 12, 13, 14. Recently, RRMM patient samples were analyzed by flow cytometry for assessment of immune population count changes, in addition to evaluation of the functionality of CD38+ immune‐cell populations, T‐cell activation, and T‐cell receptor clonality. The data revealed a previously unknown immune‐modulatory mechanism of action of daratumumab that may contribute to prolonged and deep clinical responses 15. In that study, daratumumab was found to exert immune‐modulatory effects via T‐cell induction/expansion, T‐cell activity enhancement, and reduction of immune‐suppressive cell populations. These populations included CD38+ myeloid‐derived suppressor cells, CD38+ regulatory T cells (Tregs), and CD38+ regulatory B cells.
Further characterization of daratumumab's immune‐modulatory mechanism of action is desirable. Technical limitations of flow cytometry make broad surveys of the effects of daratumumab on additional subtypes of immune‐cell populations unlikely with that technology alone 16, 17. We therefore conducted a study using cytometry by time of flight (CyTOF®) to corroborate and build upon the findings of flow cytometry 16, 18. Because CyTOF® enables the analysis of 35+ markers at the single‐cell level without the need for compensation 18, it was used to deeply characterize the effects of daratumumab on immune cell subset composition and provide insights on functionality from a single‐sample collection, moving well beyond the limitations of traditional flow cytometry. This approach was selected to increase robustness as an alternative to extrapolating conclusions from separate flow cytometry acquisitions. The objective of this study was to identify novel changes in immune‐cell phenotypes that correlate with the efficacy and depth of response observed during treatment with daratumumab in patients with RRMM.
Methods
CyTOF® Sample Sources and Staining
Full details on the study designs and patient populations included in GEN501 5 and SIRIUS 6 studies have been previously published. GEN501 Part 2 (phase 2) and SIRIUS (phase 2) were single‐arm studies of daratumumab monotherapy at 16 mg/kg in patients with heavily pre‐treated RRMM. In SIRIUS, whole‐blood (WB) samples were obtained from 32 patients at baseline and after 2 months on study. In GEN501, 5 patients provided WB and/or bone‐marrow (BM) samples at baseline, after 2 months on study, and at the end of treatment. Samples were stained using the antibody panel in Supporting Information Table S1 and acquired on a CyTOF® C5 system. Clinical response data of the GEN501 Part 2 5 and SIRIUS 6 studies were from patients who received daratumumab 16 mg/kg at the clinical cut‐off date of January 9, 2015. Ethics committees or institutional review boards at each study site approved the study protocols and the study analysis plans 5, 6. The studies were conducted in accordance with the principles of the Declaration of Helsinki and the International Conference on Harmonisation Good Clinical Practice guidelines. All patients provided written informed consent.
Data Pre‐processing and Quality Control
Following data acquisition, channel intensity was normalized using calibration beads. Data were gated using Cytobank® (www.cytobank.org; Cytobank, Inc., Santa Clara, CA, USA) and further processed using a set of custom scripts based on the flowCore package. Measured intensities for each channel were transformed using the arcsinh function with a cofactor of 5. Quality control assessment of 9 WB control samples from 9 different batches was performed using the earth mover's distance algorithm 19 and marker enrichment modeling 20. These analyses revealed no evidence of technical sources of variation that needed batch correction (Fig. 1A,B).
Figure 1.
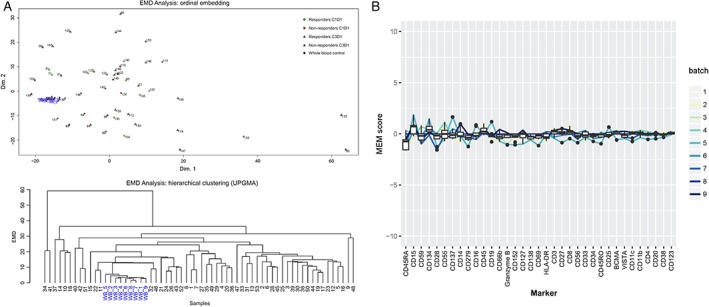
Quality control assessment using whole‐blood controls (displayed in blue) by (A) EMD analysis (ordinal embedding [43; top] and hierarchical clustering [UPGMA; bottom]) and (B) MEM revealed no evidence of technical sources of variation that required batch correction. EMD, earth mover's distance; UPGMA, unweighted pair group method with arithmetic means; MEM, marker enrichment modeling; C, cycle; D, day.
Identification of Immune‐Cell Populations
After identification of live single‐cell events, similar cellular events were grouped together using the spanning‐tree progression analysis of density normalized events (SPADE) algorithm 18, 21. To ensure that every cell population was represented, the analysis included only samples in which at least 10,000 events in the lymphocyte/monocyte population had been recorded.
Differential Analysis of Immune‐Cell Population Counts and Marker Intensities
Sample group comparisons per marker were performed based on the sample annotation for different time points (before and after treatment) and response groups (responder and non‐responder). For each sample, the mean marker intensity (MMI) was calculated for each cluster in the SPADE tree. A 2‐sided 2‐sample t‐test allowing for unequal group variance was then performed to compare the group (time/response) MMI means. This analysis yielded raw P values for all cluster/marker combinations. In case of repeated observations over time for the same subject, response group means were calculated for the fold changes (MMI difference between two time points).
Bin Analysis
Changes in the distribution of signal intensity were quantified using the following procedure: for a given population and channel of interest, the centiles of the single‐cell data were computed across all conditions and the values were used to define bins. Overlapping bins were merged when more than 1% of cells had similar signal intensity. The percentage of cells in each bin was then computed across conditions. To compare conditions, the fraction of cells in each bin for the condition of interest was compared with the total number of cells in the bin. The effect of treatment over time was visualized by plotting the cell fraction to the lower limit of intensity of each bin.
Visualization
Results of the statistical analysis of differences between conditions were visualized with a SPADE‐blend tree, where each SPADE cluster is colored with a color gradient that reflects the dimensions of raw P values to fold changes computed between marker intensities or to differences in cell population fractions observed under different conditions. To visualize trends at the single‐cell level, we used Radviz, a method in which cells are projected in two dimensions in a manner that preserves the original dimensions and enables rapid interpretation of changes within a population 22. Treatment effects on specific cell subsets were visualized using relevant channels representing different phenotypic and translational markers. Radviz shifts were used to guide manual gating and downstream statistical analysis. To explore the individual contributions of each channel and to assess the homogeneity of the response across a given cell population, we used the fan charts developed by the Bank of England 23. Briefly, we computed the centiles for each marker and each condition, and visualized the corresponding values as stacked area plots colored according to the centile to which they corresponded. The color palette is centered on the 50th centile, and color intensity decreases symmetrically with higher and lower centiles.
The Supporting Information Appendix presents the detailed methodologies used for sample staining, data pre‐processing and quality control, differential analysis of population fractions, bootstrapping P values (P*), node‐level imputation, and data visualization.
Results
CD38 Expression and Modulation in Lymphocyte Populations and Natural Killer (NK)‐Cell Reduction
To further the understanding of the immune‐modulatory activity of the CD38‐targeting antibody daratumumab observed with flow cytometry 15, WB and BM samples from RRMM patients who received single‐agent daratumumab in the SIRIUS (n = 32) and GEN501 (n = 5) studies were analyzed via CyTOF®. This platform allows for in‐depth functional protein‐level characterization at single‐cell resolution and enables multidimensional, quantitative analysis of single‐cell–level differences between patient subgroups or effects of therapeutics on cell populations and marker expression.
In alignment with earlier findings from BM samples profiled with flow cytometry 15, CyTOF® analysis showed that CD38 was expressed across various immune‐cell subpopulations, including plasma cells, NK cells, monocytes, B cells, and T cells as shown in baseline BM samples of RRMM patients (Fig. 2). Also, confirming previously reported flow cytometry findings of peripheral blood mononuclear cells from daratumumab single agent–treated RRMM patients 24, CD38 marker intensity decreased over 2 months of treatment in WB samples from SIRIUS (from Cycle 1, Day 1 to Cycle 3, Day 1; Fig. 3A). The most prominent decrease was observed for NK cells, B cells, and naïve CD4+ T cells (P* = 0.0099). Along with reduced CD38 expression, the NK‐cell population was significantly depleted (P* = 0.0099) in WB samples from SIRIUS (from Cycle 1, Day 1 to Cycle 3, Day 1; Fig. 3B, Supporting Information Fig. S1). NK cells were depleted in WB (data not shown) and BM samples from GEN501 but the NK‐cell reductions were not statistically significant from Cycle 1, Day 1 to end of treatment (Fig. 3C, Supporting Information Fig. S1). Decreases in immune‐suppressive cells, such as CD38+ Tregs, are described in greater detail below. Collectively, these findings align with flow cytometry observations performed in these clinical studies 25, 26.
Figure 2.
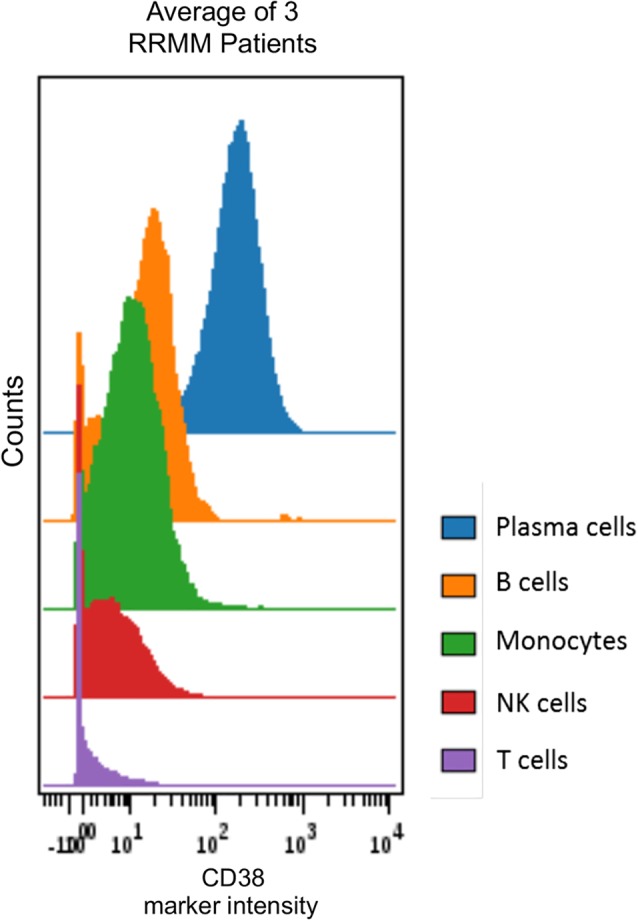
Hierarchy of CD38 expression across immune subtypes in bone‐marrow samples of 3 RRMM GEN501 patients as assessed by CyTOF®. Exported patient‐specific SPADE tree bubbles fcs files for corresponding immune‐cell populations were concatenated and subsequently visualized using Cytobank® software. CD38 marker intensity in NK, monocyte, and B‐ and T‐cell compartments. NK, natural killer; RRMM, relapsed/refractory multiple myeloma; NK, natural killer.
Figure 3.
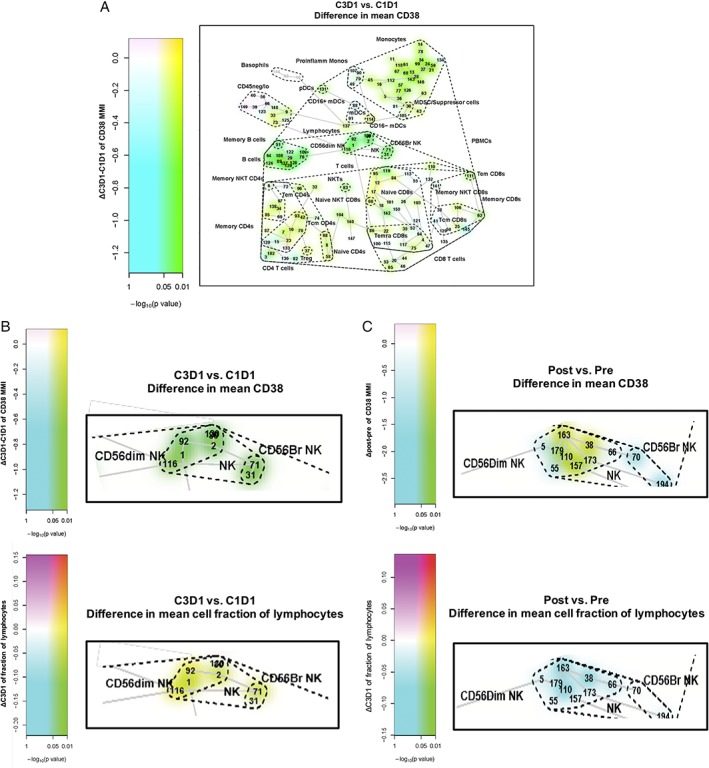
Pre‐ versus on‐ and post‐daratumumab treatment samples revealed (A) CD38 expression changes in SIRIUS whole‐blood samples across different immune cells and NK‐cell depletion in (B) SIRIUS whole‐blood and (C) GEN501 bone‐marrow samples. Data is visualized using the SPADE‐blend tree method, which involved the projection of the differential testing results (here, contrasting pre‐ versus on‐ and post‐treatment sample data) as colored highlighting of the SPADE tree clusters to maximize exploration of the statistical analysis results, simultaneously, in the samples under investigation. SPADE, spanning‐tree progression analysis of density‐normalized events; NK, natural killer; C, cycle; D, day; MMI, mean marker intensity; ProInflamm Monos, pro‐inflammatory monocytes; MDSC, myeloid‐derived suppressor cell; PBMC, peripheral blood mononuclear cell; mDC, myeloid dendritic cell; pDC, plasmacytoid dendritic cell; NKT, natural killer T cell; Tcm, central memory T cell; Tem, effector memory T cell; Temra, effector memory RA+ T cell; Treg, regulatory T cell. Nodes are colored by decrease (cyan; green if significant [raw P value]) or increase (magenta; red if significant [raw P value]).
Activated Phenotype of Remaining NK Cells
Interestingly, although NK cells were reduced upon daratumumab treatment, phenotypic changes were observed in the remaining cells of this immune population. Differential analysis and SPADE‐blend tree visualization of WB samples from SIRIUS patients after 2 months of daratumumab treatment revealed significant decreases in CD45RA (NK, P* = 0.0198; dim NK, P* = 0.0297) and significant increases in CD69 and CD127 (NK, dim NK, bright NK, P* = 0.0198; Supporting Information Fig. S2). In both SIRIUS and GEN501, a consistent trend toward decreased granzyme B and increased CD25, and increased CD137 and CD27 were observed. Similar trends of these proteins were observed in BM samples from patients in GEN501 (data not shown).
Immune‐Suppressive Populations and Basophils
To assess rare populations and their CD38+ subsets, WB samples from the SIRIUS trial were manually gated and analyzed. Beyond confirmation of the previously reported daratumumab‐mediated depletion of immune‐suppressive populations, like CD38+ Tregs (CD4+CD25+CD127−) 14 (Fig. 4A), our high‐dimensional cytometry analysis enabled the first evaluation of basophils (CD45−CD123+HLA‐DR−) under daratumumab treatment. Daratumumab reduced the percentage of CD38+ basophils after 2 months of monotherapy treatment, independent of response to treatment and the manifestation or grade of infusion‐related reactions (IRRs) at first (Fig. 4B) or any infusion (data not shown). The baseline level of CD38+ basophils was also not associated with IRRs at first infusion.
Figure 4.
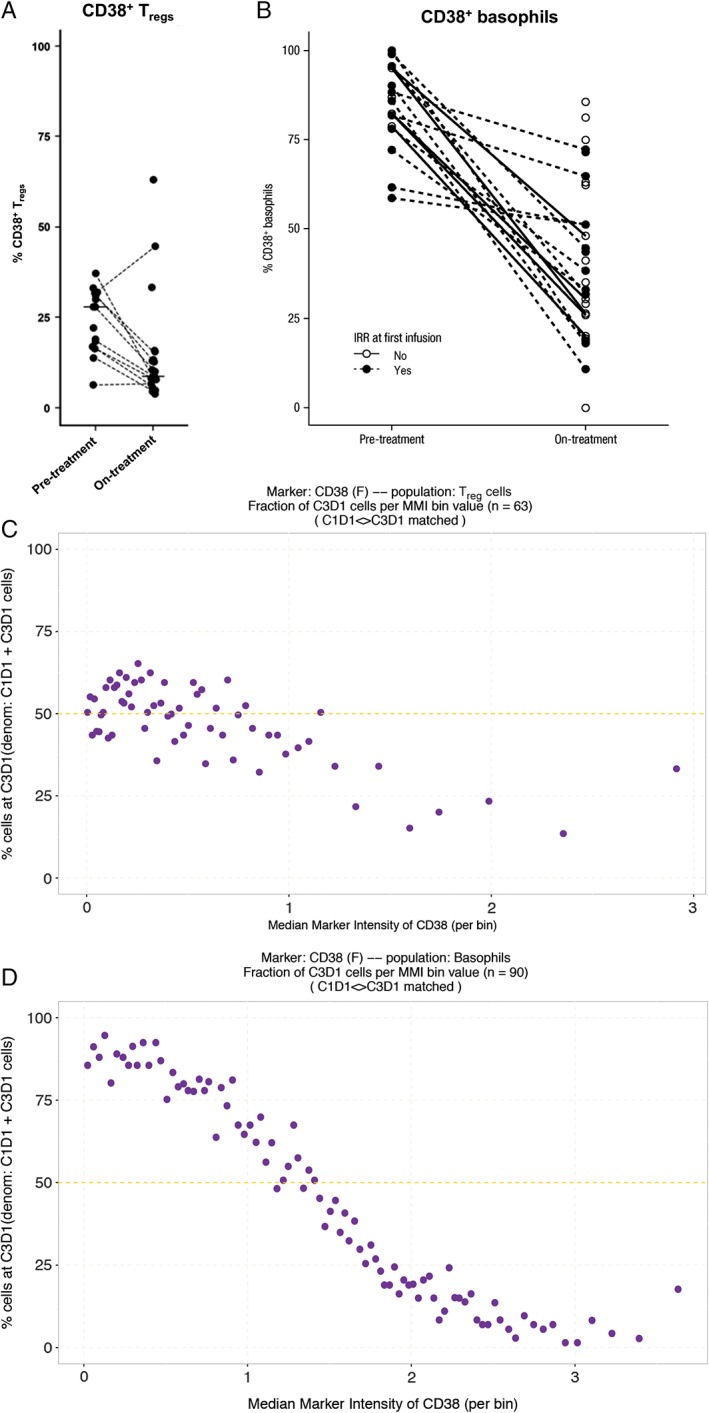
Depletion of the immune‐suppressive populations (A) CD38+ Tregs and (B) CD38+ basophils in whole‐blood samples from patients at baseline and after 2 months of daratumumab monotherapy treatment (SIRIUS). Analysis of changes in distribution of signal intensity shows a decrease in the fraction of Tregs expressing high levels of CD38, suggesting a change in the Treg population (C). For basophils (D) a shift of cell fractions from high intensity to low intensity is observed, suggesting a change in CD38 expression. Treg, regulatory T‐cell; IRR, infusion‐related reaction C, cycle; D, day; MMI, mean marker instensity. Lines indicate paired samples.
To determine whether changes in CD38 signal intensity corresponded to a change in the composition of the cell population or to a change in gene expression, we implemented a new method, the bin analysis. In short, for each population the expression data of a given marker is first summarized by binning cells using the centiles of the distribution. Then, the number of cells per bin and per time point is computed and the number of cells post‐treatment to the total number of cells in each bin is compared. The fraction of cells is constant at 50% if no change in gene expression distribution has occurred.
When changes in distribution of signal intensity were assessed with the bin analysis, the fraction of Tregs expressing low levels of CD38 was observed to have remained constant during the course of treatment (average of post‐treatment to baseline change is 50%; Fig. 4C), while the fraction of Tregs expressing high levels of CD38 was decreased (average of post‐treatment to baseline change is less than 50%; Fig. 4C). We interpret this change as shift in the Treg population, where cells expressing high levels of CD38 are depleted upon treatment with daratumumab. For basophils, however, the fraction of cells expressing low levels of CD38 was greatly increased (average of post‐treatment to baseline change is greater than 50%; Fig. 4D) while the fraction of cells expressing high levels of CD38 was decreased (average of post‐treatment to baseline change is less than 50%; Fig. 4D), representing a decrease of CD38 expression in basophils upon treatment with daratumumab.
T‐Cell Phenotyping
Distinct shifts in T‐cell phenotypes are observed in WB after 2 months of daratumumab monotherapy. Of the CD4+ and CD8+ T cells that were both present at baseline (Fig. 5A, left panel, top figures, respectively), in responders, a distinct shift toward the CD8+ population was observed upon treatment in each study (lower right panels). Because shifts in Radviz plots correspond to either an increase in signal intensity in channels in the direction of the shift, or to a decrease in signal intensity on the opposite end of the circle, we used fan plots to identify the nature of the population change over treatment, using the asinh‐transformed MMI values for selected channels (Fig. 5B). We confirmed the shift by computing the fraction of total T cells that are positive for Granzyme B, using a fixed threshold of 2 units (asinh‐transformed MMI values) for all conditions. Interestingly, this shift to CD8+ T‐cell prevalence was associated with an increase in granzyme B positivity, which is suggestive of increased killing capacity of the cytotoxic T cells 27. This observation was most pronounced in WB and BM of responders in both monotherapy studies.
Figure 5.
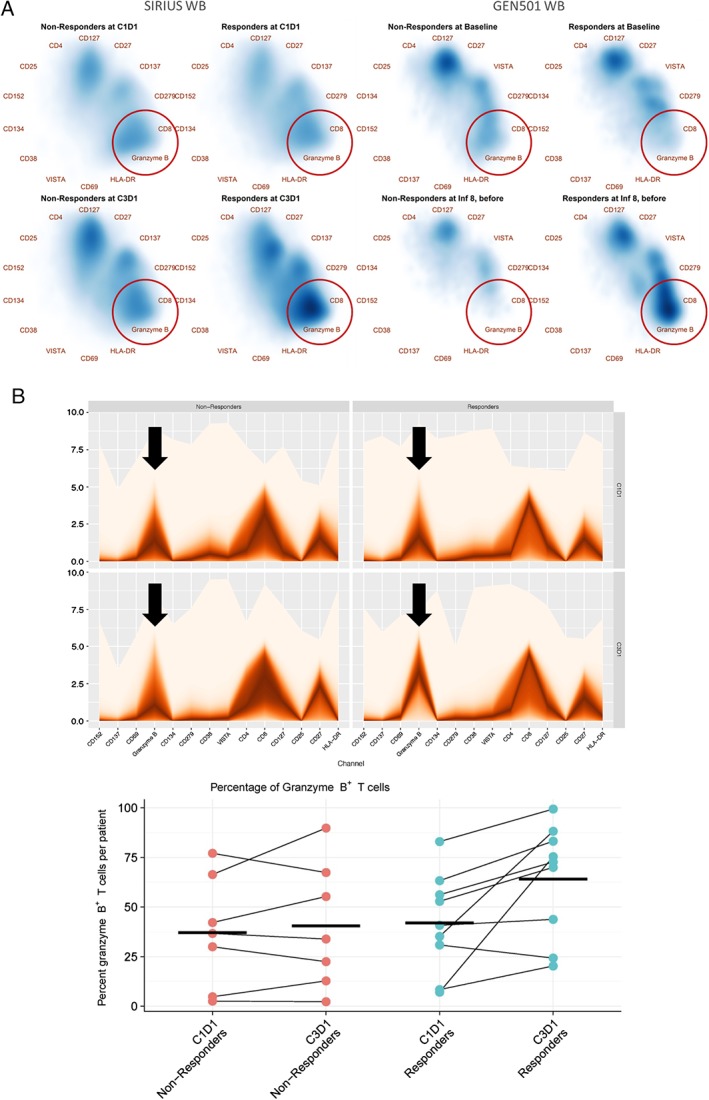
High‐dimensional algorithm–based analysis result visualization of whole blood T‐cell phenotypic changes in responders and non‐responders from SIRIUS and GEN501 after 2 months of daratumumab monotherapy. (A) Radviz density projections of T cells from SIRIUS and GEN501 studies. (B) Fan plots and scatterplots of T‐cell data from SIRIUS patients. Radviz plots of SPADE tree‐defined T‐cell populations reveal a shift to CD8 prevalence with granzyme B positivity in responders (red circles). Arrows on fan plots highlight the contribution of granzyme B to the phenotype of T cells at a given time point for patients. The analysis was based on whole‐blood samples from 32 SIRIUS patients (17 non‐responders; 15 responders) and 4 GEN501 patients (2 non‐responders; 2 responders). SPADE, spanning‐tree progression analysis of density‐normalized events; WB, whole‐blood; C, cycle; D, day; Inf, infusion.
Complementary manual gating performed on SIRIUS WB samples revealed changes in activation markers of CD8+ T cells. There was a significant increase (P = 0.017, Wilcoxon) in granzyme B production and trends for increased expression of CD69 and HLA‐DR (Fig. 6A). Modest changes were observed in markers associated with exhaustion (Fig. 6B). Following daratumumab treatment, the median percentage of PD1+ CD8+ T cells decreased from baseline in responders (from 10.2% to 8.97%) but increased in non‐responders (from 8.76% to 11.42%; Fig. 6B). However, neither change from baseline was statistically significant. Additionally, CD8+ T‐cell changes were associated with minimal alterations of the exhaustion marker CTLA4 after 2 months of daratumumab treatment.
Figure 6.
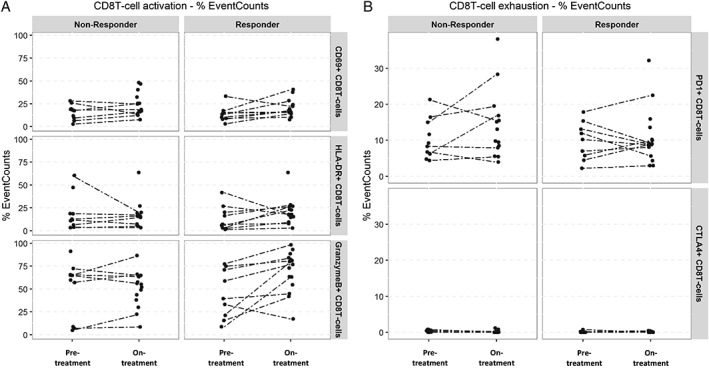
CyTOF® manual gating performed on SIRIUS whole‐blood samples, showing changes in CD8+ T‐cell (A) activation and (B) exhaustion marker changes following daratumumab treatment in responders and non‐responders. Activation marker analysis revealed a significant increase (P = 0.017) in granzyme B production among responders. Modest changes were observed in markers associated with exhaustion. Dotted lines indicate paired samples.
Discussion
Daratumumab, a human monoclonal antibody that targets CD38, has demonstrated unprecedented clinical benefit and rapid, deep responses as monotherapy and in combination with standard‐of‐care regimens in patients with RRMM 5, 6, 7, 8, 9, 10. Several studies have identified a variety of different mechanisms by which daratumumab exerts its anti‐myeloma activity 1, 12, 13, 14, 15, which likely contributes to the significant treatment benefit associated with daratumumab in the RRMM setting 5, 6, 7, 8, 9.
In another recent study, we demonstrated that daratumumab's mechanism of action includes immune‐modulatory effects, in addition to the previously established direct, on‐tumor effects 15. That study evaluated the impact of daratumumab on CD38+ immunosuppressive populations, T‐cell proliferation and activation, and T‐cell receptor clonality using WB and BM samples from patients who received daratumumab 16 mg/kg in the GEN501 study (n = 42) or the SIRIUS study (n = 106). Samples were analyzed using flow cytometry, functional assays, and T‐cell receptor beta‐chain repertoire sequencing. The study showed that daratumumab exerted immune‐modulatory effects by expanding the T‐cell population, while eliminating a population of highly immunosuppressive CD38+ Tregs, myeloid‐derived suppressor cells, and regulatory B cells, thus stimulating T‐cell effector functions 15.
We undertook the present study to complement the findings of Krejcik et al. 15, Nijhof et al. 24, and Casneuf et al. 26 assessing the effects of daratumumab on a more comprehensive profile of immune‐cell populations and immune functional characteristics. Our objective was to identify changes in immune‐cell profiles that correlated with efficacy and depth of response observed during treatment with daratumumab. Our primary analytical tool was CyTOF®, a next‐generation mass cytometry platform 18. Compared with flow cytometry, CyTOF® greatly increases the number of parameters that can be measured simultaneously per cell by eliminating cell‐dependent background signals and the need for mathematical correction of spectral overlap caused by fluorescence 18. We also introduce a novel tool for visualizing subgroup analysis results on SPADE trees by incorporating Radviz plots to better understand changes in response categories and with respect to time. Recent studies have started to incorporate the CyTOF® technology to examine immune responses to cancer therapies 28, 29, 30.
The results of our study confirm and build upon our previously reported flow cytometry findings 15. CD38 was found on plasma cells, NK cells, monocytes, B cells, and T cells in BM samples and peripheral blood mononuclear cell samples from RRMM patients. In line with Nijhof et al.’s findings in which CD38+ plasma cells were found to decrease over time 24, CyTOF® profiling shows decreased CD38 expression on many immune‐cell subtypes in both WB and BM upon daratumumab monotherapy treatment (SIRIUS, GEN501). Manual gating of samples in the SIRIUS monotherapy study revealed decreased CD38+ Tregs (all patients) after 2 months of daratumumab monotherapy, which may contribute to the depth and durability of responses associated with daratumumab 5, 6, 7, 8, 9. Furthermore, CD38+ basophils, a type of granulocytic white blood cell that releases histamine as an inflammatory reaction during an immune response and contributes to anaphylactic reactions 31, were reduced independent of response. This decrease could be due solely to phenotypic changes after daratumumab engagement and may contribute to IRRs; however, their baseline level was found not to be correlated to the presence of IRRs at first infusion. To fully elucidate this element of immunological change, functional assays with basophils in the presence of daratumumab will need to be performed to properly assess the impact for treated patients.
Furthermore, in agreement with data we presented previously 25, 26, our study has demonstrated that in addition to immune‐suppressive cells, NK cells (CD45+CD3−CD56+/dim) are also reduced in patients treated with daratumumab. However, Casneuf et al. have shown that this reduction in NK cell number in daratumumab‐treated patients does not negatively impact the efficacy of daratumumab, nor does lack of NK cells increase the susceptibility of patients to infections 25, 26. We made several novel observations when phenotypically characterizing the residual NK cells, including a significant decrease in CD45RA and increase in CD127 and CD69. While we observed trends for increased expression of CD27 in NK cells that persisted across WB and BM samples, and while there are conflicting hypotheses around the functional role of NK cells with this phenotype 32, 33, 34, 35, the observed trends toward increased signal intensity of CD137 combined with increased levels of activation markers CD69 and CD25 may suggest any compromised cytolytic activity could be offset to allow for daratumumab‐mediated antibody‐dependent cellular cytotoxicity 32. Furthermore, Krzywinska et al. showed that in hematologic malignancies, NK cells with an immature profile are still able to show cytotoxic activity 36, 37, and Wang et al. showed that post‐daratumumab NK cells are more effective at eradicating myeloma cells 38. Recently, Feng et al. have demonstrated that isatuximab, another CD38‐targeted monoclonal antibody, promotes NK‐cell activity through interferon γ induction, as evidenced by CD107a upregulation. However, it remains unclear whether the authors also observed a decrease in the levels of NK cells following isatuximab treatment 39. While these in vitro findings require further validation, collectively they highlight the immune‐modulatory role of CD38‐targeted therapies. Subsequent studies will be required to explore the functional role of this unique population of NK cells in daratumumab‐treated patients.
Of particular interest in our study are characteristics differentiating daratumumab‐treated responders from non‐responders. In an analysis of WB samples from SIRIUS and GEN501 patients, Radviz plots of SPADE tree‐defined T‐cell populations revealed a shift to CD8 prevalence with granzyme B positivity following daratumumab treatment in responders. Manual gating of SIRIUS WB samples showed that there was a significant increase in granzyme B production among responders to daratumumab. This is important to note, given that recent studies have spoken to the varied functional status of both peripheral blood and BM T cells from multiple myeloma patients 40, 41. Moreover, these findings align with work conducted by Chatterjee et al., which showed that inhibition of CD38 increased NAD+‐dependent activity of Sirt1 and enhanced the anti‐tumor effect of CD8+ T cells 42. Since the majority of our samples were from WB, we investigated this aspect of T‐cell biology for markers associated with CD8+ T‐cell exhaustion and showed that the median percentage of PD1+CD8+ T cells trended downward in responders but tended to increase in non‐responders (both not significant). Additionally, daratumumab treatment minimally affected the exhaustion marker CTLA4. It is important to note that SIRIUS and GEN501 samples were assessed at the presumed initiation of daratumumab‐mediated T‐cell expansion 15. Therefore, most patient T cells will likely have had limited interaction with target cells and thus have a high intracellular granzyme B and low checkpoint receptor expression phenotype.
In summary, CyTOF® analysis of patient samples from studies of daratumumab therapy highlight the immune‐modulatory mechanism of action of daratumumab. T‐cell changes toward a cytolytic, granzyme B+ phenotype observed in WB and BM and increased expression of the activation markers on NK cells, such as CD69, support an adaptive response in patients that may contribute to the depth of response observed in patients treated with daratumumab. Synonymous findings in WB and BM CyTOF® samples reinforce this platform's ability to allow WB surrogacy for interpretation of the immune system in multiple myeloma, which is of high value to the field. These results build upon earlier findings and advocate further utility of CyTOF® and related methodologies to gain greater insights into the mechanism of action of daratumumab monotherapy and daratumumab in combination with other immune‐modulatory agents.
Disclosure of interests
H.C.A.III, F.S., K.V.dB., T.S., J.B., Y.A., H.C., C.C., G.V., and T.C. are employees of Janssen. T.C., G.V., H.C., and A.K.S. report equity ownership in Johnson & Johnson. S.Z.U. received research funding from Onyx, Sanofi, Array BioPharma, Pharmacyclics, Takeda, Celgene, and BMS; served as a consultant for Sanofi, Takeda, Celgene, and Amgen; served on advisory committees for Onyx, Sanofi, Celgene, Skyline, Millennium, and Janssen; and served on speakers bureaus for Takeda, Celgene, and Amgen. T.P. served on advisory committees for Janssen and Genmab and received research funding from Janssen. S.L. served as a consultant for Millennium, Merck, BMS, Celgene, Novartis, Janssen, and Onyx. H.M.L. received research funding from Janssen and Genmab and served on advisory committees for Janssen. T.M. received research funding from Celgene, Janssen, and Genmab, and served on advisory committees for Janssen. N.W.C.J.vdD. received research funding from and served on advisory committees for Janssen, Celgene, BMS, and Amgen. A.K.S. is a former employee of Janssen.
Supporting information
Appendix S1: Supporting information
Appendix S2: Supporting information
Acknowledgments
The authors would like to thank Berris van Kessel‐Welmers for her support in sample operation and Tahamtan Ahmadi for oversight of these clinical studies. Editorial support was provided by Jason Jung, PhD, and Sima Patel, PhD, of MedErgy, and was funded by Janssen Global Services, LLC.
ClinicalTrials.gov Identifiers: NCT01985126 and NCT00574288.
Previously presented in part at the 58th American Society of Hematology (ASH) Annual Meeting & Exposition; December 3‐6, 2016; San Diego, California.
Literature Cited
- 1. de Weers M, Tai YT, van der Veer MS, Bakker JM, Vink T, Jacobs DCH, Oomen LA, Peipp M, Valerius T, Slootstra JW, et al. Daratumumab, a novel therapeutic human CD38 monoclonal antibody, induces killing of multiple myeloma and other hematological tumors. J Immunol 2011;186:1840–1848. [DOI] [PubMed] [Google Scholar]
- 2. Lin P, Owens R, Tricot G, Wilson CS. Flow cytometric immunophenotypic analysis of 306 cases of multiple myeloma. Am J Clin Pathol 2004;121:482–488. [DOI] [PubMed] [Google Scholar]
- 3. Santonocito AM, Consoli U, Bagnato S, Milone G, Palumbo GA, Di Raimondo F, Stagno F, Guglielmo P, Giustolisi R. Flow cytometric detection of aneuploid CD38(++) plasmacells and CD19(+) B‐lymphocytes in bone marrow, peripheral blood and PBSC harvest in multiple myeloma patients. Leuk Res 2004;28:469–477. [DOI] [PubMed] [Google Scholar]
- 4. Deaglio S, Mehta K, Malavasi F. Human CD38: A (r)evolutionary story of enzymes and receptors. Leuk Res 2001;25:1–12. [DOI] [PubMed] [Google Scholar]
- 5. Lokhorst HM, Plesner T, Laubach JP, Nahi H, Gimsing P, Hansson M, Minnema MC, Lassen U, Krejcik J, Palumbo A, et al. Targeting CD38 with daratumumab monotherapy in multiple myeloma. N Engl J Med 2015;373:1207–1219. [DOI] [PubMed] [Google Scholar]
- 6. Lonial S, Weiss BM, Usmani SZ, Singhal S, Chari A, Bahlis NJ, Belch A, Krishnan A, Vescio RA, Mateos MV, et al. Daratumumab monotherapy in patients with treatment‐refractory multiple myeloma (SIRIUS): An open‐label, randomised, phase 2 trial. Lancet 2016;387:1551–1560. [DOI] [PubMed] [Google Scholar]
- 7. Plesner T, Arkenau HT, Gimsing P, Krejcik J, Lemech C, Minnema MC, Lassen U, Laubach JP, Palumbo A, Lisby S, et al. Phase 1/2 study of daratumumab, lenalidomide, and dexamethasone for relapsed multiple myeloma. Blood 2016;128:1821–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Palumbo A, Chanan‐Khan A, Weisel K, Nooka AK, Masszi T, Beksac M, Spicka I, Hungria V, Munder M, Mateos MV, et al. Daratumumab, bortezomib, and dexamethasone for multiple myeloma. N Engl J Med 2016;375:754–766. [DOI] [PubMed] [Google Scholar]
- 9. Dimopoulos MA, Oriol A, Nahi H, San‐Miguel J, Bahlis NJ, Usmani SZ, Rabin N, Orlowski RZ, Komarnicki M, Suzuki K, et al. Daratumumab, lenalidomide, and dexamethasone for multiple myeloma. N Engl J Med 2016;375:1319–1331. [DOI] [PubMed] [Google Scholar]
- 10. Chari A, Suvannasankha A, Fay JW, Arnulf B, Kaufman JL, Ifthikharuddin JJ, Weiss BM, Krishnan A, Lentzsch S, Comenzo R, et al. Daratumumab plus pomalidomide and dexamethasone in relapsed and/or refractory multiple myeloma. Blood 2017;130:974–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DARZALEX® (daratumumab) injection, for intravenous use [package insert]. Horsham, PA: Janssen Biotech, Inc.; 2016.
- 12. Lammerts van Bueren J, Jakobs D, Kaldenhoven N, Roza M, Hiddingh S, Meesters J, Voorhorst M, Gresnigt E, Wiegman L, Buijsse AO, et al. Direct in vitro comparison of daratumumab with surrogate analogs of CD38 antibodies MOR03087, SAR650984 and Ab79. Blood 2014;124:3474. [Google Scholar]
- 13. Overdijk MB, Verploegen S, Bogels M, van Egmond M, Lammerts van Bueren JJ, Mutis T, Groen RW, Breij E, Martens AC, Bleeker WK, et al. Antibody‐mediated phagocytosis contributes to the anti‐tumor activity of the therapeutic antibody daratumumab in lymphoma and multiple myeloma. MAbs 2015;7:311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Overdijk MB, Jansen JH, Nederend M, Lammerts van Bueren JJ, Groen RW, Parren PW, Leusen JH, Boross P. The therapeutic CD38 monoclonal antibody daratumumab induces programmed cell death via Fcgamma receptor‐mediated cross‐linking. J Immunol 2016;197:807–813. [DOI] [PubMed] [Google Scholar]
- 15. Krejcik J, Casneuf T, Nijhof IS, Verbist B, Bald J, Plesner T, Syed K, Liu K, van de Donk NWCJ, Weiss BM, et al. Daratumumab depletes CD38+ immune‐regulatory cells, promotes T‐cell expansion, and skews T‐cell repertoire in multiple myeloma. Blood 2016;128:384–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, Pavlov S, Vorobiev S, Dick JE, Tanner SD. Mass cytometry: Technique for real time single cell multitarget immunoassay based on inductively coupled plasma time‐of‐flight mass spectrometry. Anal Chem 2009;81:6813–6822. [DOI] [PubMed] [Google Scholar]
- 17. Autissier P, Soulas C, Burdo TH, Williams KC. Evaluation of a 12‐color flow cytometry panel to study lymphocyte, monocyte, and dendritic cell subsets in humans. Cytometry A 2010;77:410–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bendall SC, Simonds EF, Qiu P, Amir e‐AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, et al. Single‐cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 2011;332:687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qiu P. Inferring phenotypic properties from single‐cell characteristics. PLoS One 2012;7:e37038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Diggins KE, Greenplate AR, Leelatian N, Wogsland CE, Irish JM. Characterizing cell subsets using marker enrichment modeling. Nat Methods 2017;14:275–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qiu P, Simonds EF, Bendall SC, Gibbs KD Jr, Bruggner RV, Linderman MD, Sachs K, Nolan GP, Plevritis SK. Extracting a cellular hierarchy from high‐dimensional cytometry data with SPADE. Nat Biotechnol 2011;29:886–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abraham Y, Gerrits B, Ludwig MG, Rebhan M, Gubser Keller C. Exploring glucocorticoid receptor agonists mechanism of action through mass cytometry and radial visualizations. Cytometry B Clin Cytom 2017;92:42–56. [DOI] [PubMed] [Google Scholar]
- 23. Britton E, Fisher P, Whitley J. The inflation report projections: understanding the fan chart. BEQB 1998;Q1:30–37. [Google Scholar]
- 24. Nijhof IS, Casneuf T, van Velzen JF, van Kessel B, Axel AE, Syed K, Groen RWJ, van Duin M, Sonneveld P, Minnema MC, et al. CD38 expression and complement inhibitors affect response and resistance to daratumumab therapy in myeloma. Blood 2016;128:959–970. [DOI] [PubMed] [Google Scholar]
- 25. Casneuf T, Xu XS, Adams H, III , Axel A, Verbist B, Liu K, Khan I, Ahmadi T, Yan X, Lonial S, et al. Pharmacodynamic relationship between natural killer cells and daratumumab exposure in relapsed/refractory multiple myeloma. The 21st European Hematology Association Congress; 2016.
- 26. Casneuf T, Xu XS, Adams HC, III , et al. Effects of daratumumab on natural killer cells and impact on clinical outcomes in relapsed or refractory multiple myeloma. Blood Adv 2017;1:2105–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Russell JH, Ley TJ. Lymphocyte‐mediated cytotoxicity. Annu Rev Immunol 2002;20:323–370. [DOI] [PubMed] [Google Scholar]
- 28. Hekim C, Ilander M, Yan J, Michaud E, Smykla R, Vähä‐Koskela M, Savola P, Tähtinen S, Saikko L, Hemminki A, et al. Dasatinib changes immune cell profiles concomitant with reduced tumor growth in several murine solid tumor models. Cancer Immunol Res 2017;5:157–169. [DOI] [PubMed] [Google Scholar]
- 29. Jiao S, Xia W, Yamaguchi H, Wei Y, Chen MK, Hsu JM, Hsu JL, Yu WH, Du Y, Lee HH, et al. PARP inhibitor upregulates PD‐L1 expression and enhances cancer‐associated immunosuppression. Clin Cancer Res 2017;23:3711–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carter BZ, Mak PY, Mu H, Zhou H, Mak DH, Schober W, Leverson JD, Zhang B, Bhatia R, Huang X, et al. Combined targeting of BCL‐2 and BCR‐ABL tyrosine kinase eradicates chronic myeloid leukemia stem cells. Sci Transl Med 2016;8:355ra117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Montañez MI, Mayorga C, Bogas G, Barrionuevo E, Fernandez‐Santamaria R, Martin‐Serrano A, Laguna JJ, Torres MJ, Fernandez TD, Doña I. Epidemiology, mechanisms, and diagnosis of drug‐induced anaphylaxis. Front Immunol 2017;8:614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vossen MTM, Matmati M, Hertoghs KML, Baars PA, Gent MR, Leclercq G, Hamann J, Kuijpers TW, van Lier RAW. CD27 defines phenotypically and functionally different human NK cell subsets. J Immunol 2008;180:3739–3745. [DOI] [PubMed] [Google Scholar]
- 33. Hayakawa Y, Smyth MJ. CD27 dissects mature NK cells into two subsets with distinct responsiveness and migratory capacity. J Immunol 2006;176:1517–1524. [DOI] [PubMed] [Google Scholar]
- 34. Chiossone L, Chaix J, Fuseri N, Roth C, Vivier E, Walzer T. Maturation of mouse NK cells is a 4‐stage developmental program. Blood 2009;113:5488–5496. [DOI] [PubMed] [Google Scholar]
- 35. Fu B, Tian Z, Wei H. Subsets of human natural killer cells and their regulatory effects. Immunology 2014;141:483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Krzywinska E, Cornillon A, Allende‐Vega N, Vo DN, Rene C, Lu ZY, Pasero C, Olive D, Fegueux N, Ceballos P, et al. CD45 isoform profile identifies natural killer (NK) subsets with differential activity. PLoS One 2016;11:e0150434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krzywinska E, Allende‐Vega N, Cornillon A, Vo DN, Cayrefourcq L, Panabieres C, Vilches C, Déchanet‐Merville J, Hicheri Y, Rossi JF, et al. Identification of anti‐tumor cells carrying natural killer (NK) cell antigens in patients with hematological cancers. EBioMedicine 2015;2:1364–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang Y, Zhang Y, Hughes T, Zhang J, Caligiuri MA, Benson DM, Yu J. Fratricide of NK cells in daratumumab therapy for multiple myeloma overcome by ex vivo‐expanded autologous NK cells. Clin Cancer Res 2018;24:4006–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Feng X, Zhang L, Acharya C, An G, Wen K, Qiu L, Munshi NC, Tai YT, Anderson KC. Targeting CD38 suppresses induction and function of T regulatory cells to mitigate immunosuppression in multiple myeloma. Clin Cancer Res 2017;23:4290–4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zelle‐Rieser C, Thangavadivel S, Biedermann R, Brunner A, Stoitzner P, Willenbacher E, Greil R, Jöhrer K. T cells in multiple myeloma display features of exhaustion and senescence at the tumor site. J Hematol Oncol 2016;9:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Suen H, Brown R, Yang S, Weatherburn C, Ho PJ, Woodland N, Nassif N, Barbaro P, Bryant C, Hart D, et al. Multiple myeloma causes clonal T‐cell immunosenescence: Identification of potential novel targets for promoting tumour immunity and implications for checkpoint blockade. Leukemia 2016;30:1716–1724. [DOI] [PubMed] [Google Scholar]
- 42. Chatterjee S, Daenthanasanmak A, Chakraborty P, Wyatt MW, Dhar P, Selvam SP, Fu J, Zhang J, Nguyen H, Kang I, et al. CD38‐NAD+Axis regulates immunotherapeutic anti‐tumor T cell response. Cell Metab 2017;27(1):85‐100.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Terada Y, von Luxburg U. Local ordinal embedding. Proceedings of the 31st International Conference on Machine Learning, Beijing, China; 2014.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting information
Appendix S2: Supporting information


