Abstract
Toll‐like receptors (TLRs) play critical roles in host defense after recognition of conserved microbial‐ and host‐derived components, and their dysregulation is a common feature of various inflammation‐associated cancers, including gastric cancer (GC). Despite the recent recognition that metabolic reprogramming is a hallmark of cancer, the molecular effectors of altered metabolism during tumorigenesis remain unclear. Here, using bioenergetics function assays on human GC cells, we reveal that ligand‐induced activation of TLR2, predominantly through TLR1/2 heterodimer, augments both oxidative phosphorylation (OXPHOS) and glycolysis, with a bias toward glycolytic activity. Notably, DNA microarray‐based expression profiling of human cancer cells stimulated with TLR2 ligands demonstrated significant enrichment of gene‐sets for oncogenic pathways previously implicated in metabolic regulation, including reactive oxygen species (ROS), p53 and Myc. Moreover, the redox gene encoding the manganese‐dependent mitochondrial enzyme, superoxide dismutase (SOD)2, was strongly induced at the mRNA and protein levels by multiple signaling pathways downstream of TLR2, namely JAK‐STAT3, JNK MAPK and NF‐κB. Furthermore, siRNA‐mediated suppression of SOD2 ameliorated the TLR2‐induced metabolic shift in human GC cancer cells. Importantly, patient‐derived tissue microarrays and bioinformatics interrogation of clinical datasets indicated that upregulated expression of TLR2 and SOD2 were significantly correlated in human GC, and the TLR2‐SOD2 axis was associated with multiple clinical parameters of advanced stage disease, including distant metastasis, microvascular invasion and stage, as well as poor survival. Collectively, our findings reveal a novel TLR2‐SOD2 axis as a potential biomarker for therapy and prognosis in cancer.
Keywords: gastric cancer, toll‐like receptor 2, superoxide dismutase 2, metabolism
Short abstract
What's new?
Many types of tumor cells have altered metabolism, which supports their rapid growth. Pattern‐recognition receptors such as Toll‐like receptors (TLRs) can have a broad range of effects on inflammation‐associated cancers such as gastric cancer (GC), and can influence proliferation, apoptosis, and migration. In this study, the authors identified a novel TLR2‐ SOD2 axis that promotes metabolic reprogramming of GC cells, mediated by multiple oncogenic signaling pathways. Increased expression of these proteins in gastric tumors may serve as a valuable biomarker for therapy and prognosis in GC.
Abbreviations
- ECAR
extracellular acidification rate
- OCR
oxygen consumption rate
- OXPHOS
oxidative phosphorylation.
- ROS
reactive oxygen speces
- SOD2
superoxide dismutase 2
- SRC
spare respiratory capacity
- TLR2
toll‐like receptor 2
Introduction
A defining feature of cancer cells is their capacity to employ a modified metabolic program with increased glucose uptake and glycolytic activity, even when oxygen is present (the “Warburg effect”), which facilitates both energy needs and the redirection of metabolites into biosynthesis to support vigorous tumor growth.1, 2 This aerobic glycolysis of cancers generally does not occur as a consequence of defective mitochondrial respiration, with most cancers retaining mitochondrial function to facilitate the dynamic interplay between oxidative phosphorylation (OXPHOS) and glycolysis.3 Rather, the altered glycolytic state of cancer cells is influenced by various mechanisms involving the deregulation of classic oncogenic signaling pathways, such as the phosphatidylinositol 3‐kinase (PI3K)‐mTOR axis through sensing nutrients and activation of anabolic pathways,4 oncogenic c‐Myc via transcriptional upregulation of transporters and enzymes required for glucose metabolism,5 and oncogenic Kras by multiple effects such as mediating nutrient absorption and redox homeostasis.6
Separate from these cell‐autonomous effects, extrinsic factors, such as access to nutrients and oxygen, as well as interactions with immune cells and other stromal components in the tumor microenvironment, can also contribute to the reprogrammed metabolic phenotype of cancer cells.1 In addition, recent evidence supports the notion that microbial‐derived products can induce the metabolic reprogramming of inflammatory/innate immune cells in the tumor microenvironment, thus impacting upon host defense responses such as cytokine production and phagocytosis, which can shape tumor immunity.7, 8, 9 In this respect, considering that the gut epithelium plays a central role in mucosal immunity against microbes and can be vulnerable to neoplastic transformation, the reprogramming of metabolic pathways in tumor (epithelial) and innate immune cells by microbial components could have relevance to the pathogenesis of gastrointestinal cancers. However, the molecular mechanisms by which microbial interactions with epithelial surfaces of the gastrointestinal tract can alter the metabolic state of tumor (epithelial) cells, and thus potentially contribute to tumorigenesis, remain unresolved.
Pattern recognition receptors, in particular Toll‐like receptors (TLRs), were first discovered as critical modulators of the innate immune response to a myriad of microbial (i.e. viral, bacterial, fungal) and host‐derived factors.10 Since their discovery, it has become apparent that TLRs (such as TLR2) exhibit diverse activities on the gastrointestinal tract, including maintaining the structural and functional integrity of the enteric nervous system via production of neurotrophic factors.11, 12 Over recent years, it has also emerged that TLRs can impact on a range of oncogenic activities on cancer cells within the gastrointestinal tract, including proliferation, apoptosis and migration.13 Notably, we have demonstrated a key role for TLR2 in GC, which ranks as the third most lethal cancer worldwide, and represents one of a growing number of cancers whose pathogenesis is intimately linked to dysregulation of innate immunity. Specifically, we have shown that upregulation of TLR2 promotes gastric tumorigenesis by directly augmenting gastric epithelial cell proliferation and survival, independent of inflammation, which was associated with a TLR2‐regulated gene signature enriched for anti‐apoptotic genes.14, 15 However, the potential for TLR2 or other pattern recognition receptors to direct the metabolic transcriptional programming of tumor cells is ill‐defined.
Here, we reveal that ligand‐induced TLR2 activation in human GC cells upregulated both OXPHOS and glycolysis, which was coincident with marked upregulation of the manganese‐dependent superoxide dismutase (SOD)2, a mitochondrial‐located antioxidant enzyme that regulates hydrogen peroxide (H2O2) and oxygen production from superoxide anion radicals formed by OXPHOS.16 TLR2‐induced SOD2 expression was dependent on multiple signaling pathways, namely Janus kinase (JAK)‐signal transducer and activator of transcription (STAT)3, c‐Jun N‐terminal kinase (JNK) mitogen‐activated protein kinase (MAPK), and nuclear factor kappa B (NF‐κB). Furthermore, siRNA‐mediated knock‐down of SOD2 abrogated the bioenergetic activities of TLR2 in human GC cells. Notably, tissue microarrays on clinical biopsies demonstrated a significant correlation between upregulated protein expression levels of SOD2 and TLR2 in gastric tumors, which also served as a predictor of poor patient survival. Collectively, our findings reveal a novel TLR2‐SOD2 axis as a potential biomarker of metabolic reprogramming, which could be employed for therapeutic and prognostic uses in GC, and potentially other cancers.
Materials and Methods
Cell culture
Human GC cell lines MKN1, NUGC4 (Japanese Collection of Research Bioresources Cell Bank) and AGS (American Type Culture Collection, ATCC) were cultured in Roswell Park Memorial Institute 1640 (RPMI 1640) medium supplemented with 10% fetal calf serum (FCS) and 1% L‐glutamine (Gibco). Cells were authenticated by short tandem repeat profiling (PowerPlex HS16 System kit, Promega), and were passaged for under 6 months after receipt. Cells were routinely tested for mycoplasma contamination (MycoAlert PLUS Mycoplasma Detection Kit, Lonza). AZ521, a duodenal adenocarcinoma line was characterized a Hutu 80 derivative. DNA array data generated from this line was only used as a TLR2 low expressing cell comparison for gene profiling and signaling pathway analysis. No experiment on AZ521 was conducted in our study.
Immunoblotting
The cell lysates were extracted using RIPA lysis buffer with phosphatase and protease inhibitor cocktails (ThermoFisher). The protein concentration was measured using a Bio‐Rad Protein Assay Kit. Equivalent amounts of protein (30 μg) form each cell lysate were electrophoresed on 10–12% SDS‐PAGE gels followed by transferring to PVDF membranes (Millipore). Total protein lysates were immunoblotted with antibodies against SOD2, phospho‐STAT3 (Tyr705), total STAT3, phospho‐AKT (Ser473), total AKT, phospho‐ERK1/2, total ERK1/2, phospho‐JNK, total JNK, phospho‐MAPKAPK2, total MAPKAPK2, phospho‐NF‐κB p65 (Ser536), total NF‐κB p65 (Cell Signaling Technologies), TLR2 (Santa Cruz), Tubulin (Abcam) and Actin (Sigma). Protein bands were visualized and analyzed using the Odyssey Infrared Imaging System (LI‐COR) as described previously.14 The protein expression was quantified by densitometry and normalization to actin or tubulin expression levels with Image J software.
RNA extraction and quantitative real‐time PCR
RNA was isolated using TRI reagent solution (Sigma) followed by the on‐column RNeasy mini kit and DNase treatment (Qiagen). cDNA synthesis was performed using the Transcription First Strand cDNA Synthesis Kit (Roche). Quantitative real‐time polymerase chain reaction (qPCR) was performed using the ABI 7900T PCR System (Applied Biosystems). The gene expression was normalized to the expression of housekeeping gene 18S. Relative fold changes were transformed using the comparative threshold cycle (CT) method (the 2‐ΔΔCT method) with the Sequence Detection System V2.4 software (Applied Biosystems). Primer sequences are provided in Supporting Information Table S1.
TLR activation, transfection and assay reagents
GC cells were grown in 6‐well plates in triplicate. After overnight starvation, cells were treated with phosphate‐buffered saline (PBS control), Pam3CSK4 (P3C, InvivoGen), Lipopolysaccharides from Escherichia coli O111:B4 (LPS, Sigma) or FSL‐1 (Pam2CGDPKHPKSF, InvivoGen) at the indicated concentrations after a time course. For transfection experiments, GC cells at 50–60% confluence in 6‐well plates were transiently transfected with 10 nM nontarget control or siRNA targeting SOD2 gene (Ambion) using Lipofectamine 3000 (ThermoFisher). For signaling pathway analysis, cells were pre‐treated for 1 h with either DMSO vehicle; the JAK inhibitor, CYT387 (Selleck); the JNK inhibitor, SP600125 (Selleck); the ERK inhibitor, U0126 (InvivoGen); the PI3K inhibitor, Wortmannin (Sigma); the NF‐κB inhibitor, BAY11‐7085 (Santa Cruz Biotechnology); or the P38 inhibitor, SB203580 (Selleck) at the indicated concentrations prior to stimulation with P3C or PBS. Cell viability assay in response to TLR agonists’ treatment was also conducted using the MTT assay (Invitrogen) in 96‐well plate after the manufacturer's instructions.
CRISPR‐driven gene editing
Self‐complementary oligonucleotides (Sigma‐Aldrich) used as single‐guided (sg) RNA sequences for targeting human STAT3 (Exons 3 and 4) were ligated into the LentiCRISPRv2 construct (Addgene). Lentivirus was produced by transfecting vectors into Lenti‐X/H293T cells with LentiCRISPR: psPAX2:pMD2.G at a ratio of 4:3:1. Virus was harvested 48 h after transfection, filtered and used to infect MKN1 cell cultures containing 5 μg/mL polybrene. Cells infected with nontargeting control sgRNA vector were used as negative controls. Infected cells were selected with puromycin at 0.2 μg/mL for 7 days and then isolated as a single cell clone. Each clone was amplified and validated by immunoblotting and Sanger sequencing. Cells with a complete STAT3 knockout genotype were chosen for experiments in our study, as previously described.17
Bioenergetic assays
To measure the mitochondrial and glycolytic activity in human GC cells upon TLR2 activation, we used the Seahorse XFp analyzer (Seahorse Bioscience). Cells were plated at indicated numbers (MKN1, 1.5 × 104 per well; NUGC4, 2.5 × 104 per well; AGS, 1 × 104 per well) overnight in Seahorse XFp miniplates (8 wells) with RPMI containing 10% FCS. On the day of assay, each well was confirmed sub‐confluent with comparable cell density in the assay plates, after which cells were washed and incubated in Seahorse assay medium supplemented with 1 mM pyruvate, 2 mM glutamine, and 10 mM glucose (adjust the pH to 7.4) in a 37 °C non‐CO2 incubator for 1 h prior to assay. Pharmaceutical compounds including oligomycin (1 μM), FCCP (0.2 μM for MKN1, 0.5 μM for NUGC4 and AGS), antimycin (0.5 μM) and rotenone (0.5 μM) within the Seahorse cell energy phenotype test kit were reconstituted and made to stressor mix at optimized concentration. The oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were then recorded and analyzed by Wave software (Seahorse). At least three biological independent experiments were conducted in triplicates.
ROS measurement
Total cellular ROS were measured using the ROS detection assay kit (Abcam) or ROS‐Glo™ H2O2 assay (Promega) after treatment of cells with 120 μM oxidative stress detection reagent or substrate solution, according to the manufacturer's instruction. Cells were then analyzed using a FLUOstar Omega microplate reader. The results of these experiments were normalized using the background readings from medium only wells in triplicates.
l‐lactate assay
Briefly, L‐lactate standards were firstly determined after the manufacturer's instructions (Abcam). After culture medium was collected and deproteinated, lactate production was assessed by measuring the absorbance at 450 nm and calculated using the equation obtained from the linear regression of the standard curve.
Clinical tissue microarrays
Human tissue microarrays containing gastric tumor and paired normal adjacent tissue samples from 84 GC patients undergoing surgically treatment, as well as normal gastric mucosa (n = 6) and ulcerative mucosa (n = 8) tissue biopsies, were collected between July 2006 and April 2007 at Shanghai General Hospital after written informed consent. Patients were recruited with the after requirements: met the medical diagnostic criteria of advanced stage of GC with no serious cardiopulmonary disease, and received neither chemotherapy nor radiotherapy before surgery. our study was approved by the ethics committee of Shanghai General Hospital. All GC patients were followed up until 2015, with detailed clinical and pathologic data.
Immunohistochemistry
The tissue microarray (TMA) sections were subjected to deparaffinization, rehydration and heat‐induced antigen retrieval to unmask the epitopes, and then incubated with TLR2 or SOD2 antibodies overnight. For negative control, the primary antibody was omitted and replaced by a matched isotype control IgG. For positive control, we used a metastatic human GC sample. after incubation with HRP‐conjugated secondary detection reagent and substrate (Dako), whole slides were visualized and scanned (Aperio T2 scanner). Staining intensity was independently scored by 2 pathologists blinded to the clinical data (0, no color; 1, weak; 2, moderate; 3, strong), and the percentage of positive cells (0, <5%; 1, 6–25%; 2, 26–50%; 3, 51–75%; and 4, 76–100%) was semi‐quantitatively assessed. The final scores (0–12) were then calculated by multiplying these 2 values. For analysis of clinical parameters, patients were divided into 4 subgroups based on this final score (negative, 0–2; weak, 3–4; moderate, 5–8; and strong, 9–12). Furthermore, overall survival analyses comparing patients with low expression of TLR2 or/and SOD2 (negative and weak) against those with high expression of TlR2 or/and SOD2 (moderate and strong) were conducted in this cohort.
Statistical analyses
Statistical analysis was performed using SPSS version 21.0. All data were shown from three independent experiments and presented as the mean ± SEM. Statistical comparisons were determined using student's t‐test. Pearson's Chi‐square tests were used for analysis of the associations between TLR2 and SOD2 expression as well as the clinical parameters. Univariate and multivariate Cox regression models were used to analyze independent indicators of patient prognosis. Kaplan–Meier plots and log‐rank tests were also performed for survival analysis. A value of p < 0.05 was considered statistically significant.
Results
TLR2 activation in GC cells stimulates OXPHOS and glycolysis
To assess whether TLR2 regulates metabolic changes in human GC cells, two human GC cell lines with high TLR2 expression, MKN1 and NUGC4, were selected15 (Supporting Information Fig. S1A) and subjected to cell energy phenotyping after stimulation with the TLR2 ligand, P3C for 24 h. In both cell lines, the basal oxygen consumption rate (OCR) and the OCR induced by the proton ionophore (uncoupler) FCCP (which measures maximal respiration capacity) were increased by the presence of P3C (Figs. 1 a and 1 b). Spare respiratory capacity (SRC), a measure of the ability of cells to respond to increased energy demands (i.e. the difference between basal and maximal OCR), was then calculated: P3C‐treated cells displayed a significant increase (2–3‐fold) compared to PBS‐treated control cells, suggestng an enhancement of mitochondrial respiration upon TLR2 activation (Figs. 1 c and 1 d). In addition, the basal extracellular acidification rate (ECAR), which measures acid production in cell culture media and thus represents the glycolytic rate, was significantly increased in P3C‐treated cells (Figs. 1 c and 1 d). Since the basal OCR/ECAR ratio was significantly reduced in P3C‐treated cells (Figs. 1 c and 1 d), these data indicate collectively that TLR2 activation augments both glycolysis and OXPHOS in TLR2‐high expressing cancer cells, with a greater cellular glycolytic capacity observed compared to OXPHOS. This notion is also supported by the finding that P3C treatment of the TLR2‐low expressing AGS human GC cell line has no effect on OCR, SRC, ECAR or basal OCR/ECAR (Supporting Information Fig. S1B and 1C). Furthermore, this metabolic effect appeared to be specific for TLR2 activation, predominantly through a TLR1/2 heterodimer, since lipopolysaccharide (LPS) ‐ recognized by TLR4 ‐ suppressed OCR and SRC in both MKN1 and NUGC4 cells, without significant alteration in ECAR or basal OCR/ECAR (Supporting Information Fig. S1D‐G). In addition, only minimal changes in OCR and SRC were observed in response to FSL‐1, a synthetic diacylated lipopeptide recognized by a TLR2/6 heterodimer (Supporting Information Fig. S1H‐K). We also confirmed that these metabolic alterations induced by different TLR agonists were not due to changes in cell viability or compound toxicity (Supporting Information Fig. S1L).
Figure 1.
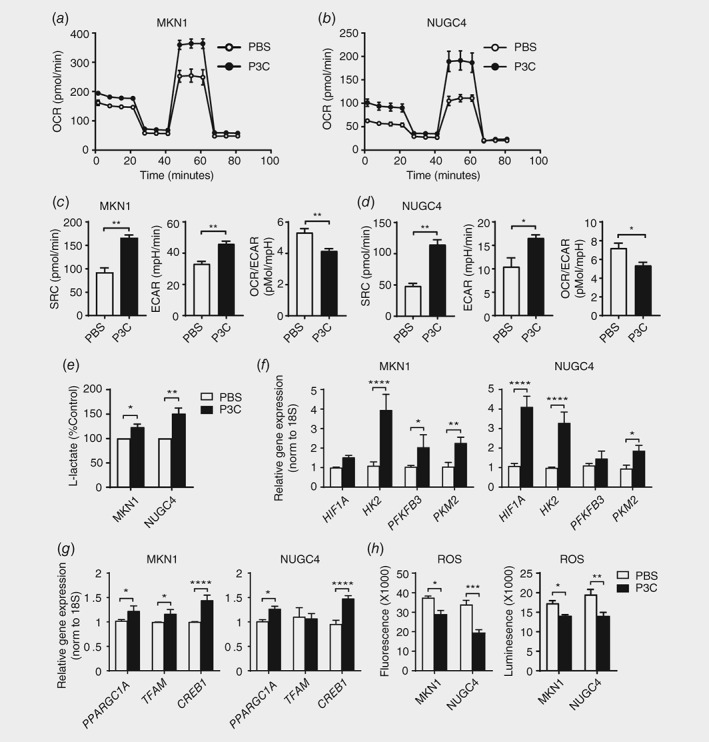
Alteration of the metabolic phenotype and ROS production in TLR2‐activated human GC cells. (a, b) Oxygen consumption rate was assayed using the Seahorse XFp analyzer (Seahorse Bioscience) in two GC cell lines, MKN1 and NUGC4, stimulated with PBS or TLR2 ligand P3C (10 μg/mL) for 24 h after sequential treatment with compounds oligomycin, FCCP and a mix of antimycin A and rotenone. (c, d) Spare respiratory capacity (SRC), basal extracellular acidification rate (basal ECAR) and basal OCR/ECAR were measured in MKN1 and NUGC4 cells. (e) Lactate production was measured in the culture medium of P3C stimulated cells compared to PBS control cells. (f, g) qPCR expression analyses of genes mediating glycolysis and oxidative phosphorylation in P3C treated MKN1 in (a) or NUGC4 cells in (b). (h) Total cellular ROS were measured using fluorescence‐based and luminescence ROS detection assay kits in two GC cell lines. Data are shown from three independent experiments in triplicates, and is presented as mean ± SEM, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
In light of the above findings, to further explore the role of ligand‐induced TLR2 activation in GC, subsequent stimulations were performed with the TLR1/2 agonist, P3C. Consistent with the above observations, the production of L‐lactate, the major lactate stereoisomer in human metabolism, was also increased in P3C‐treated cells (Fig. 1 e). Therefore, to investigate the molecular basis for the TLR2‐induced bias toward glycolysis in GC cells, we profiled the expression of numerous genes that control glycolysis. As shown in Figure 1 f, quantitative real‐time (q) PCR analyses revealed that genes encoding hypoxia‐inducible factor 1 alpha (HIF1Α), pyruvate kinase isozyme M2 (PKM2), hexokinase 2 (HK2) and 6‐phosphofructo‐2‐kinase/fructose‐2, 6‐biphosphatase 3 (PFKFR3) were significantly elevated. In particular, HK2, an enzyme that catalyzes one of the few rate limiting steps of glucose metabolism to convert glucose to glucose‐6‐phosphate,18 displayed the strongest increase (4–5‐fold) in both P3C‐treated NUGC4 and MKN1 cells (Fig. 1 f). Consistent with the elevated OXPHOS in P3C‐treated cells (compared to PBS‐treated controls), genes encoding factors that promote OXPHOS, namely peroxisome proliferator‐activated receptor gamma coactivator 1‐alpha (PPARGC1A), mitochondrial transcription factor A (TFAM) and cyclic AMP‐responsive element‐binding protein 1 (CREB1) were also increased (Fig. 1 g).
We next evaluated the cellular levels of oxidative stress in response to mitochondrial activity induced by TLR2 activation. For this purpose, real‐time total cellular reactive oxygen species (ROS) production was measured using a fluorescent, cell permeable probe, which reacts directly with a wide range of ROS. In parallel, we also assessed H2O2 levels in cell lysates by a luminescence‐based assay, since many ROS are converted to H2O2. Surprisingly, despite increased mitochondrial activity in TLR2‐activated cells, cellular ROS levels were significantly reduced in both P3C‐treated TLR2 high‐expressing MKN1 and NUGC4 cells (Fig. 1 h), whereas ROS levels were unchanged in the P3C‐treated TLR2 low‐expressing AGS cells (Supporting Information Fig. S1M).
Collectively, these observations suggest that activation of TLR2 signaling in high TLR2‐expressing GC cell lines gives rise to diverse metabolic reprogramming and cellular consequences, including augmented OXPHOS and glycolysis, increased lactate production, and increased expression of key genes in control of both bioenergetics processes. By contrast, reduced ROS production may reflect a compensatory protective mechanism that is an adaptation to microbial stimulated stress and subsequent cellular stress and metabolic changes.
SOD2 is strongly induced upon TLR2 activation in human GC cells
To dissect the intricate antioxidant defense mechanisms and identify key genes contributing to the TLR2‐induced metabolic changes in human cancer cells, we performed DNA microarrays on TLR2 high‐expressing NUGC4 and MKN1 GC cells, and as a comparison, TLR2 low‐expressing AZ521 cells. The cells were stimulated with a combination of TLR2 ligands, P3C and FSL‐1, for 6 h (GSE108345). As shown in Figure 2 a, after TLR2 ligand stimulation, the expression of 135 and 52 genes was significantly upregulated by at least 1.5‐fold in NUGC4 and MKN1 cells, respectively, of which 19 genes overlapped. Genes upregulated in NUGC4 and MKN1 cells were involved in anti‐apoptosis and chemokine activity, which mediate TLR2‐induced pro‐oncogenic properties in human GC cells.15 We also observed that the manganese‐dependent SOD2 gene, whose enzyme has key reduction–oxidation (redox) functions in scavenging ROS, was significantly augmented upon ligand‐induced TLR2 activation in MKN1 and NUGC4 cells (Fig. 2 b). By contrast, only 23 genes were uniquely increased in AZ521 cells (Fig. 2 a).
Figure 2.
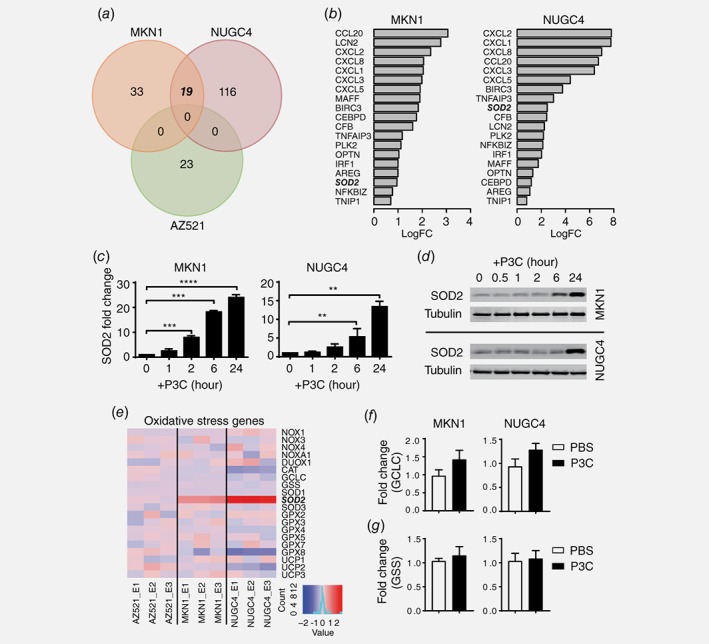
Identification of key TLR2‐regulated metabolic genes in human GC cells. (a) Venn diagram depicting significantly upregulated gene expression as determined by gene expression profiling of MKN1, NUGC4 and AZ521 cells stimulated for 6 h in triplicate with TLR2 agonists P3C and FSL‐1 together (each as 10 μg/mL), or saline control. (b) Significantly upregulated genes in both MKN1 and NUGC4 cells upon TLR2 activation are displayed ranked according to logFC change. Increased expression of SOD2 in both cell lines is indicated in bold italic text. (c) qPCR of SOD2 gene expression in MKN1 cells or NUGC4 cells after P3C stimulation (10 μg/mL) at indicated times. (d) Western blots of lysates of MKN1 and NUGC4 from (c). (e) Heatmap displaying the changes in expression of key redox genes analyzed from the gene array. (f, g) qPCR validation of selected GCLC and GSS genes in two GC cell lines treated with PBS or P3C. The expression data are presented from technical triplicates after normalization against 18S rRNA. Expression data are presented as the mean ± SEM. **p < 0.01, ***p < 0.001, ****p < 0.0001. [Color figure can be viewed at wileyonlinelibrary.com]
Consistent with the microarray data, qPCR analyses demonstrated that SOD2 mRNA levels were significantly induced within 2–6 h of P3C stimulation, and immunoblot assays confirmed elevated SOD2 protein levels at 24 h (Figs. 2 c and 2 d). In contrast, consistent with the lack of metabolic changes in TLR2 low‐expressing AGS cells, SOD2 protein expression in response to TLR2 activation was unchanged in AGS cells (Supporting Information Fig. S2A), suggesting that the upregulation of SOD2 expression is associated with TLR2 expression levels. Furthermore, in agreement with the observation of seahorse bioenergetics assay, FSL‐1 alone has no effect on SOD2 induction in TLR2 high‐expressing cell MKN1 and NUGC4 (Supporting Information Fig. S2B). As the consequences, subsequent experiments were conducted using P3C.
Interestingly, further analysis of many key redox genes19 implicated in other antioxidant systems including SOD family members, the major H2O2 scavenger catalase (CAT), glutathione peroxidase (GPX) family members, glutamate‐cysteine ligase catalytic subunit (GCLC), glutathione synthetase (GSS), NAPDH oxidase (NOXs) family members and dual oxidase1 (DUOX1), indicated that only SOD2 was uniquely upregulated in both high TLR2‐expressing GC cells we studied (Figs. 2 e– 2 g). Furthermore, gene‐set enrichment analyses (GSEA) using hallmark gene‐sets revealed that the 10 most TLR2‐responsive pathways in high TLR2‐expressing human GC cells included ROS, p53, c‐Myc, JAK‐STAT3 and Kras signaling gene networks (Supporting Information Fig. S2C), which are also implicated in respiration, ROS production and cell metabolism.20, 21, 22 Collectively, these data suggest that the expression of SOD2 is strongly induced at the transcriptional level upon TLR2 activation in high TLR2‐expressing cancer cells, and is associated with enrichment of multiple metabolic or oxidative stress related genes.
SOD2 is required for TLR2‐induced metabolic changes
To investigate the involvement of SOD2 in TLR2‐regulated metabolic alterations, we employed siRNA‐mediated knockdown of SOD2 in MKN1 and NUGC4 cell lines. Upon transfection, SOD2 expression was reduced by 70–80% at the mRNA and protein levels, and the P3C‐induced significant upregulation of SOD2 in nontargeting control (NTC) cells, which was abolished in cells transfected with SOD2 siRNA to knockdown SOD2 (Supporting Information Fig. S3A and 3B). Cells with SOD2 deficiency exhibited a significant reduction of maximal mitochondrial respiration, measured by FCCP‐stimulated OCR, compared to that of NTC cells, regardless of P3C treatment (Figs. 3 a and 3 c). Furthermore, the P3C‐induced modulation of SRC, basal ECAR and OCR/ECAR parameters, along with L‐lactate production, was abolished in SOD2 siRNA transfectants (Figs. 3 b, 3 d and 3 e). In addition, the TLR2‐induced suppression of cellular ROS levels in NTC cells (consistent with parental MKN1 and NUGC4 cells; Fig. 1 h) was reversed in SOD2‐deficient MKN1 and NUGC4 cells, whereby enhanced ROS levels were observed irrespective of P3C treatment (Fig. 3 f). Taken together, these data reveal an essential role for SOD2 in TLR2‐mediated metabolic alterations in cancer cells.
Figure 3.
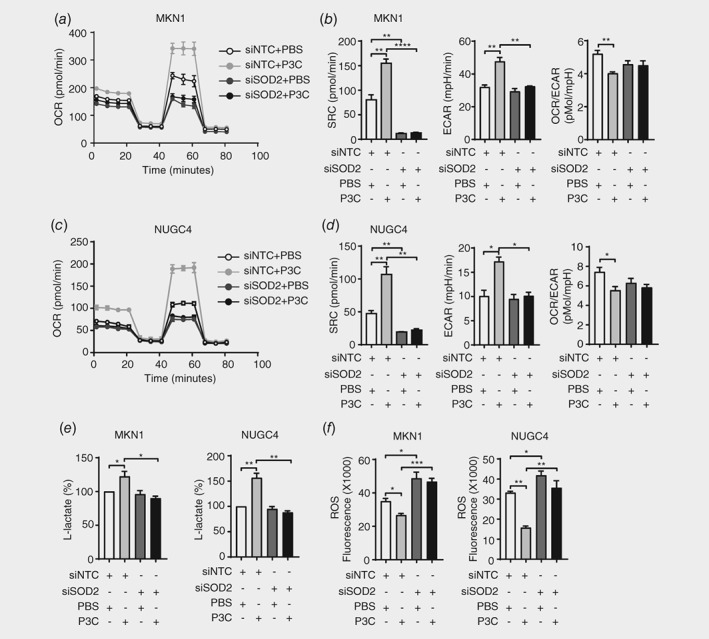
SOD2 is required for TLR2‐driven metabolic reprogramming and the limiting of ROS. (a‐d) Two GC cells lines were transiently transfected with SOD2 siRNA or non‐target control (NTC) siRNA for 48 h after 24 h P3C stimulation, then subjected to cell metabolic phenotype assay using Seahorse XFp analyzer to measure OCR (a, c), SRC, basal ECAR and basal OCR/ECAR (b, d). (e) Lactate production was measured in the culture medium of siRNA transfectants treated with P3C or PBS control. (f) Cellular ROS in siRNA transfected MKN1 from (a) and NUGC4 from (c) after P3C treatment was measured by fluorescence‐based ROS detection assay kits. Data are shown from three independent experiments, and is presented as the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
TLR2 employs multiple signaling pathways to upregulate SOD2 in cancer cells
Next we investigated the intracellular signaling mechanisms by which TLR2 induces SOD2 expression. Prior to stimulation of MKN1 and NUGC4 cells with P3C, cells were pre‐treated with a series of pharmacological inhibitors that block the activation of well‐documented TLR2‐driven signaling pathways, namely extracellular signal‐related kinase (ERK)1/2, p38 and JNK MAPKs, PI3K/AKT, JAK‐STAT3 and NF‐κB. Suppression of the JAK‐STAT3, JNK MAPK and NF‐κB cascades using CYT387, SP600125 and BAY11‐7085 inhibitors, respectively, at the indicated concentrations resulted in a significant (and comparable) inhibition of SOD2 expression (Figs. 4 a– 4 d). Further support specifically for STAT3 in mediating SOD2 expression downstream of TLR2 was provided by the observation that P3C‐induced SOD2 expression was significantly dampened in STAT3 knock‐out MKN1 cells derived by CRISPR‐Cas9‐mediated gene editing17 (Supporting Information Fig. S4A and 4B). By contrast, the blockade of p38 and ERK MAPKs, as well as PI3K/AKT, did not impair TLR2‐induced SOD2 expression in either MKN1 or NUGC4 cells (Supporting Information Fig. S4C‐E). Collectively, these results suggest that TLR2 activation utilizes multiple signaling cascades to render antioxidant protection via induction of SOD2 expression.
Figure 4.
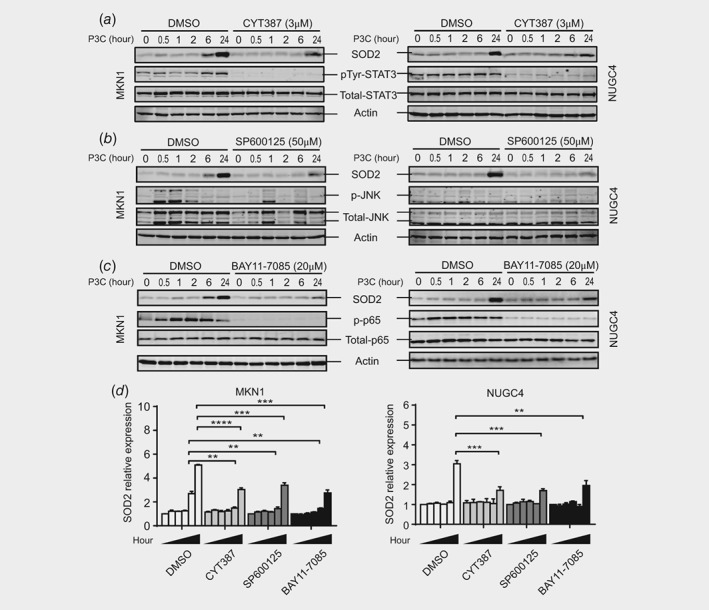
Multiple signaling pathways facilitate TLR2‐induced SOD2 expression. (a‐c) Two GC cell lines pretreated with DMSO vehicle control or specific pathway inhibitors at indicated concentrations for 1 h, were stimulated with P3C at the different time points. Cells were then lysed for analysis by Western blotting using phospho‐specific and total protein antibodies to Stat3 (a), JNK (b) and p65 (c) as well as total SOD2. (d) P3C induced SOD2 expression was determined by densitometry and normalization to actin expression levels. Data shown represent three independent experiments, and is presented as the mean ± SEM. **p < 0.01, ***p < 0.001, ****p < 0.0001.
Significant correlation between upregulated TLR2 and SOD2 expression in human GC
The clinical relevance of this novel TLR2‐SOD2 axis in GC was investigated by immunohistochemical analysis of gastric tissue microarrays (TMAs) derived from healthy individuals, or those with ulcerative gastric mucosa or cancer (tumor matched with adjacent nontumor tissues) after negative and positive controls were tested as shown in Supporting Information Figure S5. In GC patients, ∼50% of tumor‐bearing tissues displayed either strong (11%, n = 9) or moderate (38%, n = 32) cytoplasmic TLR2 staining, while no moderate or strong staining was observed in ulcerative (n = 8) or normal tissues from cancer‐free individuals (n = 6) (Figs. 5 a– 5 c). Similarly, strong (25%, n = 21) or moderate (21%, n = 18) cytoplasmic SOD2 staining was detected in half of GC tissues, while 25% of patients (n = 2) with stomach ulcers had moderate SOD2 expression. By contrast, normal tissue from healthy individuals exhibited only weak or no expression of SOD2 (Figs. 5 d–5 f). Importantly, comparison of gastric tumor versus paired adjacent nontumor tissues from patients (n = 84) also revealed that TLR2 and SOD2 expression was significantly upregulated in tumor tissues, with ∼50% (n = 41) of tumors displaying either strong or moderate TLR2 staining, whereas ∼75% of adjacent nontumor tissue presented weak or no TLR2 expression (Figs. 5 g– 5 k). Likewise, strong or moderate SOD2 expression was also observed in ∼50% of tumor tissues, while the majority (94%) of adjacent nontumor tissues exhibited no or weak expression of SOD2 (Fig. 5 l).
Figure 5.
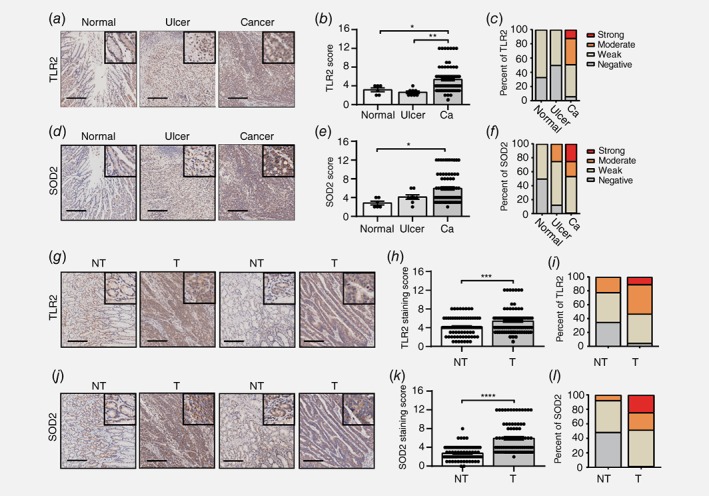
Significant correlation between TLR2 and SOD2 upregulation in human GC tissue. (a, d) Representative images of immunohistochemistry for TLR2 and SOD2 expression in normal (n = 6), ulcer (n = 8) and GC specimens (n = 84). (b, c, e and f) Immunohistochemical staining score and percentage of patients with different levels of TLR2 and SOD2 expression were visualized and determined as described in Materials and Methods. (g, j) Representative images for TLR2 and SOD2 expression in gastric tumor (T) and paired adjacent normal tissue (NT) (n = 84). (h, i, k and l) Immunohistochemical staining score and percentage of patients with different levels of TLR2 and SOD2 expression were visualized and determined as described in Materials and Methods. Scale bar, 200 μm; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. [Color figure can be viewed at wileyonlinelibrary.com]
Elevated expression of TLR2 and SOD2 is closely associated with disease progression and poor patient prognosis
We next investigated the correlation between TLR2 and SOD2 expression with key clinicopathological parameters, including metastasis, microvascular invasion, tumor size, American Joint Committee on Cancer (AJCC) stage, as well as overall patient survival (follow up time, 8.2–9 years). Notably, immunohistochemistry of tumor sections from GC patients revealed that cytoplasmic TLR2 staining steadily increased with advancing tumor stage, with highest expression in advanced stage IV tumors (Figs. 6 a and 6 b). Indeed, tumor tissues from 20% of stage III and 30% of stage IV cancer patients exhibited strong TLR2 staining, while no tumors from stage I or II cancer patients had strong TLR2 staining (Fig. 6 c). Interestingly, although TLR2 expression is well‐documented in innate immune cells,13 in tumor tissues elevated TLR2 staining was predominantly detected in the gastric epithelium rather than immune cell‐rich connective or stroma tissues (Fig. 6 a), which further supports the notion that cancer cell intrinsic TLR2 plays a major role in promoting tumorigenesis.14, 15 Furthermore, increased TLR2 expression was significantly associated with distant metastasis, microvascular invasion, tumor size, as well as AJCC stage (Supporting Information Table. S2). Importantly, analogous to our observations for TLR2, SOD2 staining also steadily and significantly increased during disease progression, with ∼70% of stage IV tumors showing strong cytoplasmic staining in the epithelium, whereas stage I tumors were devoid of any strong SOD2 staining (Figs. 6 d– 6 f). In addition, the incidence of distant metastasis markedly correlated with SOD2 expression (Supporting Information Table. S3). Most notably, Spearman analyses revealed a significant correlation between the immunostaining of TLR2 and SOD2 in tumor tissues, thus further supporting the close inter‐relationship between TLR2 and SOD2 in cancer (Fig. 6 g).
Figure 6.
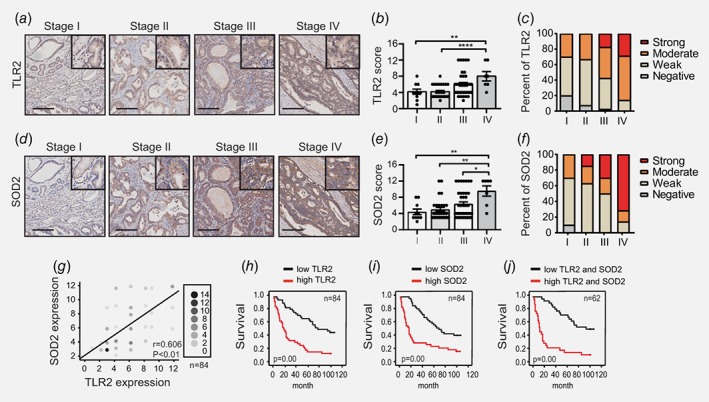
Elevated expression of TLR2 and SOD2 associates with disease progression and poor prognosis in human GC. (a, d) Representative images of immunohistochemistry for TLR2 and SOD2 expression in GC tissue in different stages of disease (n = 84). (b, c, e and f) Immunohistochemical staining score and percentage of patients with different levels of TLR2 and SOD2 expression against disease stages were visualized and determined as described in Materials and Methods, *p < 0.05, **p < 0.01, ****p < 0.0001. (g) The correlation between TLR2 and SOD2 expression was shown on the graph (n = 84). The intensity of scale indicates the repetition frequency. (h, i and j) Kaplan–Meier graphs of overall survival for GC patients stratified into either low or high expressing groups for TLR2, SOD2 or both. A log‐rank test was used for each Kaplan–Meier dataset. Total patient number and p values are indicated. [Color figure can be viewed at wileyonlinelibrary.com]
A comparative analysis of the survival of GC patients stratified as ‘high’ or ‘low’ expression for either TLR2 or SOD2 demonstrated that patients with a high expression of TLR2 or SOD2 had a significantly poorer prognosis than those with low expression (Fig. 6 h and I). We also confirmed that the combination of both TLR2 and SOD2 expression yielded a highly significant predictive value for patient survival (Fig. 6 j). Importantly, a multivariate analysis revealed that apart from conventional key clinicopathological factors such as distant metastasis, microvascular invasion and stage, SOD2 expression is an independent indicator of patient prognosis (Supporting Information Table. S4). Collectively, in support of our in vitro study, these clinical data suggest that TLR2 and SOD2 play interactive roles in GC progression and together provide a superior predictive power for patient survival.
Discussion
Emerging evidence has demonstrated that immune cells can switch from OXPHOS toward glycolysis after recognizing microbial‐derived ligands.2 Indeed, a universal increase in aerobic glycolysis has been observed in many naïve immune cells, including neutrophils, dendritic cells, and macrophages, after activation of TLRs.8, 23 More recent studies have shown that a variety of microbial components can induce metabolic changes in immune cells, the precise nature of which is determined by TLR ligand, dose and cell type.7 For example, in human monocytes P3C stimulation enhances mitochondrial respiration, which can be inhibited by the complex I inhibitor rotenone, resulting in reduced cytokine production and phagocytic capacity; this contrasts to the suppressive effect on OXPHOS by LPS.7 Studies into the relationship between immunity and metabolism are predominantly based on immune cells and there is a relative paucity of information regarding the role of the epithelium in facilitating metabolic adaptions during disease, particularly cancer. In our study, we have investigated the epithelium of the gastrointestinal tract, which serves as the first line of host defense.
Using preclinical models of GC, along with clinical data, we have previously demonstrated that among TLRs, the specific upregulation of TLR2 (but not TLR4) both enhances the ligand‐induced growth responsiveness of GC cells, and is associated with poor patient outcomes.14, 15 In this current study, we now reveal that TLR4 activation has a suppressive effect on oxidative phosphorylation, but TLR2 activation causes diverse metabolic reprogramming in human GC (epithelial) cells. In these cells, we observe enhanced OXPHOS and glycolytic activity, with a bias toward the latter, which we propose may facilitate highly proliferative cancerous clones during the later progression stage of disease, thus contributing to unfavorable survival outcomes in patients. We note that the ability of TLR2 activation to direct metabolic changes is aligned with high TLR2‐expressing cancer cell lines, namely MKN1 and NUGC4, which is a clinically‐relevant observation since TLR2 expression is augmented in >50% of human GC cases.14, 15 In this respect, it is noteworthy that both TLR agonists and antagonists have been actively exploited for their safety and therapeutic efficacy as adjuvants or monotherapy in trials for metastatic diseases.24, 25, 26 Our findings reveal that TLR2 expression levels and activation can influence the metabolic state, and thus proliferative potential of cancer cells.
It is well established that malignant cells rely on the ‘Warburg effect’ for ATP production, biosynthesis and acidification of the tumor microenvironment.27 A number of studies also report that active mitochondrial OXPHOS is a feature of human cancers: this supports anabolism and energy needs through efficient ATP synthesis rates, and also by supplying intermediates in the tricarboxylic acid (TCA) cycle that are required for biosynthesis processes.28, 29 Furthermore, in light of the heterogeneous bioenergetics profile of cancer cells, cancer metabolism can be self‐reprogrammed in response to factors in the tumor microenvironment, such as glycaemia, hypoxia, and nutrients.29, 30 In support of this notion, we provide evidence here that a synthetic molecule TLR2 ligand, P3C, can trigger metabolic changes in cancer cells by increasing both glycolysis (preferentially) and OXPHOS; this finding suggests that these two bioenergetic signatures can cooperate to support tumor progression in certain cancers. Furthermore, this metabolic effect appeared to be highly specific, predominantly through a TLR1/2 heterodimer, rather than a TLR2/6 heterodimer. It is therefore worth considering the notion that this structural‐based specificity may contribute to the diverse bioenergetics signatures of cancer cells in response to host defense responses (e.g., those triggered by microbial pathogens), thus impacting on the pathogenesis of infection‐associated cancers, such as GC. Indeed, since many experimental data for TLR2 signaling has been generated using synthetic ligands, the use of whole bacteria (e.g. H. pylori) in future studies may provide more insight into the full range of potential metabolic responses after TLR2 signaling activation.
An important consideration that arises from our study stems from analysis of gene array data from cancer cell lines treated with TLR2 agonists (Fig. 2), which revealed a pivotal alignment of specific signaling pathways and gene networks with metabolic changes that impact on oncogenic cellular functions. We identified that key oncogenic signaling pathways are involved in metabolic regulation, such as ROS, p53, JAK‐STAT3 and c‐Myc, a finding that is corroborated by previous studies suggesting that p53 and c‐Myc activation mediate ROS production associated with oxidant stress or enhanced cellular proliferation.31, 32 In addition, mitochondrial STAT3 has emerged as a critical regulator of energy metabolism and cellular ROS, as it is implicated in neoplastic transformation of Kras mutant cells.33, 34
Another key finding of our study is that SOD2, a mitochondrial redox protein, was identified as a master regulator required for metabolic reprogramming of cancer cells in response to TLR2 activation, as evidenced by inhibition of oxygen consumption rate, ECAR and lactate production upon siRNA‐mediated targeting of SOD2. This indispensable role of SOD2 in both oxidative respiration and glycolysis is highly likely attributed to SOD2 regulation of the cellular redox balance. For instance, SOD2 regulates dismutase enzymatic activity to convert superoxide generated from mitochondrial respiration to H2O2, the levels of which are closely connected to redox capability and the intracellular ROS equilibrium.35, 36 Indeed, SOD2 depletion in HEK293T cells elevates intracellular ROS, causing ultrastructural abnormalities in mitochondria and poor clonogenic fitness.37 In this respect, our observation that high TLR2‐SOD2 activity in GC cells (i.e. MKN1, NUGC4) abrogates intracellular ROS levels suggests that SOD2 may be required for detoxifying acutely elevated ROS, in order to maintain homeostasis in response to TLR2 stimuli. Conversely, SOD2 has been shown to regulate cellular redox flux by limiting the glucose consumption rate and glycolysis in mouse embryonic fibroblasts,38 while increased H2O2 release from mitochondria in SOD2‐overexpressing cancer cells promotes the metabolic shift to glycolysis by activating AMPK.39 Therefore, TLR2‐driven SOD2 induction may also enforce a more glycolytic metabolic phenotype to further limit ROS levels originating from TLR2‐induced oxidation.
SOD2 is a highly regulated nuclear‐encoded gene, whose expression can be altered at transcriptional, translational and posttranslational levels.40 Sequence‐specific transcription factors such as NF‐κB, AP1 and p53 have been shown to regulate SOD2 expression by directly binding to promoter elements or by interacting as cofactors under oxidative stress conditions.40 SOD2 has been also shown to be upregulated in A549 human lung cancer cells infected with an adenoviral vector expressing a hyperactive mutant of STAT3, STAT3‐C,41 suggesting augmented SOD2 expression in various human cancers correlates with STAT3 hyperactivation. In support of this idea, we provided evidence that ligand‐induced TLR2 activation upregulates SOD2 expression, which was partially suppressed by inhibitors targeting multiple signaling pathways, such as JAK‐STAT3, NF‐κB and JNK MAPK, which are dysregulated in many cancers, including GC.14, 42, 43, 44
In the context of cancer, the single nucleotide polymorphism SOD2 rs4880 is associated with a high risk of lymph node metastasis and poor outcomes in GC patients undergoing fluorouracil‐based chemotherapy.45, 46 Elevated SOD2 expression has also been associated with increased aggressiveness and metastasis in gastric and colorectal cancers,47, 48 which may be attributed to enhanced MMP production.49 More recently, it was reported that increased SOD2 in human breast cancer cells disrupts mitochondrial bioenergetics and enhances the ‘Warburg effect’ by activating the CaMKII‐AMPK axis and cellular H2O2 production.39 Here, our evaluation of the clinical relevance of the TLR2‐SOD2 axis using tissue microarrays revealed that similar to TLR2, upregulation of SOD2 protein expression is significantly associated with multiple clinical parameters, and is an independent indicator for patient survival. Therefore, activation of the TLR2‐SOD2 axis may foster a malignant phenotype by limiting ROS production and promoting a metabolic shift toward high OXPHOS and glycolysis.
In summary, our current study reveals that metabolic reprogramming in TLR2‐high expressing GC cells, which reflect the majority of GC cases, aligns with the induction of the redox gene, SOD2, and that the TLR2‐SOD2 axis is associated with multiple clinical parameters of high malignancy and poor survival in GC patients. By furthering our understanding of the underlying mechanisms of TLR2‐driven metabolic alterations during tumorigenesis, our study has the potential to inform better combinations of traditional chemotherapies that targeting metabolic adaptations. Furthermore, considering that metabolism and immune modulation are hallmarks of cancer, along with the fact that TLR2 is upregulated in increasing numbers of diverse cancers, it is likely that out findings here will apply to many other cancer types displaying dysregulated TLR2 activity.
Availability of data and material
The datasets supporting the conclusion of our study are available in the Gene Expression Omnibus under accession number GSE108345.
Ethics approval and consent to participate
Our study involving the use of any human tissue was approved the ethics committee of Shanghai General Hospital. Written informed consents were obtained from all patients (IRB number: 2014KY047).
Supporting information
Figure S1 Protein expression of TLR2 in a human GC cell line panel and metabolic phenotype of GC cells in response to TLR agonists.
Figure S2. TLR2 activation induced SOD2 expression in GC cells, and enrichment of key signaling pathways was identified in TLR2 activated human GC cells.
Figure S3. Inhibition of SOD2 expression using siRNA in MKN1 and NUGC4 cells.
Figure S4. Multiple signaling pathways regulate P3C‐induced SOD2 expression.
Figure S5. Analysis of clinical relevance for TLR2 and SOD2 protein expression using TMA.
Table S1. Primer sequences for SYBR green based qPCR for human genes.
Table S2. Comparison of clinical pathological profiles of the TMA cohort with low and high TLR2 expression.
Table S3. Comparison of clinical pathological profiles the TMA cohort with low and high SOD2 expression.
Table S4. Univariate and multivariate analysis of factors for patient overall survival.
Acknowledgement
The authors thank Dr. Rebecca Smith for editing the study.
Conflict of interest: The authors declare that they have no conflict of interest to declare.
Contributor Information
Brendan J. Jenkins, Email: brendan.jenkins@hudson.org.au.
Ji Kun Li, Email: jkli65975@163.com, Email: lijikunphd@163.com.
References
- 1. Vander Heiden MG, Deberardinis RJ. Understanding the intersections between metabolism and cancer biology. Cell 2017;168:657–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Palsson‐McDermott EM, O'Neill LAJ. The Warburg effect then and now: from cancer to inflammatory diseases. Bioessays 2013;35:965–73. [DOI] [PubMed] [Google Scholar]
- 3. Zong WX, Rabinowitz JD, White E. Mitochondria and cancer. Mol Cell 2016;61:667–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ben‐Sahra I, Manning BD. mTORC1 signaling and the metabolic control of cell growth. Curr Opin Cell Biol 2017;45:72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer 2016;16:749. [DOI] [PubMed] [Google Scholar]
- 6. White E. Exploiting the bad eating habits of Ras‐driven cancers. Genes Dev 2013;27:2065–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lachmandas E, Boutens L, Ratter JM, et al. Microbial stimulation of different toll‐like receptor signalling pathways induces diverse metabolic programmes in human monocytes. Nat Microbiol 2016;2:16246. [DOI] [PubMed] [Google Scholar]
- 8. Krawczyk CM, Holowka T, Sun J, et al. Toll‐like receptor–induced changes in glycolytic metabolism regulate dendritic cell activation. Blood 2010;115:4742–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mills EL, Kelly B, Logan A, et al. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell 2016;167:457–70 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kawai T, Akira S. The role of pattern‐recognition receptors in innate immunity: update on toll‐like receptors. Nat Immunol 2010;11:373–84. [DOI] [PubMed] [Google Scholar]
- 11. Brun P, Giron MC, Qesari M, et al. Toll‐like receptor 2 regulates intestinal inflammation by controlling integrity of the enteric nervous system. Gastroenterology 2013;145:1323–33. [DOI] [PubMed] [Google Scholar]
- 12. Brun P, Gobbo S, Caputi V, et al. Toll like receptor‐2 regulates production of glial‐derived neurotrophic factors in murine intestinal smooth muscle cells. Mol Cell Neurosci 2015;68:24–35. [DOI] [PubMed] [Google Scholar]
- 13. West AC, Jenkins BJ. Inflammatory and non‐inflammatory roles for toll‐like receptors in gastrointestinal cancer. Curr Pharm Des 2015;21:2968–77. [DOI] [PubMed] [Google Scholar]
- 14. Tye H, Kennedy Catherine L, Najdovska M, et al. STAT3‐driven Upregulation of TLR2 promotes gastric tumorigenesis independent of tumor inflammation. Cancer Cell 2012;22:466–78. [DOI] [PubMed] [Google Scholar]
- 15. West AC, Tang K, Tye H, et al. Identification of a TLR2‐regulated gene signature associated with tumor cell growth in gastric cancer. Oncogene 2017;36:5134–44. [DOI] [PubMed] [Google Scholar]
- 16. Kim Y, Gupta Vallur P, Phaëton R, et al. Insights into the dichotomous regulation of SOD2 in cancer. Antioxidants 2017;6:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu L, Wu D, Gao H, et al. Clinical utility of a STAT3‐regulated miRNA‐200 family signature with prognostic potential in early gastric cancer. Clin Cancer Res 2018;24:1459–72. [DOI] [PubMed] [Google Scholar]
- 18. Ahn KJ, Kim J, Yun M, et al. Enzymatic properties of the N‐ and C‐terminal halves of human hexokinase II. BMB Rep 2009;42:350–5. [DOI] [PubMed] [Google Scholar]
- 19. Ježek J, Cooper K, Strich R. Reactive oxygen species and mitochondrial dynamics: the yin and Yang of mitochondrial dysfunction and cancer progression. Antioxidants 2018;7:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gough DJ, Corlett A, Schlessinger K, et al. Mitochondrial STAT3 supports Ras‐dependent oncogenic transformation. Science 2009;324:1713–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Solaini G, Sgarbi G, Baracca A. Oxidative phosphorylation in cancer cells. Biochim Biophys Acta 1807;2011:534–42. [DOI] [PubMed] [Google Scholar]
- 22. Rodic S, Vincent MD. Reactive oxygen species (ROS) are a key determinant of cancer's metabolic phenotype. Int J Cancer 2018;142:440–8. [DOI] [PubMed] [Google Scholar]
- 23. West AP, Brodsky IE, Rahner C, et al. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature 2011;472:476–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Iribarren K, Bloy N, Buqué A, et al. Trial watch: Immunostimulation with toll‐like receptor agonists in cancer therapy. OncoImmunology 2016;5:e1088631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li JK, Balic JJ, Yu L, et al. TLR agonists as adjuvants for cancer vaccines. Adv Exp Med Biol 2017;1024:195–212. [DOI] [PubMed] [Google Scholar]
- 26. Patra MC, Choi S. Recent progress in the development of toll‐like receptor (TLR) antagonists. Expert Opin Ther Pat 2016;26:719–30. [DOI] [PubMed] [Google Scholar]
- 27. Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci 2016;41:211–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wellen KE, Thompson CB. A two‐way street: reciprocal regulation of metabolism and signalling. Nat Rev Mol Cell Biol 2012;13:270–6. [DOI] [PubMed] [Google Scholar]
- 29. Jose C, Bellance N, Rossignol R. Choosing between glycolysis and oxidative phosphorylation: a tumor's dilemma? Biochim Biophys Acta 1807;2011:552–61. [DOI] [PubMed] [Google Scholar]
- 30. Garber K. Energy deregulation: licensing tumors to grow. Science 2006;312:1158–9. [DOI] [PubMed] [Google Scholar]
- 31. Wanka C, Steinbach JP, Rieger J. Tp53‐induced glycolysis and apoptosis regulator (TIGAR) protects Glioma cells from starvation‐induced cell death by up‐regulating respiration and improving cellular redox homeostasis. J Biol Chem 2012;287:33436–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vafa O, Wade M, Kern S, et al. C‐Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function. Mol Cell 2002;9:1031–44. [DOI] [PubMed] [Google Scholar]
- 33. Garama DJ, White CL, Balic JJ, et al. Mitochondrial STAT3: powering up a potent factor. Cytokine 2016;87:20–5. [DOI] [PubMed] [Google Scholar]
- 34. Gough DJ, Marié IJ, Lobry C, et al. STAT3 supports experimental K‐RasG12D–induced murine myeloproliferative neoplasms dependent on serine phosphorylation. Blood 2014;124:2252–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miao L. St. Clair DK. Regulation of superoxide dismutase genes: implications in disease. Free Radic Biol Med 2009;47:344–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liochev SI, Fridovich I. The effects of superoxide dismutase on H2O2 formation. Free Radic Biol Med 2007;42:1465–9. [DOI] [PubMed] [Google Scholar]
- 37. Cramer‐Morales K, Heer C, Mapuskar K, et al. SOD2 targeted gene editing by CRISPR/Cas9 yields human cells devoid of MnSOD. Free Radic Biol Med 2015;89:379–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sarsour EH, Kalen AL, Xiao Z, et al. Manganese superoxide dismutase regulates a metabolic switch during the mammalian cell cycle. Cancer Res 2012;72:3807–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hart PC, Mao M, de Abreu ALP, et al. MnSOD upregulation sustains the Warburg effect via mitochondrial ROS and AMPK‐dependent signalling in cancer. Nat Commun 2015;6:6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dhar SK, St. Clair DK. Manganese superoxide dismutase regulation and cancer. Free Radic Biol Med 2012;52:2209–22. [DOI] [PubMed] [Google Scholar]
- 41. Dauer DJ, Ferraro B, Song L, et al. Stat3 regulates genes common to both wound healing and cancer. Oncogene 2005;24:3397–408. [DOI] [PubMed] [Google Scholar]
- 42. Ernst M, Najdovska M, Grail D, et al. STAT3 and STAT1 mediate IL‐11–dependent and inflammation‐associated gastric tumorigenesis in gp130 receptor mutant mice. J Clin Invest 2008;118:1727–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shibata W, Maeda S, Hikiba Y, et al. C‐Jun NH2‐terminal kinase 1 is a critical regulator for the development of gastric cancer in mice. Cancer Res 2008;68:5031–9. [DOI] [PubMed] [Google Scholar]
- 44. Sasaki N, Morisaki T, Hashizume K, et al. Nuclear factor‐κB p65 (RelA) transcription factor is constitutively activated in human gastric carcinoma tissue. Clin Cancer Res 2001;7:4136–42. [PubMed] [Google Scholar]
- 45. Xu Z, Chen Y, Gu D, et al. SOD2 rs4880 CT/CC genotype predicts poor survival for Chinese gastric cancer patients received platinum and fluorouracil based adjuvant chemotherapy. Am J Transl Res 2015;7:401–10. [PMC free article] [PubMed] [Google Scholar]
- 46. Xu Z, Zhu H, Luk JM, et al. Clinical significance of SOD2 and GSTP1 gene polymorphisms in Chinese patients with gastric cancer. Cancer 2012;118:5489–96. [DOI] [PubMed] [Google Scholar]
- 47. Toh Y, Kuninaka S, Oshiro T, et al. Overexpression of manganese superoxide dismutase mRNA may correlate with aggressiveness in gastric and colorectal adenocarcinomas. Int J Oncol 2000;17:107–12. [DOI] [PubMed] [Google Scholar]
- 48. Malafa M, Margenthaler J, Webb B, et al. MnSOD expression is increased in metastatic gastric cancer. J Surg Res 2000;88:130–4. [DOI] [PubMed] [Google Scholar]
- 49. Nelson KK, Ranganathan AC, Mansouri J, et al. Elevated Sod2 activity augments matrix metalloproteinase expression:evidence for the involvement of endogenous hydrogen peroxide in regulating metastasis. Clin Cancer Res 2003;9:424–32. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Protein expression of TLR2 in a human GC cell line panel and metabolic phenotype of GC cells in response to TLR agonists.
Figure S2. TLR2 activation induced SOD2 expression in GC cells, and enrichment of key signaling pathways was identified in TLR2 activated human GC cells.
Figure S3. Inhibition of SOD2 expression using siRNA in MKN1 and NUGC4 cells.
Figure S4. Multiple signaling pathways regulate P3C‐induced SOD2 expression.
Figure S5. Analysis of clinical relevance for TLR2 and SOD2 protein expression using TMA.
Table S1. Primer sequences for SYBR green based qPCR for human genes.
Table S2. Comparison of clinical pathological profiles of the TMA cohort with low and high TLR2 expression.
Table S3. Comparison of clinical pathological profiles the TMA cohort with low and high SOD2 expression.
Table S4. Univariate and multivariate analysis of factors for patient overall survival.
Data Availability Statement
The datasets supporting the conclusion of our study are available in the Gene Expression Omnibus under accession number GSE108345.


