Abstract
The essential liver exocrine and endocrine functions require a precise spatial arrangement of the hepatic lobule consisting of the central vein, portal vein, hepatic artery, intrahepatic bile duct system, and hepatocyte zonation. This allows blood to be carried through the liver parenchyma sampled by all hepatocytes and bile produced by the hepatocytes to be carried out of the liver through the intrahepatic bile duct system composed of cholangiocytes. The molecular orchestration of multiple signaling pathways and epigenetic factors is required to set up lineage restriction of the bipotential hepatoblast progenitor into the hepatocyte and cholangiocyte cell lineages, and to further refine cell fate heterogeneity within each cell lineage reflected in the functional heterogeneity of hepatocytes and cholangiocytes. In addition to the complex molecular regulation, there is a complicated morphogenetic choreography observed in building the refined hepatic epithelial architecture. Given the multifaceted molecular and cellular regulation, it is not surprising that impairment of any of these processes can result in acute and chronic hepatobiliary diseases. To enlighten the development of potential molecular and cellular targets for therapeutic options, an understanding of how the intricate hepatic molecular and cellular interactions are regulated is imperative. Here, we review the signaling pathways and epigenetic factors regulating hepatic cell line-ages, fates, and epithelial architecture.
1. Hepatic specification
1.1. Establishment of hepatic competence by pioneer factors
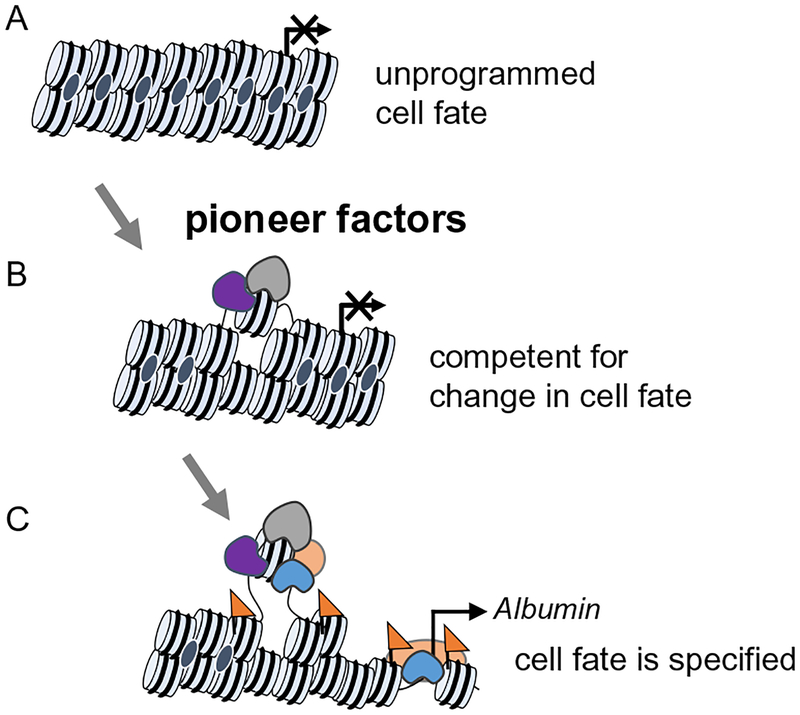
Multipotent progenitors establish competence to differentiate into specific lineages but not others. Embryonic foregut endoderm cells have not expressed liver-specific genes yet, but the isolated foregut progenitors retain the competence to respond to liver inductive signals (Gualdi et al., 1996). This result proposes that the molecular nature of competence must be cell intrinsic, such as the combined pattern of transcription factors and chromatin states. The transcription factors Forkhead Box A1 (FoxA1) and FoxA2 are expressed in foregut endoderm cells, and conditional deletion of both genes in foregut endoderm fails to induce hepatic genes, such as albumin (Alb) and transthyretin, in response to fibroblast growth factor 2 (Fgf2) signaling (Lee, Friedman, Fulmer, & Kaestner, 2005). This study provides genetic evidence that FoxA provides the hepatic competence to respond to the inductive cue. The Alb enhancer is pre-occupied by FoxA as well as Gata Binding Protein (Gata) factors in foregut endoderm cell chromatin before the onset of Alb expression (Bossard & Zaret, 1998; Gualdi et al., 1996). FoxA and Gata belong to a special class of transcription factors known as “pioneer factors” that have unique ability to engage closed and silent chromatin converting it to an open and permissive chromatin state (Fig. 1) (Iwafuchi-Doi & Zaret, 2016). A genome-wide study using mouse embryonic stem cell (ESC)-derived endoderm and early hepatic cells further support that FoxA genes function as pioneer factors in hepatic differentiation. FoxA preoccupies promoters and enhancers of silent hepatic genes before their differentiation is directed to hepatic cells (Xu et al., 2012). Taken together, pioneer factors FoxA and Gata first engage silent chromatin in foregut endoderm to impart the competence for hepatic fate. Given that different pioneer factors also work in other developmental contexts (Iwafuchi-Doi & Zaret, 2014), the studies of hepatic competence provide general principles by which multipotent progenitors gain the competence to differentiate into specific lineages but not others.
Fig. 1.
Initial chromatin targeting of pioneer factor and subsequent events. (A) Closed chromatin configuration in either a low signal or repressed state. (B) Pioneer factors (gray shape) locally open gene regulatory regions in chromatin, referred to as a “poised” state. This enables cooperative binding with other transcription factors (purple shape) and establishes competence to respond to signals. (C) An active chromatin state is established (active histone marks, orange triangles). Additional transcription factors (orange and blue shapes) are recruited promoting cooperative and stable binding.
1.2. The onset of hepatic morphogenesis
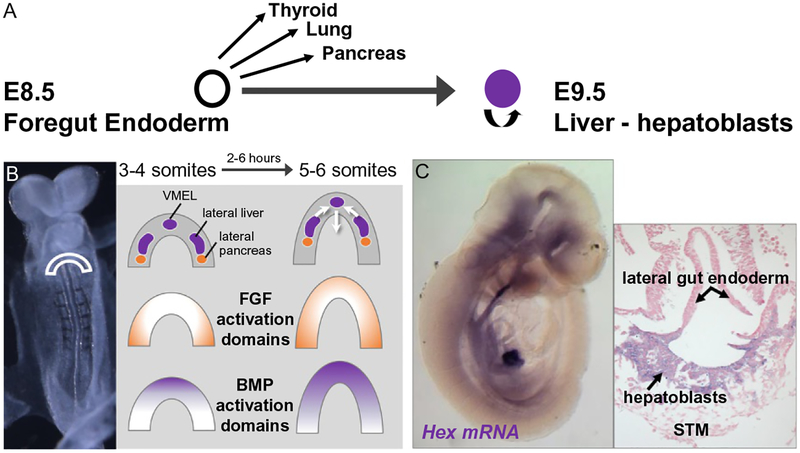
The definitive endoderm is established during gastrulation and invaginates at the anterior end to generate the foregut, which ultimately gives rise to liver, pancreas, lung, and thyroid (Fig. 2). At mouse embryonic day 8.0 of gestation (E8.0), liver progenitors arise from paired lateral domains in the ventral foregut as well as from a small population in the ventral midline endodermal lip (Fig. 2) (Angelo, Guerrero-Zayas, & Tremblay, 2012; Tremblay & Zaret, 2005). At E8.5, morphogenetic movements help close off the foregut, where the paired lateral domains and ventral midline endodermal lip cells merge to create the hepatic endoderm, composed of cells known as hepatoblasts. Shortly after hepatic specification (E8.5–E9.0), the hepatic endoderm thickens as the cells transition from a columnar epithelium to a pseudostratified epithelium (Bort, Signore, Tremblay, Martinez Barbera, & Zaret, 2006). Between E9.0 and E9.5, hepatoblasts delaminate from the epithelium and migrate into the adjacent septum transversum mesenchyme to form the nascent liver bud (Fig. 2) (Bort et al., 2006) (reviewed in Ober & Lemaigre, 2018).
Fig. 2.
Specification of the foregut endoderm. (A) During embryogenesis, the thyroid, lung, liver, and ventral pancreas arise by the budding of diverticula from the ventral foregut endoderm that contains multipotent progenitor cells. (B) Hepatic and pancreatic organogenesis requires complex and temporally precise FGF and BMP signaling. At the 3–4 somite stage of mouse development, FGF target genes are activated in lateral endoderm, whereas BMP targets are activated in the ventral midline endodermal lip (VMEL). Therefore, hepatic domains initially reside in a region of high BMP activity at the ventral midline endodermal lip and high FGF activity at the lateral endoderm.(C) Once specified, the Hex-positive hepatoblasts then delaminate from the epithelium and migrate into the septum transversum (STM).
1.3. Hepatic specification by signals from adjacent mesoderm tissues
Hepatic fate is specified by combinatorial signaling through a progressive series of reciprocal tissue interactions between the endoderm epithelium and nearby mesoderm during morphogenetic movements of foregut closure (Fig. 2). At early somite stages, Fgf from the cardiac mesoderm and bone morphogenetic protein (Bmp) from septum transversum mesenchyme cells coordinately specify the hepatic fate in the ventral foregut endoderm and suppress the pancreas program (Deutsch, Jung, Zheng, Lora, & Zaret, 2001; Jung, Zheng, Goldfarb, & Zaret, 1999; Rossi, Dunn, Hogan, & Zaret, 2001). As a downstream regulator of Fgf signaling, mitogen-activated protein kinase (Mapk) pathway is activated first in the lateral hepatic progenitors, and then in the ventral midline endodermal lip progenitors about 1h later (Calmont et al., 2006). In contrast, downstream of Bmp signaling, mothers against decapentaplegic Drosophila homolog (Smad)1, 5, 8 are phosphorylated first in the ventral midline endodermal lip progenitors and then in the lateral hepatic progenitors about 1h later (Wandzioch & Zaret, 2009). The effect of the different order of Fgf and Bmp signaling on gene expression and cellular function remains to be determined.
After foregut closure, the hepatoblasts proliferate and migrate into the surrounding mesenchyme, interacting to form a bud that becomes vascularized. Loss-of-function studies in mice have demonstrated that kinase insert domain receptor (Kdr) which encodes the vascular endothelial growth factor receptor 2 (Vegfr2) is required for hepatic outgrowth, but not hepatic specification (Matsumoto, Yoshitomi, Rossant, & Zaret, 2001). Because of the early embryonic lethality observed in homozygous Kdr mutants, an embryonic explant culture system was used to assess the expansion of the albumin-positive hepatic cells. The liver bud was isolated at E9.5 and put into culture for 72h. The outgrowth of the hepatic endoderm was specifically affected in homozygous Kdr mutants, in contrast to the growth of the surrounding fibroblast cells or the initial expression of early liver genes in the endoderm. Additionally, the hepatic outgrowth observed in the explant cultures, induced by the endogenous endothelial cells in the wild-type and heterozygous explants, suggests that an intact vasculature and embryo are not necessary for hepatic outgrowth (Matsumoto et al., 2001).
With the advent of single-cell RNA sequencing and the ability to generate hepatic endoderm from human-induced pluripotent stem cells (iPSCs), a more thorough evaluation can be performed to determine the influence mesenchymal and endothelial cells impart on hepatic differentiation. Single-cell RNA sequencing of three-dimensional liver bud organoids reconstituted from human iPSC-derived hepatic endoderm, mesenchymal stem cells and endothelial cells established that the transcriptome states of the cells comprising the liver bud organoids more closely resemble the single-cell transcriptomes of human fetal hepatic cells isolated from samples at gestation weeks 10.5 and 17.5 than human adult liver (Camp et al., 2017; Takebe et al., 2015). In silico receptor-ligand pairing of the single-cell transcriptomes identified potential inter-lineage signaling mechanisms between specific cell lineages by means of complementary receptor-ligand pairs. The pairings implicated signaling pathways such as tumor necrosis factor (Tnf), Fgf, Janus tyrosine kinase/signal transducer, and activator of transcription (Jak/Stat), nuclear factor kappa-light-chain-enhancer of activated B cells (Nf-κB), hypoxia-inducible factor (Hif), and Vegf suggesting extensive crosstalk between hepatic, endothelial, and mesenchymal lineages (Camp et al., 2017). Using chemical inhibitors to investigate whether any of these signaling pathways influence the ratio of hepatic endoderm to endothelial cell composition of the liver bud organoids identified Nf-κB, Fgf, insulin-like growth factor (Igf), Jak, and Vegf as important (Camp et al., 2017). Focusing on the hepatic endoderm and endothelial cell interaction, chemical Kdr/Vegfr2 inhibitor treatment of human liver bud organoids or mouse ESC-derived hepatic endoderm co-cultured with endothelial cells resulted in impaired endothelial sprouting and hepatic differentiation (Camp et al., 2017; Han et al., 2018), independent of liver bud organoid self-condensation driven by mesenchymal cells (Camp et al., 2017; Takebe et al., 2015). As expected, since the majority of hepatic endoderm does not express Kdr/Vegfr2, the transcriptome of cultures consisting solely of human iPSC-derived hepatic endoderm treated with chemical Kdr/Vegfr2 inhibitors were not changed (Camp et al., 2017; Goldman et al., 2013). Because cultures consisting of only human iPSC-derived hepatic endoderm do express some mature hepatocyte markers, but their overall transcriptome is more similar to mouse E8.5–E10 liver, the conclusion that the endoderm-endothelial interaction is not required for initial hepatic specification is further supported (Camp et al., 2017).
The molecular regulation required for the in vivo process of forming a compact liver bud is unknown. However, in vitro studies strongly point to a requirement for mesenchymal cells. Using the liver bud organoid system, all possible combinations of human iPSC-derived hepatic endoderm, mesenchymal stem cells, and endothelial cells were tested for the critical cell type driving the dynamic collective movement of cells into a liver bud organoid (Takebe et al., 2015). The required cell lineage was identified as the mesenchymal stem cell component. Cell-cell contact appears to be required because conditioned medium generated by mesenchymal stem cells was not sufficient to drive collective cell movement and liver bud organoid formation (Takebe et al., 2015). Live imaging studies imply that the self-condensation dynamic is not based on active cell migration but rather on cell contraction through stress fibers, similar to mechanical contraction of viscoelastic body physical models (Shinozawa, Yoshikawa, & Takebe, 2016). During embryonic gastrulation, inward displacement of cell-cell junctions is driven by myosin II, a motor protein responsible cell contraction (Bertet, Sulak, & Lecuit, 2004; Pouille, Ahmadi, Brunet, & Farge, 2009; Shindo & Wallingford, 2014). Similarly, collective movement of cells to form the liver bud organoid requires myosin II. Self-condensation of liver bud organoids is antagonized by the treatment with blebbistatin, a myosin II ATPase inhibitor (Takebe et al., 2015). In total, these results suggest that mesenchymal stem cell-based traction force produced by the actomyosin cytoskeleton is central to the directed and drastic movements of cells observed in the formation of the liver bud organoid. Many in vivo experiments are needed to confirm whether these cell interactions are required for liver bud formation in a living organism.
The liver bud expansion intrinsically requires the divergent homeobox genes hematopoietically expressed homeobox (Hex) and prospero homeobox protein 1 (Prox1). Hex is the earliest gene known to be expressed and required for differentiation of hepatocytes (Fig. 2) (Keng et al., 2000; Martinez Barbera et al., 2000). At E9.5, Hex homozygous mutants form a ventral endoderm thickening, but no outgrowth of the specified hepatoblasts is observed. Similarly, Prox1 homozygous mutants exhibit a smaller liver and absence of hepatoblast migration due to an increase of E-cadherin expression and a failure to breakdown the laminin-rich membrane surrounding the liver bud. There are currently no known specific molecular markers associated with the function of initial endoderm thickening (Costa, Kalinichenko, Holterman, & Wang, 2003).
1.4. Chromatin basis of fate choice for liver versus pancreas
As discussed above, pioneer factors can open chromatin and establish permissive states for gene activation, but are alternative genes repressed? Small cell number chromatin immunoprecipitation (ChIP) studies using ventral foregut endoderm cells from mouse embryos identified a different “pre-pattern” of the chromatin states at hepatic versus pancreatic regulatory elements (Xu et al., 2011). In the ventral foregut, the pancreatic regulatory elements contained active (diacetylated histone 3 lysine 9 and 14 [H3K9acK14ac]) and repressed (trimethylated histone 3 lysine 27 [H3K27me3]) histone modifications, but the liver regulatory elements were devoid of these marks. During hepatic specification, the liver regulatory elements gain H3 acetylation, but regulatory elements of pancreatic genes retain the “bivalent” chromatin state in hepatoblasts. When enhancer of zeste homolog 2 (Ezh2), a key methyltransferase for H3K27me3 modification and catalytic subunit of polycomb repressive complex, was deleted in mouse foregut endoderm cells, pancreas specification was promoted, and the liver bud became smaller (Xu et al., 2011). Thus, the repressed chromatin state can modulate the liver versus pancreas fate choice by suppressing the pancreas lineage. More comprehensive, genome-wide studies reveal dynamic H3K27me3 patterns during endodermal lineages progression (van Arensbergen et al., 2010; Xie et al., 2013; Xu et al., 2014). It is important to understand how stage- and lineage-specific gene loss or gain of H3K27me3 pre-patterns a specific lineage in multipotent progenitor cells.
2. Hepatic architecture
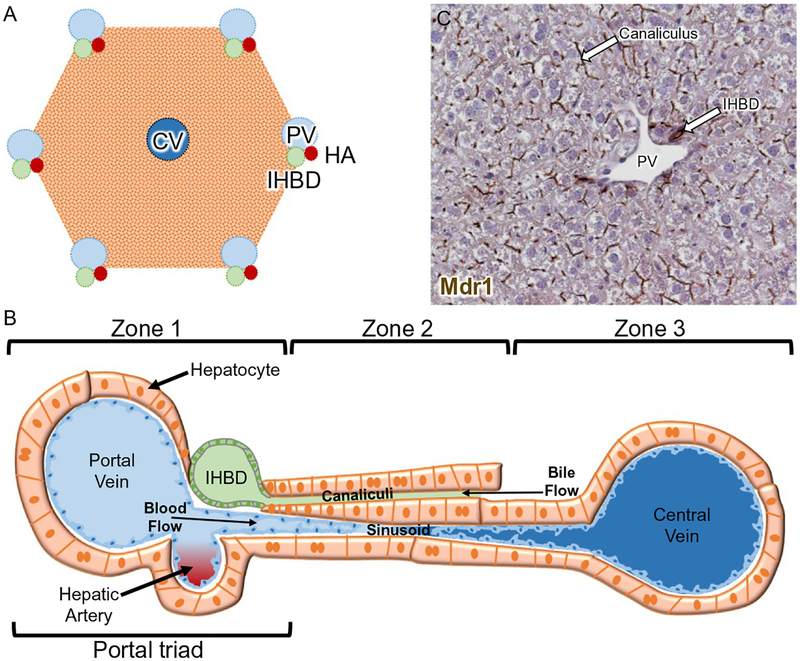
The hepatic lobule is an approximate hexagonal or pentagonal shape with a portal vein at each vertex and a central vein branch in the middle (Fig. 3) (Desmet, 2011). The portal vein (PV), hepatic artery (HA), and intrahepatic bile duct (IHBD) branches are arranged in close association at the vertices in structures known as portal triads. The hierarchical, branched structures of the PV, HA, and IHBD bifurcate approximately 17–20 times in human forming smaller and smaller branches (Crawford, 2002).
Fig. 3.
Hepatic architecture. (A) Schematic of the approximate hexagonal or pentagonal hepatic lobule. (B) One radius of the approximate hexagonal or pentagonal hepatic lobule. Hepatocyte cords run along the radius of the lobule between portal veins (PV) and central veins (CV), and are arranged into three zones: zone 1 near the PV, zone 2, and zone 3 near the CV. Bile is secreted from hepatocytes into the canalicular channels and transported to the intrahepatic bile ducts (IHBD). The sinusoidal capillaries carry oxygenated blood from the PV and hepatic artery (HA) past the hepatocytes to the CV.(C) Localization of multidrug resistance (Mdr1) protein encoded by ATP binding cassette subfamily B member 1 (ABCB1) on the apical hepatocyte canalicular membrane and apical surface of cholangiocytes comprising IHBDs in mouse liver.
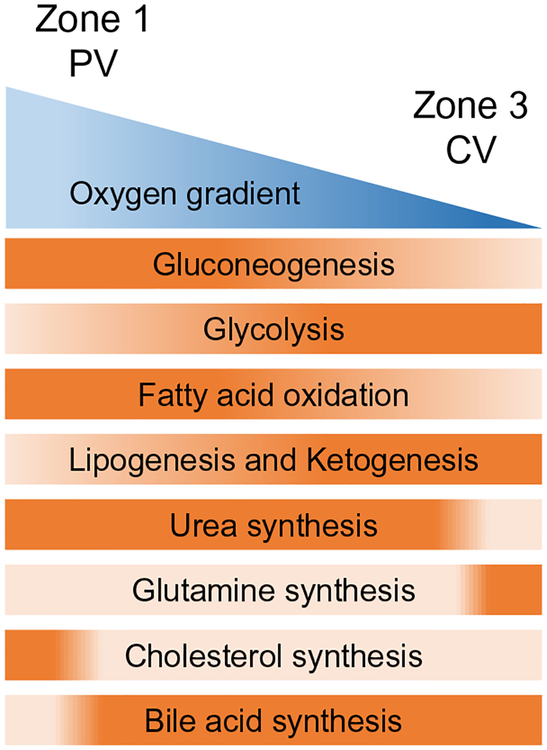
Hepatocytes are arranged in cords to fill the space between the central vein (CV) and PV. Collectively, hepatocytes perform a wide variety of tasks, including metabolic, detoxification, synthetic, and immunologic (Bhatia, Underhill, Zaret, & Fox, 2014). These functions are segregated between different zonal subpopulations of hepatocytes enabling various metabolic pathways to run in parallel (Fig. 3). Zone 1 is the periportal zone (closest to PV), zone 2 is the intermediate zone, and zone 3 is the pericentral zone (closest to CV). Zone 1 hepatocytes specialize in gluconeogenesis, fatty acid oxidation, urea synthesis, and cholesterol synthesis while zone 3 hepatocytes specialize in glycolysis, lipogenesis, ketogenesis, glutamine synthesis, and bile acid synthesis (Kietzmann, 2017; Torre, Perret, & Colnot, 2011).
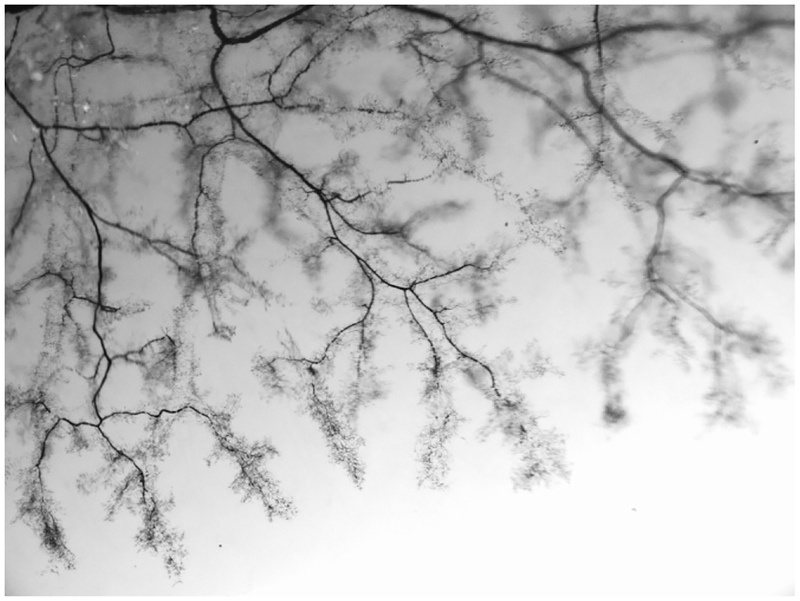
The assembly of hepatocytes as a polarized epithelium is critical for them to perform their exocrine and endocrine functions. Extending along the hepatocyte basal side are the sinusoids. Sinusoids are capillary-like structures that connect and permit blood flow from the PV and HA to the CV. Extending along the hepatocyte apical side are the bile canalicular channels. Canaliculi form a connected network extending throughout the parenchymal epithelium, allowing passive transport of bile produced by the hepatocytes (Gissen & Arias, 2015; Keppler, 2017). The multidrug resistance P-glycoprotein (Mdr1 or ATP binding cassette subfamily B member 1, ABCB1) encodes a large transmembrane efflux pump that decorates the apical membrane of hepatocytes (i.e., canalicular membrane) and cholangiocytes (i.e., bile duct epithelial cells) (Fig. 3) (Boyer, 2013; Fickert & Wagner, 2017). Bile flows from the canaliculi to the canals of Hering that are conduits lined partly by hepatocytes and partly by cholangiocytes (Fig. 3). The canals of Hering are eventually connected to ductules, the smallest ramification of the IHBD system (Roskams et al., 2004). All sizes of ducts are composed entirely of cholangiocytes that establish an apicobasal epithelial polarity and form the connected IHBD system. The small peripheral IHBDs merge into fewer, larger ducts, until finally a single hilar duct carries the bile out of the liver and transports it into the gallbladder for storage and ultimately into the intestine to aid in digestion. The IHBD system relies on its intricate three-dimensional structure to access all of the hepatocytes and effectively clear bile out of the liver (Fig. 4).
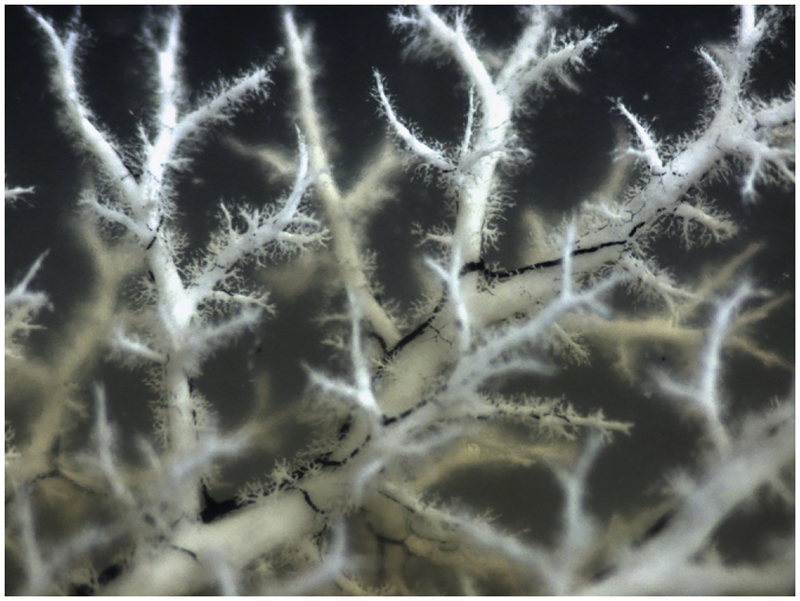
Fig. 4.
Hierarchical branching architecture of the intrahepatic bile duct (IHBD) system. Retrograde ink injections into the left lobe of the mouse liver IHBD system. Liver lobe cleared with benzyl benzoate and benzyl alcohol (BABB) solution to visualize hierarchical structure.
3. Hepatocyte cell fate decision
3.1. Molecular regulation of hepatocyte differentiation
Hepatocyte-specific transcription factors are already expressed in bipotential hepatoblasts whether they are entering the hepatocyte or cholangiocyte lineage transcriptional program (Gerard, Tys, & Lemaigre, 2017). During mid-gestation until after birth, specified hepatocytes undergo a process of postnatal differentiation where they adopt the physiological functions and morphology associated with the adult liver. During the last weeks of gestation, inhibition of glycolytic enzymes is coupled with the rise in gluconeogenic enzyme levels, reflecting maturation of the liver from a primarily glycolytic role in the first trimesters to a gluconeogenic role before birth (Devi, Habeebullah, & Gupta, 1992b). Additionally, there is an age-dependent reduction in hepatocyte membrane fluidity with liver maturation. The decreased hepatocyte membrane fluidity is suggested to be due to a decrease of lipid content, an increase in plasma membrane cholesterol, and a progressive reduction in the lipid:protein ratio throughout the prenatal period and in adult human liver (Devi, Gupta, & Habeebullah, 1992a). Finally, hepatocyte maturation is assessed by the expression and activity of phase-I drug metabolizing enzymes, including cytochrome P450 (CYPs), to modify drugs and environmental toxins. The absence of most phase-I enzyme expression at birth is thought to be responsible for the substantial pharmacokinetic differences and toxicity between newborns and adults (Hart, Cui, Klaassen, & Zhong, 2009; Peng et al., 2013, 2012; Sadler et al., 2016).
3.1.1. Models for postnatal hepatocyte differentiation
During the process of human and mouse postnatal differentiation, the expression of hepatic specific genes is regulated by a progressively complex transcriptional network deemed the six key master regulators or liver-enriched factors: hepatocyte nuclear factor 4a (Hnf4a), Hnf3b/FoxA2, Hnf1a, Hnf6/Onecut 1(OC1), Hnf1b, and liver receptor homolog 1 (LRH1)/nuclear receptor subfamily 5 group A member 2 (Nr5a2) in mouse and HNF4A, HNF3B/FOXA2, HNF1A, HNF6/OC1, CAMP-responsive element binding protein 1 (CREB1), and upstream transcription factor 1 (USF1) in human (Kyrmizi et al., 2006; Odom et al., 2006, 2004; White, Brestelli, Kaestner, & Greenbaum, 2005). It is unclear how master regulators, expressed in both embryonic hepatoblasts and adult hepatocytes, coordinate the gene expression changes that are necessary for hepatocyte zonal architecture and mature hepatocyte cell function.
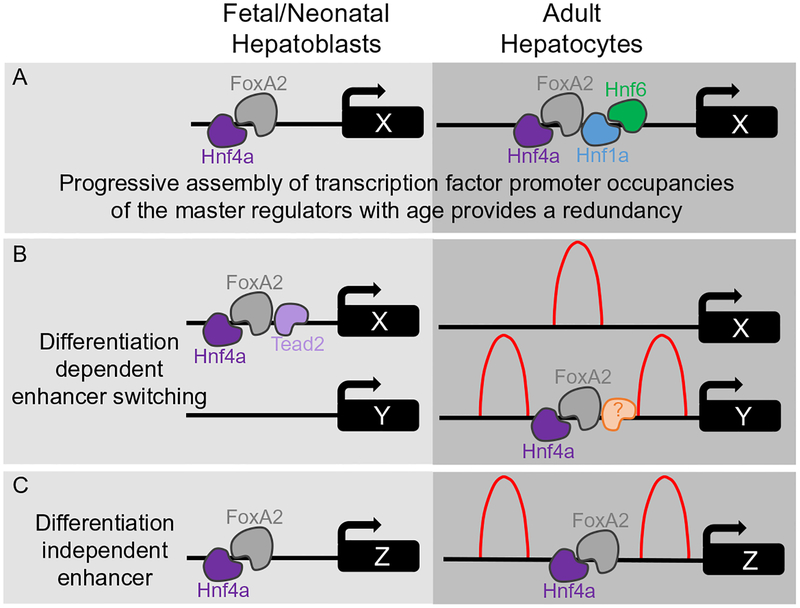
Published studies have identified a dichotomy in the severity of pheno-type and number of differentially expressed hepatic genes that is dependent upon whether key master regulators such as Hnf4a and FoxA2 are deleted at embryonic versus adult stages of liver development (Battle et al., 2006; Bochkis et al., 2008; Hayhurst, Lee, Lambert, Ward, & Gonzalez, 2001; Kyrmizi et al., 2006; Lee et al., 2005; Li, Schug, Tuteja, White, & Kaestner, 2011; Parviz et al., 2003; Sund et al., 2000). Two explanations have been provided for why embryonic deletion causes more severe pheno-types and greater changes in gene expression (Fig. 5). First, the complex transcription factor promoter occupancies of the master regulators at adult postnatal ages provides a redundancy to maintain postnatal hepatocyte gene expression, stabilizing the transcription factor network and potentially demonstrating synergistic interdependence (Fig. 5) (Hayhurst et al., 2001; Sund et al., 2000). This is based on protein-DNA interactions (i.e., chromatin immunoprecipitation followed by PCR of enhancer regions or sequencing) demonstrating that the number and complexity of transcription factor interactions on enhancer regions increase as hepatocyte maturation proceeds. There is a correlative rise in the expression of the key master regulators and each was found to occupy the gene regulatory regions of each other increasing occupation with age (Kyrmizi et al., 2006; Odom et al., 2006).
Fig. 5.
Mechanistic models of hepatocyte differentiation. (A) Schematic demonstrating progressive assembly of transcription complexes. Fetal/neonatal hepatoblasts have less complicated transcription factor complexes associated at enhancers of the same genetic locus compared to adult hepatocytes. (B) Schematic of differentiation-dependent enhancers. Example demonstrates that the enhancer at genetic locus X is occupied in fetal/neonatal hepatoblasts but not occupied in adult hepatocytes, and the enhancer at genetic locus Y is not occupied in fetal/neonatal hepatoblasts but is occupied in adult hepatocytes. (C) Schematic of a differentiation-independent enhancer. Example demonstrates that the enhancer at genetic locus Z is similarly occupied in both fetal/neonatal hepatoblasts and adult hepatocytes. (B and C) Representation of H3K4me1 average enrichment profiles of binding (red peaks) at differentiation-dependent and -independent enhancers in adult hepatocytes. Distinct binding patterns (bimodal and monomodal distribution) are present, depending on whether FoxA2 and Hnf4a are bound to enhancers.
Second, different gene targets and/or enhancers are regulated by the similar transcription factor complexes in embryonic versus adult time points (Fig. 5) (Alder et al., 2014). Hnf4a and FoxA2 interact with thousands of enhancer regions in a differentiation-dependent manner during hepatic development. In fact, 55% of Hnf4a and 60% of FoxA2 binding sites are uniquely occupied in either the embryonic hepatoblasts or adult hepatocytes, suggesting that these sites are regulated in a differentiation-dependent manner (Alder et al., 2014). Differentiation-dependent enhancer switching would enable key master regulators to perform distinct roles in both hepatoblasts and hepatocytes. This switch of DNA-binding sites is distinct from situations in which a transcription factor binds only high-affinity sites in one cell type. A motif-finding algorithm oPOSSUM was used to identify hepatoblast-enriched motifs bound by Hnf4a and FoxA2 (Alder et al., 2014). This analysis identified known DNA-binding motifs for key master regulators such as Hnf1a and Hnf1b validating the approach of motif analysis as these master regulators are known to be bound within the same peaks as Hnf4a and FoxA2. Surprisingly, this analysis also identified a motif for TEA domain transcription factor (Tead) whose transcriptional activity is dependent on interactions with yes-associated protein 1 (Yap1), a downstream component of the Hippo signaling pathway (Alder et al., 2014; Wu, Liu, Zheng, Dong, & Pan, 2008; Zhang et al., 2008). To determine whether the presence of Tead2 and Yap1 can influence the recruitment of Hnf4a and FoxA2 to differentiation-dependent enhancers, a Yap1 inducible transgenic mouse model was expressed in the liver in response to doxycycline (Alder et al., 2014; Dong et al., 2007). No changes of Hnf4a and FoxA2 were observed at differentiation-independent enhancers, those found occupied in both hepatoblasts and hepatocytes. However, detection of Hnf4a and FoxA2 occupancy was decreased at adult hepatocyte-specific differentiation-dependent enhancers upon ectopic expression of Yap1 (Alder et al., 2014). Hippo signaling is a critical regulator of liver size (Camargo et al., 2007; Dong et al., 2007). Overexpression of Yap1 results in an increase of liver size by fourfold (Camargo et al., 2007; Dong et al., 2007). Hippo pathway activity results in phosphorylation and inactivation of the transcriptional coactivator Yap1 (Oka, Mazack, & Sudol, 2008; Zhao et al., 2007), and in vivo inactivation of Hippo signaling is sufficient to dedifferentiate adult hepatocytes into cells resembling hepatoblast characteristics, expressing both hepatocyte and cholangiocyte markers (Yimlamai et al., 2014). Taken together, Hippo signaling may affect hepatocyte differentiation through regulating the pool of nuclear Yap1 and thereby influence a temporal switch of DNA sites bound by Hnf4a and FoxA2 at differentiation-dependent enhancers via direct interaction with Yap1 or indirectly activating a repressor that masks adult hepatocyte enhancer regions in hepatoblasts.
3.1.2. Potential epigenetic regulators of enhancer switching
If enhancer switching is true at a more global level versus a few specific targets that recruit additional cofactors during hepatocyte postnatal differentiation, like Tead2 during hepatoblast stages (Alder et al., 2014), then it begs the question—how is occupation of transcriptional factors such as Hnf4a and FoxA2 at different targets regulated given that most of the master regulators are expressed in both embryonic and adult liver? Generally, enhancers exist in a primed state that is characterized by the presence of monomethylated histone 3 lysine 4 (H3K4me1) prior to acetylated histone 3 lysine 27 (H3K27ac) and enhancer activation (Calo & Wysocka, 2013). Indeed, differentiation-dependent enhancers in mouse and human ESCs are pre-marked by H3K4me1 (Creyghton et al., 2010; Rada-Iglesias et al., 2011; Zentner, Tesar, & Scacheri, 2011). Although H3K4me1 is promoted to be linked to enhancer activity, its presence at an enhancer is probably more accurately defined as providing a window of opportunity for activation, facilitating nucleosomal accessibility, and/or pioneer factor binding (Iwafuchi-Doi & Zaret, 2014). Adult liver ChIP sequencing data reveal that differentiation-independent (embryonic and adult) and -dependent (adult only) enhancer regions bound by Hnf4a and/or FoxA2 have the highest levels of H3K4me1 at each side of the central transcription factor binding site position (Fig. 5), exhibiting a bimodal distribution pattern of H3K4me1 at active enhancer regions (Alder et al., 2014; Hoffman et al., 2010). In contrast, differentiation-dependent embryonic sites that are not bound by Hnf4a and/or FoxA2 in adult liver show the highest levels of H3K4me1 in the central transcription factor binding site position (Fig. 5), exhibiting a monomodal distribution pattern of H3K4me1 at inactive enhancer regions (Alder et al., 2014; Hoffman et al., 2010). Conversely, enrichment patterns of the repressive histone modifications, H3K27me3, and trimethylated histone 3 lysine 9 (H3K9me3), are not predominately found at differentiation-dependent embryonic sites in adult liver as might be expected if this type of mechanism is used to exclude binding of transcription factors via chromatin compaction (Alder et al., 2014). In fact, only 3% H3K27me3 and 7% H3K9me3 of the differentiation-dependent embryonic enhancers were enriched in adult liver. The polycomb repressive complex protein Ezh2 expression is the highest in hepatoblasts at E9.5 and decreases after E13.5 with age, suggesting that Ezh2 downregulation is required for liver maturation, contrasting functional results have been obtained from in vivo loss of function and ex vivo knockdown studies (Aoki et al., 2010; Koike et al., 2014). In vivo inducible deletion of Ezh2 in a non-lineage-specific manner starting at E10.5 and analyzed at E18.5 by global gene expression analysis demonstrated that progression of differentiation into functional hepatocytes is impaired by loss of Ezh2 (Koike et al., 2014). In contrast, ex vivo short hairpin RNA-mediated knockdown of Ezh2 from isolated E14.5 hepatoblasts promoted differentiation into hepatocytes based on increased expression of key master regulators or liver-enriched factors (Aoki et al., 2010). Further studies are required to determine whether non-hepatic lineage cells influence the phenotype of Ezh2 loss or Ezh2 loss at discrete time windows impacts the phenotype and transcriptional landscape as hepatoblasts differentiate into hepatocytes. Nonetheless, analysis of publicly available DNase I hypersensitivity data confirms that 60% of the differentiation-dependent embryonic sites remain accessible in adult liver (Alder et al., 2014). Therefore, differentiation-dependent enhancer switching is not simply explained by polycomb-mediated chromatin compaction.
3.1.3. Potential role of alternative histones
In addition to transcriptional networks and covalent modification on his-tones, incorporation of histone variants can regulate gene transcription. H2A.Z, a highly conserved variant of the histone H2A, is known to be associated with nucleosomes adjacent to the transcription start sites and linked to dynamic changes in gene expression (Bargaje et al., 2012). The differentiation of embryonic stem cells to endoderm/hepatic progenitors is regulated by FoxA2 binding to nucleosomal DNA on H2A.Z-containing nucleosomes, followed by FoxA2 and H2A.Z recruitment nucleosome disassembly complexes to enable nucleosome depletion and cell differentiation (Li et al., 2012), implicating chromatin remodeling as a regulator of hepatic lineage-specific gene regulation during embryonic stem cell differentiation. The in vivo data supporting incorporation of histone variants remain to be generated for hepatoblast cell fate decisions and postnatal differentiation of hepatocyte and cholangiocyte identities.
3.2. Molecular regulation of hepatocyte zonation
Hepatic metabolic zonation and further delineation of differentiated hepatocyte functional subsets begin in the first weeks after birth (Jungermann & Katz, 1989). Single-cell RNA sequencing of mouse hepatocytes highlights that half of the detected transcripts are not randomly expressed (3496 of 7277 genes) (Halpern et al., 2017). This emphasizes that liver zonation is a highly regulated process. However, only 25% (884 of 3496 genes) of all zonated genes are regulated by Wnt/beta-catenin (Benhamouche et al., 2006; Halpern et al., 2017; Sekine, Lan, Bedolli, Feng, & Hebrok, 2006), and an additional 9% (298 genes) are suggested to be regulated by hypoxia, Ras-dependent signaling, and pituitary hormones (Halpern et al., 2017). Therefore, the regulation of many zonally localized hepatocyte genes remains to be uncovered (Fig. 6).
Fig. 6.
Zonal hepatocyte functions. Dark orange color indicates more activity for hepatocyte functions.
3.2.1. Wnt/beta-catenin signaling pathway
As stated above, the main signaling pathway involved in setting up and maintaining hepatocyte zonation is canonical Wnt/beta-catenin. To activate canonical Wnt signaling, Wnt ligands bind to frizzled cell surface receptors and co-receptors, low-density lipoprotein receptor-related protein 5 (LRP5) and LRP6 (Pinson, Brennan, Monkley, Avery, & Skarnes, 2000; Tamai et al., 2000), blocking the beta-catenin destruction complex thereby allowing beta-catenin to accumulate and be transported into the nucleus. In the nucleus, beta-catenin acts as a coactivator for the transcription of Wnt target genes by binding to transcription factors from the T-cell factor (TCF) and lymphoid enhancer factor (Lef) family. In the absence of Wnt ligands, frizzled and LRP receptors are inactive, and the destruction complex including adenomatous polyposis coli (Apc) and Axin act as a scaffold, recruiting newly synthesized cytoplasmic beta-catenin for phosphorylation and targeted ubiquitin-mediated proteasomal degradation (Kretzschmar & Clevers, 2017). The R-spondins and their leucine-rich repeat containing G protein-coupled receptors, (LGR)4 and LGR5 (de Lau et al., 2011; Glinka et al., 2011; Ruffner et al., 2012), act as agonists by opposing the degradation of beta-catenin. Active R-spondin-Lgr complexes bind and inactivate the transmembrane E3 ubiquitin ligases zinc and ring finger 3 (Znrf3) and ring finger protein 43 (Rnf43) permitting prolonged stabilization of beta-catenin (Hao et al., 2012; Koo et al., 2012).
In vivo beta-catenin stabilization is both necessary and sufficient for expression of the pericentral enzyme glutamine synthetase in mouse liver (Fig. 6). Inducible loss of beta-catenin in hepatocytes results in all hepatocytes portraying a more periportal phenotype and lack of zone 3 hepatocytes expressing glutamine synthetase (Gougelet et al., 2014). Additional support comes from liver-specific deletion of the Wnt co-receptors (Lrp5 and 6) and hepatocyte-specific deletion of the Wnt agonists (Lgr4 and 5), resulting in the lack of liver zonation and failure of hepatocytes to express zone 3 metabolic genes (Planas-Paz et al., 2016; Yang et al., 2014). In contrast, stabilization of beta-catenin due to liver-specific deletion of Apc, a negative regulator of beta-catenin stabilization, induces a zone 3 hepatocyte pheno-type and glutamine synthetase expression throughout the hepatic lobule (Benhamouche et al., 2006).
How Wnt activity is restricted or activated spatially in pericentral or zone 3 hepatocytes are unclear. The expression of Wnt ligands has been found in several hepatic cell types: hepatocytes, cholangiocytes, endothelial cells, liver sinusoidal endothelial cells (i.e., LSECs), stellate cells, and liver resident macrophages (i.e., Kupffer cells) (Zeng et al., 2007). Cell lineage-specific deletion of Wntless, a putative G protein-coupled receptor that transports all Wnts intracellularly for secretion, was used to determine which hepatic cell type is responsible for providing Wnt ligands for establishing and maintaining hepatic zonation (Yang et al., 2014). The results indicate that hepatocytes, cholangiocytes, and macrophages are not the source of Wnt ligands for beta-catenin activation and hepatic zonation. The loss of Wntless from endothelial cells using Tie2-Cre results in embryonic lethality, preventing assessment of proper hepatic zonation. However, screening all 19 Wnts by in situ hybridization identified that central vein (CV) endothelial cells specifically express Wnt2 and Wnt9b (Wang, Zhao, Fish, Logan, & Nusse, 2015). Finally, using VE-cadherin-CreERT2 to conditionally induce deletion of Wntless in endothelial cells resulted in loss of zone 3 pericentral expression of glutamine synthetase (Wang et al., 2015). Therefore, these data support a model where CV endothelial cells provide the source of Wnt ligands to activate the Wnt pathway in the pericentral zone 3.
3.2.2. Hedgehog signaling pathway
Another signaling pathway found to be important for maintaining liver zonation is hedgehog (Hh). To activate canonical Hh signaling, the ligands Sonic, Indian, and Desert hedgehog (Shh, Ihh, and Dhh) interact with the patched (Ptch1 and Ptch2) receptors, removing their inhibition of the co-receptor smoothened (Smo). Active Smo then triggers the nuclear localization and activation of the glioma-associated oncogene transcription factors (Gli1, 2, and 3) by preventing the conversion of Gli2 and Gli3 into transcriptional repressors (Petrov, Wierbowski, & Salic, 2017). In vivo conditional mouse hepatocyte-specific deletion of Smo at 8 weeks of age results in steatosis within 5 weeks (Matz-Soja et al., 2016). Interestingly, the lipid accumulation was observed in zone 1 hepatocytes near the PV. In correlation with the lipid accumulation in zone 1, upon Smo hepatocyte deletion, upregulation of the lipogenic transcription factors sterol regulatory element binding protein (Srebp) and peroxisome proliferator activator receptor (Ppar) as well as the enzyme fatty acid synthase (Fasn) is observed in zone 1. Normally these lipogenic genes are expressed in zone 3 to perform their liponeogenic function of generating fat by converting carbohydrates into fatty acids (Fig. 6). However, the ectopic expression of these lipogenic genes potentially drives ectopic liponeogenesis in zone 1 where hepatocytes do not possess the ability to use the generated lipid. This idea is mechanistically feasible, as Srebp1 does have binding sites located at −7000 and −500bp from the transcriptional start of the Fasn gene (Amemiya-Kudo et al., 2002; Morishita, Mochizuki, & Goda, 2014), and overexpression of Srebp1c is sufficient to upregulate Fasn gene expression in hepatocytes (Dentin et al., 2004). In confirmation of a Hh role in hepatocyte zonation, knockdown of Gli transcription factors in hepatocyte cultures results in similar phenotypic characteristics as observed with Smo deletion (Matz-Soja et al., 2016). Importantly, the changes in lipid metabolic and liver zonation in mouse hepatocytes deficient for Smo are independent of changes in cholesterol biosynthesis, glycogen content, and glycolysis, which suggests a specific role for Hh signaling as these processes encompass general and zonal hepatocyte functions.
3.2.3. Other key signaling molecules involved in hepatocyte zonation
Two additional key molecules mediate hepatic zonation. Hnf4a loss-of-function results in liver with a normal pericentral zone 3 expression of glutamine synthetase, but glutamine synthetase is expanded to be expressed in zone 1 periportal hepatocytes (Stanulovic et al., 2007). Data support a model where Hnf4a and beta-catenin compete for binding to the DNA-binding cofactor transcription factor (Tcf) (Gougelet et al., 2014). Thereby, Hnf4a/Tcf drives zone 1 gene expression and beta-catenin/Tcf drives zone 3 gene expression. Thus, Hnf4a opposes Wnt signaling to promote the expression of zone 1 periportal hepatocyte genes and inhibit the expression of zone 3 pericentral hepatocyte genes. Glucagon loss-of-function mice also appear to have normal pericentral zone 3 expression of glutamine synthetase, but glutamine synthetase is expanded to be expressed in zone 2 and a gradient toward zone 1 periportal hepatocytes (Cheng et al., 2018). Glucagon is secreted from the pancreatic alpha-cells and transported to the liver through the PV to the CV in order to support glucose homeostasis by stimulating hepatic gluconeogenesis in zone 1 hepatocytes. Interestingly, glucagon infusion appears to modulate expression of many Wnt/beta-catenin target genes. The effects are most pronounced in zone 1 hepatocytes where 28% of all genes are activated by glucagon and inhibited by Wnt/beta-catenin (Cheng et al., 2018). Therefore, the counter concentration gradients of glucagon and Wnt ligands may maintain the liver metabolic zonation.
Finally, liver zonation is influenced by oxygen and reactive oxygen species (ROS) however certain genes and functions within the different hepatocyte zones diverge in their level of sensitivity to oxygen modulation. Genes involved in glucose and drug metabolism are more readily influenced by blood flow and oxygen tension, while genes involved in ammonia detoxification and glutamine synthesis have a more stable and defined expression pattern in the face of oxygen manipulations (Kietzmann, 2017). While it is clear that oxygen can influence hepatocyte zonation, an in vivo mechanism through which the regulation occurs and the importance of oxygen pressure during developmental zone establishment and homeostasis remains unknown. Data support the idea that Wnt/beta-catenin and the hedgehog pathway can be modulated by hypoxia and hypoxia-inducible transcription factors (Hif). In vitro studies using colon cancer cell lines demonstrated that hypoxic conditions reduced levels of Apc via a Hif1a-dependent mechanism (Newton, Kenneth, Appleton, Nathke, & Rocha, 2010). Hif1a represses the Apc gene by binding to a hypoxia responsive element within the Apc promoter. In contrast, Apc can mediate repression of Hif1a, but simultaneously requires low levels of beta-catenin and nuclear factor κB (Newton et al., 2010). Further, data support, at minimum, a parallel activity of hedgehog and Hif to induce the expression of Fasn. Hypoxic induction in adult mice can induce a rapid hedgehog response observed through expression of the ligand Shh and evidence of activity through expression of Ptch1 in multiple organs including the liver (Bijlsma et al., 2009). Also, Srebp1 expression is upregulated and has the potential to subsequently bind to the promoter of Fasn activating its expression as similarly demonstrated for hedgehog signaling (Bijlsma et al., 2009). Therefore, the dynamic interplay between oxygen levels and signaling pathways may influence liver zonation and contribute to modulations of hepatic physiological function.
3.2.4. Hepatocyte zonation is dynamic
It is important to note that hepatocyte zonation is dynamic. The liver performs its exocrine and endocrine functions by responding to nutrition levels, drugs, environmental toxin intake, and levels of hormone and other blood borne factors that result in changes of gene expression patterns. Therefore, gradients of morphogenesis, such as Wnt, Hh, and hormones, in concert with oxygen, induce and restrict gene expression in different subsets of hepatocytes located in different zones of the hepatic lobule to segregate hepatocyte function. Given the innate ability of the liver to regenerate, it is this flexibility that allows hepatocytes of different zonal origins to effectively evade and adeptly adapt and replace damaged hepatocytes (Planas-Paz et al., 2016).
4. Cholangiocyte cell fate decision and intrahepatic bile duct morphogenesis
4.1. Intrahepatic bile duct formation
Mammalian organs with a branched, multicellular epithelial network, such as the lung, kidney, pancreas, and salivary gland form tubes by processes involving wrapping of an epithelial sheet, branching via subdivision or proliferation from an existing cellular compartment, or cavitation of a cylindrical cluster of cells (Baer, Chanut-Delalande, & Affolter, 2009). In contrast to these organs, the liver forms a branched IHBD network through a multistep process including specification of cholangiocytes and subsequent morphogenesis of the specified cholangiocytes into a tube (Lemaigre, 2010; Ober & Lemaigre, 2018; Tanimizu & Mitaka, 2017). This alternative process of tubulogenesis could theoretically bestow the potential of the liver to continually generate a connected biliary system coordinated with a normal enlarging parenchymal mass and enable the unique ability of the liver to regenerate after injury (Desmet, 2011). A contrast is observed in organs where the process of tubulogenesis uses terminally differentiated cells such as alveoli, glomeruli, and beta cells from which to build tubes with very little potential for regeneration.
4.1.1. Cholangiocyte specification
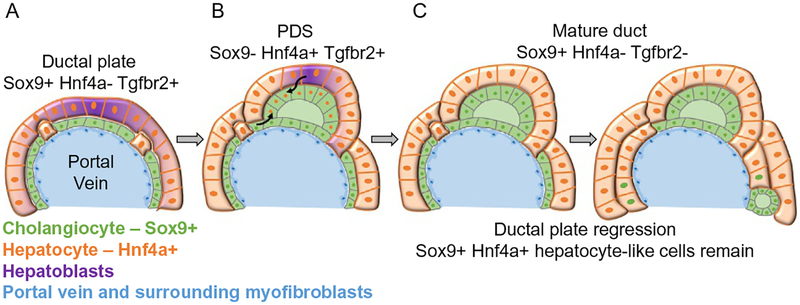
IHBD architectural formation is a highly complex and regulated process that occurs in a coordinated fashion along the portal vein (PV) network to form a connected IHBD branched network that intimately follows the PV branched network (Fig. 7). The first step in IHBD formation is cholangiocyte specification where hepatoblasts enter into the cholangiocyte transcriptional program detected by expression of specific cytokeratins (e.g., CK19) and the transcription factor Sry-related HMG box 9 (Sox9) (Fig. 8). Two-dimensional section analysis of human liver samples along with examination of experimental genetic mouse models exhibiting abnormal IHBD development have provided critical information regarding IHBD formation (Desmet, 1992; Raynaud et al., 2011; Terada, 2017; Van Eyken et al., 1988; Vestentoft et al., 2011). The cells that contribute to the IHBD structure are a subpopulation of the bipotential hepatoblasts in close proximity to the myofibroblasts surrounding PVs. Around E11–E14 in mice and 7–10 weeks of gestation in humans, a subpopulation of hepatoblasts forms a temporary structure appearing as a band of potential cholangiocytes, termed the ductal plate, encircling the PVs (Antoniou et al., 2009; Terada, 2017; Van Eyken et al., 1988; Vestentoft et al., 2011).
Fig. 7.
Three-dimensional association of the portal vein (PV) system (white) and the intrahepatic bile duct (IHBD) system (black). Retrograde ink injections into the left lobe of the mouse liver PV and IHBD system. Liver lobe cleared with benzyl benzoate and benzyl alcohol (BABB) solution to visualize hierarchical structure.
Fig. 8.
Schematic of temporal cholangiocyte specification and morphogenesis process.(A) Hepatoblasts that are in close association with the portal vein (PV) myofibroblasts begin to express Sox9 and enter into the cholangiocyte transcriptional program in addition to repressing the hepatocyte program. These tightly associated Sox9+ cells form a temporary structure termed the ductal plate as they appear to form a cover surrounding the PV. (B) Primitive ductal structures (PDS) or luminal structures surrounded by asymmetrical gene expressing cells begin to form. These are luminal spaces are surrounded by Sox9+ Hnf4a−Tgfbr2− cells on the PV side and Sox9− Hnf4a+ Tgfbr2+ cells on the parenchymal side. The origin of these cells is uncertain. They may arise from the parenchymal hepatoblast-like cells or cells contributing from the initially formed ductal plate (black arrows). (C) Symmetrical ductal structures or mature ducts are formed of Sox9+ Hnf4a− Tgfbr2− cells encircling the luminal structure. The remaining ductal plate cells that are not incorporated into an intrahepatic bile duct (IHBD) regress back to Sox9+ Hnf4a+ hepatocytes-like cells.
Hepatoblasts entering into the cholangiocyte program not only induce cholangiocyte-specific genes and repress hepatocyte genes but also undergo growth arrest during the specification process (Carpentier et al., 2011). There is evidence that growth arrest can initiate cholangiocyte specification, but it is not sufficient in all scenarios of genetic induced growth arrest. For example, loss of Prox1 or T-Box transcription factor 3 (Tbx3) in mice results in growth arrest and increased commitment of hepatoblasts to the cholangiocyte program (Ludtke, Christoffels, Petry, & Kispert, 2009; Seth et al., 2014; Suzuki, Sekiya, Buscher, Izpisua Belmonte, & Taniguchi, 2008). The implicated regulator of growth arrest is p19ARF due to its over-expression in the absence of Tbx3 and known role as a tumor suppressor (Suzuki et al., 2008). In contrast, absence of the polycomb repressive complex protein Ezh2 leads to an increase of cyclin-dependent kinase inhibitor (Cdkn2A)/p19ARF and reduced hepatoblast proliferation, but without inducing the cholangiocyte program even though hepatocyte differentiation was inhibited (Koike et al., 2014). Consequently, there is a genetic context dependency that plays into the balance between proliferation and growth arrest, as well as initiation and repression of specific cell lineage transcriptional programs ultimately regulating the hepatoblast biopotential cell fate decision.
4.1.2. Transient primitive ductal structure as part of tubulogenesis
Tubulogenesis begins around E15–E17 in mice and 11–15 gestational weeks in humans (Antoniou et al., 2009; Terada, 2017; Van Eyken et al., 1988; Vestentoft et al., 2011). This morphogenesis step delineates the structure surrounding a forming lumen as either a primitive ductal structure or a mature duct (Fig. 8). Transient asymmetrical primitive ductal structures are observed in mice and humans (Raynaud et al., 2011). The primitive ductal structure is composed of two distinct cell types as distinguished by the presence or absence of marker expression (Sox9, Hnf4a, and transforming growth factor beta receptor type 2 [Tgfbr2]) compared to a mature duct. The primitive ductal structure is asymmetrical; cells on the PV side of the lumen express the marker Sox9, compared to cells on the parenchymal side that express Hnf4a and Tgfbr2. A mature duct is symmetrical, composed of cells expressing Sox9 (Antoniou et al., 2009; Raynaud et al., 2011). Additionally, the PV side of primitive ductal structure displays higher levels of E-cadherin expression and is in contact with laminin presumably produced from the PV myofibroblasts. Upon symmetrical mature duct formation, the lumen displays equal levels of E-cadherin expression and is surrounded by extracellular matrix and myofibroblasts (Antoniou et al., 2009). Detailed analysis using immunostaining suggests that there is a radial progression of differentiation—mature cells on the PV side of the lumen promote differentiation of the neighboring less mature cells on the parenchymal side of the lumen (Antoniou et al., 2009; Zong et al., 2009). However, it remains unclear if cells from the ductal plate move and contribute to the less mature cells of the forming lumen or more hepatoblasts are recruited to contribute to the lumen (Fig. 8). Given the expression of Hnf4a in the less mature cells on the parenchymal side of the primitive ductal structure, the more plausible explanation is that hepatoblasts are recruited to contribute to the forming lumen. Nevertheless, sandwich cultures of hepatoblasts suggest that mono-layers of progenitors can fold up to form tubular structures as observed in a wrapping process of tubulogenesis (Tanimizu, Miyajima, & Mostov, 2009).
Finally, the ductal plate cells that remain unincorporated into an IHBD then either undergo apoptosis (Terada & Nakanuma, 1995) or turn off some of the cholangiocyte markers and contribute to periportal hepatocytes or the canal of Hering (Carpentier et al., 2011). Lineage tracing Sox9-positive cells in mouse strongly suggests that the unincorporated cholangiocytes transdifferentiate into hepatocytes without apoptosis or proliferation (Fig. 8) (Carpentier et al., 2011). If the unincorporated ductal plate cells do not receive or are unresponsive to the proper signals, they may contribute to ductal plate malformations as observed in patients with autosomal recessive polycystic kidney disease (ARPKD), congenital hepatic fibrosis, autosomal dominant polycystic kidney disease (ADPKD), Caroli’s disease, Caroli’s syndrome, and von Meyenburg complexes (Desmet, 1992; Huppert, 2011; Raynaud et al., 2011). Thus, there is a high level of coordination that must regulate sequential tubulogenesis and regression of the ductal plates along PVs within the three-dimensional space of the liver to connect the entire IHBD system to the extrahepatic ductal system. This indicates that careful orchestration of signals between epithelial and mesenchymal cells is required to guide IHBD formation (Gerard et al., 2017; Lemaigre, 2010).
4.1.3. Three-dimensional view of tubulogenesis
Due to the fact that the liver forms the IHBD system after the hepatic mass has already begun to expand and the lobes of the liver have formed, it is difficult to visualize and record the morphogenesis. Very interesting biology has been revealed using technical advancements to describe rendered IHBD three-dimensional models through quantitative measures and to visualize the IHBD three-dimensional architecture through retrograde ink injections in combination with tissue clearing (Figs. 4 and 7) (Takashima, Terada, Kawabata, & Suzuki, 2015; Tanimizu et al., 2016).
To map cholangiocyte specification and morphogenesis at a high resolution in three-dimension, serial sections of mouse mid-gestational to adult liver samples were immunostained with CK19 and digital three-dimensional reconstruction of images was performed. Using quantitative morphometric analyses, length, number of branch points, and distance from the PV were analyzed. Information garnered from the high resolution three-dimensional maps suggests that clusters of CK19-positive cholangiocytes are specified individually or in clusters rather than a cell layer as deduced from two-dimensional analyses (Takashima et al., 2015). As IHBD development proceeds, the clusters of CK19-positive cholangiocytes are further increased and form dense networks encircling the PV (Takashima et al., 2015). Thus, the ductal plate structure assumed to be a cell layer of specified cholangiocytes surrounding the PV may instead be a three-dimensional dense plexus-like network of cholangiocytes (Fig. 9). Finally, the network of CK19-positive cells surrounding the PV decreases with age as specific segments of the network become larger diameter CK19-positive tubes extending parallel to the PV, but at a greater distance away from the PV than when the CK19-positive tubes were initially formed (Takashima et al., 2015).
Fig. 9.
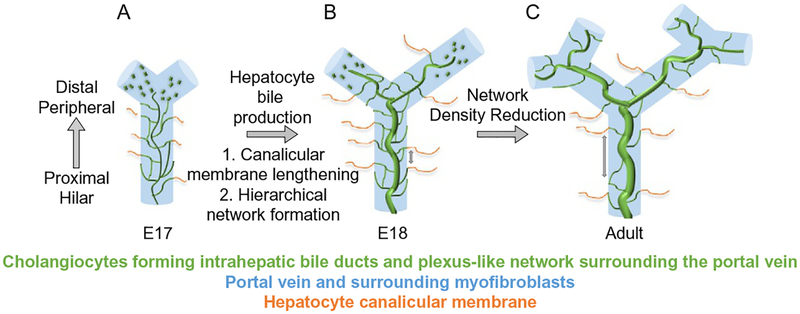
Three-dimensional model of intrahepatic bile duct formation (IHBD) formation.(A) At the beginning of IHBD development, cholangiocytes are specified in the region adjacent to the portal vein (PV) system and are quickly incorporated into a dense homogeneous network that is communicating with the extrahepatic bile duct. (B) Upon hepatocyte bile production, secretion, and canalicular membrane lengthening, the homogenous network begins to reorganize into a hierarchical network between mouse E17 and E18. (C) As the liver parenchyma expands, new IHBDs are generated peripherally and the distance between the fine network structures surrounding the PV increases.
To provide a global view of IHBD formation, a retrograde injection of carbon ink into the luminal space from the common bile duct was performed at time points between mid-gestation and 1 week after birth followed by clearing the liver tissue to expose the ink filled IHBD structure (Kaneko, Kamimoto, Miyajima, & Itoh, 2015; Tanimizu et al., 2016). Ink fills the continuous luminal space of IHBDs but does not leak into the PV or the hepatic parenchyma (Kaneko et al., 2015). Immunostaining used in combination with the ink injection detects the correlation between cholangiocyte specification and communicating luminal IHBD structures. The main findings are consistent with three-dimensional reconstruction, however a greater appreciation for the process of cholangiocyte morpho-genesis and IHBD formation is revealed. Remodeling of specified cholangiocytes into IHBDs starts at the larger PVs at the hilar/proximal regions and based on serial sections and three-dimensional visualization moves toward the peripheral/distal region of liver following the PV system. Therefore, all steps of biliary morphogenesis progress in a hilumto-periphery direction, allowing several stages of IHBD formation to be analyzed within a single liver during hepatogenesis (Fig. 9) (Antoniou et al., 2009; Tanimizu et al., 2016; Van Eyken et al., 1988).
Newly committed cholangiocytes are specified peripherally and are quickly incorporated into a plexus-like network of communicating luminal structures encircling the PV (Fig. 9). In mice, the homogenous communicating network begins to be rearranged into a hierarchical network between E17 and E18. The timing of the hierarchical arrangement correlates with lengthening of the bile canalicular network, hepatocyte excretion of bilirubin into bile, and presence of bile in the intestine (Tanimizu et al., 2016). Using the multidrug resistance-associated protein 2 (Mrp2) inhibitor benzbromarone to block the unidirectional efflux of bile from the hepatocyte canalicular membrane (thereby decreasing the flow of bile between mouse E16 and E18), the structural rearrangement and formation of the hierarchical network of IHBDs were inhibited (Colombo, Armstrong, Duan, & Rioux, 2012; Tanimizu et al., 2016). Additionally, co-localization between liver kinase B1 (Lkb1 encoded by Stk11), a serine/threonine kinase, and E-cadherin has been described at adherens junctions of kidney and intestinal polarized epithelial cell lines in culture (Sebbagh, Santoni, Hall, Borg, & Schwartz, 2009). Lkb1 is an evolutionary conserved serine/threonine protein kinase implicated in a wide range of cellular functions including inhibition of cellular proliferation, regulation of cellular polarity, and metabolism (Salvi & DeMali, 2018). Loss of Lkb1 at mid-gestation in hepatoblasts results in jaundice, loss of protein localization at the bile canalicular membrane, and inability of specified cholangiocytes to undergo formation of the IHBD hierarchical network (Woods et al., 2011). Lkb1-deficient cholangiocytes are able to generate primitive ductal structure but fail to form mature symmetrical ducts (Just et al., 2015). These results indicate that formation of a hepatocyte canalicular membrane and bile flow drives the structural transition of IHBDs from the homogenous tubular plexus-like network into the mature hierarchical network (Fig. 9). This model proposes that any communicating luminal duct that is part of the homogeneous network and connected to a bile canaliculus has the potential to receive more bile secreted from hepatocytes compared to the other communicating lumena and thereby its luminal space may enlarge. Whether the luminal enlargement occurs through localized proliferation of cholangiocytes, movement and incorporation of nearby cholangiocytes, or stretching and enlargement of cholangiocyte cell size remains to be experimentally determined.
After birth, a homogenous network surrounding the PV is still visible in the liver periphery of mice until 1 week of age (Tanimizu et al., 2016). The incomplete IHBD architecture at birth, also exists in humans where the IHBD system is still forming during the first years of life (Van Eyken et al., 1988). Therefore, the process by which the IHBD system forms allows for progressive assembly of a communicating IHBD architecture coincident with the enlargement of the liver during normal growth in childhood. The final IHBD hierarchical architecture consists of large ducts running along PVs and small channels forming a mesh-like network around PVs. The IHBD intricate structure is formed by a very complicated specification and movement of cells that somehow know their place in the three-dimensional space to form connections to drain bile out of the liver. Therefore, the model for IHBD formation can be refined as follows:(1) cholangiocyte specification initially occurs as discontinuous clusters of cells surrounding the PV, (2) specified cholangiocytes rapidly undergo morphogenesis to form a homogeneous communicating luminal network of small ducts, and (3) the homogeneous IHBD network is rearranged into a hierarchical IHBD system coincident with hepatocyte bile production and secretion.
4.2. Molecular regulation of intrahepatic bile duct formation
Cholangiocyte specification and tubulogenesis require regulated input from various signaling pathways (Fig. 10). Specification of cholangiocytes and the process of tubulogenesis to form IHBDs take place in the vicinity of the PV, suggesting that local cues drive differentiation of hepatoblasts toward cholangiocytes and control the formation of primitive ductal structures. Notch and Tgfb are the two main signaling pathways known to supply localized ligand expression and receptor activation within the PV region.
Fig. 10.
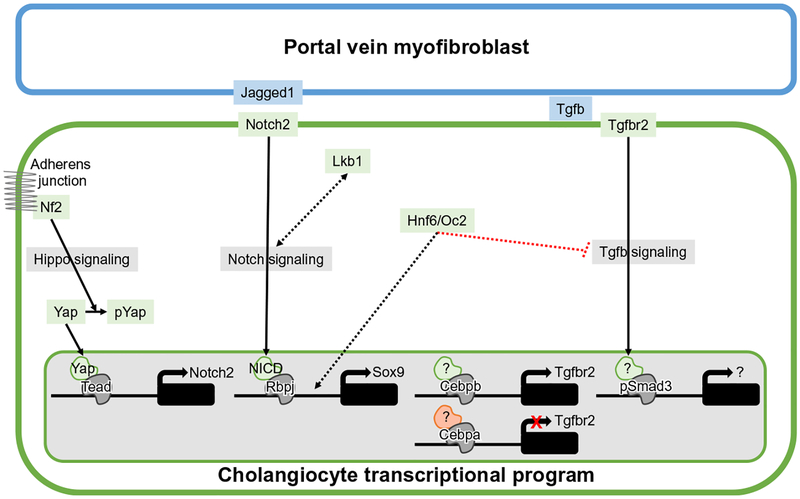
Mechanistic model of cholangiocyte differentiation. Basic schematic of the regulatory nodes and pathways involved in setting up the cholangiocyte transcriptional program. Green outlined cell represents cholangiocyte and blue outlined cell represents portal vein (PV) myofibroblasts. Evidence of functional requirement indicated by solid lines and unknown level of interaction is indicated by dotted lines. Unidentified binding partners/coactivators or targets during cholangiocyte differentiation indicated by “?”. Double-headed arrow indicates mutual cross regulation between Lkb1 and Notch signaling.
4.2.1. Notch signaling pathway
In humans, mutations in both Jagged1 (Jag1) and Notch2, a Notch pathway ligand and receptor, respectively, cause Alagille syndrome (ALGS1 and ALGS2) (Li et al., 1997; McDaniell et al., 2006; Oda et al., 1997). ALGS is an autosomal dominant disorder that is primarily characterized by neonatal jaundice, cholestasis, and paucity of intrahepatic bile ducts (IHBDs), and patients with ALGS also may have characteristic appearance of facial structure along with abnormalities of heart, eye, skeleton, vasculature, kidney, and pancreas (Emerick et al., 1999). Based on the ALGS hepatic phenotype, the primary pathway that has been implicated in IHBD formation is Notch signaling. In general, Notch ligands and receptors are both membrane-bound and participate in juxtacrine signaling between adjacent cells. Ligand binding triggers activation of the Notch receptor through a series of proteolytic events, culminating in a gamma-secretase, presenilin-dependent, proteolytic release of the Notch intracellular domain (NICD) from the membrane. NICD translocates to the nucleus where it associates with recombination signal binding protein for immunoglobulin kappa J region (Rbpj) to activate transcription of downstream targets. Therefore, the Notch signaling pathway is likely the main determinant that positions the proximal spatial localization of the PV and IHBD systems.
During cholangiocyte specification, myofibroblasts surrounding the PV express the Notch ligand Jag1 and most hepatic cells express Notch2 (Hofmann et al., 2010; Kodama, Hijikata, Kageyama, Shimotohno, & Chiba, 2004; Tanimizu & Miyajima, 2004; Zong et al., 2009). Importantly, cell lineage requirement studies have confirmed that Jag1 is required in the PV myofibroblasts, but not the endothelium (Hofmann et al., 2010) or hepatic epithelium (Loomes et al., 2007). Homozygous deletion of Jag1 in smooth muscle protein 22 alpha (SM22a)-positive cells, including PV myofibroblasts, leads to defective bile duct development resulting in jaundice, liver failure, and paucity of IHBDs. CK19-positive cells are detected surrounding the PV, but they are unable to undergo efficient tubulogenesis. Notably, initial cholangiocyte specification based on CK19 expression and the formation of the first layer of ductal plate appears to be independent of the PV myofibroblast supplied Jag1. However, Jag1-dependent signaling from the PV myofibroblast is essential for tubulogenesis and expansion of the mesenchymal cell population (Hofmann et al., 2010), all supporting a model whereby Jag1 interacts with Notch receptors expressed on bipotential hepatoblasts to activate the Notch pathway and regulate cholangiocyte specification and/or morphogenesis.
However, Jag1 expression is not only detected in the PV myofibroblasts but also in differentiating cholangiocytes (Zong et al., 2009), suggesting that Jag1-Notch activation within the differentiating cholangiocytes may contribute to cholangiocyte specification and/or tubulogenesis. Newly differentiated Jag1-positive cholangiocytes may activate the Notch receptors in adjacent cells. Therefore, hepatoblasts positioned between Jag1-positive cholangiocytes and near PV myofibroblasts would receive stronger induction signals than those outside the homogeneous IHBD network. Despite the observed expression data, initial in vivo studies investigating the requirement of Jagged1 in hepatoblasts suggests that Jag1 is not required in the hepatic epithelium for IHBD formation (Loomes et al., 2007).
Work with cultured hepatoblasts indicates that constitutive Notch activation represses expression of hepatocyte genes but induces expression of biliary genes, although no morphological indications of tubular structures were formed (Tanimizu & Miyajima, 2004). In vivo, when the ligand-independent constitutively active intracellular domain of Notch1 or Notch2(i.e., NICD1 or NICD2) is expressed in hepatoblasts or hepatocytes, either is sufficient to promote cholangiocyte specification (Jeliazkova et al., 2013; Sparks, Huppert, Brown, Washington, & Huppert, 2010; Tchorz et al., 2009; Zong et al., 2009). Notably, not only is NICD1 able to induce cholangiocyte specification but also cholangiocyte morphogenesis, increasing the three-dimensional density of communicating IHBDs as assessed by retrograde resin injections (Sparks et al., 2010; Sparks, Perrien, Huppert, Peterson, & Huppert, 2011).
Inhibiting all canonical Notch activity by in vivo hepatoblast-specific deletion of Rbpj lead to a visible decrease of ductal plate cells and specified cholangiocytes (Sparks et al., 2010; Zong et al., 2009). Alternatively, using DAPT, a gamma-secretase inhibitor, to block all canonical Notch signaling between postnatal day 2 and postnatal day 6 in mice (when clusters of cholangiocytes are being specified and undergo tubulogenesis to form a communicating IHBD network), a deficiency of specified cholangiocytes to participate in generating the communicating network of IHBDs results (Tanimizu et al., 2016). Although, in the presence of DAPT, cholangiocytes were able to establish the apicobasal polarity (Tanimizu et al., 2016). Additionally, homozygous deletion of Notch2 in hepatoblasts using a mid-gestational Cre driver containing mouse albumin regulatory elements and the alpha-fetoprotein enhancers (i.e., Alfp-Cre, Kellendonk, Opherk, Anlag, Schutz, & Tronche, 2000) results in an absence of embryonic cholangiocyte specification (Falix et al., 2014). This result clearly indicates that Notch2 is the fundamental hepatic Notch receptor involved in cholangiocyte specification.
Inhibiting the expression of individual Notch receptors using a midgestational Cre driver containing the promoter and an upstream enhancer of the rat albumin gene (i.e., Alb-Cre, Postic & Magnuson, 2000), demonstrated that Notch2, but not Notch1 homozygous deletion results in tubulogenesis defects, but not clear cholangiocyte specification defects with regard to cell number (Geisler et al., 2008; Lozier, McCright, & Gridley, 2008; Sparks et al., 2010). The resulting phenotype is persistent unremodeled CK-positive cholangiocytes. These findings suggest that a reduction in Notch activity results in precocious differentiation, not proliferation, of ductal plate cells that are unable to either receive cues for tubulogenesis or for elimination if they are not incorporated into an IHBD. Therefore, although these specified cholangiocytes appear normal based on current cholangiocyte marker expression, they clearly are unable to complete morphogenesis. All results indicate that the Notch-deficient cholangiocytes are not correctly specified. This phenotype is consistent with what is observed in Jag1 haploinsufficient mice and when Jag1 is deleted from the myofibroblasts using the SM22-Cre transgenic line (Hofmann et al., 2010; Thakurdas et al., 2016). Therefore, it remains unknown if the observed tubulogenesis defects are due to incorrect cholangiocyte specification or whether Notch directly regulates gene targets important for tubulogenesis in the context of IHBD formation.
Sox9, one of the first indications of cells entering the cholangiocyte transcriptional program is a Notch target gene (Zong et al., 2009). There are 10 consensus Rbpj DNA-binding sites in the Sox9 promoter. Two cis-regulatory DNA-binding sites close to the Sox9 are verified to be bona fide Rbpj sites by ChIP PCR (Zong et al., 2009), suggesting that a specific level of Notch activity may be necessary to induce the correct level of Sox9 expression for correct cholangiocyte specification. Additionally, crosstalk between Lkb1 and Notch signaling may regulate tubulogenesis. Phenotypically, the phenotypes between loss of Lkb1 and Notch pathway components are very similar with jaundice and defects in bile duct tubulogenesis. Similarities between liver transcriptomes were analyzed using microarrays comparing the differential genes lists between liver-specific Rbpj loss of function to controls and liver-specific Lkb1 loss of function compared to controls. Parallels were found in 20–25% of the differential gene lists. Ingenuity pathway analysis highlighted metabolic pathways as the main signatures driving the similarity between LKB1 and Notch gene datasets (Just et al., 2015). Whether these are the genes driving the similar tubulogenesis defect remain unknown, especially given the role of Lkb1 in hepatocyte canalicular membrane formation. However, epistatic studies in vitro and in vivo reveal mutual cross regulation between Lkb1 and Notch signaling without clear evidence that one is epistatic to the other (Just et al., 2015).
4.2.2. Tgfb signaling pathway
Upon Tgfb ligand binding to the extracellular domain of Tgfbr2 a conformational change is elicited, resulting in the phosphorylation and activation of Tgfbr1. Tgfbr1 phosphorylates Smad2 or Smad3 on two serine residues within their C-terminus enabling binding to Smad4 to form heteromeric Smad complexes that enter the nucleus and initiate gene transcription (Vander Ark, Cao, & Li, 2018). In the liver, the mRNA of both Tgfb2 and Tgfb3 ligands are predominantly expressed by PV myofibroblasts while Tgfb1 is produced throughout the liver parenchyma (Antoniou et al., 2009). Tgfb activity assessed using a transgenic mouse line harboring an activin/Tgfb reporter consisting of 12 Smad-consensus binding sites upstream of enhanced green fluorescent protein is present in a portal to central gradient at E12.5 (Clotman et al., 2005). Therefore, the predominant spatial activation of the Tgfb pathway in the region of the PV suggests a role for Tgfb in regulating hepatoblast competency to become cholangiocytes and/or inducing cholangiocyte specification.
The dynamic expression pattern of Tgfbr2 suggests that the Tgfb pathway also plays a role in tubulogenesis. Immunostaining of Tgfb2 and Tgfb3 demonstrates binding specifically to the parenchymal side of primitive ductal structure coincident with localization of Tgfbr2 on the parenchymal side (Antoniou et al., 2009). At the onset of cholangiocyte specification, Tgfbr2 is expressed in all cells of the ductal plate. However, as stated above, during primitive ductal structure formation Tgfbr2 is specifically localized on cells surrounding the forming lumen on the parenchymal side and is absent on the side nearest the PV. In mature ducts, all cholangiocytes are negative for Tgfbr2 expression (Antoniou et al., 2009). Although speculative, this model suggests that Tgfb successively induces cholangiocyte specification and formation of the transient ductal plate and primitive ductal structure, then is downregulated.
Several functional sufficiency studies support the premise that Tgfb signaling plays a role in cholangiocyte specification. The first used E12.5 fetal liver explants treated with either Tgfb ligand added to the media or a bead supplying Tgfb ligand demonstrated an increase of cholangiocyte marker mRNA and induction of a local gradient of cholangiocyte cytokeratin markers (Clotman et al., 2005). Second, treating an E14.5 hepatoblast cell line (i.e., BMEL) in culture with any of the three Tgfb ligands (Tgfb1, Tgfb2, or Tgfb3) reduces the mRNA expression of hepatocyte markers and increases the mRNA expression of cholangiocyte markers (Antoniou et al., 2009). Coincident with the increase of cholangiocyte markers is the decrease of Tgfbr2 expression, indicating that activation of the Tgfb pathway ultimately prevents further activation. Finally, using human ESC generated hepatoblasts, treatment with Tgfbr2, Tgfb1, or Tgfb2, but not Tgfb3 increases mRNA expression of cholangiocyte markers and decreases mRNA expression of hepatocyte markers in vitro (Takayama et al., 2014), supporting the ability of the Tgfb pathway to induce cholangiocyte specification in the context of functional Notch signaling.
A clear in vivo requirement for Tgfb signaling in the process of IHBD formation is lacking. In vitro knockdown of Tgfbr2 in human ESC generated hepatoblasts show a decrease in mRNA expression of cholangiocyte markers in response to exogenous stimulus for cholangiocyte differentiation (Takayama et al., 2014). Further, the presence of small molecule Tgfb inhibitors in an in vitro epithelial cyst forming assay, using a hepatoblast cell line (i.e., HPPL), reduced the number of formed cysts (Tanimizu, Miyajima, & Mostov, 2007), suggesting that Tgfb signaling is required for cholangiocyte specification and tubulogeneis. One in vivo study, treating wild-type pregnant female mice with an anti-Tgfb neutralizing antibody at E10.5 and analysis performed at E14.5 show a qualitative reduction in the cholangiocyte cytokeratin-positive cells around the PV (Clotman et al., 2005). However, deletion of Tgfbr2 specifically in hepatoblasts using a mid-gestational Cre driver (i.e., Alb-Cre, Postic & Magnuson, 2000) does not result in an IHBD phenotype (Schaub et al., 2018). Therefore, the Tgfb pathway may only be required temporally at the very beginning of cholangiocyte specification either in the process of setting up competency or direct regulation of cholangiocyte target genes or has a role in a non-hepatoblast cell lineage.
The dynamic expression pattern of Tgfbr2 and the potential temporal requirement of the Tgfb pathway imply a high level of regulation. Indeed, persistent Tgfbr2 expression is associated with delayed transition from primitive ductal structure to mature bile duct formation observed when Sox9 is deleted specifically from hepatoblasts at mid-gestation (Antoniou et al., 2009; Poncy et al., 2015). Part of the phenotypic delay of mature bile duct formation may also be attributed to persistent expression of CCAAT enhancer-binding protein alpha (Cebpa) on both the portal and parenchymal side of the primitive ductal structure in Sox9-deficient liver. Deletion of Cebpa specifically in hepatoblasts at mid-gestation results in preferential differentiation of cholangiocytes rather than hepatocytes (Yamasaki et al., 2006). Interestingly, Tgfbr2 is negatively and positively regulated by Cebpa and Cebpb, respectively (Takayama et al., 2014). ChIP experiments demonstrate that Cebpa and Cebpb are recruited to the same DNA-binding site in the Tgfbr2 promoter region in cells differentiated to the hepatocyte-like or cholangiocyte-like fate from human ESCs, respectively (Takayama et al., 2014). Further supporting the model that Cebpa and Cebpb oppositely regulates Tgfbr2 promoter activity, reporter assays of the Tgfbr2 promoter and hepatoblast differentiation assays all verify that forced Cebpb expression activates expression of Tgfbr2 and phenotypically induces cholangiocyte-like cells from human ESC generated hepatoblasts and isolated E13.5 mouse hepatoblasts (Takayama et al., 2014). Therefore, the ratio of Cebpb to Cebpa appears to govern the hepatoblast cell fate decision by negatively and positively regulating the expression of Tgfbr2.
The Onecut (OC) family of transcription factors also impacts Tgfb activity. Hnf6 (i.e., OC1) and OC2 are transcription factors that bind to DNA as monomers utilizing their cut- and homeo-DNA-binding domains. A critical threshold of Hnf6 and OC2 dosage has been proposed for regulating hepatoblast differentiation. Too little or too much can result in premature biliary differentiation where hepatoblasts express both hepatocyte and cholangiocyte markers (Lemaigre, 2009). In adult mice, Hnf6 and OC2 are detected in both hepatocytes and cholangiocytes and therefore levels of Hnf6 may remain critical in maintaining adult hepatic cell fates (Lemaigre, 2009). Loss of Hnf6 and/or OC2 results in ectopic Tgfb activity. Changes to the portal to central gradient of Tgfb activity is detected, presumably allowing formation of intermediate cells and formation of primitive ductules within the parenchyma instead of restricting the lineage commitment in the vicinity of the PV (Clotman et al., 2005, 2002). The change in the portal to central gradient of Tgfb activity may be instigated through the observed stimulation of Tgfbr2 expression by Hnf6 and/or OC2 direct transcriptional regulation or reduced expression of the Activin/Tgfb inhibitors follistatin and/or alpha2-macroglobulin (Clotman et al., 2005). However, no evidence of Hnf6 binding to the Tgfbr2 promoter and directly regulating transcription in ChIP assays has been found (Takayama et al., 2014). Hnf6 can bind to the Sox9 promoter, but whether this is in response to Tgfb signaling or is recruited independently is unknown (Alder et al., 2014; Laudadio et al., 2012). These findings allude to indirect regulation of Tgfb by the Onecut family of transcription factors and imply a positive and negative feedback loop involving the Cebp family of transcription factors.
4.2.3. Other pathways with Notch-dependent and -independent mechanisms of IHBD formation
Although Notch signaling is the main pathway involved in IHBD formation, the emergence of Notch-independent cholangiocytes has been demonstrated in experimental mouse models (Falix et al., 2014; Walter, Vanderpool, Cast, & Huppert, 2014). In these models, mice initially lack peripheral IHBDs at neonatal stages, leading to severe cholestasis, necrotic liver injury, and liver fibrosis (Falix et al., 2014; Vanderpool et al., 2012). However, de novo IHBD formation occurs in both mouse models after weaning (Falix et al., 2014; Walter et al., 2014), allowing reversal of cholestasis and normalization of liver histology by postnatal day 120 (Schaub et al., 2018; Walter et al., 2014). The ductular reactive structures that emerge in these mutant mice resemble those seen in human, chronically injured livers containing intermediate cells expressing both hepatocyte and cholangiocyte markers. Hepatocytes not only have the ability to self-duplicate but also have the remarkable capability of assuming cholangiocyte identity in injured rodent and human liver (Ernst, Spinner, Piccoli, Mauger, & Russo, 2007; Fabris et al., 2007; Fukuda et al., 2004; Limaye et al., 2008; Michalopoulos, Barua, & Bowen, 2005; Schaub et al., 2018; Sekiya & Suzuki, 2014; Tanimizu, Nishikawa, Ichinohe, Akiyama, & Mitaka, 2014; Tarlow et al., 2014; Yanger et al., 2013; Yimlamai et al., 2014). Because the Notch-independent cholangiocytes emerge in the PV region, the most likely candidate for an alternative signaling pathway is the Tgfb pathway. Hepatocyte lineage tracing confirms that the de novo formed IHBDs are hepatocyte-derived and require Tgfbr2 (Schaub et al., 2018). Hence, the Tgfb pathway can compensate for the loss of Notch in the process of hepatocyte-to-cholangiocyte transdifferentiation but is unable to compensate for loss of Notch during hepatoblast-to-cholangiocyte differentiation. The targets of Tgfbr2/pSmad3 leading to activation of the cholangiocyte transcriptional program in hepatocytes are unknown. Even given this ability of Tgfb signaling, Notch activation appears to be more efficient to induce hepatocyte-to-cholangiocyte transdifferentiation than Tgfb (Sparks et al., 2010; Zong et al., 2009).
One other pathway has demonstrated the clear ability to influence the cholangiocyte fate in a Notch-dependent mechanism. Increased Yap activity induces hepatoblast and hepatocyte conversion to a cholangiocyte-like fate although the potential of these cells to undergo morphogenesis and form de novo IHBDs has not been demonstrated (Yimlamai et al., 2014). Similarly, the loss of the upstream tumor suppressor of the Hippo pathway, Neurofibromin 2 (Nf2), leads to increased Yap activity and generation of hepatoblast conversion to a cholangiocyte-like fate (Zhang et al., 2010). These phenotypes are very reminiscent of the induced hepatoblast and hepatocyte conversion to cholangiocytes upon NICD1 and NICD2 hepatic expression in mice (Jeliazkova et al., 2013; Sparks et al., 2010; Tchorz et al., 2009; Zong et al., 2009). Based on ChIP PCR confirmation of TEAD4 binding to a Notch2 promoter region, the resulting phenotype of increased Yap activity is thought to be a Notch2-dependent mechanism with Notch2 being a direct transcriptional target of the Hippo pathway effector Yap (Yimlamai et al., 2014). Epistatic studies have confirmed this model by reducing the Notch signaling activity through hepatoblast-specific deletion of Notch2. The Nf2 phenotype of increased cholangiocyte differentiation is suppressed while rescuing the paucity of IHBDs observed by midgestational hepatoblast-specific deletion of Notch2 (Wu et al., 2017). These results indicate that Notch signaling is one of the downstream effectors of the Hippo signaling pathway in regulating IHBD development (Fig. 10).
5. Conclusions
Future studies investigating the signaling pathways and epigenetic factors required for lineage restriction of the bipotential hepatoblast into the hepatocyte and cholangiocyte lineages, and further refinement of cell fate heterogeneity within each cell lineage for essential functional diversity of the hepatic epithelium remain critical for enlightening the disease processes of acute and chronic hepatobiliary diseases as well as the development of potential molecular and cellular targets for therapeutic options. The liver is alone among solid organs in its ability to regenerate full mass and function following extensive acute injury or partial resection (Huppert & Campbell, 2016). However, the reason for functional failure of the liver to regenerate mass or replace specific hepatic fate heterogeneity in most cases is unknown. Mouse studies using genetic reporters to trace the hepatocyte or cholangiocyte lineage reveal no evidence of contribution from a reserve stem cell population for liver homeostasis or liver regeneration (Malato et al., 2011; Raven et al., 2017; Schaub et al., 2018; Schaub, Malato, Gormond, & Willenbring, 2014; Yimlamai et al., 2014). Instead, during the liver injury or regenerative process, cells originating from the hepatocyte or cholangiocyte cell lineage can be found in different states of cell identity, transitioning to and from the hepatocyte and cholangiocyte lineage (Raven et al., 2017; Schaub et al., 2018). This highlights the amazing plasticity of adult hepatic epithelial cells. Still, we do not know whether all the signaling pathways and epigenetic factors used during hepatogenesis are the same molecular factors exploited during regeneration. Nevertheless, knowledge of molecular and cellular hepatogenesis regulators gives us a solid starting point. Discovery of the molecular regulators that bestow the ability of hepatic cell fates to be dynamic in response to injury, nutrition levels, drugs, environmental toxin intake, and levels of hormone, and other blood borne factors are vital to lessen the need for liver transplantation and inform potential pluripotent stem cell-derived hepatocyte and cholangiocyte cell replacement therapies.
References
- Alder O, Cullum R, Lee S, Kan AC, Wei W, Yi Y, et al. (2014). Hippo signaling influences HNF4A and FOXA2 enhancer switching during hepatocyte differentiation. Cell Reports, 9, 261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemiya-Kudo M, Shimano H, Hasty AH, Yahagi N, Yoshikawa T, Matsuzaka T, et al. (2002). Transcriptional activities of nuclear SREBP−1a, −1c, and −2 to different target promoters of lipogenic and cholesterogenic genes. Journal of Lipid Research, 43, 1220–1235. [PubMed] [Google Scholar]
- Angelo JR, Guerrero-Zayas MI, & Tremblay KD (2012). A fate map of the murine pancreas buds reveals a multipotent ventral foregut organ progenitor. PLoS One, 7, e40707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou A, Raynaud P, Cordi S, Zong Y, Tronche F, Stanger BZ, et al. (2009). Intrahepatic bile ducts develop according to a new mode of tubulogenesis regulated by the transcription factor SOX9. Gastroenterology, 136, 2325–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki R, Chiba T, Miyagi S, Negishi M, Konuma T, Taniguchi H, et al. (2010). The polycomb group gene product Ezh2 regulates proliferation and differentiation of murine hepatic stem/progenitor cells. Journal of Hepatology, 52, 854–863. [DOI] [PubMed] [Google Scholar]
- Baer MM, Chanut-Delalande H, & Affolter M (2009). Cellular and molecular mechanisms underlying the formation of biological tubes. Current Topics in Developmental Biology, 89, 137–162. [DOI] [PubMed] [Google Scholar]
- Bargaje R, Alam MP, Patowary A, Sarkar M, Ali T, Gupta S, et al. (2012). Proximity of H2A.Z containing nucleosome to the transcription start site influences gene expression levels in the mammalian liver and brain. Nucleic Acids Research, 40, 8965–8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle MA, Konopka G, Parviz F, Gaggl AL, Yang C, Sladek FM, et al. (2006). Hepatocyte nuclear factor 4alpha orchestrates expression of cell adhesion proteins during the epithelial transformation of the developing liver. Proceedings of the National Academy of Sciences of the United States of America, 103, 8419–8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamouche S, Decaens T, Godard C, Chambrey R, Rickman DS, Moinard C, et al. (2006). Apc tumor suppressor gene is the “zonation-keeper” of mouse liver. Developmental Cell, 10, 759–770. [DOI] [PubMed] [Google Scholar]
- Bertet C, Sulak L, & Lecuit T (2004). Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation. Nature, 429, 667–671. [DOI] [PubMed] [Google Scholar]
- Bhatia SN, Underhill GH, Zaret KS, & Fox IJ (2014). Cell and tissue engineering for liver disease. Science Translational Medicine, 6(245), 245sr2 10.1126/scitranslmed.3005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlsma MF, Groot AP, Oduro JP, Franken RJ, Schoenmakers SH, Peppelenbosch MP, et al. (2009). Hypoxia induces a hedgehog response mediated by HIF-1alpha. Journal of Cellular and Molecular Medicine, 13, 2053–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkis IM, Rubins NE, White P, Furth EE, Friedman JR, & Kaestner KH (2008). Hepatocyte-specific ablation of Foxa2 alters bile acid homeostasis and results in endoplasmic reticulum stress. Nature Medicine, 14, 828–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bort R, Signore M, Tremblay K, Martinez Barbera JP, & Zaret KS (2006). Hex homeobox gene controls the transition of the endoderm to a pseudostratified, cell emergent epithelium for liver bud development. Developmental Biology, 290, 44–56. [DOI] [PubMed] [Google Scholar]
- Bossard P, & Zaret KS (1998). GATA transcription factors as potentiators of gut endoderm differentiation. Development, 125, 4909–4917. [DOI] [PubMed] [Google Scholar]
- Boyer JL (2013). Bile formation and secretion. Comprehensive Physiology, 3, 1035–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calmont A, Wandzioch E, Tremblay KD, Minowada G, Kaestner KH, Martin GR, et al. (2006). An FGF response pathway that mediates hepatic gene induction in embryonic endoderm cells. Developmental Cell, 11, 339–348. [DOI] [PubMed] [Google Scholar]
- Calo E, & Wysocka J (2013). Modification of enhancer chromatin: What, how, and why? Molecular Cell, 49, 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, et al. (2007). YAP1 increases organ size and expands undifferentiated progenitor cells. Current Biology, 17, 2054–2060. [DOI] [PubMed] [Google Scholar]
- Camp JG, Sekine K, Gerber T, Loeffler-Wirth H, Binder H, Gac M, et al. (2017). Multilineage communication regulates human liver bud development from pluripotency. Nature, 546, 533–538. [DOI] [PubMed] [Google Scholar]
- Carpentier R, Suner RE, van Hul N, Kopp JL, Beaudry JB, Cordi S, et al. (2011). Embryonic ductal plate cells give rise to cholangiocytes, periportal hepatocytes, and adult liver progenitor cells. Gastroenterology, 141, 1432–1438. e1431–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Kim SY, Okamoto H, Xin Y, Yancopoulos GD, Murphy AJ, et al. (2018). Glucagon contributes to liver zonation. Proceedings of the National Academy of Sciences of the United States of America, 115, E4111–E4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clotman F, Jacquemin P, Plumb-Rudewiez N, Pierreux CE, Van der Smissen P, Dietz HC, et al. (2005). Control of liver cell fate decision by a gradient of TGF beta signaling modulated by Onecut transcription factors. Genes & Development, 19, 1849–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clotman F, Lannoy VJ, Reber M, Cereghini S, Cassiman D, Jacquemin P, et al. (2002). The Onecut transcription factor HNF6 is required for normal development of the biliary tract. Development, 129, 1819–1828. [DOI] [PubMed] [Google Scholar]
- Colombo F, Armstrong C, Duan J, & Rioux N (2012). A high throughput in vitro mrp2 assay to predict in vivo biliary excretion. Xenobiotica, 42, 157–163. [DOI] [PubMed] [Google Scholar]
- Costa RH, Kalinichenko VV, Holterman AX, & Wang X (2003). Transcription factors in liver development, differentiation, and regeneration. Hepatology, 38, 1331–1347. [DOI] [PubMed] [Google Scholar]
- Crawford JM (2002). Development of the intrahepatic biliary tree. Seminars in Liver Disease, 22, 213–226. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. (2010). Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America, 107, 21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lau W, Barker N, Low TY, Koo BK, Li VS, Teunissen H, et al. (2011). Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature, 476, 293–297. [DOI] [PubMed] [Google Scholar]
- Dentin R, Pegorier JP, Benhamed F, Foufelle F, Ferre P, Fauveau V, et al. (2004). Hepatic glucokinase is required for the synergistic action of ChREBP and SREBP-1c on glycolytic and lipogenic gene expression. The Journal of Biological Chemistry, 279, 20314–20326. [DOI] [PubMed] [Google Scholar]
- Desmet VJ (1992). Congenital diseases of intrahepatic bile ducts: Variations on the theme“ductal plate malformation” Hepatology, 16, 1069–1083. [DOI] [PubMed] [Google Scholar]
- Desmet VJ (2011). Ductal plates in hepatic ductular reactions. Hypothesis and implications. II. Ontogenic liver growth in childhood. Virchows Archiv, 458, 261–270. [DOI] [PubMed] [Google Scholar]
- Deutsch G, Jung J, Zheng M, Lora J, & Zaret KS (2001). A bipotential precursor population for pancreas and liver within the embryonic endoderm. Development, 128, 871–881. [DOI] [PubMed] [Google Scholar]
- Devi BG, Gupta PD, & Habeebullah CM (1992). Changes in membrane fluidity during human liver development. Biochemistry International, 28, 41–49. [PubMed] [Google Scholar]
- Devi BG, Habeebullah CM, & Gupta PD (1992). Glycogen metabolism during human liver development. Biochemistry International, 28, 229–237. [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, et al. (2007). Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell, 130, 1120–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerick KM, Rand EB, Goldmuntz E, Krantz ID, Spinner NB, & Piccoli DA (1999). Features of Alagille syndrome in 92 patients: Frequency and relation to prognosis. Hepatology, 29, 822–829. [DOI] [PubMed] [Google Scholar]
- Ernst LM, Spinner NB, Piccoli DA, Mauger J, & Russo P (2007). Interlobular bile duct loss in pediatric cholestatic disease is associated with aberrant cytokeratin 7 expression by hepatocytes. Pediatric and Developmental Pathology, 10, 383–390. [DOI] [PubMed] [Google Scholar]
- Fabris L, Cadamuro M, Guido M, Spirli C, Fiorotto R, Colledan M, et al. (2007). Analysis of liver repair mechanisms in Alagille syndrome and biliary atresia reveals a role for notch signaling. The American Journal of Pathology, 171, 641–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falix FA, Weeda VB, Labruyere WT, Poncy A, de Waart DR, Hakvoort TB, et al. (2014). Hepatic Notch2 deficiency leads to bile duct agenesis perinatally and secondary bile duct formation after weaning. Developmental Biology, 396, 201–213. [DOI] [PubMed] [Google Scholar]
- Fickert P, & Wagner M (2017). Biliary bile acids in hepatobiliary injury—What is the link? Journal of Hepatology, 67, 619–631. [DOI] [PubMed] [Google Scholar]
- Fukuda K, Sugihara A, Nakasho K, Tsujimura T, Yamada N, Okaya A, et al. (2004). The origin of biliary ductular cells that appear in the spleen after transplantation of hepatocytes. Cell Transplantation, 13, 27–33. [DOI] [PubMed] [Google Scholar]
- Geisler F, Nagl F, Mazur PK, Lee M, Zimber-Strobl U, Strobl LJ, et al. (2008). Liver-specific inactivation of Notch2, but not Notch1, compromises intrahepatic bile duct development in mice. Hepatology, 48, 607–616. [DOI] [PubMed] [Google Scholar]
- Gerard C, Tys J, & Lemaigre FP (2017). Gene regulatory networks in differentiation and direct reprogramming of hepatic cells. Seminars in Cell & Developmental Biology, 66, 43–50. [DOI] [PubMed] [Google Scholar]
- Gissen P, & Arias IM (2015). Structural and functional hepatocyte polarity and liver disease. Journal of Hepatology, 63, 1023–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinka A, Dolde C, Kirsch N, Huang YL, Kazanskaya O, Ingelfinger D, et al. (2011). LGR4 and LGR5 are R-spondin receptors mediating Wnt/beta-catenin and Wnt/PCP signalling. EMBO Reports, 12, 1055–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman O, Han S, Sourisseau M, Dziedzic N, Hamou W, Corneo B, et al. (2013). KDR identifies a conserved human and murine hepatic progenitor and instructs early liver development. Cell Stem Cell, 12, 748–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gougelet A, Torre C, Veber P, Sartor C, Bachelot L, Denechaud PD, et al. (2014). T-cell factor 4 and beta-catenin chromatin occupancies pattern zonal liver metabolism in mice. Hepatology, 59, 2344–2357. [DOI] [PubMed] [Google Scholar]
- Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, & Zaret KS (1996). Hepatic specification of the gut endoderm in vitro: Cell signaling and transcriptional control. Genes & Development, 10, 1670–1682. [DOI] [PubMed] [Google Scholar]
- Halpern KB, Shenhav R, Matcovitch-Natan O, Toth B, Lemze D, Golan M, et al. (2017). Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature, 542, 352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Tan C, Ding J, Wang J, Ma’ayan A, & Gouon-Evans V (2018). Endothelial cells instruct liver specification of embryonic stem cell-derived endoderm through endothelial VEGFR2 signaling and endoderm epigenetic modifications. Stem Cell Research, 30, 163–170. [DOI] [PubMed] [Google Scholar]
- Hao HX, Xie Y, Zhang Y, Charlat O, Oster E, Avello M, et al. (2012). ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature, 485, 195–200. [DOI] [PubMed] [Google Scholar]
- Hart SN, Cui Y, Klaassen CD, & Zhong XB (2009). Three patterns of cytochrome P450 gene expression during liver maturation in mice. Drug Metabolism and Disposition, 37, 116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayhurst GP, Lee YH, Lambert G, Ward JM, & Gonzalez FJ (2001). Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Molecular and Cellular Biology, 21, 1393–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman BG, Robertson G, Zavaglia B, Beach M, Cullum R, Lee S, et al. (2010). Locus co-occupancy, nucleosome positioning, and H3K4me1 regulate the functionality of FOXA2-, HNF4A-, and PDX1-bound loci in islets and liver. Genome Research, 20, 1037–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann JJ, Zovein AC, Koh H, Radtke F, Weinmaster G, & Iruela-Arispe ML (2010). Jagged1 in the portal vein mesenchyme regulates intrahepatic bile duct development: Insights into Alagille syndrome. Development, 137, 4061–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert SS (2011). A new set of classifications for ductal plate malformations. Hepatology,53, 1795–1797. [DOI] [PubMed] [Google Scholar]
- Huppert SS, & Campbell KM (2016). Emerging advancements in liver regeneration and organogenesis as tools for liver replacement. Current Opinion in Organ Transplantation, 21, 581–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, & Zaret KS (2014). Pioneer transcription factors in cell reprogramming. Genes & Development, 28, 2679–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, & Zaret KS (2016). Cell fate control by pioneer transcription factors.Development, 143, 1833–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeliazkova P, Jors S, Lee M, Zimber-Strobl U, Ferrer J, Schmid RM, et al. (2013). Canonical Notch2 signaling determines biliary cell fates of embryonic hepatoblasts and adult hepatocytes independent of Hes1. Hepatology, 57, 2469–2479. [DOI] [PubMed] [Google Scholar]
- Jung J, Zheng M, Goldfarb M, & Zaret KS (1999). Initiation of mammalian liver development from endoderm by fibroblast growth factors. Science, 284, 1998–2003. [DOI] [PubMed] [Google Scholar]
- Jungermann K, & Katz N (1989). Functional specialization of different hepatocyte populations. Physiological Reviews, 69, 708–764. [DOI] [PubMed] [Google Scholar]
- Just PA, Poncy A, Charawi S, Dahmani R, Traore M, Dumontet T, et al. (2015). LKB1 and Notch pathways interact and control biliary morphogenesis. PLoS One, 10, e0145400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko K, Kamimoto K, Miyajima A, & Itoh T (2015). Adaptive remodeling of the biliary architecture underlies liver homeostasis. Hepatology, 61, 2056–2066. [DOI] [PubMed] [Google Scholar]
- Kellendonk C, Opherk C, Anlag K, Schutz G, & Tronche F (2000). Hepatocyte-specific expression of Cre recombinase. Genesis, 26, 151–153. [DOI] [PubMed] [Google Scholar]
- Keng VW, Yagi H, Ikawa M, Nagano T, Myint Z, Yamada K, et al. (2000). Homeobox gene Hex is essential for onset of mouse embryonic liver development and differentiation of the monocyte lineage. Biochemical and Biophysical Research Communications, 276, 1155–1161. [DOI] [PubMed] [Google Scholar]
- Keppler D (2017). Progress in the molecular characterization of hepatobiliary transporters. Digestive Diseases, 35, 197–202. [DOI] [PubMed] [Google Scholar]
- Kietzmann T (2017). Metabolic zonation of the liver: The oxygen gradient revisited. RedoxBiology, 11, 622–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama Y, Hijikata M, Kageyama R, Shimotohno K, & Chiba T (2004). The role of notch signaling in the development of intrahepatic bile ducts. Gastroenterology, 127, 1775–1786. [DOI] [PubMed] [Google Scholar]
- Koike H, Ouchi R, Ueno Y, Nakata S, Obana Y, Sekine K, et al. (2014). Polycomb group protein Ezh2 regulates hepatic progenitor cell proliferation and differentiation in murine embryonic liver. PLoS One, 9, e104776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo BK, Spit M, Jordens I, Low TY, Stange DE, van de Wetering M, et al. (2012). Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature, 488, 665–669. [DOI] [PubMed] [Google Scholar]
- Kretzschmar K, & Clevers H (2017). Wnt/beta-catenin signaling in adult mammalian epithelial stem cells. Developmental Biology, 428, 273–282. [DOI] [PubMed] [Google Scholar]
- Kyrmizi I, Hatzis P, Katrakili N, Tronche F, Gonzalez FJ, & Talianidis I (2006). Plasticity and expanding complexity of the hepatic transcription factor network during liver development. Genes & Development, 20, 2293–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudadio I, Manfroid I, Achouri Y, Schmidt D, Wilson MD, Cordi S, et al. (2012). A feedback loop between the liver-enriched transcription factor network and miR-122 controls hepatocyte differentiation. Gastroenterology, 142, 119–129. [DOI] [PubMed] [Google Scholar]
- Lee CS, Friedman JR, Fulmer JT, & Kaestner KH (2005). The initiation of liver development is dependent on Foxa transcription factors. Nature, 435, 944–947. [DOI] [PubMed] [Google Scholar]
- Lemaigre FP (2009). Mechanisms of liver development: Concepts for understanding liver disorders and design of novel therapies. Gastroenterology, 137, 62–79. [DOI] [PubMed] [Google Scholar]
- Lemaigre FP (2010). Molecular mechanisms of biliary development. Progress in MolecularBiology and Translational Science, 97, 103–126. [DOI] [PubMed] [Google Scholar]
- Li Z, Gadue P, Chen K, Jiao Y, Tuteja G, Schug J, et al. (2012). Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell, 151, 1608–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Krantz ID, Deng Y, Genin A, Banta AB, Collins CC, et al. (1997). Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nature Genetics, 16, 243–251. [DOI] [PubMed] [Google Scholar]
- Li Z, Schug J, Tuteja G, White P, & Kaestner KH (2011). The nucleosome map of the mammalian liver. Nature Structural & Molecular Biology, 18, 742–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limaye PB, Alarcon G, Walls AL, Nalesnik MA, Michalopoulos GK, Demetris AJ, et al. (2008). Expression of specific hepatocyte and cholangiocyte transcription factors in human liver disease and embryonic development. Laboratory Investigation, 88, 865–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomes KM, Russo P, Ryan M, Nelson A, Underkoffler L, Glover C, et al. (2007). Bile duct proliferation in liver-specific Jag1 conditional knockout mice: Effects of gene dosage. Hepatology, 45, 323–330. [DOI] [PubMed] [Google Scholar]
- Lozier J, McCright B, & Gridley T (2008). Notch signaling regulates bile duct morpho-genesis in mice. PLoS One, 3, e1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke TH, Christoffels VM, Petry M, & Kispert A (2009). Tbx3 promotes liver bud expansion during mouse development by suppression of cholangiocyte differentiation. Hepatology, 49, 969–978. [DOI] [PubMed] [Google Scholar]
- Malato Y, Naqvi S, Schurmann N, Ng R, Wang B, Zape J, et al. (2011). Fate tracing of mature hepatocytes in mouse liver homeostasis and regeneration. The Journal of Clinical Investigation, 121, 4850–4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Barbera JP, Clements M, Thomas P, Rodriguez T, Meloy D, Kioussis D, et al. (2000). The homeobox gene Hex is required in definitive endodermal tissues for normal forebrain, liver and thyroid formation. Development, 127, 2433–2445. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Yoshitomi H, Rossant J, & Zaret KS (2001). Liver organogenesis promoted by endothelial cells prior to vascular function. Science, 294, 559–563. [DOI] [PubMed] [Google Scholar]
- Matz-Soja M, Rennert C, Schonefeld K, Aleithe S, Boettger J, Schmidt-Heck W, et al. (2016). Hedgehog signaling is a potent regulator of liver lipid metabolism and reveals a GLI-code associated with steatosis. eLife, 5, e13308 10.7554/eLife.13308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniell R, Warthen DM, Sanchez-Lara PA, Pai A, Krantz ID, Piccoli DA, et al. (2006). NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the notch signaling pathway. American Journal of Human Genetics, 79, 169–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalopoulos GK, Barua L, & Bowen WC (2005). Transdifferentiation of rat hepatocytes into biliary cells after bile duct ligation and toxic biliary injury. Hepatology, 41, 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita S, Mochizuki K, & Goda T (2014). Bindings of ChREBP and SREBP1, and histone acetylation around the rat liver fatty acid synthase gene are associated with induction of the gene during the suckling-weaning transition. Journal of Nutritional Science and Vitaminology (Tokyo), 60, 94–100. [DOI] [PubMed] [Google Scholar]
- Newton IP, Kenneth NS, Appleton PL, Nathke I, & Rocha S (2010). Adenomatous polyposis coli and hypoxia-inducible factor-1{alpha} have an antagonistic connection. Molecular Biology of the Cell, 21, 3630–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober EA, & Lemaigre FP (2018). Development of the liver: Insights into organ and tissue morphogenesis. Journal of Hepatology, 68, 1049–1062. [DOI] [PubMed] [Google Scholar]
- Oda T, Elkahloun AG, Pike BL, Okajima K, Krantz ID, Genin A, et al. (1997). Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nature Genetics, 16, 235–242. [DOI] [PubMed] [Google Scholar]
- Odom DT, Dowell RD, Jacobsen ES, Nekludova L, Rolfe PA, Danford TW, et al. (2006). Core transcriptional regulatory circuitry in human hepatocytes. Molecular Systems Biology, 2, 2006.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, et al. (2004). Control of pancreas and liver gene expression by HNF transcription factors. Science, 303, 1378–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T, Mazack V, & Sudol M (2008). Mst2 and Lats kinases regulate apoptotic function of Yes kinase-associated protein (YAP). The Journal of Biological Chemistry, 283, 27534–27546. [DOI] [PubMed] [Google Scholar]
- Parviz F, Matullo C, Garrison WD, Savatski L, Adamson JW, Ning G, et al. (2003). Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nature Genetics, 34, 292–296. [DOI] [PubMed] [Google Scholar]
- Peng L, Cui JY, Yoo B, Gunewardena SS, Lu H, Klaassen CD, et al. (2013). RNA-sequencing quantification of hepatic ontogeny of phase-I enzymes in mice. Drug Metabolism and Disposition, 41, 2175–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Yoo B, Gunewardena SS, Lu H, Klaassen CD, & Zhong XB (2012). RNA sequencing reveals dynamic changes of mRNA abundance of cytochromes P450 and their alternative transcripts during mouse liver development. Drug Metabolism and Disposition, 40, 1198–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov K, Wierbowski BM, & Salic A (2017). Sending and receiving hedgehog signals.Annual Review of Cell and Developmental Biology, 33, 145–168. [DOI] [PubMed] [Google Scholar]
- Pinson KI, Brennan J, Monkley S, Avery BJ, & Skarnes WC (2000). An LDL-receptor-related protein mediates Wnt signalling in mice. Nature, 407, 535–538. [DOI] [PubMed] [Google Scholar]
- Planas-Paz L, Orsini V, Boulter L, Calabrese D, Pikiolek M, Nigsch F, et al. (2016). The RSPO-LGR4/5-ZNRF3/RNF43 module controls liver zonation and size. Nature Cell Biology, 18, 467–479. [DOI] [PubMed] [Google Scholar]
- Poncy A, Antoniou A, Cordi S, Pierreux CE, Jacquemin P, & Lemaigre FP (2015). Transcription factors SOX4 and SOX9 cooperatively control development of bile ducts. Developmental Biology, 404, 136–148. [DOI] [PubMed] [Google Scholar]
- Postic C, & Magnuson MA (2000). DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis, 26, 149–150. [DOI] [PubMed] [Google Scholar]
- Pouille PA, Ahmadi P, Brunet AC, & Farge E (2009). Mechanical signals trigger myosin II redistribution and mesoderm invagination in Drosophila embryos. Science Signaling, 2, ra16. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, & Wysocka J (2011). A unique chromatin signature uncovers early developmental enhancers in humans. Nature, 470, 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven A, Lu WY, Man TY, Ferreira-Gonzalez S, O’Duibhir E, Dwyer BJ, et al. (2017). Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature, 547, 350–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud P, Tate J, Callens C, Cordi S, Vandersmissen P, Carpentier R, et al. (2011). A classification of ductal plate malformations based on distinct pathogenic mechanisms of biliary dysmorphogenesis. Hepatology, 53, 1959–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskams TA, Theise ND, Balabaud C, Bhagat G, Bhathal PS, Bioulac-Sage P, et al. (2004). Nomenclature of the finer branches of the biliary tree: Canals, ductules, and ductular reactions in human livers. Hepatology, 39, 1739–1745. [DOI] [PubMed] [Google Scholar]
- Rossi JM, Dunn NR, Hogan BL, & Zaret KS (2001). Distinct mesodermal signals, including BMPs from the septum transversum mesenchyme, are required in combination for hepatogenesis from the endoderm. Genes & Development, 15, 1998–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffner H, Sprunger J, Charlat O, Leighton-Davies J, Grosshans B, Salathe A, et al. (2012). R-Spondin potentiates Wnt/beta-catenin signaling through orphan receptors LGR4 and LGR5. PLoS One, 7, e40976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler NC, Nandhikonda P, Webb-Robertson BJ, Ansong C, Anderson LN, Smith JN, et al. (2016). Hepatic cytochrome P450 activity, abundance, and expression throughout human development. Drug Metabolism and Disposition, 44, 984–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi AM, & DeMali KA (2018). Mechanisms linking mechanotransduction and cell metabolism. Current Opinion in Cell Biology, 54, 114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub JR, Huppert KA, Kurial SNT, Hsu BY, Cast AE, Donnelly B, et al. (2018). De novo formation of the biliary system by TGFbeta-mediated hepatocyte trans-differentiation. Nature, 557, 247–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub JR, Malato Y, Gormond C, & Willenbring H (2014). Evidence against a stem cell origin of new hepatocytes in a common mouse model of chronic liver injury. Cell Reports, 8, 933–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebbagh M, Santoni MJ, Hall B, Borg JP, & Schwartz MA (2009). Regulation of LKB1/STRAD localization and function by E-cadherin. Current Biology, 19, 37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine S, Lan BY, Bedolli M, Feng S, & Hebrok M (2006). Liver-specific loss of beta-catenin blocks glutamine synthesis pathway activity and cytochrome p450 expression in mice. Hepatology, 43, 817–825. [DOI] [PubMed] [Google Scholar]
- Sekiya S, & Suzuki A (2014). Hepatocytes, rather than cholangiocytes, can be the major source of primitive ductules in the chronically injured mouse liver. The American Journal of Pathology, 184, 1468–1478. [DOI] [PubMed] [Google Scholar]
- Seth A, Ye J, Yu N, Guez F, Bedford DC, Neale GA, et al. (2014). Prox1 ablation in hepatic progenitors causes defective hepatocyte specification and increases biliary cell commitment. Development, 141, 538–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo A, & Wallingford JB (2014). PCP and septins compartmentalize cortical actomyosin to direct collective cell movement. Science, 343, 649–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozawa T, Yoshikawa HY, & Takebe T (2016). Reverse engineering liver buds through self-driven condensation and organization towards medical application. Developmental Biology, 420, 221–229. [DOI] [PubMed] [Google Scholar]
- Sparks EE, Huppert KA, Brown MA, Washington MK, & Huppert SS (2010). Notch signaling regulates formation of the three-dimensional architecture of intrahepatic bile ducts in mice. Hepatology, 51, 1391–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks EE, Perrien DS, Huppert KA, Peterson TE, & Huppert SS (2011). Defects in hepatic Notch signaling result in disruption of the communicating intrahepatic bile duct network in mice. Disease Models & Mechanisms, 4, 359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanulovic VS, Kyrmizi I, Kruithof-de Julio M, Hoogenkamp M, Vermeulen JL, Ruijter JM, et al. (2007). Hepatic HNF4alpha deficiency induces periportal expression of glutamine synthetase and other pericentral enzymes. Hepatology, 45, 433–444. [DOI] [PubMed] [Google Scholar]
- Sund NJ, Ang SL, Sackett SD, Shen W, Daigle N, Magnuson MA, et al. (2000). Hepatocyte nuclear factor 3beta (Foxa2) is dispensable for maintaining the differentiated state of the adult hepatocyte. Molecular and Cellular Biology, 20, 5175–5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Sekiya S, Buscher D, Izpisua Belmonte JC, & Taniguchi H (2008). Tbx3 controls the fate of hepatic progenitor cells in liver development by suppressing p19ARF expression. Development, 135, 1589–1595. [DOI] [PubMed] [Google Scholar]
- Takashima Y, Terada M, Kawabata M, & Suzuki A (2015). Dynamic three-dimensional morphogenesis of intrahepatic bile ducts in mouse liver development. Hepatology, 61, 1003–1011. [DOI] [PubMed] [Google Scholar]
- Takayama K, Kawabata K, Nagamoto Y, Inamura M, Ohashi K, Okuno H, et al. (2014). CCAAT/enhancer binding protein-mediated regulation of TGFbeta receptor 2 expression determines the hepatoblast fate decision. Development, 141, 91–100. [DOI] [PubMed] [Google Scholar]
- Takebe T, Enomura M, Yoshizawa E, Kimura M, Koike H, Ueno Y, et al. (2015). Vascularized and complex organ buds from diverse tissues via mesenchymal cell-driven condensation. Cell Stem Cell, 16, 556–565. [DOI] [PubMed] [Google Scholar]
- Tamai K, Semenov M, Kato Y, Spokony R, Liu C, Katsuyama Y, et al. (2000). LDL-receptor-related proteins in Wnt signal transduction. Nature, 407, 530–535. [DOI] [PubMed] [Google Scholar]
- Tanimizu N, Kaneko K, Itoh T, Ichinohe N, Ishii M, Mizuguchi T, et al. (2016). Intrahepatic bile ducts are developed through formation of homogeneous continuous luminal network and its dynamic rearrangement in mice. Hepatology, 64, 175–188. [DOI] [PubMed] [Google Scholar]
- Tanimizu N, & Mitaka T (2017). Epithelial morphogenesis during liver development. Cold Spring Harbor Perspectives in Biology, 9, a027862 10.1101/cshperspect.a027862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimizu N, & Miyajima A (2004). Notch signaling controls hepatoblast differentiation by altering the expression of liver-enriched transcription factors. Journal of Cell Science, 117, 3165–3174. [DOI] [PubMed] [Google Scholar]
- Tanimizu N, Miyajima A, & Mostov KE (2007). Liver progenitor cells develop cholangiocyte-type epithelial polarity in three-dimensional culture. Molecular Biology of the Cell, 18, 1472–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimizu N, Miyajima A, & Mostov KE (2009). Liver progenitor cells fold up a cell monolayer into a double-layered structure during tubular morphogenesis. Molecular Biology of the Cell, 20, 2486–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimizu N, Nishikawa Y, Ichinohe N, Akiyama H, & Mitaka T (2014). Sry HMG box protein 9-positive (Sox9+) epithelial cell adhesion molecule-negative (EpCAM−) biphenotypic cells derived from hepatocytes are involved in mouse liver regeneration. The Journal of Biological Chemistry, 289, 7589–7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarlow BD, Pelz C, Naugler WE, Wakefield L, Wilson EM, Finegold MJ, et al. (2014). Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell Stem Cell, 15, 605–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchorz JS, Kinter J, Muller M, Tornillo L, Heim MH, & Bettler B (2009). Notch2 signaling promotes biliary epithelial cell fate specification and tubulogenesis during bile duct development in mice. Hepatology, 50, 871–879. [DOI] [PubMed] [Google Scholar]
- Terada T (2017). Human ductal plate and its derivatives express antigens of cholangiocellular, hepatocellular, hepatic stellate/progenitor cell, stem cell, and neuroendocrine lineages, and proliferative antigens. Experimental Biology and Medicine (Maywood, N.J.), 242, 907–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada T, & Nakanuma Y (1995). Detection of apoptosis and expression of apoptosis-related proteins during human intrahepatic bile duct development. The American Journal of Pathology, 146, 67–74. [PMC free article] [PubMed] [Google Scholar]
- Thakurdas SM, Lopez MF, Kakuda S, Fernandez-Valdivia R, Zarrin-Khameh N, Haltiwanger RS, et al. (2016). Jagged1 heterozygosity in mice results in a congenital cholangiopathy which is reversed by concomitant deletion of one copy of Poglut1 (Rumi). Hepatology, 63, 550–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre C, Perret C, & Colnot S (2011). Transcription dynamics in a physiological process: Beta-catenin signaling directs liver metabolic zonation. The International Journal of Biochemistry & Cell Biology, 43, 271–278. [DOI] [PubMed] [Google Scholar]
- Tremblay KD, & Zaret KS (2005). Distinct populations of endoderm cells converge to generate the embryonic liver bud and ventral foregut tissues. Developmental Biology, 280, 87–99. [DOI] [PubMed] [Google Scholar]
- van Arensbergen J, Garcia-Hurtado J, Moran I, Maestro MA, Xu X, Van de Casteele M, et al. (2010). Derepression of polycomb targets during pancreatic organ-ogenesis allows insulin-producing beta-cells to adopt a neural gene activity program. Genome Research, 20, 722–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eyken P, Sciot R, Callea F, Van der Steen K, Moerman P, & Desmet VJ (1988). The development of the intrahepatic bile ducts in man: A keratin-immunohistochemical study. Hepatology, 8, 1586–1595. [DOI] [PubMed] [Google Scholar]
- Vander Ark A, Cao J, & Li X (2018). TGF-beta receptors: In and beyond TGF-beta signaling. Cellular Signalling, 52, 112–120. [DOI] [PubMed] [Google Scholar]
- Vanderpool C, Sparks EE, Huppert KA, Gannon M, Means AL, & Huppert SS (2012). Genetic interactions between hepatocyte nuclear factor-6 and Notch signaling regulate mouse intrahepatic bile duct development in vivo. Hepatology, 55, 233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestentoft PS, Jelnes P, Hopkinson BM, Vainer B, Mollgard K, Quistorff B, et al. (2011). Three-dimensional reconstructions of intrahepatic bile duct tubulogenesis in human liver. BMC Developmental Biology, 11, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter TJ, Vanderpool C, Cast AE, & Huppert SS (2014). Intrahepatic bile duct regeneration in mice does not require Hnf6 or Notch signaling through Rbpj. The American Journal of Pathology, 184, 1479–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandzioch E, & Zaret KS (2009). Dynamic signaling network for the specification of embryonic pancreas and liver progenitors. Science, 324, 1707–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Zhao L, Fish M, Logan CY, & Nusse R (2015). Self-renewing diploidAxin2(+) cells fuel homeostatic renewal of the liver. Nature, 524, 180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White P, Brestelli JE, Kaestner KH, & Greenbaum LE (2005). Identification of transcriptional networks during liver regeneration. The Journal of Biological Chemistry, 280, 3715–3722. [DOI] [PubMed] [Google Scholar]
- Woods A, Heslegrave AJ, Muckett PJ, Levene AP, Clements M, Mobberley M, et al. (2011). LKB1 is required for hepatic bile acid transport and canalicular membrane integrity in mice. The Biochemical Journal, 434, 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Liu Y, Zheng Y, Dong J, & Pan D (2008). The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Developmental Cell, 14, 388–398. [DOI] [PubMed] [Google Scholar]
- Wu N, Nguyen Q, Wan Y, Zhou T, Venter J, Frampton GA, et al. (2017). The Hippo signaling functions through the Notch signaling to regulate intrahepatic bile duct development in mammals. Laboratory Investigation, 97, 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie R, Everett LJ, Lim HW, Patel NA, Schug J, Kroon E, et al. (2013). Dynamic chromatin remodeling mediated by polycomb proteins orchestrates pancreatic differentiation of human embryonic stem cells. Cell Stem Cell, 12, 224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu CR, Cole PA, Meyers DJ, Kormish J, Dent S, & Zaret KS (2011). Chromatin “prepattern” and histone modifiers in a fate choice for liver and pancreas. Science, 332, 963–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu CR, Li LC, Donahue G, Ying L, Zhang YW, Gadue P, et al. (2014). Dynamics of genomic H3K27me3 domains and role of EZH2 during pancreatic endocrine specification. The EMBO Journal, 33, 2157–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Lu X, Chen EZ, He Z, Uyunbilig B, Li G, et al. (2012). Genome-wide roles of Foxa2 in directing liver specification. Journal of Molecular Cell Biology, 4, 420–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- .Suppression of C/EBPalpha expression in periportal hepatoblasts may stimulate biliary cell differentiation through increased Hnf6 and Hnf1b expression. Development, 133, 4233–4243. [DOI] [PubMed] [Google Scholar]
- Yang J, Mowry LE, Nejak-Bowen KN, Okabe H, Diegel CR, Lang RA, et al. (2014). Beta-catenin signaling in murine liver zonation and regeneration: A Wnt-Wnt situation! Hepatology, 60, 964–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanger K, Zong Y, Maggs LR, Shapira SN, Maddipati R, Aiello NM, et al. (2013). Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes & Development, 27, 719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yimlamai D, Christodoulou C, Galli GG, Yanger K, Pepe-Mooney B, Gurung B, et al. (2014). Hippo pathway activity influences liver cell fate. Cell, 157, 1324–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng G, Awan F, Otruba W, Muller P, Apte U, Tan X, et al. (2007). Wnt’er in liver: Expression of Wnt and frizzled genes in mouse. Hepatology, 45, 195–204. [DOI] [PubMed] [Google Scholar]
- Zentner GE, Tesar PJ, & Scacheri PC (2011). Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Research, 21, 1273–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Bai H, David KK, Dong J, Zheng Y, Cai J, et al. (2010). The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Developmental Cell, 19, 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ren F, Zhang Q, Chen Y, Wang B, & Jiang J (2008). The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Developmental Cell, 14, 377–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, et al. (2007). Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes & Development, 21, 2747–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Y, Panikkar A, Xu J, Antoniou A, Raynaud P, Lemaigre F, et al. (2009). Notch signaling controls liver development by regulating biliary differentiation. Development, 136, 1727–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Further reading
- Yanger K, Knigin D, Zong Y, Maggs L, Gu G, Akiyama H, et al. (2014). Adult hepatocytes are generated by self-duplication rather than stem cell differentiation. Cell Stem Cell, 15, 340–349. [DOI] [PMC free article] [PubMed] [Google Scholar]