Abstract
The mass migration that occurred during 2009–2013 and after the insurgency in northeastern Nigeria could have increased malaria incidence and Plasmodium falciparum genetic diversity in North Central Nigeria. To determine P. falciparum sequence diversity in this region, we screened 282 samples collected in regional clinics during 2015–2018 for Plasmodium spp. and, with positive samples, determined P. falciparum infection complexity and allele diversity using PCR. Of 34 P. falciparum–positive samples, 39 msp1, 31 msp2, and 13 glurp alleles were detected, and 88% of infections were polyclonal. We identified trimorphic and dimorphic allele combinations in a high percentage of samples, indicative of a high infection complexity in the study population. High genetic diversity is a catalyst for the evolution of drug-resistant alleles. Improved measures (e.g., better drug quality, diagnostics) are needed to control P. falciparum transmission and reduce the potential for the emergence of drug resistance in Nigeria.
Keywords: genotypes, genetic, diversity, allele, complexity, infection, Plasmodium falciparum, nested PCR, malaria, polyclonal, resistance, population displacement, parasites, Nigeria, insurgency, Federal Capital Territory, Abuja, Plateau, Nasarawa, Kaduna, Kogi, glurp, msp1, msp2
Malaria is endemic in Nigeria; its national prevalence among children 6–59 months of age is 27% (1). The World Health Organization World Malaria Report 2017 indicated that Nigeria accounted for 27% of the global malaria cases and 30% of the deaths in 2016 (2). The insurgency that occurred during 2009–2013 in northeastern Nigeria has been marked as one of the key factors responsible for driving this increased number of malaria cases and deaths; the insurgency disrupted the healthcare system, and up to 2.1 million persons were displaced (3–6). Added to the insurgency burden were communal conflicts between internally displaced persons (IDPs) and locals, which predominantly occurred in the states of Plateau, Benue, Taraba, Kaduna, and Nasarawa (3). Published reports established that 3 patterns of IDP movement transpired in Nigeria: migration to North Central Nigeria (defined as the states of Kwara, Niger, Nasarawa, Plateau, Benue, and Kaduna, and the Federal Capital Territory), economic migration from rural to urban areas, and secondary displacement of host communities because of communal conflicts (5).
The population displacement that occurred in North Central Nigeria was likely a key factor affecting the epidemiology of malaria transmission in this region. A high level of new clones of Plasmodium falciparum probably would have been introduced into the host communities of towns and cities experiencing influxes of IDPs. Immigration of infected persons into new areas can increase the rate of malaria transmission (7) and might subsequently result in infections of higher complexity and pathogens of more extensive genetic diversity in the host communities. In other malaria-endemic areas, population displacement, political unrest, and insurgency have been found to affect malaria epidemiology (8–10).
Meiotic recombination of P. falciparum parasites in Anopheles sp. mosquitoes has been proposed as the origin of the generation of novel alleles leading to new P. falciparum strains, and this cycle is likely to continue as long as there are vectors, parasites, and human hosts (11–13). Treatment failures occur more often in patients infected with higher numbers (>3 vs. <3) of P. falciparum strains (14,15). Drug resistance and treatment failures are envisaged to be among the challenges to achieving elimination of malaria in Nigeria (16,17). Ajayi et al. (18) reported 3 cases of artemisinin-based combination treatment failure of P. falciparum infection that were later adequately cleared with quinine. However, health practitioners and professionals in Nigeria have determined that the failure to clear parasites and resolve clinical disease after drug treatment is the result of many factors (drug nonpotency, incorrect diagnosis, noncompliance with dosing regimen duration, use of substandard drugs, and drug interactions). These reports spurred us to investigate the complexity of P. falciparum infections and P. falciparum genetic diversity (allele frequencies) in the North Central region of Nigeria.
Merozoite surface protein 1 (msp1), merozoite surface protein 2 (msp2), and glutamate-rich protein (glurp) gene-based studies of P. falciparum genetic diversity and infection complexity have been extensively carried out in other parts of sub-Saharan Africa, but only a few studies have taken place in regions of Nigeria (19–24). Research is scant on the genetic diversity of P. falciparum in North Central Nigeria, and this knowledge is critical for the implementation of successful control measures (13). Earlier reports by Jelinek et al. (25) and Meyer et al. (12) showed that increased genetic diversity of circulating malaria parasites in a population increases the potential for the selection of drug resistance. In our study, we investigated P. falciparum genetic diversity and the complexity of P. falciparum infections by assessing msp1, msp2, and glurp allele frequencies and genetic diversity in densely populated areas of North Central Nigeria.
Materials and Methods
Study Design
During August 2015–April 2018, whole blood samples for this study were collected from patients treated at 6 randomly selected healthcare centers in 5 locations of North Central Nigeria: Jos (Plateau State), Karu (Nasarawa State), Kafanchan (Kaduna State), Lokoja (Kogi State), and Abuja (Federal Capital Territory) (Figure 1). We chose these locations because previously published reports indicated that IDP movement occurred in these areas during the peak periods of insurgency, communal conflicts, and economic migration. After collection, whole blood samples were stored at 4°C until we performed PCR analyses at the Department of Biochemistry in the Faculty of Medical Sciences, University of Jos (Jos, Nigeria). Our target was to analyze >50 representative archived whole blood samples from each of the 6 healthcare centers. We received permission to collect and analyze these samples from the National Code for Health Research Ethics Committee.
Figure 1.
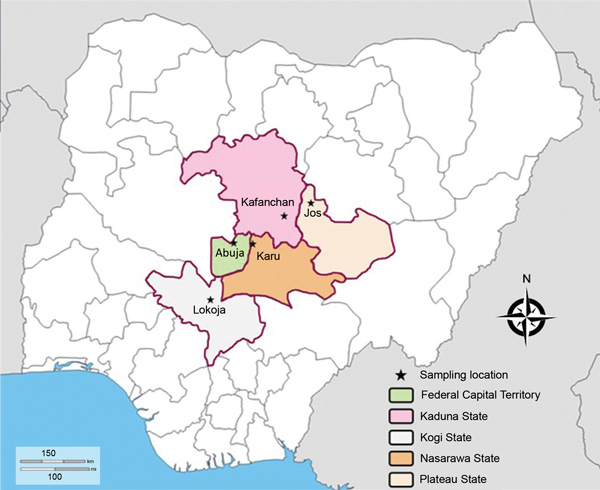
Sampling site locations in study of genetic diversity of Plasmodium falciparum spp., North Central Nigeria, 2015–2018. The Jabi region was the sampling site in Abuja.
PCR Screening for P. falciparum
We characterized P. falciparum isolates using the nested PCR methods of Snounou et al. (26) as modified by Singh et al. (27). These modifications included the use of a different genus-specific primer combination (rPLU-1 with rPLU-5 instead of rPLU-5 with rPLU-6) and the use of the nest-1 PCR product as the template for the genus-specific (nest-2) PCR. The primers for these PCRs (Table 1) were designed by using the Plasmodium small subunit rRNA genes (29) and synthesized by Inqaba Biotec (https://www.inqababiotec.co.za). We performed the nest-1 PCR, then the nest-2 PCR (to identify the samples positive for Plasmodium spp.), and then the P. falciparum species–specific PCR.
Table 1. Primer sequences used for PCRs to screen and genotype samples collected in study of genetic diversity of Plasmodium falciparum parasites, North Central Nigeria, 2015–2018*.
| PCR description | Primer name or type: sequence, 5′→3′ | Size, bp | Reference |
|---|---|---|---|
| Nest-1 |
rPLU1: TCAAAGATTAAGCCATGCAAGTGA | 620 |
(27) |
| rPLU5: CCTGTTGTTGCCTTAAACTCC | |||
| Nest-2, genus-specific PCR |
rPLU3: TTTTTATAAGGATAACTACGGAAAAGCTGT | 240 |
(27) |
| rPLU4: TACCCGTCATAGCCATGTTAGGCCAATACC | |||
|
Plasmodium falciparum species–specific PCR |
rFAL1: TTAAACTGGTTTGGGAAAACCAAATATATT | 205 |
(27) |
| rFAL2: ACACAATGAACTCAATCATGACTACCCGTC | |||
|
msp1, primary reaction |
Forward: CTAGAAGCTTTAGAAGATGCAGTATTG | Variable |
(28) |
| Reverse: CTTAAATAGTATTCTAATTCAAGTGGATCA | |||
|
K1
|
Forward: AAATGAAGAAGAAATTACTACAAAAGGTGC | Variable |
(28) |
| Reverse: GCTTGCATCAGCTGGAGGGCTTGCACCAGA | |||
|
MAD20
|
Forward: AAATGAAGGAACAAGTGGAACAGCTGTTAC | Variable |
(28) |
| Reverse: ATCTGAAGGATTTGTACGTCTTGAATTACC | |||
|
RO33
|
Forward: TAAAGGATGGAGCAAATACTCAAGTTGTTG | Variable |
(28) |
| Reverse: CATCTGAAGGATTTGCAGCACCTGGAGATC | |||
|
msp2, primary reaction |
Forward: ATGAAGGTAATTAAAACATTGTCTATTATA | Variable |
(28) |
| Reverse: CTTTGTTACCATCGGTACATTCTT | |||
|
FC27
|
Forward: AATACTAAGAGTGTAGGTGCARATGCTCCA | Variable |
(28) |
| Reverse: TTTTATTTGGTGCATTGCCAGAACTTGAAC | |||
| IC/3D7 |
Forward: AGAAGTATGGCAGAAAGTAAKCCTYCTACT | Variable |
(28) |
| Reverse: GATTGTAATTCGGGGGATTCAGTTTGTTCG | |||
|
glurp, primary reaction |
Forward: TGAATTTGAAGATGTTCACACTGAAC | Variable |
(28) |
| Reverse: GTGGAATTGCTTTTTCTTCAACACTAA | |||
| glurp | Forward: TGTTCACACTGAACAATTAGATTTAGATCA | Variable | (28) |
| Reverse: GTGGAATTGCTTTTTCTTCAACACTAA |
*glurp, glutamate-rich protein; msp1, merozoite surface protein 1; msp2, merozoite surface protein 2.
The nest-1 reaction mix contained 5 μL of DNA template, 25 μL of One Taq Quick-Load 2× Master Mix (New England BioLabs, https://www.neb.com), 1 μL of each primer (10 μmol/L rPLU1 and rPLU5), and 18 μL nuclease-free water. We used the following cycling program for the nest-1 PCR: initial denaturation at 94°C for 4 min; 35 cycles of 94°C for 30 s, 55°C for 60 s, and 72°C for 60 s; and a final extension at 72°C for 4 min. The nest-2 reaction mix was the same as the nest-1 reaction mix, except that we used 2 μL of the nest-1 PCR product as the template and different primers (rPLU3 and rPLU4). The P. falciparum species–specific reaction mix was the same as the nest-2 PCR reaction mix, except that we used different primers (rFAL1 and rFAL2). The cycling programs we used for the nest-2 and P. falciparum species–specific PCRs were the same as the one used for the nest-1 PCR, except that we used different annealing temperatures (62°C for the nest-2 PCR and 58°C for the Plasmodium species–specific PCR). We performed these amplifications (as well as the genotyping PCR described in the next section) in the GeneAmp 9700 (Applied Biosystems, https://www.thermofisher.com).
Genotyping by Nested PCR
With P. falciparum–positive samples, we performed primary and genotype-specific PCRs for msp1, msp2, and glurp according to the modified method of Snounou and Färnert (28). The primary PCRs amplified the block 2 variable region (for msp1), block 3 central repeat region (for msp2), or the glutamate-rich protein region (for glurp). These regions have repetitive segments and motifs that are variable in length among strains; this variability occurs because of the intragenic recombination in Anopheles mosquitoes. PCRs targeting these regions amplify sequences of different lengths, which are the basis of clonality in P. falciparum. The primary reagent mixture included 5 μL of genomic DNA extract; 25 μL One Taq Quick-Load 2× Master Mix; 1.0 μL of each of the msp1, msp2, or glurp first reaction primers (10 μmol/L) (Table 1); and 14 μL nuclease-free water. We performed amplifications using an initial denaturation at 95°C for 5 min, followed by 25 cycles of denaturation at 94°C for 1 min, annealing at 58°C for 2 min, and extension at 72°C for 2 min and then a final extension at 72°C for 5 min. We used the products of the primary PCRs as the template for the subsequent genotype-specific PCRs, which were used to determine msp1 (K1, MAD20, RO33), msp2 (FC-27, IC/3D7), and glurp allele type. The reaction mix and conditions we used for the genotype-specific amplifications were the same as the ones we used for the primary amplification, except that the primers (Table 1), annealing temperature (61°C), and DNA template (2 μL) were different.
Statistical Analysis
We used GraphPad Prism version 7.03 (https://www.graphpad.com) for allele frequency graph construction. We also used this tool to compare allele frequencies by paired t test and analyze the polyclonal distribution of allele families.
Results
P. falciparum Nested PCR
We obtained 282 archived whole blood samples from the 6 healthcare centers participating in the study. Many of the samples originated from patients who visited the centers for general medical checkups, genotyping, and screening for hepatitis viruses. Various clinical laboratory tests were performed on the collected blood samples as requested by the medical doctors for determining diagnosis or as requested by the patient.
Of 282 blood samples, 54 were positive for a Plasmodium species (240-bp PCR product) by nested PCR and 34 samples were positive for P. falciparum (205-bp PCR product; Figure 2). The estimated prevalence of P. falciparum among positive samples was 63% (34/54), unusually low compared with the national P. falciparum prevalence of ˃95% in 2015 (1). However, this finding represents a limited number of samples and cannot truly reflect the national average. The estimated prevalence of 37% for non–P. falciparum malaria species can probably be attributed to the circulation of >2% of the other Plasmodium species that infect humans.
Figure 2.
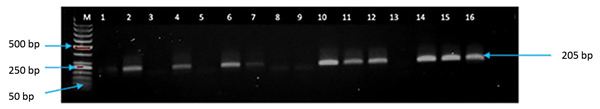
Screening PCR results of persons with Plasmodium falciparum parasite infections, North Central Nigeria, 2015–2018. Lane M, 50-bp DNA marker (ThermoFisher Scientific, https://www.thermofisher.com); lanes 1–4, archived blood samples from Nisa Premier Hospital (Jabi, Federal Capital Territory, Nigeria); lanes 5–15, archived blood samples from Kogi Specialist Hospital (Lokoja, Kogi State, Nigeria); lane 16, positive control. Samples positive for P. falciparum had a PCR product size of 205 bp.
Allele Diversity and Complexity of Infections
In genotyping PCRs, 32 persons were positive for msp1, 29 for msp2, and 31 for glurp. We found more genetic variation (i.e., more alleles) for the msp1 gene than the msp2 or glurp genes. We detected a total of 39 msp1 (16 K1, 13 MAD20, and 10 RO33), 31 msp2 (16 FC27 and 15 IC/3D7) and 13 glurp clones in P. falciparum–positive blood samples (Tables 2, 3). A high percentage of the population had evidence of polyclonal infections; >2 different-sized PCR products (i.e., >2 alleles) were present on agarose gel for the msp1, msp2, or glurp genes in 88% of the samples. Similar studies conducted previously in Nigeria (mainly in the southwestern part) during 2004–2014 showed less genetic diversity (Table 3). In those studies, the total number of clones detected were 4 for K1, 2 for MAD20, and 4 for RO33 of the msp1 gene and 9 for FC27 and 4 for IC/3D7 of the msp2 gene. Polyclonal P. falciparum infections were more prevalent in our study (multiplicity of infection [MOI], i.e., parasite clones per sample, 2.4) than in the previous reports (MOIs 1.1 and 1.4; Table 3).
Table 2. Alleles of genes detected in Plasmodium falciparum isolates, North Central Nigeria, 2015–2018*.
| Allele family |
No. positive by PCR |
No. alleles |
Allele size range, bp |
Mean MOI† |
% Polyclonal |
|---|---|---|---|---|---|
| msp1 | 32 | ||||
| K1 | 32 | 16 | 100–916 | 2.63 | 68 |
| MAD20 | 21 | 13 | 100–1,200 | 2.38 | 73 |
|
RO33
|
22 |
10 |
100–1,100 |
2.23 |
64 |
| msp2 | 29 | ||||
| FC27 | 17 | 16 | 150–1,130 | 2.52 | 53 |
|
IC/3D7
|
28 |
15 |
300–1,200 |
2.21 |
68 |
| glurp | 31 | 13 | 200–1,550 | 1.03 | 23 |
*glurp, glutamate-rich protein; MOI, multiplicity of infection; msp1, merozoite surface protein 1; msp2, merozoite surface protein 2. †Defined as number of parasite clones per sample.
Table 3. Distribution of msp 1, msp 2, and glurp clones detected in previous studies conducted in southwestern Nigeria, 2004–2014, and North Central Nigeria, 2015–2018*.
| Study | Region (state or territory) | No. samples | No. alleles† |
Mean MO‡ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
msp1
|
msp2
|
glurp | ||||||||
| K1 | MAD20 | R033 | FC27 | IC/3D7 | ||||||
| Happi et al. (30) | Southwestern (Ogun) | 47 | 4 | 2 | 4 | 9 | 4 | 5 | – | |
| Olasehinde et al. (31) | Southwestern (Ogun) | 100 | 4 | 3 | 1 | 3 | 3 | – | 1.1 | |
| Oyebola et al. (32) | Southwestern (Lagos) | 100 | 3 | 2 | 1 | 3 | 3 | – | 1.4 | |
| Bamidele Abiodun et al. (33) | Southwestern (Lagos) | 78 | 2 | 2 | 1 | – | – | – | 1.4 | |
| This study | North Central (Plateau, Nasarawa, Kogi, Kaduna, FCT) | 54 | 16 | 13 | 10 | 16 | 15 | 13 | 2.4 | |
*FDT, Federal Capital Territory; glurp, glutamate-rich protein; MOI, multiplicity of infection; msp1, merozoite surface protein 1; msp2, merozoite surface protein 2. †Total number alleles found, regardless of size. ‡Defined as number of parasite clones per sample.
In our study, fragment sizes were 100–1,200 bp for msp1, 150–1,200 bp for msp2, and 200–1,550 bp for glurp (Figures 3,4,5). The highest number of clones seen for a single allele in a single sample was 8 (with 8 clear bands of the FC27 allele); this sample came from a patient in Kafanchan. A number of samples contained only 1 clone: 8 samples contained a single clone of K1, 8 a single clone of RO33, 6 a single clone of MAD20, 7 a single clone of FC27, and 9 a single clone of IC/3D7.
Figure 3.
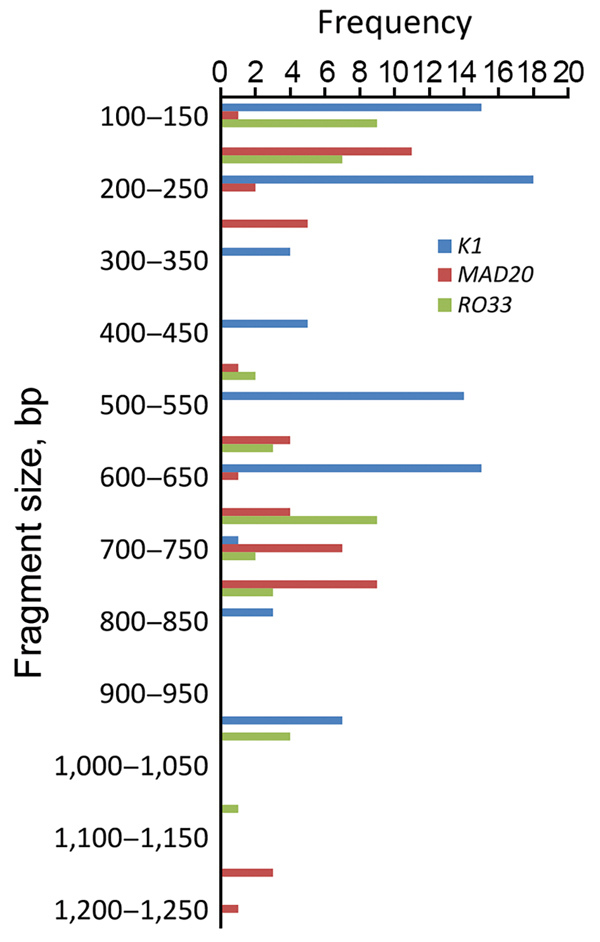
Allele frequency of msp1 in persons with Plasmodium falciparum infection, North Central Nigeria, 2015–2018. The K1 allele of size 200–250 bp was detected at the highest frequency (n = 18). The next highest detected were the MAD20 allele of fragment size 150–200 bp (n = 11) and the RO33 alleles of fragment sizes 100–150 bp (n = 9) and 650–700 bp (n = 9).
Figure 4.
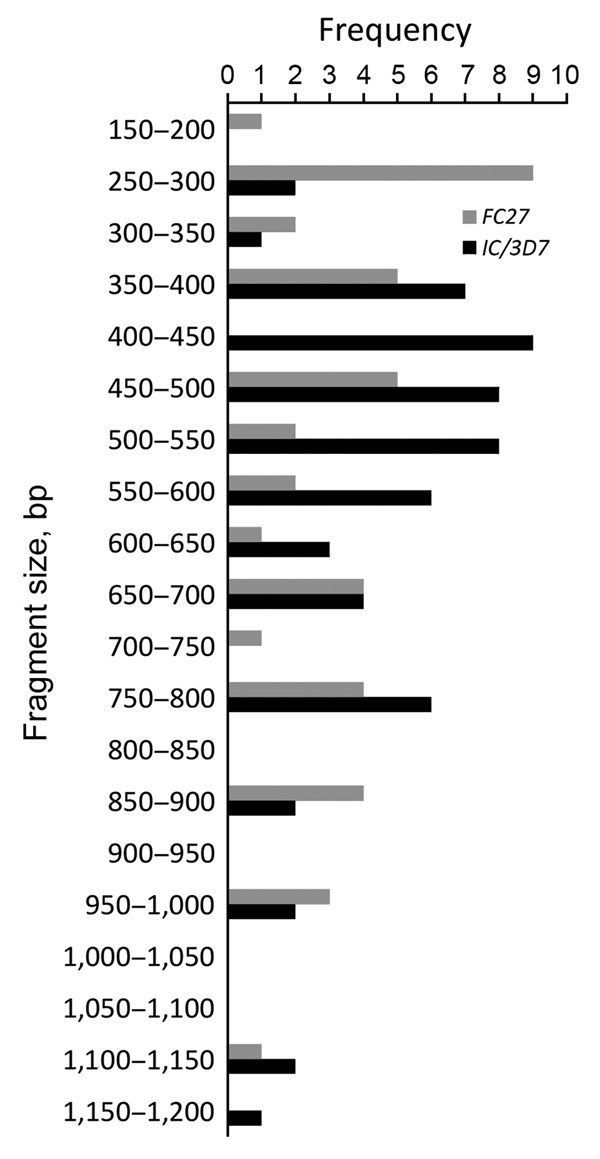
Allele frequency of msp2 in persons with Plasmodium falciparum infection, North Central Nigeria, 2015–2018. The most frequently detected alleles were the FC27 allele of size 250–300 bp (n = 9) and the IC/3D7 allele of size 400–450 bp (n = 9).
Figure 5.
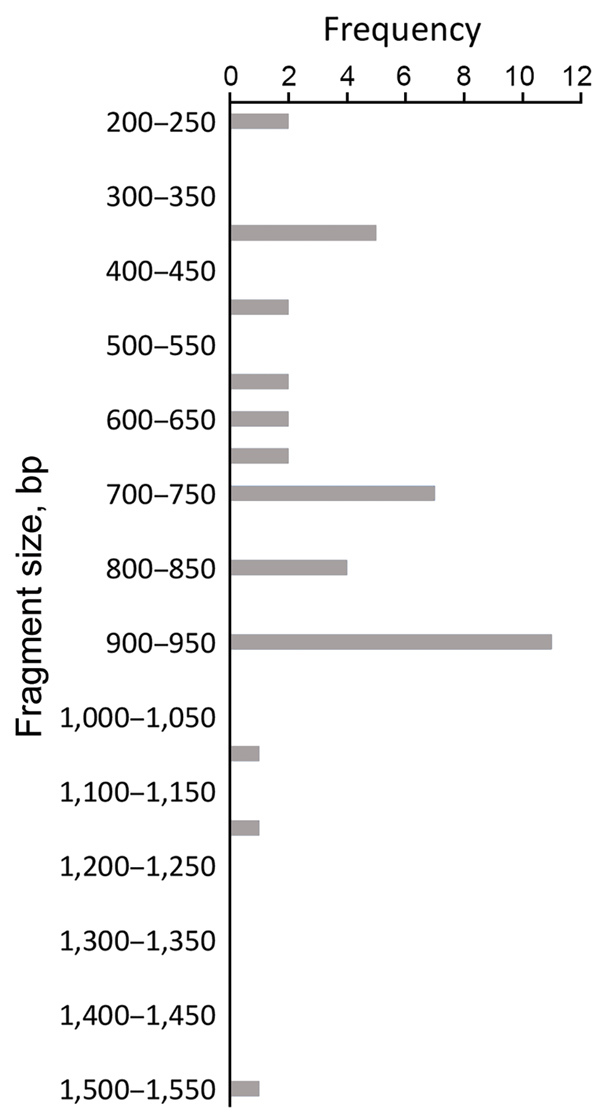
Allele frequency of glurp in persons with Plasmodium falciparum infection, North Central Nigeria, 2015–2018. The allele with the highest frequency (n = 11) was 900–950 bp in size.
The combinations of msp1 family alleles observed were K1, MAD20, and RO33 (63%, 20/32); K1 and MAD20 (3%, 1/32); K1 and RO33 (6%, 2/32); and MAD20 and RO33 (3%, 1/32). Infections were predominantly trimorphic (63%), and fewer were dimorphic (12%); 25% were monomorphic. For the msp2 allele family, dimorphic infections (FC27 + IC/3D7) occurred in 50% of the population; 35% of the population was positive for only IC/3D7 and 15% for only FC27. Many patients had infections with high MOIs. The mean MOI per allele family was 2.4 for msp1 and msp2 and 1.0 for glurp (Table 2). The wide occurrence of trimorphic and dimorphic allele combinations is indicative of high complexity of infection in the study population.
We assessed the frequency of the occurrence of each allele in the population. We categorized clones into molecular weight groups differing by 50 bp for clear discrimination from other clones and elimination of errors that would result from estimating the molecular weight on agarose gels. For the msp1 allele family, the K1 allele of size 200−250 bp was detected at the highest frequency (n = 18), followed by the MAD20 allele of fragment size 150–200 bp (n = 11) and the RO33 alleles of fragment sizes 100–150 bp (n = 9) and 650–700 bp (n = 9) (Figure 3). For the msp2 allele family, the most frequently detected alleles were the FC27 allele of size 250–300 bp (n = 9) and the IC/3D7 allele of size 400–450 bp (n = 9) (Figure 4). The glurp allele with the highest frequency (n = 11) was 900–950 bp in size (Figure 5).
We did not attempt to compare the distribution patterns of the allele families by location because the number of P. falciparum–positive samples in some locations was low. However, an analysis of the seasonal patterns of diversity showed that 60% of the infections that occurred during the dry season (November–March) and 50% of those that occurred during the wet season (April–October) were polyclonal. Paired t tests showed that the temporal distribution of polyclonal infections and the frequency of clones were statistically significant (p<0.05). During the dry season, the person infected with the highest number of clones (8 clones) was sampled, as well as others infected with 4–5 clones. The finding that polyclonal infections were more common in the dry season was unexpected. However, the increased transmission during this period can be explained by the open gutter and sewage systems present in urban areas of Nigeria; this factor has led to the mosquitoes that potentiate malaria transmission being prevalent year-round.
Discussion
In this prospective cross-sectional study, we investigated P. falciparum genetic diversity and the complexity of P. falciparum infection in North Central Nigeria because a high level of genetic diversity and infection complexity was a likely aftermath of the influx of IDPs into the region. We hypothesized that a high level of genetic diversity would be found in the region partly as a result of immigration of P. falciparum–infected IDPs into the region. High genetic diversity of malaria parasites circulating in a population could serve as a risk factor for genetic recombination and the generation of novel alleles (11–13). We found that 63% of the Plasmodium spp.–positive samples were positive for P. falciparum. Our PCR screening results cannot be used as an estimate of the prevalence of P. falciparum in North Central Nigeria, but this finding has revealed that non–P. falciparum malaria species are circulating in this region and infecting humans. The finding of 37% positivity for non–P. falciparum Plasmodium spp. is high compared with previous reports of low-level prevalence (0.4%–6.3%) in other parts of Nigeria (30–33). An epidemiologic study of non–P. falciparum malaria species circulating in the North Central region would be vital for national malaria control. Treatment failures reported by some authors (16,17) in Nigeria could be related to incorrect diagnosis and hence inappropriate drug administration. Artemisinin-based combination therapy is recommended for the treatment of P. falciparum malaria, but chloroquine plus primaquine is the first-line regimen for P. vivax malaria (34).
We detected the msp1, msp2, and glurp alleles in most patients positive for P. falciparum parasites, and these alleles had high genetic variability. Many samples from infected patients contained multiple alleles, and the genetic variation was more extensive with the msp1 and msp2 allele families than with the glurp allele. Overall, 88% of the population had evidence of polyclonal infection with >1 of the allelic forms. We discovered 10–16 msp1 clones and 15–16 msp2 clones. This finding does not corroborate results of similar previous studies from southwestern Nigeria, in which 1–4 msp1 clones and 3–9 msp2 clones were found (Table 3). We detected a total of 39 msp1, 31 msp2, and 13 glurp alleles, which is quite high compared with the reported values in other regions of Nigeria (30,32) and other countries of West Africa (21). Infections with pathogens harboring high numbers of alleles could influence the emergence of resistance (35), considering that, in this scenario, multiple alleles are exposed to the same chemotherapeutic agents. The high level of trimorphic, dimorphic, and monomorphic infections in the population is evidence of extensive genetic diversity. Our work revealed a higher frequency of occurrence of polyclonal infections and infections with P. falciparum parasites of multiple genotypes, as well as infections of higher MOIs, compared with previous studies (30,33,36).
The question yet to be answered, however, is whether polyclonal infection is a frequent occurrence in Nigeria. The frequent occurrence of this phenomenon in a single geographic area has been determined to be an early indicator for the emergence of alleles associated with drug resistance (11,35). The high number of polyclonal infections seen in North Central Nigeria could be attributed to a high level of genetic recombination and high evolutionary pressure on the msp1, msp2, and glurp genes in this region, which could partially be attributed to the high level of movement into the North Central region (the economic hub of the nation) for commercial reasons. Drug resistance of P. falciparum parasites is a highly complex mechanism involving multiple genes, pfk13 being a gene responsible (37). Work by Oboh et al. (36) and Ebenebe et al. (37) indicated that in 2018 drug resistance of malaria pathogens to artemisinin-based combination therapy was not an immediate public health threat for southwestern Nigeria. Nonetheless, a nationwide study should be performed to comprehensively evaluate for the presence of P. falciparum drug resistance genes with techniques such as next-generation sequencing and genome walking (38,39). Oboh et al. (36) were able to detect synonymous mutations (not validated AA mutations of the pfk13 gene), such as the nucleotide change from CCG to CCA in the pfk13 gene in P. falciparum from southwestern Nigeria, and called for close monitoring of parasites in Nigeria. A nationwide study should also be conducted to comprehensively determine the genetic diversity of P. falciparum and complexity of infections in Nigeria. Determining the genetic diversity of malaria parasites in the North Central region, as well as for the entire nation of Nigeria in general, is needed for the design of a national treatment policy, vaccine development, and immunogenicity studies.
The frequency of polyclonal infections in southwestern Nigeria during 2004–2014 can be described as low and not extensive compared with the level discovered in this study. The period of this study (2015–2018) falls within peak periods of communal conflicts in Nigeria (2009–2017), which resulted in the displacement of persons in the North Central region (40,41). Our findings suggest a relationship between the high number of polyclonal infections discovered in North Central Nigeria and the movements of IDPs into the region, although a previous study of the msp2 genotype by Oyedeji et al. (42) in Lafia (Nasarawa State) showed a high level of genetic diversity in 2005–2006. A high level of introduction of new P. falciparum clones could have occurred in the host communities in the towns and cities that experienced IDP influxes.
A major limitation of this study is the lack of a comparison of the genetic diversity of Plasmodium spp. in patients in other regions of the country. However, previous studies conducted in southwestern Nigeria provided a baseline for comparison to determine whether the level of Plasmodium spp. genetic diversity discovered in this study was different and thus potentially associated with the movement of IDPs. The correlation would have been more comprehensive if we had also included comparison groups from other regions and specific IDP camps in our investigation.
In summary, this study revealed a high level of P. falciparum genetic diversity and infection complexity in North Central Nigeria and suggested that this complexity might be a result of the insurgency and movement of IDPs that occurred in Nigeria during 2009–2017. A deliberate effort is needed to control malaria and eliminate the risk for the evolution of resistance alleles across Nigeria, but particularly in this region of Nigeria, which is the economic hub of the nation and at highest risk for distributing new strains worldwide. The information we generated on Plasmodium spp. epidemiology and genetic diversity could serve as a source in a database to be used for policy development on nationwide malaria control and intervention programs for Nigeria. Our work also led to the establishment of a laboratory capable of performing PCR methods to evaluate malaria epidemiology in the region, which could be used for future comprehensive malaria control measures.
Acknowledgments
We sincerely thank Bitrus Matawal for his valuable assistance in securing the national code for ethics approval to conduct this healthcare research. We also appreciate the medical laboratory scientists in the selected healthcare centers who provided aliquots of archived blood samples analyzed in this study.
This study was sponsored by the Biotechnology Development Center of the University of Jos.
Biography
Dr. Yakubu is a research fellow at the Biotechnology Division of the National Veterinary Research Institute Vom in Jos, Nigeria. He specializes in PCR methodology and diagnostics and has research interests in DNA barcoding, vaccinology, and protein biotechnology.
Footnotes
Suggested citation for this article: Yakubu B, Longdet IY, Tony HJ, Davou DT, Obishakin E. High-complexity Plasmodium falciparum infections, North Central Nigeria, 2015–2018. Emerg Infect Dis. 2019 Jul [date cited]. https://doi.org/10.3201/eid2507.181614
References
- 1.National Malaria Elimination Programme, National Population Commission, and National Bureau of Statistics, Federal Republic of Nigeria; ICT International. Nigeria malaria indicator survey 2015. 2016. Aug [cited 2018 Oct 18]. https://dhsprogram.com/pubs/pdf/MIS20/MIS20.pdf
- 2.World Health Organization. World malaria report 2017. Geneva: The Organization; 2017. [cited 2018 Oct 18]. https://www.who.int/malaria/publications/world-malaria-report-2017
- 3.United Nations Office for the Coordination of Humanitarian Affairs. Situation report no.1, 2014. 2014. [cited 2018 Oct 18]. http://www.unocha.org/nigeria
- 4.Aribodor DN, Ugwuanyi IK, Aribodor OB. Challenges to achieving malaria elimination in Nigeria. Am J Public Health Res. 2016;4:38–41. [Google Scholar]
- 5.Internal Displacement Monitoring Centre. GRID 2016. Global report on internal displacement. 2016. May [cited 2018 Oct 18]. http://www.internal-displacement.org/globalreport2016
- 6.Emmanuelar I. Insurgency and humanitarian crisis in northern Nigeria: the case of Boko Haram. Afr J Pol Sci Int Relat. 2015;9:284–96. 10.5897/AJPSIR2015.0789 [DOI] [Google Scholar]
- 7.Martens P, Hall L. Malaria on the move: human population movement and malaria transmission. Emerg Infect Dis. 2000;6:103–9. 10.3201/eid0602.000202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenwood B, Mutabingwa T. Malaria in 2002. Nature. 2002;415:670–2. 10.1038/415670a [DOI] [PubMed] [Google Scholar]
- 9.Gu W, Killeen GF, Mbogo CM, Regens JL, Githure JI, Beier JC. An individual-based model of Plasmodium falciparum malaria transmission on the coast of Kenya. Trans R Soc Trop Med Hyg. 2003;97:43–50. 10.1016/S0035-9203(03)90018-6 [DOI] [PubMed] [Google Scholar]
- 10.Osorio L, Todd J, Pearce R, Bradley DJ. The role of imported cases in the epidemiology of urban Plasmodium falciparum malaria in Quibdó, Colombia. Trop Med Int Health. 2007;12:331–41. 10.1111/j.1365-3156.2006.01791.x [DOI] [PubMed] [Google Scholar]
- 11.Conway DJ, Roper C, Oduola AMJ, Arnot DE, Kremsner PG, Grobusch MP, et al. High recombination rate in natural populations of Plasmodium falciparum. Proc Natl Acad Sci U S A. 1999;96:4506–11. 10.1073/pnas.96.8.4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer CG, May J, Arez AP, Gil JP, Do Rosario V. Genetic diversity of Plasmodium falciparum: asexual stages. Trop Med Int Health. 2002;7:395–408. 10.1046/j.1365-3156.2002.00875.x [DOI] [PubMed] [Google Scholar]
- 13.Kiwanuka GN. Genetic diversity in Plasmodium falciparum merozoite surface protein 1 and 2 coding genes and its implications in malaria epidemiology: a review of published studies from 1997-2007. J Vector Borne Dis. 2009;46:1–12. [PubMed] [Google Scholar]
- 14.Lee SA, Yeka A, Nsobya SL, Dokomajilar C, Rosenthal PJ, Talisuna A, et al. Complexity of Plasmodium falciparum infections and antimalarial drug efficacy at 7 sites in Uganda. J Infect Dis. 2006;193:1160–3. 10.1086/501473 [DOI] [PubMed] [Google Scholar]
- 15.Kyabayinze DJ, Karamagi C, Kiggundu M, Kamya MR, Wabwire-Mangen F, Kironde F, et al. Multiplicity of Plasmodium falciparum infection predicts antimalarial treatment outcome in Ugandan children. Afr Health Sci. 2008;8:200–5. [PMC free article] [PubMed] [Google Scholar]
- 16.Amodu OK, Adeyemo AA, Ayoola OO, Gbadegesin RA, Orimadegun AE, Akinsola AK, et al. Genetic diversity of the msp-1 locus and symptomatic malaria in south-west Nigeria. Acta Trop. 2005;95:226–32. 10.1016/j.actatropica.2005.06.017 [DOI] [PubMed] [Google Scholar]
- 17.Amodu OK, Oyedeji SI, Ntoumi F, Orimadegun AE, Gbadegesin RA, Olumese PE, et al. Complexity of the msp2 locus and the severity of childhood malaria, in south-western Nigeria. Ann Trop Med Parasitol. 2008;102:95–102. 10.1179/136485908X252340 [DOI] [PubMed] [Google Scholar]
- 18.Ajayi NA, Ukwaja KN. Possible artemisinin-based combination therapy-resistant malaria in Nigeria: a report of three cases. Rev Soc Bras Med Trop. 2013;46:525–7. 10.1590/0037-8682-0098-2013 [DOI] [PubMed] [Google Scholar]
- 19.Ferreira MU, Kaneko O, Kimura M, Liu Q, Kawamoto F, Tanabe K. Allelic diversity at the merozoite surface protein-1 (MSP-1) locus in natural Plasmodium falciparum populations: a brief overview. Mem Inst Oswaldo Cruz. 1998;93:631–8. 10.1590/S0074-02761998000500013 [DOI] [PubMed] [Google Scholar]
- 20.Ferreira MU, Ribeiro WL, Tonon AP, Kawamoto F, Rich SM. Sequence diversity and evolution of the malaria vaccine candidate merozoite surface protein-1 (MSP-1) of Plasmodium falciparum. Gene. 2003;304:65–75. 10.1016/S0378-1119(02)01180-0 [DOI] [PubMed] [Google Scholar]
- 21.Mwingira F, Nkwengulila G, Schoepflin S, Sumari D, Beck H-P, Snounou G, et al. Plasmodium falciparum msp1, msp2 and glurp allele frequency and diversity in sub-Saharan Africa. Malar J. 2011;10:79. 10.1186/1475-2875-10-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manske M, Miotto O, Campino S, Auburn S, Almagro-Garcia J, Maslen G, et al. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature. 2012;487:375–9. 10.1038/nature11174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irawati N, Jamsari, Wirasti Y. Genetic diversity of merozoite surface protein-1 in Plasmodium falciparum field isolates from a mountain and coastal area in West Sumatera, Indonesia. J Pharm Biomed Sci. 2013;30:1061–4. [Google Scholar]
- 24.Mawili-Mboumba DP, Mbondoukwe N, Adande E, Bouyou-Akotet MK. Allelic diversity of msp1 gene in Plasmodium falciparum from rural and urban areas of Gabon. Korean J Parasitol. 2015;53:413–9. 10.3347/kjp.2015.53.4.413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jelinek T, Kilian AH, Westermeier A, Pröll S, Kabagambe G, Nothdurft HD, et al. Population structure of recrudescent Plasmodium falciparum isolates from western Uganda. Trop Med Int Health. 1999;4:476–80. 10.1046/j.1365-3156.1999.00428.x [DOI] [PubMed] [Google Scholar]
- 26.Snounou G, Viriyakosol S, Zhu XP, Jarra W, Pinheiro L, do Rosario VE, et al. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61:315–20. 10.1016/0166-6851(93)90077-B [DOI] [PubMed] [Google Scholar]
- 27.Singh B, Bobogare A, Cox-Singh J, Snounou G, Abdullah MS, Rahman HA. A genus- and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am J Trop Med Hyg. 1999;60:687–92. 10.4269/ajtmh.1999.60.687 [DOI] [PubMed] [Google Scholar]
- 28.Snounou G, Fӓrnert A. Genotyping of Plasmodium falciparum parasites by PCR. In: Moll K, Ljungstrӧm I, Perlmann H, Scherf A, Wahlgren M, editors. Methods in malaria research. Manassas (VA): American Type Culture Collection; 2008. p. 238–42 [cited 2018 Oct 18]. https://ki.se/sites/default/files/methods_in_malaria_research.pdf [Google Scholar]
- 29.Waters AP, McCutchan TF. Rapid, sensitive diagnosis of malaria based on ribosomal RNA. Lancet. 1989;1:1343–6. 10.1016/S0140-6736(89)92800-6 [DOI] [PubMed] [Google Scholar]
- 30.Happi CT, Gbotosho GO, Sowunmi A, Falade CO, Akinboye DO, Gerena L, et al. Molecular analysis of Plasmodium falciparum recrudescent malaria infections in children treated with chloroquine in Nigeria. Am J Trop Med Hyg. 2004;70:20–6. 10.4269/ajtmh.2004.70.20 [DOI] [PubMed] [Google Scholar]
- 31.Olasehinde GI, Yah CS, Singh R, Ojuronbge OO, Ajayi AA, Valecha N, et al. Genetic diversity of Plasmodium falciparum field isolates from south western Nigeria. Afr Health Sci. 2012;12:355–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oyebola MK, Idowu ET, Olukosi YA, Iwalokun BA, Agomo CO, Ajibaye OO, et al. Genetic diversity and complexity of Plasmodium falciparum infections in Lagos, Nigeria. Asian Pac J Trop Biomed. 2014;4(Suppl 1):S87–91. 10.12980/APJTB.4.2014C1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bamidele Abiodun I, Oluwadun A, Olugbenga Ayoola A, Senapon Olusola I. Plasmodium falciparum merozoite surface proein-1 polymorphisms among asymptomatic sickle cell anemia patients in Nigeria. Acta Med Iran. 2016;54:44–53. [PubMed] [Google Scholar]
- 34.Sam Wobo SO, Adekunle NO, Adeleke MA, Dedeke GA, Oke OA, Abimbola WA, et al. Epidemiological factors in prevalence of malaria parasites in primary health facilities attendants, Ogun State, Nigeria. Malar Chemother Control Elimin. 2014;3:1000111. [Google Scholar]
- 35.Cui L, Mharakurwa S, Ndiaye D, Rathod PK, Rosenthal PJ. Antimalarial drug resistance: literature review and activities and findings of the ICEMR network. Am J Trop Med Hyg. 2015;93(Suppl):57–68. 10.4269/ajtmh.15-0007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oboh MA, Ndiaye D, Antony HA, Badiane AS, Singh US, Ali NA, et al. Status of artemisinin resistance in malaria parasite Plasmodium falciparum from molecular analysis of kelch13 gene in southwestern Nigeria. BioMed Res Int. 2018;2018:2305062. 10.1155/2018/2305062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ebenebe JC, Ntadom G, Ambe J, Wammanda R, Jiya N, Finomo F, et al. ; For The Antimalarial Therapeutic Efficacy Monitoring Group National Malaria Elimination Programme The Federal Ministry Of Health Abuja Nigeria. Efficacy of artemisinin-based combination treatments of uncomplicated falciparum malaria in under-five year-old Nigerian children ten years following adoption as first line antimalarials. Am J Trop Med Hyg. 2018;99:649–64. 10.4269/ajtmh.18-0115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flannery EL, Wang T, Akbari A, Corey VC, Gunawan F, Bright AT, et al. Next-generation sequencing of Plasmodium vivax patient sample shows evidence of direct evolution in drug-resistance genes. ACS Infect Dis. 2015;1:367–79. 10.1021/acsinfecdis.5b00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nag S, Dalgaard MD, Kofoed PE, Ursing J, Crespo M, Andersen LO, et al. High throughput resistance profiling of Plasmodium falciparum infections based on custom dual indexing and Illumina next generation sequencing-technology. Sci Rep. 2017;7:2398. 10.1038/s41598-017-02724-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.International Organization for Migration. Displacement tracking matrix (DTM) round 12 report-October 2016. 2016. Oct [cited 2018 Oct 18]. https://reliefweb.int/report/nigeria/displacement-tracking-matrix-dtm-round-12-report-october-2016
- 41.International Committee of the Red Cross. Internal displacement in northeastern Nigeria. Kampala Convention. Supra note 6. Article 5(1). 2016 Dec [cited 2018 Oct 18]. http://www.icrc.org/where-we-work/africa/nigeria
- 42.Oyedeji SI, Awobode HO, Anumudu C, Kun J. Genetic diversity of Plasmodium falciparum isolates from naturally infected children in north-central Nigeria using the merozoite surface protein-2 as molecular marker. Asian Pac J Trop Med. 2013;6:589–94. 10.1016/S1995-7645(13)60102-9 [DOI] [PubMed] [Google Scholar]


