Abstract
Hepatocellular carcinoma (HCC) is one of the most fatal cancers with common features of invasion and metastasis. Recent evidence indicate that the long noncoding RNA NORAD is a potential oncogene and is significantly upregulated in several cancers. However, the general biological role and clinical value of NORAD in HCC remains unknown. Here, NORAD expression was measured in 29 paired tumor and paratumor tissues via quantitative real‐time polymerase chain reaction (qPCR). The effects of NORAD on HCC cell malignant potential were investigated via NORAD overexpression and knockdown both in vitro and in vivo. The mechanism of competitive endogenous RNAs (ceRNAs) was acquired and identified by bioinformatics analyses and luciferase assays. Moreover, the impact of NORAD level on the transforming growth factor β (TGF‐β) pathway was further determined by qPCR. We found that HCC tissues had a high level of NORAD compared with the paratumor tissues, and NORAD upregulation was associated with the shorter overall survival of patients with HCC. Furthermore, NORAD overexpression was demonstrated to promote HCC cell migration and invasion. Mechanically, NORAD might function as a ceRNA to regulate miR‐202‐5p, which served as a tumor‐suppressing microRNA via the TGF‐β pathway. We address that NORAD has a tumor‐promoting effect in HCC and describes a novel mechanism whereby NORAD regulates the TGF‐β pathway as a ceRNA of Homo sapiens (hsa)‐miR‐202‐5p.
Keywords: HCC, invasion and metastasis, miR‐202‐5p, NORAD, TGF‐β
1. INTRODUCTION
Liver cancer is one of the most fatal and prevalent cancers in the world (Bray et al., 2018). Hepatocellular carcinoma (HCC) represents 80% of all primary liver neoplasms and has a complex etiology with different contributing factors (Zhou et al., 2018), for example, the chronic infections with hepatitis B and C viruses, alcohol consumption, cirrhosis, and aflatoxin B1 exposure (Singal & El‐Serag, 2015; Zhou et al., 2018). Over the past decades, a large number of genes and multiple pathways involved in this disease have been disclosed to be the cellular and molecular biology of HCC (Zucman‐Rossi et al., 2015). Despite the intense efforts, therapeutic progress has not been significantly improved and the benefit of HCC patient is limited. Thus, the further and complete understanding of the mechanism of this disease is still an urgent need.
MicroRNAs (miRNAs) are small noncoding RNAs, which is involved in posttranscriptional RNA regulation (Hammond, 2015). Mechanistically, miRNAs bind to target transcripts via sequence complementarity to reduce the certain transcripts expression (Afonso‐Grunz & Muller, 2015; Hammond, 2015). Now, miRNA‐mediated regulation has been revealed to be one of the most common posttranscriptional regulatory mechanisms in eukaryotes and involved in a variety of cellular processes and diseases, including cancer (Hammond, 2015; Mendell & Olson, 2012; Stroynowska‐Czerwinska, Fiszer, & Krzyzosiak, 2014). In addition, alterations in the miRNA balance can lead to dysfunction of the tumor‐suppressor genes and/or oncogenes, which further induces the process of tumor initiation and progression (Wahid, Khan, & Kim, 2014), including HCC (Mao & Wang, 2015; Morishita & Masaki, 2015). For example, miR‐122 is the most abundant miRNA in normal liver, whereas it is almost totally silenced in HCC, and loss of miR‐122 frequently promotes tumorigenesis and tumor progression (Thakral & Ghoshal, 2015). However, the potential oncogenic role of miRNA in HCC remains to be fully elucidated (Mizuguchi, Takizawa, Yoshida, & Uchida, 2016). For example, miR‐21 and miR‐224 are usually upregulated in HCC compared with healthy liver, but their functions in HCC initiation and/or progression is still poorly understood (Mizuguchi et al., 2016).
Long noncoding RNAs (lncRNAs) can function as competing endogenous RNAs (ceRNAs) or RNA sponges, thus interacting with miRNAs in a way that can sequester miRNAs and reduce their regulatory effect on target messenger RNAs (mRNAs; Y. Gao et al., 2014; Legnini, Morlando, Mangiavacchi, Fatica, & Bozzoni, 2014). For example, the lncRNA Unigene56159 acts as a ceRNA of miR‐140‐5p in HCC cells (Lv et al., 2016). The lncRNA HOTAIR may act as a ceRNA to force a sink for miR‐331‐3p, thus resulting in the derepression of erb‐b2 receptor tyrosine kinase 2 (HER2) and promoting the proliferation, migration, and invasion of gastric carcinoma cells (Liu et al., 2014). NORAD is one of the star lncRNAs because it is highly conserved, abundantly expressed, and important for DNA protection and chromosomal stability (Munschauer et al., 2018). NORAD is highly expressed in the cytoplasm and significantly upregulated by hypoxia in epithelial cells (Tichon et al., 2016). The dysregulation of NORAD has been discovered in several malignant diseases (Huo et al., 2018; Kawasaki et al., 2018; H. Li et al., 2017; Q. Li et al., 2018; Zhang et al., 2018). However, the roles and underlying mechanisms have not been clarified.
In the present study, we demonstrate that NORAD is upregulated and promotes invasion and metastasis of HCC. Moreover, elevated NORAD expression in HCC tissues was associated with the poor prognosis of patients. Mechanistically, NORAD may function as a ceRNA of miR‐202‐5p to regulate the expression of transforming growth factor β receptors (TGFBRs), thereby plays an oncogenic role in the pathogenesis of HCC.
2. METHODS
2.1. Cell lines and cell culture
The human HCC cell lines SMMC‐7721, Huh7, PLC/PRF/5, and Hep3B (obtained from Liver Cancer Institute, Zhongshan Hospital, Fudan University, Shanghai, China) were used in the study. The Hep3B cells were cultured in minimum Eagle's medium (Gibco, Grand Island, NY) and SMMC‐7721, Huh7, and PLC/PRF/5 cells were cultured in Dulbecco's modified Eagle's medium (Gibco, Grand Island, NY) containing 10% fetal bovine serum (Gibco, Grand Island, NY) supplemented with 100 IU/ml penicillin and 100 µg/ml streptomycin. All cell lines were incubated at 37°C in a humidified atmosphere with 5% CO2. All other supplies were purchased from Corning (Grand Island, NY).
2.2. Quantitative real‐time polymerase chain reaction and western blot analysis
Quantitative real‐time polymerase chain reaction (qPCR) and western blot analysis were performed as previously described (Peng et al., 2017). Reverse transcription was used to detect miRNA expression, and miRNAs were amplified using stem‐loop primers (RiboBio, Guangzhou, China). U6 small nucleolar RNA was used as the endogenous control. All PCR assays were repeated three times. The qPCR primers were as follows: TGFBR1: forward 5′‐TTCTTCATGTGTTCCTGTAGCT‐3′, reverse 5′‐TGACTAGCA ACAAGTCAGGATT‐3′, TGFBR2: forward 5′‐TTCTTCATGTGTTCCTGTAGCT‐3′, reverse 5′‐TGACTAGCAACAAGTCAGGATT‐3′; GAPDH (glyceraldehyde‐3‐phosphate dehydrogenase): forward 5′‐TGCCAAATATGACATCAAGAA‐3′, reverse 5′‐GGAGTGGGTG TCGTCGCTGTTG‐3′; NORAD forward 5′‐AAGCTGCTCTCAACTCCACC‐3′, reverse 5′‐GGACGTATCGCTTCCAGAGG‐3′. All western blot assays were repeated three times. Antibody used were listed below: anti‐phospho‐Smad2/Smad3 (p‐smad2/3; bs‐8853R; Bioss, Beijing, China), anti‐Smad2/3 (8685s; Cell Signaling Technology, Danvers, MA), anti‐GAPDH (10494‐1‐AP; Proteintech, Wuhan, China).
2.3. Luciferase reporter gene assay
pGL3 plasmid (E1761; Promega, Madison, WI) containing the luciferase reporter gene was used. The NORAD and TGF‐β receptors sequences containing wild‐type or mutated miR‐202‐5p binding sites were synthesized respectively and inserted into pGL3 luciferase vector. The constructed plasmids were cotransfected with miR‐202‐5p mimics into HCC cell lines by Lipofectamine 2000 (Thermo Fisher Scientific, Inc., Waltham, MA). The luciferase assay was performed according to the manufacturer's protocol.
2.4. Transfection and clone selection
Lentiviral‐mediated pLKO‐short hairpin RNA NORAD and GV208‐NORAD were from Genebiochem (Shanghai, China). Empty vectors were used as the negative control. The lentiviral vectors were transfected into HCC cells using polybrene according to the manufacturer's instructions. Infected HCC cells were selected against puromycin for 7 days before use in the assays.
2.5. Patients and follow‐up
Tissues from 95 patients with HCC who underwent tumor resection between January 2006 and May 2016 were obtained at Zhongshan Hospital (Shanghai, China). Tissues were stored in −80°C ultra‐low temperature freezer. Patents were collected randomly and none of the patients had received any chemotherapy or radiation treatment before the surgery. The Ethics Committee has examined and certified the study, all informed written consents were obtained from the patients.
2.6. Cell viability, colony formation, cell migration, and matrigel invasion assays
The cell migration and colony formation assays were performed as previously described (P.T. Gao et al., 2018; Peng et al., 2017). The Cell Counting Kit‐8 (YEASON, Shanghai, China) and Matrigel invasion assays (BD Bioscience, Franklin Lakes, NJ) were performed as the manufacturers’ instructions.
2.7. Tumor xenograft assay
Previously constructed HCC cells were used to establish subcutaneous xenograft tumor models in nude mice (4–6 weeks old). Tumor growth was monitored weekly, and fresh subcutaneous tumor tissues were cut into 1 × 1 mm pieces and orthotopic implanted in healthy nude mice (4–6 weeks old). Mice were observed carefully and killed after 6 weeks.
2.8. Bioinformatics analysis
The potential miRNA binding sites within NORAD and the messenger RNA binding sites within miR‐202‐5p were predicted using computer‐aided algorithms from starBase (http://starbase.sysu.edu.cn; J. H. Li, Liu, Zhou, Qu, & Yang, 2014) and TargetScan (http://www.targetscan.org; Agarwal, Bell, Nam, & Bartel, 2015), The Cancer Genome Atlas (TCGA) data were analyzed using Gene Expression Profiling Interactive Analysis (http://gepia.cancer‐pku.cn/; Tang et al., 2017).
2.9. Statistical analysis
Statistical analysis was performed with SPSS 19.0 software (SPSS, Chicago, IL). Values are expressed as the mean and standard deviation. The Student t test and one‐way analysis of variance were used for comparisons. Cumulative recurrence and survival rates were analyzed using the Kaplan–Meier method and the log‐rank test. Independent prognostic factors were analyzed using Cox's proportional hazards regression model. p < 0.05 was considered statistically significant.
3. RESULTS
3.1. NORAD overexpressed in HCC tissues and elevated NORAD expression correlated with the poor prognosis of HCC patients
First, NORAD levels were detected in 29 HCC and their corresponding paratumor tissues by qPCR. NORAD expression was significantly increased in 62.1% (18/29) of tumor compared to the matched paratumor tissues (p < 0.01; Figure 1a). Moreover, we found that high level of NORAD was positively correlated with the large tumor size (p = 0.003) and HbsAg positive rate (p = 0.004; Table 1). To investigate the relation between NORAD expression and the prognosis of HCC patients, the 95 HCC patients were grouped into NORADhigh and NORADlow according to NORAD expression. Kaplan–Meier analysis revealed that the patients in NORADhigh group had shorter overall survival (OS) rate and higher recurrence rate than those of the patients in NORADlow group. Importantly, the log‐rank test identified the NORAD expression was an independent predictor of OS and postoperative recurrence in patients with HCC (Figure 1c,d; Table 2). These results indicate that high level of NORAD participates in HCC progression.
Figure 1.
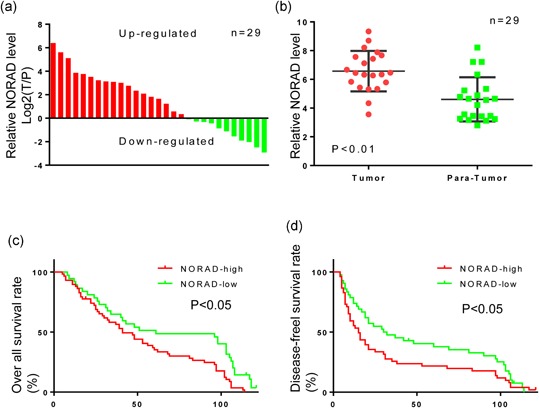
Long noncoding RNA NORAD is highly expressed in hepatocellular carcinoma (HCC) tumor tissues and associated with poor prognosis in HCC patients. (a) Relative expression levels of NORAD/GAPDH were calculated by formula log2 (T/P) in 29 paired HCC tissues. Values of Y axis which were greater than zero indicated higher expression in tumor. (b) Relative expression levels of NORAD/GAPDH were presented in tumor and normal (paratumor) groups. Paired t test was performed and p < 0.01. (c,d) Kaplan–Meier estimator of overall survival and disease‐free survival. p values were calculated by the log‐rank test. p < 0.05 in c, d. GAPDH: glyceraldehyde 3‐phosphate dehydrogenase [Color figure can be viewed at wileyonlinelibrary.com]
Table 1.
Correlation between NORAD and clinicopathological characteristics in 95 HCC patients
| Number of patients | |||
|---|---|---|---|
| Variables | NORADhigh | NORADlow | p Value |
| Sex | |||
| Female | 7 | 8 | 0.255 |
| Male | 51 | 29 | |
| Year | |||
| <52 | 37 | 17 | 0.095 |
| ≥52 | 21 | 20 | |
| Hepatic cirrhosis | |||
| Yes | 56 | 30 | 0.026 |
| No | 2 | 7 | |
| HbsAg | |||
| Positive | 43 | 36 | 0.004 |
| Negative | 15 | 1 | |
| HCV | |||
| Positive | 0 | 1 | 1 |
| Negative | 57 | 37 | |
| AFP | |||
| <20 | 25 | 13 | 0.522 |
| ≥20 | 33 | 24 | |
| Tumor size (cm) | |||
| <5 | 25 | 28 | 0.003 |
| ≥5 | 33 | 9 | |
| Tumor number | |||
| Single | 50 | 28 | 0.272 |
| Multiple | 8 | 9 | |
| Tumor encapsulation | |||
| Complete | 28 | 17 | 0.837 |
| None | 30 | 20 | |
| Tumor differentiation | |||
| I + II | 37 | 27 | 0.379 |
| III + IV | 21 | 10 | |
| TNM stage | |||
| I | 38 | 28 | 0.364 |
| II + III | 20 | 9 | |
Note. p < 0.01 was considered significant.
AFP: α‐feto protein; HBsAg: hepatitis B surface antigen; HCC: hepatocellular carcinoma; HCV: hepatitis C virus; TNM: tumor, node, and metastases.
Table 2.
Univariate and multivariate analyses of factors associated with overall survival
| Multivariate | ||||
|---|---|---|---|---|
| Factors | Univariate p value | HR | 95% Cl | p Value |
| Sex (female vs. male) | 0.044 | NA | ||
| Age (years) (≥53 vs.<53) | 0.441 | NA | ||
| Liver cirrhosis (yes vs. no) | 0.376 | NA | ||
| HBsAg (positive vs. negative) | 0.587 | NA | ||
| HCV (positive vs. negative) | 0.138 | NA | ||
| Serum AFP, ng/mL (≥20 vs.<20) | 0.876 | NA | ||
| Tumor encapsulation (yes vs. no) | 0.121 | NA | ||
| Tumor differentiation (III/IV vs. I/II) | 0.061 | NA | ||
| Tumor number (multiple vs. single) | 0.399 | NA | ||
| Tumor size (diameter, cm; ≥5 vs. <5) | 0.039 | NS | ||
| TNM stage (I/II vs. III/IV) | 0.854 | NA | ||
| NORAD expression (high vs. low) | 0.005 | 1.763 | 1.104–2.815 | 0.018 |
Note. Cox proportional hazards regression model. P < 0.05 was considered significant.
AFP: α‐feto protein; CI: confidence interval; HBsAg: hepatitis B surface antigen; HCV: hepatitis C virus; HR: hazard ratio; NA: not adopted; NS: not significant; TNM: tumor node metastasis.
3.2. NORAD overexpression promoted HCC cell proliferation, invasion, and metastasis in vivo and in vitro
The above results indicated that elevated NORAD expression might involve in HCC invasion and metastasis. Thus, we measured NORAD expression in four HCC cell lines (Figure 2a), and further established cell models with NORAD knockdown and overexpression (Figure 2b). Then the variety of cell phenotype influenced by NORAD expression was determined, and we found that high level of NORAD induced cell proliferation, enhanced the colony formation, as well as promoted the HCC cell migration and invasion (Figure 2c–f). In vivo, subcutaneous tumor models were constructed using NORAD knockdown HCC cell line and its control (Figure 2h). Tumor weights of ctrl group were significantly greater than NORAD knockdown group. Fresh subcutaneous tumor tissues were orthotopic implanted to new nude mice to construct orthotopic tumor models. After 42 days’ incubation, 4/5 (80%) mice of in the control group presented with lung metastases under serial section with H&E staining, whereas no mice in the NORAD knockdown group with lung metastases (Figure 2i). Here, we conclude that the high level of NORAD do serve as a promoter in HCC.
Figure 2.
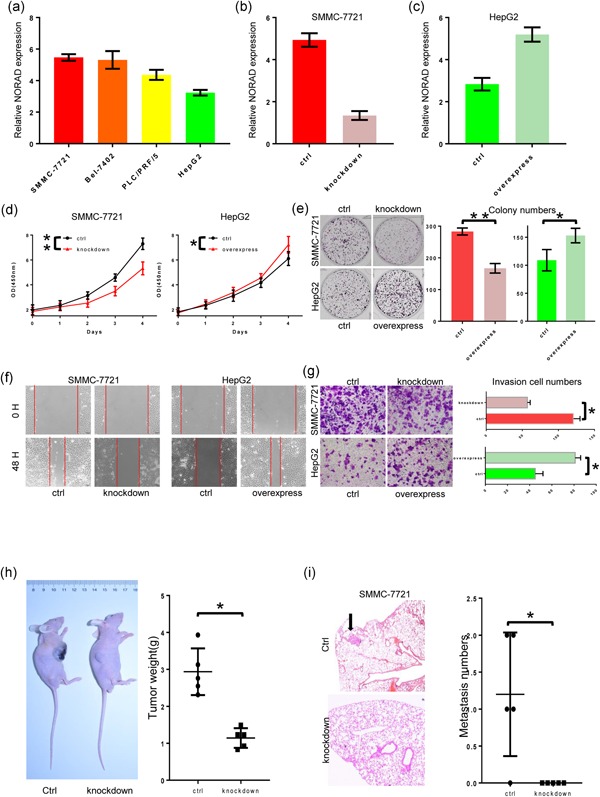
NORAD acts as a tumor enhancer in hepatocellular carcinoma (HCC) both in vitro and in vivo. (a) NORAD expression was tested in different HCC cell lines. (b) Knockdown of NORAD in SMMC‐7721 cells was detected. (c) Overexpression of NORAD in HepG2 cells was detected. (d) The Cell Counting Kit‐8 assay was carried out in the two stable cell lines and their control cells. (e) Colony formation assays were carried out in the two stable cell lines and their control cells. (f) Cell migration assays (wound healing experiment) were performed in those cell lines. (g) Transwell assays (Matrigel invasion assay) were performed in those cell lines. (h) Subcutaneous xenograft tumor models in nude mice and statistic of tumor weights. (i) Lung metastasis model in nude mice and statistic of metastasis numbers. *Statistical significance, p < 0.05; **Statistical significance, p < 0.01. [Color figure can be viewed at wileyonlinelibrary.com]
3.3. NORAD activated the TGF‐β pathway as a ceRNA of miR‐202‐5p
To explore the molecular mechanism of NORAD in HCC progression, we used starBase and TargetScan bioinformatics analysis to predict its’ target and revealed three putative miRNA response elements of miR‐202‐5p in NORAD and the 3′ untranslated regions (3′‐UTRs) of TGFBR1 and TGFBR2 (Figure 3a). To confirm whether the TGFBRs and NORAD were target RNAs of miR‐202‐5p in HCC cells, we conducted luciferase reporter assays after transfecting Huh7 cells containing synthetic miR‐202‐5p mimic with the wild‐type or mutated miR‐202‐5p target sequences in NORAD and the TGFBR1 and TGFBR2 3′‐UTRs. We found that the miR‐202‐5p mimic reduced luciferase activity in the cells transfected with the wild‐type sequences but had no effect on luciferase activity in cells transfected with the mutant sequences (Figure 3b,c). These data indicate that miR‐202‐5p may be a target of NORAD, and NORAD might promote HCC progression by the TGF‐β pathway.
Figure 3.
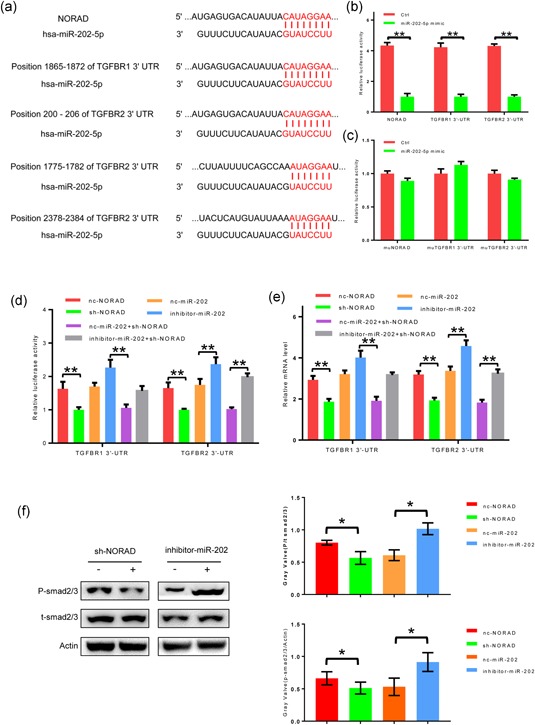
miR‐202‐5p, targeted by NORAD, inhibit TGFBR1 and TGFBR2 in hepatocellular carcinoma (HCC) cell lines. (a) Targets prediction of NORAD and miR‐202‐5p. (b,c) miR‐202‐5p mimic and its control were added in HCC cells which were transfected by wild‐type or mutated NORAD, TGFBR1 3′‐UTR and TGFBR2 3′‐UTR. The histogram indicates the luciferase values measured 48 hr after transfection. (d) NORAD knockdown, miR‐202‐5p inhibitor and their controls were added in HCC cells which contained TGFBR1 3′‐UTR and TGFBR2 3′‐UTR. The histogram indicates the luciferase values measured 48 hr after transfection. (e) Messenger RNA levels of TGFBR1 and TGFBR2 were measured after NORAD knockdown, miR‐202‐5p inhibitor and their controls were added. (f) Phosphorylation level of Smad2/3 were tested by western blot analysis. *Statistical significance, p < 0.05; **Statistical significance, p < 0.01. TGFBR: transforming growth factor β receptors; UTR: untranslated region [Color figure can be viewed at wileyonlinelibrary.com]
As mentioned above, NORAD mediated the sequestration of miR‐202‐5p, and miR‐202‐5p sponges TGFBRs. To discuss whether NORAD‐mediated miR‐202‐5p sequestration could regulate the TGF‐β pathway, we measured the TGFBRs luciferase activity in SMMC‐7721 cells. NORAD knockdown reduced the TGFBRs luciferase activity, which miR‐202‐5p inhibitor partially rescued (Figure 3d); these results were confirmed at mRNA level (Figure 3e). Further, the phosphorylation levels of smad2/3, which was considered as downstream of TGFBRs were measured. NORAD knockdown reduced the phosphorylation levels of smad2/3 and inhibition of miR‐202‐5p made the opposite effect (Figure 3f). These data demonstrate that NORAD functions as a molecular inhibitor of miR‐202‐5p, then indirectly upregulated TGFBRs expression.
3.4. MiR‐202‐5p expression was higher in paratumor tissues and correlated with better OS
Here, we further determined miR‐202‐5p expression in 29 HCC and their corresponding paratumor tissues by qPCR. As showed in Figure 4a, the paratumor tissues had higher miR‐202‐5p expression than that in HCC tissues (p < 0.01). Accordingly, the 95 patients were grouped into miR‐202‐5phigh and miR‐202‐5plow according to miR‐202‐5p expression, and the consequential Kaplan–Meier analysis indicated that HCC patients in miR‐202‐5phigh group had higher OS rate than the patients in miR‐202‐5plow group (Figure 4b).
Figure 4.
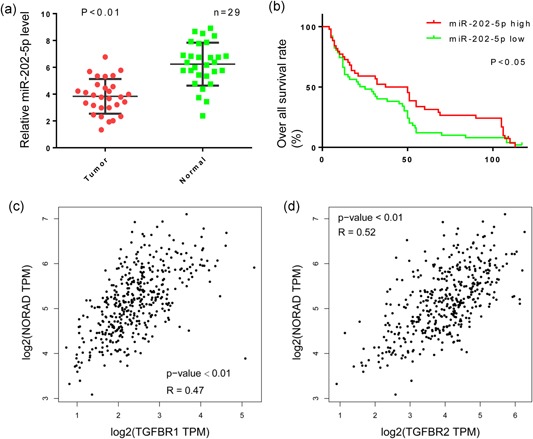
MiR‐202‐5p expression is associated with better prognosis in hepatocellular carcinoma patients and TGFBR1/TGFBR2 expression. (a) Relative expression levels of miR‐202‐5p/GAPDH were presented in tumor and normal(paratumor) groups. Paired t test was performed and p < 0.01. (b,d) Kaplan−Meier estimator of overall survival. p values were calculated by the log‐rank test. p < 0.05. (c,d) Correlation of miR‐202‐5p and TGFRB1/2 were tested in data from The Cancer Genome Atlas. GAPDH: glyceraldehyde 3‐phosphate dehydrogenase; TGFBR: transforming growth factor β receptors [Color figure can be viewed at wileyonlinelibrary.com]
3.5. Positive correlation between NORAD and TGFBRs in HCC tissues
Additionally, we also examined TCGA data to determine whether NORAD regulates TGFBRs expression in tumor tissue. A scatter plot of NORAD and TGFBRs mRNA expression in the HCC tissues revealed a positive correlation between NORAD and the TGFBRs (Figures 4c,d and 5).
Figure 5.
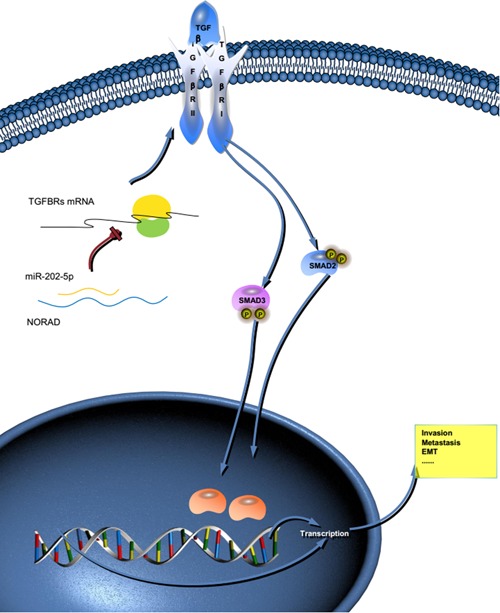
NORAD/miR‐202‐5p/TGFBRs axis regulates hepatocellular carcinoma (HCC) progression. Schematic diagram shows that NORAD increases HCC invasion and metastasis via sponging miR‐202‐5p, a negative regulator of transforming growth factor β pathway. mRNA: messenger RNA; TGBFR: transforming growth factor β receptors [Color figure can be viewed at wileyonlinelibrary.com]
4. DISCUSSION
HCC is a fatal disease in which the majority of patients experience recurrence and metastasis (Bray et al., 2018; Tang, 2001; Tang et al., 1999). However, its mechanism is poorly understood. lncRNAs acting as ceRNAs or natural miRNA sponges are important posttranscriptional regulators of gene expression, which communicate with and coregulate each other by competing for binding to shared miRNAs. Understanding this novel RNA crosstalk will shed light on gene regulatory networks and human development and disease (Afonso‐Grunz & Muller, 2015; Huarte, 2015; Mao & Wang, 2015; Mendell & Olson, 2012). Here, we showed that lncRNA NORAD overexpressed in HCC tissues, and elevated NORAD positively related to the poor prognosis and high recurrence rate of HCC patients. By NORAD interference and overexpression, we demonstrated that NORAD functioned as a promoter in HCC development both in vitro and in vivo. Mechanically, high level of NORAD upregulated TGFBRs expression via functioning as a ceRNA of miR‐202‐5p. Thus, we identified a novel NORAD/miR‐202‐5p/TGFBRs axis in HCC progression.
NORAD is highly conserved and abundant, with expression levels of approximately 500–1,000 copies/cell (Tichon et al., 2016). Particularly, NORAD loss of function has been reported to induce chromosomal instability and triggers dramatic aneuploidy in karyotypically stable cell lines. In addition, NORAD has been discovered to directly regulate both ploidy and chromosomal stability by sequestering the PUMILIO proteins, which repress key factors in mitosis, DNA repair, and DNA replication (Munschauer et al., 2018; Tichon et al., 2016). Moreover, NORAD was revealed to be a cytoplasmic multivalent PUMILIO‐binding platform that acted as a negative regulator of PUMILIO activity (Tichon et al., 2016). Now, NORAD was reported to promote tumor cell growth and proliferation in several types of cancer (Huo et al., 2018; Kawasaki et al., 2018; H. Li et al., 2017; Q. Li et al., 2018). However, studies on NORAD in HCC are lacking, and existing studies have not been able to shed much light on its mechanism. Previously, miR‐202‐5p was found to influence the TGF‐β pathway by targeting SMAD2/3 transport in pancreatic cancer, and NORAD inhibited miR‐202‐5p in colorectal cancer (H. Li et al., 2017; Zhang et al., 2018). Here, we further reported a novel axis, thus, NORAD could act as a ceRNA to modulate miR‐202‐5p expression, which also could target the 3′‐UTRs of TGFBRs to regulate the TGF‐β pathway. Importantly, we partially validated this axis in clinical sample. Thus, we displayed a definite axis initiated by NORAD, which indicated an oncogenic role of NORAD in HCC.
5. CONCLUSION
This study defines a novel mechanism in which the lncRNA NORAD exerts a tumor‐promoting effect in HCC through a NORAD/miR‐202‐5p/TGF‐β axis.
Yang X, Cai J-B, Peng R, et al. The long noncoding RNA NORAD enhances the TGF‐β pathway to promote hepatocellular carcinoma progression by targeting miR‐202‐5p. J Cell Physiol. 2019;234:12051–12060. 10.1002/jcp.27869
Contributor Information
Guo‐Ming Shi, Email: shi.guoming@zs-hospital.sh.cn.
Jia Fan, Email: fan.jia@zs-hospital.sh.cn.
References
REFERENCES
- Afonso‐Grunz, F. , & Müller, S. (2015). Principles of miRNA‐mRNA interactions: Beyond sequence complementarity. Cellular and Molecular Life Science, 72(16), 3127–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal, V. , Bell, G. W. , Nam, J. W. , & Bartel, D. P. (2015). Predicting effective microRNA target sites in mammalian mRNAs. eLife, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray, F. , Ferlay, J. , Soerjomataram, I. , Siegel, R. L. , Torre, L. A. , & Jemal, A. (2018). Global Cancer Statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians, 68, 394–424. [DOI] [PubMed] [Google Scholar]
- Gao, P. T. , Ding, G. Y. , Yang, X. , Dong, R. Z. , Hu, B. , Zhu, X. D. , … Huang, C. (2018). Invasive potential of hepatocellular carcinoma is enhanced by loss of selenium‐binding protein 1 and subsequent upregulation of CXCR4. American Journal of Cancer Research, 8(6), 1040–1049. [PMC free article] [PubMed] [Google Scholar]
- Gao, Y. , Wu, F. , Zhou, J. , Yan, L. , Jurczak, M. J. , Lee, H. Y. , … Huang, Y. (2014). The H19/let‐7 double‐negative feedback loop contributes to glucose metabolism in muscle cells. Nucleic Acids Research, 42(22), 13799–13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, S. M. (2015). An overview of microRNAs. Advanced Drug Delivery Reviews, 87, 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte, M. (2015). The emerging role of lncRNAs in cancer. Nature Medicine, 21(11), 1253–1261. [DOI] [PubMed] [Google Scholar]
- Huo, H. , Tian, J. , Wang, R. , Li, Y. , Qu, C. , & Wang, N. (2018). Long non‐coding RNA NORAD upregulate SIP1 expression to promote cell proliferation and invasion in cervical cancer. Biomedicine & Pharmacotherapy, 106, 1454–1460. [DOI] [PubMed] [Google Scholar]
- Kawasaki, N. , Miwa, T. , Hokari, S. , Sakurai, T. , Ohmori, K. , Miyauchi, K. , … Koinuma, D. (2018). Long noncoding RNA NORAD regulates transforming growth factor‐beta signaling and epithelial‐to‐mesenchymal transition‐like phenotype. Cancer Prevention Research, 109(7), 2211–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnini, I. , Morlando, M. , Mangiavacchi, A. , Fatica, A. , & Bozzoni, I. (2014). A feedforward regulatory loop between HuR and the long noncoding RNA linc‐MD1 controls early phases of myogenesis. Molecular Cell, 53(3), 506–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Wang, X. , Wen, C. , Huo, Z. , Wang, W. , Zhan, Q. , … Shen, B. (2017). Long noncoding RNA NORAD, a novel competing endogenous RNA, enhances the hypoxia‐induced epithelial‐mesenchymal transition to promote metastasis in pancreatic cancer. Molecular Cancer, 16(1), 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. H. , Liu, S. , Zhou, H. , Qu, L. H. , & Yang, J. H. (2014). starBase v2.0: Decoding miRNA‐ceRNA, miRNA‐ncRNA and protein‐RNA interaction networks from large‐scale CLIP‐Seq data. Nucleic Acids Research, 42(Database issue), D92–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q. , Li, C. , Chen, J. , Liu, P. , Cui, Y. , Zhou, X. , … Zu, X. (2018). High expression of long noncoding RNA NORAD indicates a poor prognosis and promotes clinical progression and metastasis in bladder cancer. Urologic Oncology, 36(6), 310–315. [DOI] [PubMed] [Google Scholar]
- Liu, X. , Sun, M. , Nie, F. , Ge, Y. , Zhang, E. , Yin, D. , … Wang, Z. (2014). Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR‐331‐3p in gastric cancer. Molecular Cancer, 13, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv, J. , Fan, H. , Zhao, X. , Lv, P. , Fan, J. , Zhang, Y. , … Tang, H. (2016). Long non‐coding RNA Unigene56159 promotes epithelial‐mesenchymal transition by acting as a ceRNA of miR‐140‐5p in hepatocellular carcinoma cells. Cancer Letters, 382(2), 166–175. [DOI] [PubMed] [Google Scholar]
- Mao, B. , & Wang, G. (2015). MicroRNAs involved with hepatocellular carcinoma (Review). Oncology Reports, 34(6), 2811–2820. [DOI] [PubMed] [Google Scholar]
- Mendell, J. T. , & Olson, E. N. (2012). MicroRNAs in stress signaling and human disease. Cell, 148(6), 1172–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi, Y. , Takizawa, T. , Yoshida, H. , & Uchida, E. (2016). Dysregulated miRNA in progression of hepatocellular carcinoma: A systematic review. Hepatology Research, 46(5), 391–406. [DOI] [PubMed] [Google Scholar]
- Morishita, A. , & Masaki, T. (2015). miRNA in hepatocellular carcinoma. Hepatology Research, 45(2), 128–141. [DOI] [PubMed] [Google Scholar]
- Munschauer, M. , Nguyen, C. T. , Sirokman, K. , Hartigan, C. R. , Hogstrom, L. , Engreitz, J. M. , … Lander, E. S. (2018). The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature, 561(7721), 132–136. [DOI] [PubMed] [Google Scholar]
- Peng, R. , Huang, X. , Zhang, C. , Yang, X. , Xu, Y. , & Bai, D. (2017). Overexpression of UHRF2 in intrahepatic cholangiocarcinoma and its clinical significance. OncoTargets and Therapy, 10, 5863–5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singal, A. G. , & El‐Serag, H. B. (2015). Hepatocellular carcinoma from epidemiology to prevention: Translating knowledge into practice. Clinical Gastroenterology and Hepatology, 13(12), 2140–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroynowska‐Czerwinska, A. , Fiszer, A. , & Krzyzosiak, W. J. (2014). The panorama of miRNA‐mediated mechanisms in mammalian cells. Cellular and Molecular Life Science, 71(12), 2253–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Z. , Li, C. , Kang, B. , Gao, G. , Li, C. , & Zhang, Z. (2017). GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Research, 45(W1), W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Z. , Zhou, X. , Lin, Z. , Yang, B. , Ma, Z. , Ye, S. , … Zhou, J. (1999). Surgical treatment of hepatocellular carcinoma and related basic research with special reference to recurrence and metastasis. Chinese Medical Journal, 112(10), 887–891. [PubMed] [Google Scholar]
- Tang, Z. Y. (2001). Hepatocellular carcinoma‐‐cause, treatment and metastasis. World Journal of Gastroenterology, 7(4), 445–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakral, S. , & Ghoshal, K. (2015). miR‐122 is a unique molecule with great potential in diagnosis, prognosis of liver disease, and therapy both as miRNA mimic and antimir. Current Gene Therapy, 15(2), 142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichon, A. , Gil, N. , Lubelsky, Y. , Havkin solomon, T. , Lemze, D. , Itzkovitz, S. , … Ulitsky, I. (2016). A conserved abundant cytoplasmic long noncoding RNA modulates repression by Pumilio proteins in human cells. Nature Communications, 7, 12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahid, F. , Khan, T. , & Kim, Y. Y. (2014). MicroRNA and diseases: Therapeutic potential as new generation of drugs. Biochimie, 104, 12–26. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Li, X. Y. , Hu, P. , & Ding, Y. S. (2018). LncRNA NORAD contributes to colorectal cancer progression by inhibition of miR‐202‐5p. Oncology Research, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Sun, H. C. , Wang, Z. , Cong, W. M. , Wang, J. H. , Zeng, M. S. , … Fan, J. (2018). Guidelines for diagnosis and treatment of primary liver cancer in China (2017 Edition). Liver Cancer, 7(3), 235–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucman‐Rossi, J. , Villanueva, A. , Nault, J. C. , & Llovet, J. M. (2015). Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology, 149(5), 1226–1239. [DOI] [PubMed] [Google Scholar]


