Abstract
Collectively, the completion of the Human Genome Project and subsequent development of high-throughput next-generation sequencing methodologies have revolutionized genomic research. However, the rapid sequencing and analysis of thousands upon thousands of human exomes and genomes has taught us that most genes, including those known to cause heritable cardiovascular disorders such as long QT syndrome, harbor an unexpected background rate of rare, and presumably innocuous, non-synonymous genetic variation. In this Review, we aim to reappraise the genetic architecture underlying both the acquired and congenital forms of long QT syndrome by examining how the clinical phenotype associated with and background genetic variation in long QT syndrome-susceptibility genes impacts the clinical validity of existing gene-disease associations and the variant classification and reporting strategies that serve as the foundation for diagnostic long QT syndrome genetic testing.
Keywords: Arrhythmia, Genetic testing, Genetic variation, Long QT syndrome, Sudden cardiac death
Introduction
Long QT syndrome (LQTS) is a genetically and phenotypically heterogeneous disorder of cardiac repolarization characterized clinically by prolongation of the heart rate-corrected QT interval (QTc) on the 12-lead electrocardiogram (ECG) and an increased propensity for torsadogenic syncope/seizures and sudden cardiac death (SCD) [1,2]. Following the sentinel discovery of the canonical LQTS-susceptibility genes (KCNQ1, KCNH2, and SCN5A) in 1995 and 1996 [3–5]and completion of the Human Genome Project in 2003, rapid advances in deoxyribonucleic acid (DNA) sequencing technology allowed for the discovery of new Mendelian disease-susceptibility genes, including most of the 14 minor LQTS-susceptibility genes [6–12], at a dizzying pace.
However, the high-throughput DNA sequencing and subsequent analysis of tens-to-hundreds of thousands human exomes and genomes have exposed the stark reality that one in eight protein-encoding base pairs hosts a genetic variant and many Mendelian disease-susceptibility genes, including the canonical LQTS-susceptibility genes, are surprisingly tolerant to non-synonymous genetic variation [13–15]. Unfortunately, this burden of rare non-synonymous background genetic variants not only complicates variant interpretation [16–18], but for some minor LQTS-susceptibility genes, also calls into question the veracity of longstanding gene-disease associations.
In this Review, we (i) detail current paradigms regarding the molecular and cellular underpinnings of LQTS, (ii) reappraise the veracity of the QT phenotype associated with specific minor LQTS subtypes, (iii) examine ongoing attempts to classify and curate the clinical validity of existing gene-disease associations, and (iv) assess the limitations of current variant interpretation and reporting standards in an effort to highlight how a critical reappraisal of LQTS genetic architecture can help preserve, and hopefully enhance, the clinical validity of diagnostic LQTS genetic testing. To assist those readers with limited genetics/genomics backgrounds or those in need of a quick refresher, a glossary of terminology used throughout this Review is included in the online supplementary material.
Long QT syndrome: cellular mechanisms, genetic underpinnings, and prevailing paradigms
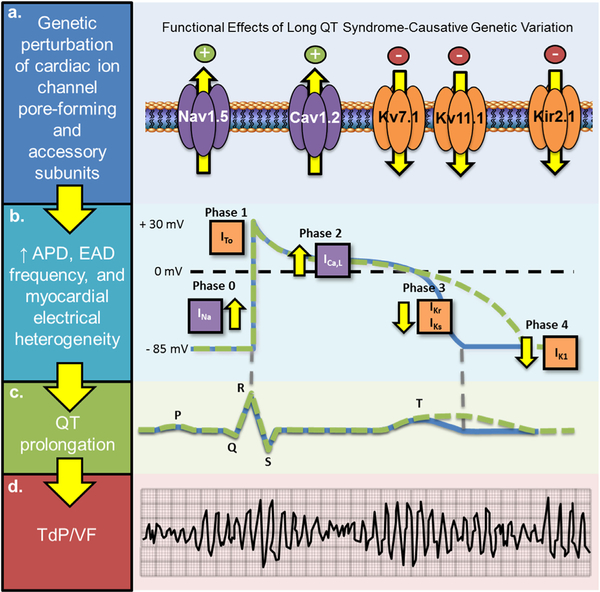
The electromechanical function of the heart is dependent on the coordinated activation and inactivation of inward depolarizing [sodium (Na+) and calcium (Ca2+)] and outward repolarizing [potassium (K+)] currents that underlie the five major phases of the cardiac action potential (Fig. 1) [19,20]. Genetic or acquired defects that accentuate depolarizing Na+ and Ca2+ currents (INa and ICa,L) or attenuate repolarizing K+ currents (IKs, IKr, and IK1) (Fig. 1a) can prolong the ventricular cardiac action potential (Fig. 1b), as reflected by a prolonged QT interval on the surface 12-lead ECG (Fig. 1c). Current joint expert consensus guidelines define a prolonged QTc value as >450 ms in males and >460 ms in females [21]. However, ~5–10% of individuals within the general population have a QTc > 460 ms on screening ECG [22]. As such, sex-specific 99.5th percentile QTc values (>460 ms for prepubertal males and females, >470 ms for postpubertal males, and >480 ms for postpubertal females) are used commonly as a cutoff to identify those who might benefit from cLQTS genetic testing [23]. Practically, a baseline QTc value ≥ 500 ms is considered definitely abnormal and if observed, in the absence of one or more QT prolonging risk factors, should compel strong clinical suspicion for congenital LQTS (cLQTS) and a class I recommendation to proceed with LQTS genetic testing [23].
Fig. 1.
Current paradigm for the molecular and cellular basis of congenital long QT syndrome. (a) Genetic variation in pore-forming α-subunits or accessory β-subunits that results in cardiac potassium channel (KCNQ1-encoded Kv7.1, KCNH2-encoded Kv11.1, and KCNJ2-encoded Kir2.1) loss-of-function, cardiac sodium channel (SCN5A-encoded Nav1.5) gain-of-function, or cardiac L-type calcium channel (CACNA1C-encoded Cav1.2) gain-of-function. (b) A genetically-mediated increase in outward depolarizing currents (purple channels/boxes) and/or inward repolarizing currents (orange channels/boxes) prolongs the action potential duration (APD) generating the substrate needed for an increased frequency of early after depolarizations (EADs). (c) An increase in APD manifests as heart-rate corrected QT (QTc) interval prolongation on 12-lead surface electrocardiogram. (d) EAD-triggered action potentials can precipitate torsades de pointes (TdP) which may degenerate into ventricular fibrillation. Adapted from Giudicessi et al. [69] with permission. Copyright © 2018, Wiley. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Regardless of the underlying mechanism of QT prolongation [acquired/drug-induced LQTS (aLQTS) vs. cLQTS], increased cardiomyocyte refractoriness generates an electrical substrate that can give rise to frequent early afterdepolarizations (EADs) mediated by re-activation of L-type Ca2+ and sodium-calcium exchange currents during phase 2 and 3 of the cardiac action potential. Triggered activity from focal EADs coupled with EAD augmented electrical heterogeneity in adjacent regions of myocardium likely generates the electrical substrate needed for the initiation, propagation, and maintenance of torsades de pointes (TdP), the hallmark form of self-sustaining polymorphic ventricular tachycardia observed in patients with LQTS (Fig. 1d) [24].
During the early-to-mid 1990s, a series of now classical linkage-analysis studies identified the KCNQ1 -encoded Kv 7.1 K+ channel, the KCNH2-encoded Kv 11.1/hERG K+ channel, and the SCN5A- encoded Nav 1.5 Na+ channel pore-forming α-subunits as genetic substrates for congenital LQTS [3–5]. Among clinically definitive LQTS cases (i.e. QTc ≥ 480 ms and “Schwartz score” ≥ 3.5) [25], ~75% harbor a pathogenic variant in one of the three canonical LQTS-susceptibility genes [KCNQ1 (~35%), KCNH2 (~30%), and SCN5A (~10%)] (Table 1) [26]. An additional ~5–10% of LQTS cases, including patients that may display prolonged QTc values as part of distinct multisystem syndromes [27–29] such as Timothy syndrome (TS1 and CACNA1C), Andersen-Tawil Syndrome (ATS1 and KCNJ2), and Triadin Knockout Syndrome (TKOS and TRDN), are anticipated to harbor a pathogenic variant in one of at least 14 additional “minor” LQTS-susceptibility genes that have been published in the literature to date (Table 1).
Table 1.
Current paradigms regarding the genetic basis of non-syndromic and multisystem LQTS subtypes.
| Gene | LQTS subtype | Historical conventiona | OMIM | Protein | Functional effect | Mode of inheritance | Frequency | Ref. |
|---|---|---|---|---|---|---|---|---|
| Non-syndromic long QT syndrome (major) | ||||||||
| KCNQ1 | LQT1 | LQT1 | 192500 | Kv7.1 | Reduced IKs | AD; AR | ~30–35% | [4,70] |
| KCNH2 | LQT2 | LQT2 | 613688 | Kv11.1 | Reduced IKr | AD | ~25–30% | [3] |
| SCN5A | LQT3 | LQT3 | 603830 | Nav1.5 | Increased INa | AD | ~5–10% | [5] |
| Non-syndromic long QT syndrome (minor) | ||||||||
| AKAP9b | AKAP9-LQTS | LQT11 | 611820 | Yotiao | Reduced IKs | AD | <1% | [11] |
| CACNA1C | CACNA1C-LQTS | LQT8 | N/A | Cav1.2 | Increased ICa,L | AD | ~1–2% | [71] |
| CALM1 | CALM1-LQTS | LQT14 | 616247 | Calmodulin 1 | Increased ICa,L (defective CDI) | Sporadic | ~1–2% | [72] |
| CALM2 | CALM2-LQTS | LQT15 | 616249 | Calmodulin 2 | Increased ICa,L (defective CDI) | Sporadic | ~1% | [72] |
| CALM3 | CALM3-LQTS | LQT16 | 114183 | Calmodulin 3 | Likely increased ICa,L (defective CDI) | Sporadic | <1% | [73] |
| CAV3 | CAV3-LQTS | LQT9 | 611818 | Caveolin 3 | Increased INa | AD | <1% | [8] |
| KCNE1 | KCNE1-LQTS | LQT5 | 613695 | MinK | Reduced IKs | AD | <1% | [6] |
| KCNE2b | KCNE2-LQTS | LQT6 | 613693 | MiRP1 | Reduced IKr | AD | <1% | [7] |
| KCNJ5b | KCNJ5-LQTS | LQT13 | 613485 | Kir3.4 | Reduced IK,Ach | AD | <1% | [12] |
| SCN4Bb | SCN4B-LQTS | LQT10 | 611819 | Nav1.5 /β4-subunit | Increased INa | AD | <1% | [9] |
| SNTA1b | SNTA1-LQTS | LQT12 | 612955 | Syntrophin-α1 | Increased INa | AD | <1% | [10] |
| Cardiac-only Timothy syndrome | ||||||||
| CACNA1C | COTS | N/A | N/A | Cav1.2 | Increased ICa,L | AD | ~1% | [74] |
| Jervell and Lange–Nielson syndrome | ||||||||
| KCNQ1 | JLNS1 | JLNS1 | 220400 | Kv7.1 | Reduced IKs | AR | Very rare | [75] |
| KCNE1 | JLNS2 | JLNS2 | 612347 | MinK | Reduced IKs | AR | Very rare | [76] |
| Ankyrin-B syndrome | ||||||||
| ANK2c,b | ABS | LQT4 | 600919 | Ankyrin B | Aberrant ion channel/transporter localization | AD | <1% | [35] |
| Andersen–Tawil Syndrome | ||||||||
| KCNJ2C | ATS1 | LQT7 | 170390 | Kir2.1 | Reduced IK1 | AD | <1% | [28] |
| Timothy syndrome | ||||||||
| CACNA1C | TS1 | LQT8 | 601005 | Cav1.2 | Increased ICa,L (slowed VDI) | Sporadic; AD mosaicism | Very rare | [29,77] |
| Triadin knockout syndrome | ||||||||
| TRDN | TKOS | N/A | 615441 | Triadin | Likely increased ICaL (disruption of CRU) | AR; sporadic | ~2% | [27] |
Abbreviations: AD, autosomal dominant; AR, autosomal recessive; ABS, Ankyrin-B syndrome; ATS, Andersen Tawil syndrome; CDI, calcium-dependent inactivation; COTS, cardiac-only Timothy syndrome; CRU, calcium release unit; LQTS, long-QT syndrome; TS, Timothy syndrome, and VDI, voltage-dependent inactivation.
Due to potential demotion of some genes previously alleged to be LQTS-causative, the authors strongly recommend limiting the use of historical numerical conventions to KCNQ1 (LQT1), KCNH2 (LQT2), and SCN5A (LQT3).
Genes that already have or likely will receive limited or disputed evidence gene designations.
Genes that merit removal from diagnostic LQTS genetic testing panels due to lack of true association with QT prolongation.
A case of mistaken identity? Reappraising the QT phenotype associated with Andersen Tawil syndrome (ATS) and ankyrin-B syndrome (ABS)
Disease severity in most forms of LQTS, including the degree of QT prolongation, is influenced strongly by the phenomena of incomplete penetrance and variable expressivity. However, since the sentinel clinical description of both ATS [30] and Ankyrin-B syndrome (ABS) [31], subsequent evidence suggests that individuals with both disorders rarely display baseline QTc values consistent with cLQTS. As such, the following paragraphs briefly examine if ATS and ABS still merit designation as bona fide LQTS subtypes and whether KCNJ2 or ANK2 should remain on diagnostic LQTS genetic testing panels.
ATS was first described in 1971 as a clinical triad of periodic paralysis, ventricular ectopy/arrhythmia, and variable developmental abnormalities of the head, face, and limbs [30]. Due to the presence of prominent U waves and resulting QT-U interval prolongation on 12-lead ECG, ATS was labelled, initially and erroneously, as a multisystem form of cLQTS and the ATS-causative KCNJ2 gene given the historical designation as LQT7 [32].
However, once the abnormal U wave is excluded, ATS patients almost always display normal QT intervals (<440 ms) [33]. Furthermore, the burden of complex ventricular ectopy at rest and biventricular tachycardia (a rare subtype of polymorphic ventricular tachycardia observed in catecholaminergic polymorphic ventricular tachycardia, but not LQTS) observed in ATS are highly uncharacteristic of LQTS [33]. As such, it is difficult to find substantive evidence to support the ongoing use of the historical LQT7 convention to describe ATS or the inclusion of KCNJ2 on diagnostic LQTS genetic testing panels, other than to catch the rare case of ATS incorrectly diagnosed as LQTS due to a failure to recognize the multisystem disorder’s distinct clinical and electrocardiographic profile.
Unlike ATS, ABS is a cardiac-only disorder characterized clinically by a range of variably present arrhythmia phenotypes including sinus node dysfunction, supraventricular and ventricular arrhythmias, and SCD [34]. Interestingly, overt QT prolongation (average QTc 490 ± 30 ms) was a characteristic of affected individuals in the multigenerational French pedigree that led to the first description of ABS (historically termed LQT4) and the discovery of the locus containing ANK2 [31,35]. However, subsequent studies have demonstrated that baseline QTc prolongation is at best inconsistently observed in ABS [34,36,37]. Like ATS, this initial discrepancy may be caused, in part, by the inclusion of prominent U waves/sinusoidal T-U abnormalities in QTc calculations. As a result, QTUc values may appear markedly prolonged in ABS, but true QTc values typically reside in the normal-to-borderline range as more commonly reported [34,36,37].
That said, the recent description of a novel ANK2 variant, p.Ser646Phe-ANK2, that appears to be more consistently associated with true QTc prolongation (average QTc 475 ± 40 ms) [38] and the prior association of common ANK2 variants with QTc duration in the general population [39] support a role for ANK2 in cardiac repolarization/LQTS pathogenesis. As such, it would be premature, at least on the basis of clinical phenotype alone, to call for the removal of ANK2 from diagnostic LQTS genetic testing panels. However, when taken in context of (i) the weak QT phenotype inconsistently observed in ABS [34,36,37], (ii) failure of subsequent larger genome-wide association studies (GWAS) to reproduce the association between ANK2 and QTc duration in the general population [40,41], and (iii) public exome data presented in the ensuing sections, there is clearly impetus to reappraise the role of ANK2 in the pathogenesis of monogenic cLQTS.
Minor LQTS-susceptibility genes: time for a critical reappraisal?
In the 15 years following the release of the completed human genome, advances in sequencing technology facilitated the discovery of new disease-susceptibility genes in scenarios (i.e. singletons and small pedigrees) not feasible with classical linkage analysis. As a result, nearly half of the gene-disease associations in existence today [42], including the majority of the minor LQTS-susceptibility genes (Table 1) [6–12], were discovered over the last ~10 years using largely hypothesis-driven, mutational analysis of biologically plausible genes and their gene-encoded proteins. However, many of these genes were discovered in an era before the burden of background non-synonymous genetic variation was appreciated fully. As such, the strength of evidence behind existing gene-disease associations is widely variable. In the following paragraphs, we explore the minor non-syndromic LQTS-susceptibility genes as a prototype to examine ongoing efforts to assess the validity of gene-disease associations and detail the potential impact of these efforts on clinical LQTS genetic testing.
Reappraising the non-syndromic minor LQTS-susceptibility genes
Due to a relative paucity of gene- and variant-level evidence, virtually every novel ultra-rare missense variant in a nonsyndromic, minor LQTS-susceptibility gene (Table 1) is likely to receive, at best, a variant of uncertain significance (VUS) designation. Not suprisingly, the identification of an ambiguous VUS in so-called weak or preliminary evidence genes is rarely clinically informative, increases the risk of genetic test misinterpretation, and ultimately can result in potentially harmful diagnostic miscues. In an effort to improve the validity of genes included on clinical genetic tests, the National Institutes of Health funded Clinical Genome Resource (ClinGen) has developed a semi-quantitative framework designed to standardize how the strength of genetic and functional evidence supporting a given gene-disease association is assessed systematically [43].
Although official ClinGen curations for the majority of LQTS-and other inherited cardiac channelopathy-susceptibility genes are eagerly awaited, independent application of the open access ClinGen gene-disease association framework suggests that 6/9 (66.7%) of the minor non-syndromic LQTS-susceptibility genes will receive (ANK2, KCNE2, KCNJ5/ and SNTA1) or have already received (AKAP9 and SCN4B) [43] disputed-evidence gene or limited-evidence gene designations (Table 3) [43].
Table 3.
Preliminary reappraisal of the evidence supporting gene-disease associations for non-syndromic LQTS minor genes.
| Gene | ExAC missense constraint Z scorea | ClinGen genetic evidence score (# of qualifying variants)b | ClinGen experimental evidence scoreb | Anticipated ClinGen classification (total score)c | Ref. |
|---|---|---|---|---|---|
| AKAP9 | −1.03 | 0.5 (1) | 3 | Limited evidence (3.5) | [11,43,86,87] |
| ANK2 | +1.06 | 0.5 (1) | 5 | Limited evidence (5.5) | [34,35,38,45] |
| CACNA1C | +6.41 | 5.0 (10)d | 2.5d | Moderate evidence (7.5) | [71,78–80,88] |
| CAV3 | +1.19 | 7 (3) | 3 | Moderate evidence (10) | [8,89] |
| KCNE1 | −0.46 | 3.0 (6) | 5.5 | Moderate evidence (8.5) | [6,76,81–84,90–92] |
| KCNE2 | −0.23 | 0 (0) | 5 | Disputed evidence (5) | [7,44,48,93–99] |
| KCNJ5 | +1.46 | 0 (0) | 3.5 | Disputed evidence (3.5) | [12,46,100] |
| SCN4B | −0.13 | 0.5 (1) | 3 | Limited evidence (3.5) | [9,101,102] |
| SNTA1 | +0.77 | 0.5 (1) | 3 | Limited evidence (3.5) | [10,85] |
Abbreviations: ClinGen, Clinical Genome Resource; and ExAC, exome aggregation consortium.
A positive Z score indicates fewer missense variants than expected (intolerant to genetic variation) whereas a negative Z score indicates more missense variants where observe than expected (tolerant to genetic variation).
Denotes author calculations using the open-access evidence-based framework proposed and in use by ClinGen curators. Only protein-truncating or functionally characterized missense variants present in ≤ 10 individuals in gnomAD were considered as qualifying variants.
Gene-disease associations are classified according to the ClinGen clinical validity matrix as limited (1–6 points), moderate (7–11 points), strong (12–18 points), and definitive (12–18 points with replication over time) on the basis of the sum of genetic (0–12 points) and experimental (0–6 points) evidence. Currently, official ClinGen curations are only available for AKAP9 (limited evidence) and SCN4B (limited evidence).
Genetic and functional data (cellular electrophysiology, model organism, etc.) related to multisystem Timothy syndrome or the complex cardiac phenotype observed in cardiac-only Timothy syndrome were not included in these calculations.
For AKAP9, SCN4B, and SNTA1, where only a single qualifying functional ultra-rare missense variant is present in the literature (Table 2), there is simply not enough genetic evidence to elevate these genes beyond a limited-evidence gene regardless of the strength of experimental evidence available (Table 3). Of note, the putative LQTS-causative variants within both AKAP9 and SNTA1 used to establish their initial gene-disease associations are present, albeit at extremely low frequency (<10 out of >10 0,00 0 individuals), in pubic exomes (Table 2). Although this finding rightfully questions the nature of AKAP9 and SNTA’s association with penetrant mongogenic cLQTS, the identification of additional LQTS-associated variants, more sophisticated functional characterization of existing and newly discovered variants such as using patient-specific and their variant-corrected induced pluripotent stem cell-derived cardiac cell lines to assess both necessity and sufficiency of an implicated genetic variant, and development of publication avenues that encourage the pursuit and reporting of these findings is needed urgently to determine if AKAP9, SCN4B/ and SNTA1 are candidates for elevation from a limited-evidence gene to a bona fide LQTS-susceptibility gene or further demotion to a disputed-evidence gene.
Table 2.
Functional rare variants in non-syndromic minor LQTS-susceptibility genes reported in seminal gene-disease association studies.
| Gene | Variant | De novo status | Number of segregations | gnomAD minor allele frequency | In vitro phenotype | Ref. |
|---|---|---|---|---|---|---|
| AKAP9 | p.Ser1570Leu | No | 1 | 2/239262 | LQT1-like IKs LOF | [11] |
| ANK2 | p.Ser646Phe | No | 14 | Absenta | Cytosolic Ca2+ accumulation | [38] |
| p.Glu1458Gly | No | 17 | 143/276786 | Cytosolic Ca2+ accumulation | [35,45] | |
| p.Thr3744Asn | No | N/A | 176/276914 | Cytosolic Ca2+ accumulation | [45] | |
| p.Leu3740Ile | No | N/A | 877/276944 | Cytosolic Ca2+ accumulation | [45] | |
| p.Arg3906Trp | No | N/A | 293/27830 | Cytosolic Ca2+ accumulation | [45] | |
| p.Glu3931Lys | No | N/A | 762/276920 | Cytosolic Ca2+ accumulation | [45] | |
| CACNA1C | p.Ala28Thr | No | 2 | 7/246970 | ICa,L GOF | [78] |
| p.Ala582Asp | No | N/A | Absent | ICa,L GOF | [79] | |
| p.Leu762Phe | No | N/A | Absent | ICa,L GOF | [80] | |
| p.Pro857Arg | No | 3 | 1/245530 | ICa,L GOF | [71] | |
| p.Arg860Gly | No | N/A | Absent | ICa,L GOF | [78] | |
| p.Ile1166Thr | No | N/A | Absent | ICa,L GOF | [78] | |
| p.Ile1166Val | No | 3 | Absent | ICa,L GOF | [78] | |
| p.Ile1475Met | No | 1 | Absent | ICa,L GOF | [78] | |
| p.Glu1496Lys | No | N/A | Absent | ICa,L GOF | [78] | |
| p.Gly1783Cys | No | N/A | Absent | ICa,L GOF | [79] | |
| CAV3 | p.Phe97Cys | Yes | N/A | Absent | LQT3-like late INa GOF | [8] |
| p.Ser141Arg | Yes | N/A | Absent | LQT3-like late INa GOF | [8] | |
| KCNE1 | p.Val47Phe | No | N/A | Absent | LQT1-like IKs LOF | [81] |
| p.Leu51His | No | N/A | Absent | LQT1-like IKs LOF | [81] | |
| p.Gly52Arg | No | 5 | Absent | LQT1-like IKs LOF | [82] | |
| p.Ser74Leu | No | 1 | 5/277126 | LQT1-like IKs LOF | [6] | |
| p.Asp76Asn | No | 2 | 19/277116 | LQT1-like IKs LOF | [6,81] | |
| p.Tyr81Cys | No | N/A | 2/246202 | LQT1-like IKs LOF | [83] | |
| p.Trp87Arg | No | N/A | Absent | LQT1-like IKs LOF | [81] | |
| p.Val109Ile | No | 1 | 39/276906 | LQT1-like IKs LOF | [84] | |
| KCNE2 | p.Thr10Met | No | N/A | 63/277186 | LQT2-like IKr LOF | [44,48] |
| p.Met54Thr | No | N/A | 66/227224 | LQT2-like IKr LOF | [7] | |
| p.Ile57Thr | No | N/A | 266/277246 | LQT2-like IKr LOF | [7] | |
| KCNJ5 | p.Gly387Arg | No | 11 | 47/255978 | IKACh LOF | [12] |
| SCN4B | p.Leu179Phe | No | 1 | Absent | LQT3-like late INa GOF | [9] |
| SNTA1 | p.Ala257Gly | No | N/A | 500/276612 | LQT3-like late INa GOF | [85] |
| p.Aal390Val | No | N/A | 6/276632 | LQT3-like late INa GOF | [10] |
Abbreviations: GOF, gain-of-function, LOF, loss-of-function; and LQTS, long QT syndrome.
Public exome databases such as ExAC and gnomAD do not contain individuals from the Gitxsan First Nations community. The minor allele frequency for this variant in this unique founder population is not established.
Already, the body of evidence for some of the other minor non-syndromic LQTS genes compel their demotion (Table 3). In a recent analysis, those variants in KCNE2-encoded MinK-related peptide 1 (MiRP1) initially deemed to be KCNE2-LQTS (historically designated as LQT6)-causative [7] were (i) collectively observed in 1.4% of public exomes, (ii) not found to co-segregate with a LQTS phenotype in either the literature or a large multicenter cohort of KCNE2 rare variant-positive subjects, and (iii) rarely associated with QT prolongation, TdP, or SCD in the absence of a secondary stressor (electrolyte abnormalities, QT prolonging medications, etc.) [44]. This collective evidence led to the conclusion that rare loss-of-function KCNE2 variants confer an underlying arrhythmia-susceptiblity that may manifest as QT prolongation, TdP, and SCD when exacerbated by additional environmental and/or genetic risk factors, but likely do not cause bona fide cLQTS in isolation [44]. As such, KCNE2 appears poised to receive a disputed-evidence gene designation when the official ClinGen gene-disease association classifications are released (Table 3).
Similarly, a majority of the ANK2 -encoded ankyrin-B loss-of-function variants previously deemed to be ABS-causative [34,35,45] are observed at unacceptably high minor allele frequencies (MAF) in public exomes (Table 2). Furthermore, as discussed above, over time, it has become increasingly clear that QT prolongation is not a consistent feature of the cardiac phenotype observed in ABS [36]. Although the disruption of multiple ion channels/transporters and resulting arrhythmia-susceptibility conferred by loss-of-function ANK2 variants is not in question, the high frequency of putative ABS-causative variants in public exomes and nature of the ABS cardiac phenotype certainly question the ongoing designation of ANK2 as a self-sufficient cLQTS-susceptibility gene. That said, the aforementioned recent discovery of a loss-of-function ANK2 variant (p.Ser646Phe-ANK2) in the Gitxsan First Nations founder population that appears to be more consistently associated with QT prolongation and is absent from public exomes may save ANK2 from receiving a disputed-evidence gene designation (Table 3) [38].
Lastly, a single trafficking-defective variant, p.Gly387Arg-KCNJ5, in the KCNJ5-encoded G-protein-coupled inward rectifier potassium channel subtype 4 (GIRK4; Kir3.4) that partially conducts the acetylcholine-induced potassium current (IiKACh) current has been linked to cLQTS through a multi-generational Chinese pedigree [12]. On one hand, KCNJ5 represents one of the only nonsyndromic, minor LQTS-susceptibility genes to be discovered by classical linkage analysis [12]. On the o present in 47/9424 (0.005%) East Asian public exomes/genomes and evidence to support a definitive role for IKACh in human ventricular repolarization is lacking currently [46]. Furthermore, a closer analysis of the clinical phenotype of p.Gly387Arg-KCNJ5-positive individuals revealed that overt QT prolongation was only observed in individuals with additional non-modifiable QT prolongation risk factors such as heart failure, bradycardia, and diabetes [47]. Therefore, it is not clear whether KCNJ5 functions as a self-sufficient but weakly penetrant cLQTS-susceptibility gene or more likely confers an underlying arrhythmia-susceptibility that can manifest as QT prolongation in the presence of multiple additional QT prolongation risk factors. Regardless, in light of the aformentioned population frequency data (Table 2) and lingering questions regarding the role of IKACh in ventricular repolarization, it would come as little suprise if KCNJ5 receives a disputed-evidence gene designation (Table 3).
While AIKAP9, ANK2, KCNE2, KCNJ5, SCN4B, and SNTA1 appear headed for demotion to either limited- or disputed-evidence gene status, it is important to remember that our understanding of LQTS genetic architecture is not static. A gene demoted today in the setting of a prevailing monogenic paradigm may well find new life tomorrow as our understanding of the oligogenic/polygenic nature of LQTS continues to evolve. As such, the temptation to simply lock these genes up and throw away the key must be resisted and the fluid nature of gene-disease association assessment must be embraced.
Genetic testing panels: is bigger really better?
From the onset of clinical genetic testing for LQTS and other Mendelian disorders, the mindset regarding the size of genetic testing panels has largely followed a “bigger is better” mentality. As new disease-susceptibility genes were discovered, the race between clinical laboratories to rapidly expand their respective diagnostic genetic testing panels was on. In recent years, a natural extension of this “bigger is better” mantra has been the development of massive pan-disease diagnostic genetic testing panels (i.e. pan-arrhythmia, pan-cardiac, etc.) that often encompass hundreds of genes with highly variable gene-disease association strengths [18]. When combined with an inherently high rate of background genetic variation, the use of these large and relatively non-specific genetic testing panels has become a recipe for the identification of an enormous number of VUS destined for so-called “genetic purgatory” [17]. Consequently, the unintentional mishandling of VUS has resulted in diagnostic miscues that range in severity from the misguided initiation of predictive cascade screening to the inappropriate implantation of prophylactic implantable cardioverterdefibrillators [16,17].
As those genes ultimately designated as disputed-evidence genes and limited-evidence genes (i.e. genes of uncertain significance or GUS) are likely to also contribute a substantial number of VUS, there are bound to be calls to remove some of the minor LQTS-susceptibility genes designated officially as either disputed/limited-evidence genes from genetic testing panels. However, the assumption that variants residing within such a gene cannot impact clinical decision making may be flawed.
For example, unless current estimates of LQTS/LQTS subtype prevalence represent vast underestimates, functional rare variants in KCNE2 (i.e. p.Thr10Met-KCNE2, p.Met54Thr-KCNE2, and p.Ile57Thr-KCNE2) [44,48] are simply observed too frequently in public exomes to support even a weakly penetrant, but selfsufficient, role in the pathogenesis of a LQTS subtype widely believed to account for ≤1% of LQTS cases [26,36,49]. However, each of these KCNE2 rare variants have been observed in individuals who exhibited a prolonged QT phenotype, including se vere manifestations such as TdP and SCD, in the presence of additional endogenous and exogenous risk factors [44]. Thus, despite an uncertain role in the pathogenesis of fully penetrant monogenic cLQTS, it still remains possible that rare, functionally pro-arrhythmic, variants in an otherwise disputed/limited-evidence gene (from a monogenetic disease-causing perspective) can potentially contribute to oligogenic/polygenic LQTS or predispose to acquired forms of LQTS. Therefore, some variants in these genes have the potential to impact clinical decision-making in some clinical scenarios.
This begs the question of how to ensure that ordering healthcare providers are made aware of the presence of potentially clinically-impactful, functional rare variants termed functional risk alleles that may contribute to oligogenic, polygenic, or acquired forms of LQTS residing within genes believed to have a limited or disputed role in penetrant monogenic LQTS without needlessly increasing the number of novel rare VUSs. At first glance, the removal of disputed-evidence genes and perhaps even limited-evidence genes from diagnostic genetic testing panels might be the best solution. However, as our understanding of the genetic architecture underlying LQTS continues to evolve, we may quickly discover that the removal of some limited/disputed-evidence genes has resulted in a genetic test that now fails to convey/capture an individual’s true genetic predilection for disease. As such, once the formal ClinGen classifications for LQTS and other SCD-predisposing cardiovascular disorders are released, it will be up to research community at large to develop more definitive means of assessing the potential pathogenic contributions of the minor LQTS-susceptibility genes that have been demoted, including potential oligogenic/polygenic contributions, as well as determining formal criteria for the fluid inclusion of genes on clinical genetic tests of all shapes and sizes.
Historical LQTS conventions: farewell old friend?
Unlike genes, which are granted an official name and symbol by a formal committee [the Human Genome Organization (HUGO) Gene Nomenclature Committee (HGNC)], there is and never was a gold standard by which the genetics and genomics community was expected to adhere when naming genetic disorders. The result has been a hodge-podge of naming conventions: (i) major sign of the disorder (e.g. LQTS), (ii) responsible genetic/biochemical defect (e.g. Ankyrin-B syndrome, ABS), (iii) surname(s) of the disorder’s discover(s) (e.g. Andersen and Tawil Syndrome, ATS, Jervell and Lange-Nielsen Syndrome, JLNS, and Timothy Syndrome, TS), and (iv) the adoption of a rigid numerical approach (i.e. LQT1, LQT2, etc.) to the naming of disease subtypes.
With the likely demotion of previously published monogenic cLQTS-causative genes due to either (i) contradictory clinical evidence (KCNJ2/LQT7), (ii) genetic evidence (AKAP9/LQT11, KCNE2/LQT6, KCNJ5/LQT13, SCN4B/LQT10, and SNTA1/LQT12), or (iii) a combination of both (ANK2/LQT4), gaps in the classical/historical LQT1–17 subtype nomenclature are bound to emerge. As such, the time to reappraise existing LQTS nomenclature and institute substantive changes has never been better.
To this end, the adoption of a system that allows the canonical, irrefutable, and common disease-susceptibility genes to retain their historical LQT1–3 designations and then assigns a gene-derived convention for the minor non-syndromic LQTS-susceptibility genes (i.e. CACNA1C-LQTS) is preferable. Such a system (i) communicates directly the gene/minor LQTS subtype in question, (ii) eliminates the need to recall additional/unnecessary associations (LQT15 is caused by what again?), and (iii) allows genes to fluidly enter or exit as the strength of evidence and/or prevailing paradigms dictate regarding LQTS genetic architecture.
Reappraisal of variant interpretation and reporting strategies in diagnostic LQTS genetic testing
As a result of inter-laboratory heterogeneity in the classification and reporting of genetic variants, the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) released updated variant classification and reporting standards in 2015 [50]. In response, most commercial genetic testing companies, including the majority of those that perform diagnostic LQTS genetic testing, have adopted variant classification and reporting strategies that closely align with the 2015 ACMG/AMP guidelines. However, these general “one-size-fits-all” guidelines were not intended to address the intricacies and idiosyncrasies associated with genetic testing for specific disorders such as LQTS [51]. As such, the following paragraphs examine specific limitations of the 2015 ACMG/AMP guidelines that have already negatively impacted diagnostic LQTS genetic testing.
Potentially pro-arrhythmic common genetic variants (functional risk alleles): missing in action or not clinically actionable?
Unfortunately, the 2015 ACMG/AMP guidelines specifically avoided guidance on how or whether common genetic variants, that may modify cLQTS phenotypic severity and/or serve as genetic risk factors for aLQTS, should be classified and incorporated into diagnostic LQTS genetic testing reports. In turn, this lack of guidance gave those clinical genetic testing laboratories, that adopted ACMG/AMP-based schemes, the latitude to interpret (or misinterpret) the 2015 guidelines in a manner that has resulted in the exclusion of common variants with the potential to impact clinical decision making (Table 4) from diagnostic LQTS genetic testing reports [51].
Table 4.
Pro-arrhythmic common variants with the potential to impact clinical decision making.
| Variant | Overall MAF (ExAC) |
African MAF (ExAC) |
Functional Evidence | Epidemiologic evidence | SIFT/PolyPhen prediction | Current ACMG classification | Amended ACMG classification | Refs. |
|---|---|---|---|---|---|---|---|---|
| p.Ser1103Tyr-SCN5A | 0.8% | 10% | LQT3-like INa GOF | Over-represented in aLQTS, SCD in SHD, and SIDS | Possibly damaging/deleterious | Benign | Functional risk allelea | [103,104] |
| p.Asp85Asn-KCNE1 | 0.9% | 0.2% | LQT1/2-like IKs and/or IKr LOF | Over-represented in aLQTS | Benign/deleterious | Likely benign | Functional risk allelea | [105–108] |
Abbreviations: ACMG, American College of Medical Genetics and Genomics; aLQTS, acquired long QT syndrome; ExAC, Exome Aggregation Consortium; GOF, gain-of-function; IK r , rapid component of the delayed-rectifier potassium current; INa , inward depolarizing sodium current; LOF, loss-of-function; LQTS, long QT syndrome; MAF, minor allele frequency; SCD, sudden cardiac death; SHD, structural heart disease; SIDS, sudden infant death syndrome.
If included on genetic testing variants, these variants should be listed under a distinct “Functional Risk Allele” or “Other Reportable” category with a disclaimer such as “This variant is NOT a self-sufficient LQTS-causative mutation in isolation. However, in the setting of either disease-causative mutations or acquired QT aggravating risk factors, the presence of this particular variant can potentially increase the patient’s risk of a potentially life threatening ventricular arrhythmia.”
As recently reviewed in detail elsewhere [51], the population frequency of common variants such as p.Asp85Asn-KCNE1 and p.Ser1103Tyr-SCN5A (Table 4) easily exceed cLQTS prevalence. However, these variants have substantial epidemiologic and experimental evidence to suggest they are capable of generating a circumstance-dependent, pro-arrhythmic state in the setting of QT prolonging medications, electrolyte abnormalities, structural heart disease, and repolarization reserve-deficient genetic backgrounds [51]. Thus, these common variants are likely not benign as frequently classified (Table 4). Rather, they represent a newly defined class of variants termed “functional risk alleles” capable of mimicking the sequelae of ultra-rare penetrant cLQTS-causative pathogenic variants, including conferring susceptibility for TdP and SCD, under specific conditions [51]. Therefore, the practice of withholding or limiting information regarding the presence of so-called functional risk alleles to a supplement that must be requested by healthcare providers has likely resulted in LQTS genetic tests that fail to accurately reflect their patient’s true genetic risk of SCD-predisposing disease.
As such, consideration should be given to reporting those variants that meet the epidemiologic (i.e. odds ratio >5 in case-control studies) and experimental (i.e. well-established functional evidence such as in vitro cellular electrophysiology data) to be classified as functional risk alleles (Table 4) under a distinct “Other Reportable” category with an accompanying disclaimer such as “This variant is not a self-sufficient, disease-causative mutation in isolation. However, in the setting of either disease-causative mutations or acquired QT-aggravating risk factors, the presence of this risk allele can potentially increase the patient’s risk of a potentially life threatening ventricular arrhythmia” [51].
This approach differentiates functional risk alleles from truly pathogenic/likely pathogenic variants and ambiguous VUS and limits the risk of misinterpretation and downstream diagnostic mis-cues. Furthermore, this approach highlights how a critical reappraisal of current variant classification and reporting strategies can improve the state of diagnostic LQTS genetic testing by ensuring patients and healthcare providers are informed of functional risk alleles and afforded every opportunity to institute the simple interventions [i.e. QT prolonging drug avoidance (www.crediblemeds. org), hydration/potassium supplementation, judicious use of antipyretics, etc.] needed to mitigate the small but increased risk of SCD that the presence of functional risk alleles may confer [51].
LQTS-lite: reappraising how weakly penetrant “pathogenic” genetic variants are classified and reported
Residing somewhere between functional risk alleles (Table 4) and penetrant ultra-rare cLQTS-causative pathogenic variants (an LQT1-causative variant for example) is a class of functional rare variants deemed previously to be LQTS-causative, but whose frequency in public exomes exceed expected cLQTS subtype prevalence (1:7,143 for LQT1, 1:10,000 for LQT2, 1:25,0 00 for LQT3, and 1:250,0 0 0 for minor LQTS subtypes). In a heterozygous state, these variants, which are perhaps best exemplified by Finnish (p.Arg176Trp-KCNH2 and p.Leu552Ser-KCNH2) [52] and Norwegian/Swedish (p.Arg518Ter-KCNQ1) [53] founder variants, are associated with a subclinical/concealed LQTS phenotype consisting of borderline QTc prolongation (~450–460 ms) and low SCD risk. Functionally, these rare variants result in haploinsufficiency (p.Arg518Ter-KCNQ1 and all truncating KCNQ1 variants for that matter) or confer modest reductions in IKr current density (p.Arg176Trp-KCNH2 and p.Leu552Ser-KCNH2) [54–57]. Furthermore, the pro-arrhythmic potential of these variants is further highlighted by their frequent contribution to the severe LQTS phenotype observed in JLNS/compound heterozygosity [53,56] and/or association with drug-induced LQTS/TdP risk [58]. As such, these weakly penetrant, but functional, rare variants represent well- known examples of an emerging class of LQTS-causative variants, that includes many KCNQ1 prematurely truncating nonsense and frame-shift variants [59], which collectively result in a latent and more common form of LQTS perhaps best referred to as “LQTSLite”.
Based on public exome-derived population frequencies, LQTSLite-causative variants are anticipated to be amongst those variants most frequently encountered during diagnostic LQTS genetic testing [13]. Although classification of LQTS-Lite-causative variants as “pathogenic variants” or “likely pathogenic variants” is appropriate, these weakly penetrant variants highlight the fact that current ACMG/AMP guidelines were not designed to communicate blatant differences in the strength/penetrance of pathogenic variants. As a result of this “all-or-nothing” approach, healthcare providers with little experience with genetic heart diseases may subject extremely low risk asymptomatic individuals with LQTS-Lite-causative variants to overly aggressive treatment on the basis of a “positive” genetic test. As such, the best management strategies for patients with LQTS-Lite-causative variant(s) and the development of variant classification and reporting mechanisms capable of differentiating between pathogenic variants with intrinsically lower/higher risk and/or penetrance represent areas in need of ongoing reappraisal.
LQTS genetic architecture: truly a monogenic disease?
The number of minor LQTS-susceptibility gene variants observed at unacceptably high frequency in public exomes (Table 2) suggests that the role many of these genes play in LQTS pathogenesis may be more limited, or alternatively more complicated, than believed initially. When viewed in the context of pro-arrhythmic common variants/functional risk alleles (Table 4) and weakly pen etrant LQTS-Lite-causative rare variants in the canonical LQTS-susceptibility genes, it appears that the spectrum of genetic variants capable of contributing to the genetic architecture of LQTS (Fig. 2) may be far more complicated than envisioned previously.
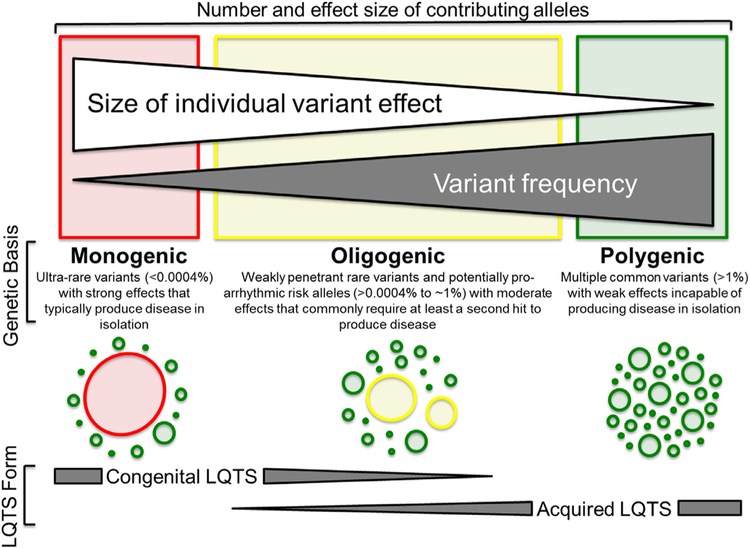
Fig. 2.
Spectrum of genetic variation underlying the acquired and congenital forms of long QT syndrome. At the severe (red) end of the spectrum are ultra-rare congenital long QT syndrome (cLQTS)-causative pathogenic variants with strong effects on gene function. In the middle of the spectrum (yellow) are weakly penetrant rare variants previously labelled as cLQTS-causative, but whose minor allele frequencies in public exomes exceed anticipated cLQTS/cLQTS subtype prevalence, and comparatively more common functional risk alleles (e.g. p.Asp85Asn-KCNE1) that exhibit moderate effects on gene function and rarely produce overt QT prolongation in the absence of additional endogenous or exogenous hits to cardiac repolarization and therefore result in so-called LQTS-Lite. Lastly, at the benign (green) end of the spectrum are common variants with weak effects on gene function, largely discovered through large genome wide association studies of QT prolongation in the general population, that are incapable of producing disease in isolation, but have been shown to result in acquired LQTS (aLQTS) risk when multiple QT-prolonging common variants are present within the genome of an individual with additional endogenous and exogenous QT prolonging risk factors. In recognition that the genetic basis of aLQTS and cLQTS is variable, dark gray triangles denote the spectrum of genetic variation underlying both LQTS forms. Adapted from Giudicessi and Ackerman [60] with permission. Copyright © 2013, Elsevier. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
On the severe end of the LQTS genetic spectrum are ultra-rare, self-sufficient pathogenic/likely pathogenic variants that substantially reduce cardiac repolarization reserve and underlie relatively penetrant monogenic cLQTS (Fig. 2). Due to the strong negative selective pressure exerted against these variants, they are likely rarely, if ever, observed at frequencies that exceed the estimated prevalence of cLQTS in the general population [1:2,500 or a minor allele frequency (MAF) < 0.00 04%] [60].
The opposite (benign) end of the LQTS genetic spectrum features common genetic variants (MAF > 1%), often identified through large-scale genome wide association studies associated with QT interval duration in the general population (Fig. 2) [40,41,61]. Although benign in isolation, the aggregate effect of multiple QT-influencing common genetic variants, as assessed by weighted-effect genetic risk scores, are associated with interindividual variability in baseline QTc duration [62] and the exaggerated QTc response/TdP risk following exposure to a medication with known QT prolonging potential [63] observed in acquired/drug-induced LQTS (Fig. 2). In theory, the burden of QT-influencing, common genetic variants within an individual’s genetic background (i.e. repolarization reserve rich vs. deficient) could also contribute to the phenomena of incomplete penetrance and variable expressivity observed in many LQTS pedigrees as well as the ~20% of LQTS cases that remain genotype-negative.
Lastly, in the middle of the LQTS genetic variation spectrum are relatively rare and comparatively more common genetic variants (MAF ranging from >0.00 04 to < 1%) that we have designated herein as either LQTS-Lite-causative rare variants or func tional risk alleles, respectively (Fig. 2). These variants result in mild-to-moderate reductions in cardiac repolarization reserve that may manifest as borderline or transient QT prolongation, but rarely cause LQTS-triggered syncope/seizures or SCD in the absence of additional QT prolonging genetic variants or endogenous/exogenous risk factors. As such, these variants may serve as primary drivers of so-called oligogenic cLQTS when present in genetic backgrounds that contain other QT-related genetic modifiers such as common variants in NOS1AP [64,65], the 3′untranslated region of KCNQ1 [66], and perhaps an increased burden of those common variants shown to influence QTc duration in health. Furthermore, LQTS-Lite-causative variants within this class likely explain, in part, the observation that functional rare variants previously associated with cLQTS are observed at unexpectedly high frequency in public exome/genome databases [67].
As the dust settles: practical implications for the clinician
As outlined in the preceding sections, the stage is set for several paradigm shifts, notably the possible demotion/reclassification of ~40% of LQTS-susceptibility genes and the transition away from a rigid monogenic model of disease, with the potential to radically alter our view of the genetic architecture underlying LQTS and other heritable cardiovascular disorders. For those charged with the care of patients with LQTS, whether it be on a daily basis or once in a blue moon, it is anticipated that this Review will generate a litany of questions, and perhaps some appropriate anxiety, regarding the current role of genetics in the diagnosis, risk-stratification, and management of LQTS.
Once formal ClinGen gene-disease association classifications are released, it will take time for (i) the field to reach a consensus regarding if/how variants in demoted/reclassified genes are reported and (ii) clinical laboratories to implement these recommendations. As genetic testing is, and should remain, a class I recommendation for LQTS [23], clinicians should continue to utilize the current generation of diagnostic LQTS genetic tests. However, ordering healthcare providers should assure they are receiving a complete genetic risk profile on every patient by requesting the supplemental information needed to determine if functional risk alleles are present and handling variants identified in genes that are poised for demotion/reclassification with extra caution and skepticism.
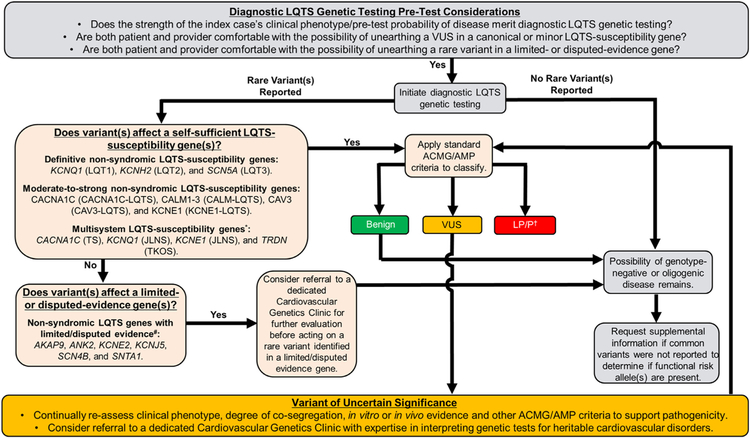
During this transition period, clinicians may find it helpful to utilize a tiered approach, independent from commercial classification and reporting schemes, to prioritize the analysis of diagnostic LQTS genetic testing results (Fig. 3). Under such an approach, findings in the canonical [KCNQ1 (LQT1), KCNH2 (LQT2), and SCN5A (LQT3)] and moderate-to-strong evidence minor LQTS-susceptibility genes (CACNA1C-LQTS, CALM1/2/3-LQTS, CAV3-LQTS, KCNE1-LQTS, and TRDN-LQTS/TKOS) are prioritized and represent the area where generalists should focus their efforts. Until the dust settles, it is recommended that patients with either a VUS or variants labelled, previously or actively, as pathogenic/likely pathogenic in limited/disputed evidence genes be referred to dedicated Cardiovascular Genetics Clinics to decrease the risk of diagnostic miscues. Furthermore, any vexing questions related to LQTS genetic testing, including appropriateness of cascade screening, VUS resolution, and the possibility of oligogenic/polygenic disease, are best addressed in a similar setting.
Fig. 3.
A rational tiered approach to LQTS genetic testing initiation, variant interpretation, and ongoing re-assessment of variants of unknown/uncertain significance (VUS). Light gray boxes denote basic considerations pertaining to the initiation of LQTS genetic testing. Light orange boxes denote a tiered approach to the assessment of rare variants in LQTS-susceptibility genes with variable gene-disease association evidence strength. Gold boxes denote basic considerations pertaining to the identification of a rare VUS in self-sufficient LQTS-susceptibility genes that currently lacks sufficient evidence to classify as either benign or pathogenic. *Due to the lack of a true QT prolongation phenotype, the authors recommend against the routine inclusion of KCNJ2 on diagnostic LQTS testing panels. # Although the self-sufficient role of these genes in LQTS pathogenesis requires further investigation, specific rare variants in these genes may still confer arrhythmia-susceptibility and/or contribute to an oligogenic/polygenic form of LQTS. Referral to a dedicated Cardiovascular Genetics Clinic is recommended.† Current ACMG/AMP guidelines are not equipped to assess or communicate the strength of likely pathogenic/pathogenic variants. Outside of beta-blocker use, management of low-risk asymptomatic/concealed individuals should not be made on genetic test results alone. Abbreviations: ACMG, American College of Medical Genetics and Genomics; AMP, Association for Molecular Pathology; JLNS, Jervell and Lange-Nielson syndrome; LP, likely pathogenic; LQTS, long QT syndrome; P, pathogenic; TKOS, triadin knock-out syndrome; TS, Timothy syndrome; and VUS, variant of uncertain significance. Adapted from Giudicessi et al. [51] with permission. Copyright © 2018, Lippincott Williams & Wilkins with permission. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Conclusion
As illustrated in this Review, the unexpectedly high rate of background genetic variation in the canonical and minor LQTS-susceptibility genes is beginning to re-shape our understanding of the genetic architecture underlying LQTS. In short order, a number of the minor LQTS-susceptibility genes, previously believed to be responsible for ~5–10% of non-syndromic LQTS cases, could be demoted to either limited-evidence or disputed-evidence gene status and, at best, be relegated to roles as oligogenic/polygenic contributors. Meanwhile, the presence of functional and potentially clinically impactful genetic variation in LQTS-susceptibility genes whose population frequencies easily exceed overall LQTS and/or LQTS subtype prevalence exposes inherent limitations in our current variant classification and reporting strategies. Unfortunately, these issues are not unique to LQTS and are anticipated to impact all heritable cardiovascular disorders, including the aortopathies, cardiomyopathies, and the other cardiac channelopathies, to some degree. As these issues continue to challenge current paradigms, it is paramount that the field continually reappraise whether (i) a larger than anticipated number of LQTS cases, including many currently labelled as minor gene-positive or genotype-negative, are actually oligogenic or polygenic in nature, (ii) previous estimates of cLQTS prevalence are indeed accurate, and (iii) existing variant classification and reporting strategies accurately reflect the full spectrum of genetic variation that may contribute to LQTS pathogenesis and therefore convey an individual’s true genetic risk of disease. Hopefully, ongoing efforts, to address these and other pressing issues (i.e. “the VUS crisis”), will continue to improve the state of diagnostic LQTS genetic testing and ensure that patients have access to accurate genotype-guided approaches to the diagnosis, risk-stratification, and clinical management of this potentially fatal, but highly treatable, genetic disorder [68].
Supplementary Material
Acknowledgments
Funding sources: This work was supported by the Windland Smith Rice Sudden Comprehensive Sudden Cardiac Death Program (to Dr. Ackerman). Dr. Giudicessi thanks the Mayo Clinic Cardiovascular Diseases Fellowship and Clinician Investigator Training Programs for fostering an outstanding environment for physician-scientist training.
Abbreviations:
- ABS
ankyrin-B syndrome
- ACMG
American College of Medical Genetics and Genomics
- aLQTS
acquired/drug-induced LQTS
- AMP
Association for Molecular Pathology
- Ca2+
calcium
- ClinGen
Clinical Genome Resource
- cLQTS
congenital LQTS
- DNA
deoxyribonucleic acid
- EAD
early afterdepolarization
- ECG
electrocardiogram
- GWAS
genome-wide association study
- I KACh
acetylcholine- induced potassium current
- K+
potassium
- LQTS
long QT syndrome
- MAF
minor allele frequency
- Na+
sodium
- QTc
heart rate-corrected QT interval
- SCD
sudden cardiac death
- TdP
torsades de pointes
- VUS
variant of uncertain significance
Footnotes
Conflict of interest statement: Dr. Ackerman is a consultant for Audentes Therapeutics, Boston Scientific, Gilead Sciences, Invitae, Medtronic, MyoKardia, and St. Jude Medical. From 2004 to 2016, M.J.A. and Mayo Clinic received sales-based royalties from Transgenomic for their FAMILION-LQTS and FAMILION-CPVT genetic tests. Currently, M.J.A. and Mayo Clinic have royalty/equity relationships (without remuneration so far) with AliveCor, Blue Ox Health, and StemoniX. However, none of these entities participated in this study. Drs. Giudicessi and Wilde declare no conflicts.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tcm.2018.03.003.
References
- [1].Schwartz PJ, Ackerman MJ, George AL Jr, Wilde AA. Impact of genetics on the clinical management of channelopathies. J Am Coll Cardiol 2013;62(3):169–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Giudicessi JR, Ackerman MJ. Calcium revisited: new insights into the molecular basis of long-QT syndrome. Circ Arrhythm Electrophysiol 2016;9(7):e002480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Curran ME, Splawski I, Timothy KW, Vincent GM, Green ED, Keating MT. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell 1995;80(5):795–803, [DOI] [PubMed] [Google Scholar]
- [4].Wang Q, Curran ME, Splawski I, Burn TC, Millholland JM, VanRaay TJ, et al. Positional cloning of a novel potassium channel gene: KVLQT1 mutations cause cardiac arrhythmias. Nat Genet 1996;12(1):17–23, [DOI] [PubMed] [Google Scholar]
- [5].Wang Q, Shen J, Splawski I, Atkinson D, Li Z, Robinson JL, et al. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell 1995;80(5):805–11, [DOI] [PubMed] [Google Scholar]
- [6].Splawski I, Tristani-Firouzi M, Lehmann MH, Sanguinetti MC, Keating MT, Mutations in the hminK gene cause long QT syndrome and suppress IKs function. Nat Genet 1997;17(3):338–40, [DOI] [PubMed] [Google Scholar]
- [7].Abbott GW, Sesti F, Splawski I, Buck ME, Lehmann MH, Timothy KW, et al. MiRP1 forms IKr potassium channels with HERG and is associated with cardiac arrhythmia. Cell 1999;97(2):175–87, [DOI] [PubMed] [Google Scholar]
- [8].Vatta M, Ackerman MJ, Ye B, Makielski JC, Ughanze EE, Taylor EW, et al. Mutant caveolin-3 induces persistent late sodium current and is associated with long-QT syndrome. Circulation 2006;114(20):2104–12. [DOI] [PubMed] [Google Scholar]
- [9].Medeiros-Domingo A, Kaku T, Tester DJ, Iturralde-Torres P, Itty A, Ye B, et al. SCN4B-encoded sodium channel beta4 subunit in congenital long-QT syndrome. Circulation 2007;116(2):134–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ueda K, Valdivia C, Medeiros-Domingo A, Tester DJ, Vatta M, Farrugia G, et al. Syntrophin mutation associated with long QT syndrome through activation of the nNOS-SCN5A macromolecular complex. Proc Natl Acad Sci USA 2008;105(27):9355–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chen L, Marquardt ML, Tester DJ, Sampson KJ, Ackerman MJ, Kass RS. Mutation of an A-kinase-anchoring protein causes long-QT syndrome. Proc Natl Acad Sci USA 2007;104(52):20990–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yang Y, Yang Y, Liang B, Liu J, Li J, Grunnet M, et al. Identification of a Kir3.4 mutation in congenital long QT syndrome. Am J Hum Genet 2010;86(6):872–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ackerman MJ, Tester DJ, Jones GS, Will ML, Burrow CR, Curran ME. Ethnic differences in cardiac potassium channel variants: implications for genetic susceptibility to sudden cardiac death and genetic testing for congenital long QT syndrome. Mayo Clin Proc 2003;78(12):1479–87. [DOI] [PubMed] [Google Scholar]
- [14].Ackerman MJ, Splawski I, Makielski JC, Tester DJ, Will ML, Timothy KW, et al. Spectrum and prevalence of cardiac sodium channel variants among black, white, Asian, and Hispanic individuals: implications for arrhythmogenic susceptibility and Brugada/long QT syndrome genetic testing. Heart Rhythm 2004;1(5):600–7. [DOI] [PubMed] [Google Scholar]
- [15].Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536(7616):285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ackerman JP, Bartos DC, Kapplinger JD, Tester DJ, Delisle BP, Ackerman MJ. The promise and peril of precision medicine: phenotyping still matters most. Mayo Clin Proc 2016. doi: 10.1016/j.mayocp.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ackerman MJ. Genetic purgatory and the cardiac channelopathies: Exposing the variants of uncertain/unknown significance issue. Heart Rhythm 2015;12(11):2325–31. [DOI] [PubMed] [Google Scholar]
- [18].Giudicessi JR, Kullo IJ, Ackerman MJ. Precision cardiovascular medicine: state of genetic testing. Mayo Clin Proc 2017;92(4):642–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Nerbonne JM, Kass RS. Molecular physiology of cardiac repolarization. Physiol Rev 2005;85(4):1205–53. [DOI] [PubMed] [Google Scholar]
- [20].Giudicessi JR, Ackerman MJ. Potassium-channel mutations and cardiac ar- rhythmias-diagnosis and therapy. Nat Rev Cardiol 2012;9(6):319–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Rautaharju PM, Surawicz B, Gettes LS. AHA/ACCF/HRS recommendations for the standardization and interpretation of the electrocardiogram part IV: the ST segment, T and U waves, and the QT interval a scientific statement from the American Heart Association electrocardiography and arrhythmias committee, Council on Clinical Cardiology; the American College of Cardiology Foundation; and the Heart Rhythm Society. Circulation 2009;119(10) E241–EE50. [DOI] [PubMed] [Google Scholar]
- [22].Taggart NW, Haglund CM, Tester DJ, Ackerman MJ. Diagnostic miscues in congenital long-QT syndrome. Circulation 2007;115(20):2613–20. [DOI] [PubMed] [Google Scholar]
- [23].Ackerman MJ, Priori SG, Willems S, Berul C, Brugada R, Calkins H, et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA). Heart Rhythm 2011;8(8):1308–39. [DOI] [PubMed] [Google Scholar]
- [24].Weiss JN, Garfinkel A, Karagueuzian HS, Chen PS, Qu ZL. Early afterdepolarizations and cardiac arrhythmias. Heart Rhythm 2010;7(12):1891–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Giudicessi JR, Ackerman MJ. Genotype- and phenotype-guided management of congenital long QT syndrome. Curr Probl Cardiol 2013;38(10):417–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Tester DJ, Will ML, Haglund CM, Ackerman MJ, Compendium of cardiac channel mutations in 541 consecutive unrelated patients referred for long QT syndrome genetic testing. Heart Rhythm 2005;2(5):507–17. [DOI] [PubMed] [Google Scholar]
- [27].Altmann HM, Tester DJ, Will ML, Middha S, Evans JM, Eckloff BW, et al. Homozygous/compound heterozygous triadin mutations associated with autosomal-recessive long-QT syndrome and pediatric sudden cardiac arrest: elucidation of the triadin knockout syndrome. Circulation 2015;131(23):2051–60. [DOI] [PubMed] [Google Scholar]
- [28].Plaster NM, Tawil R, Tristani-Firouzi M, Canun S, Bendahhou S, Tsunoda A, et al. Mutations in Kir2.1 cause the developmental and episodic electrical phenotypes of Andersen’s syndrome. Cell 2001;105(4):511–19, [DOI] [PubMed] [Google Scholar]
- [29].Splawski I, Timothy KW, Sharpe LM, Decher N, Kumar P, Bloise R, et al. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell 2004;119(1):19–31. [DOI] [PubMed] [Google Scholar]
- [30].Andersen ED, Krasilnikoff PA, Overvad H. Intermittent muscular weakness, extrasystoles, and multiple developmental anomalies. A new syndrome? Acta Paediatr Scand 1971;60(5):559–64, [DOI] [PubMed] [Google Scholar]
- [31].Schott JJ, Charpentier F, Peltier S, Foley P, Drouin E, Bouhour JB, et al. Mapping of a gene for long QT syndrome to chromosome 4q25–27. Am J Hum Genet 1995;57(5):1114–22. [PMC free article] [PubMed] [Google Scholar]
- [32].Tristani-Firouzi M, Jensen JL, Donaldson MR, Sansone V, Meola G, Hahn A, et al. Functional and clinical characterization of KCNJ2 mutations associated with LQT7 (Andersen syndrome). J Clin Invest 2002;110(3):381–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhang L, Benson DW, Tristani-Firouzi M, Ptacek LJ, Tawil R, Schwartz PJ, et al. Electrocardiographic features in Andersen-Tawil syndrome patients with KCNJ2 mutations: characteristic T-U-wave patterns predict the KCNJ2 genotype. Circulation 2005;111(21):2720–6. [DOI] [PubMed] [Google Scholar]
- [34].Mohler PJ, Splawski I, Napolitano C, Bottelli G, Sharpe L, Timothy K, et al. A cardiac arrhythmia syndrome caused by loss of ankyrin-B function. Proc Natl Acad Sci USA 2004;101(24):9137–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Mohler PJ, Schott JJ, Gramolini AO, Dilly KW, Guatimosim S, duBell WH, et al. Ankyrin-B mutation causes type 4 long-QT cardiac arrhythmia and sudden cardiac death. Nature 2003;421(6923):634–9, [DOI] [PubMed] [Google Scholar]
- [36].Sherman J, Tester DJ, Ackerman MJ. Targeted mutational analysis of ankyrin-B in 541 consecutive, unrelated patients referred for long QT syndrome ge netic testing and 200 healthy subjects. Heart Rhythm 2005;2(11):1218–1223 [DOI] [PubMed] [Google Scholar]
- [37].Ichikawa M, Aiba T, Ohno S, Shigemizu D, Ozawa J, Sonoda K, et al. Pheno typic variability of ANK2 mutations in patients with inherited primary arrhythmia syndromes. Circ J 2016;80(12):2435–42. [DOI] [PubMed] [Google Scholar]
- [38].Swayne LA, Murphy NP, Asuri S, Chen L, Xu X, McIntosh S, et al. Novel vari ant in the ANK2 membrane-binding domain is associated with ankyrin-B syndrome and structural heart disease in a First Nations population with a highrate of long QT syndrome. Circ Cardiovasc Genet 2017;10(1):e001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sedlacek K, Stark K, Cunha SR, Pfeufer A, Weber S, Berger I, et al. Common genetic variants in ANK2 modulate QT interval: results from the KORA study. Circ Cardiovasc Genet 2008;1(2):93–9. [DOI] [PubMed] [Google Scholar]
- [40].Pfeufer A, Sanna S, Arking DE, Muller M, Gateva V, Fuchsberger C, et al. Com mon variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat Genet 2009;41(4):407–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Newton-Cheh C, Eijgelsheim M, Rice KM, de Bakker PI, Yin X, Estrada K, et al. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat Genet 2009;41(4):399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Chong JX, Buckingham KJ, Jhangiani SN, Boehm C, Sobreira N, Smith JD, et al. The genetic basis of Mendelian phenotypes: discoveries, challenges, and opportunities. Am J Hum Genet 2015;97(2):199–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Strande NT, Riggs ER, Buchanan AH, Ceyhan-Birsoy O, DiStefano M, Dwight SS, et al. Evaluating the clinical validity of gene-disease associations:an evidence-based framework developed by the clinical genome resource. Am J Hum Genet 2017;100(6):895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Roberts JD, Krahn AD, Ackerman MJ, Rohatgi RK, Moss AJ, Nazer B,et al. Loss-of-function KCNE2 variants: true monogenic culprits of long-QT syndrome or proarrhythmic variants requiring secondary provocation? Circ Arrhythm Electrophysiol 2017;10(8):e005282. [DOI] [PubMed] [Google Scholar]
- [45].Mohler PJ, Le Scouarnec S, Denjoy I, Lowe JS, Guicheney P, Caron L,et al. Defining the cellular phenotype of “ankyrin-B syndrome” variants: hu man ANK2 variants associated with clinical phenotypes display a spectrum of activities in cardiomyocytes. Circulation 2007;115(4):432–41. [DOI] [PubMed] [Google Scholar]
- [46].Beckmann C, Rinne A, Littwitz C, Mintert E, Bosche LI, Kienitz MC, et al. G protein-activated (GIRK) current in rat ventricular myocytes is masked by constitutive inward rectifier current (I(K1)). Cell Physiol Biochem 2008;21(4):259–68. [DOI] [PubMed] [Google Scholar]
- [47].Wang F, Liu J, Hong L, Liang B, Graff C, Yang Y, et al. The phenotype character istics of type 13 long QT syndrome with mutation in KCNJ5 (Kir3.4-G387R). Heart Rhythm 2013;10(10):1500–6. [DOI] [PubMed] [Google Scholar]
- [48].Gordon E, Panaghie G, Deng L, Bee KJ, Roepke TK, Krogh-Madsen T, et al. A KCNE2 mutation in a patient with cardiac arrhythmia induced by auditory stimuli and serum electrolyte imbalance. Cardiovasc Res 2008;77(1):98–106. [DOI] [PubMed] [Google Scholar]
- [49].Kapplinger JD, Tester DJ, Salisbury BA, Carr JL, Harris-Kerr C, Pollevick GD, et al. Spectrum and prevalence of mutations from the first 2,500 consecutive unrelated patients referred for the FAMILION long QT syndrome genetic test. Heart Rhythm 2009;6(9):1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomicsand the Association for Molecular Pathology. Genet Med 2015;17(5):405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Giudicessi JR, Roden DM, Wilde AAM, Ackerman MJ. Classification and reporting of potentially proarrhythmic common genetic variation in long QT syndrome genetic testing. Circulation 2018;137(6):619–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lahtinen AM, Havulinna AS, Noseworthy PA, Jula A, Karhunen PJ, Perola M, et al. Prevalence of arrhythmia-associated gene mutations and risk of sudden cardiac death in the Finnish population. Ann Med 2013;45(4):328–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Winbo A, Stattin EL, Nordin C, Diamant UB, Persson J, Jensen SM, et al. Phenotype, origin and estimated prevalence of a common long QT syndrome mutation: a clinical, genealogical and molecular genetics study including Swedish R518X/KCNQ1 families. BMC Cardiovasc Disord 2014;14:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Fodstad H, Bendahhou S, Rougier JS, Laitinen-Forsblom PJ, Barhanin J,Abriel H, et al. Molecular characterization of two founder mutations causing long QT syndrome and identification of compound heterozygous patients. Ann Med 2006;38(4):294–304. [DOI] [PubMed] [Google Scholar]
- [55].Lahti AL, Kujala VJ, Chapman H, Koivisto AP, Pekkanen-Mattila M, Kerkela E, et al. Model for long QT syndrome type 2 using human iPS cells demonstrates arrhythmogenic characteristics in cell culture. Dis Model Mech 2012;5(2):220–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Piippo K, Laitinen P, Swan H, Toivonen L, Viitasalo M, Pasternack M, et al. Homozygosity for a HERG potassium channel mutation causes a severe form of long QT syndrome: identification of an apparent founder mutation in the Finns. J Am Coll Cardiol 2000;35(7):1919–25. [DOI] [PubMed] [Google Scholar]
- [57].Anderson CL, Kuzmicki CE, Childs RR, Hintz CJ, Delisle BP, January CT. Large-s cale mutational analysis of Kv11.1 reveals molecular insights into type 2 long QT syndrome. Nat Commun 2014;5:5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Lehtonen A, Fodstad H, Laitinen-Forsblom P, Toivonen L, Kontula K, Swan H. Further evidence of inherited long QT syndrome gene muta tions in antiarrhythmic drug-associated torsades de pointes. Heart Rhythm 2007;4(5):603–7. [DOI] [PubMed] [Google Scholar]
- [59].Ruwald MH, Xu Parks X, Moss AJ, Zareba W, Baman J, McNitt S, et al. Stop–codon and C-terminal nonsense mutations are associated with a lower risk of cardiac events in patients with long QT syndrome type 1. Heart Rhythm 2016;13(1):122–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Giudicessi JR, Ackerman MJ. Determinants of incomplete penetrance and variable expressivity in heritable cardiac arrhythmia syndromes. Transl Res 2013;161(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Arking DE, Pulit SL, Crotti L, van der Harst P, Munroe PB, Koopmann TT, et al. Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nat Genet 2014;46(8):826–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Noseworthy PA, Havulinna AS, Porthan K, Lahtinen AM, Jula A, Karhunen PJ, et al. Common genetic variants, QT interval, and sudden cardiac death in a Finnish population-based study. Circ Cardiovasc Genet 2011;4(3):305–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Strauss DG, Vicente J, Johannesen L, Blinova K, Mason JW, Weeke P, et al. Common genetic variant risk score is associated with drug-induced QT prolongation and torsade de pointes risk: a pilot study. Circulation 2017;135(14):1300–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Crotti L, Monti MC, Insolia R, Peljto A, Goosen A, Brink PA, et al. NOS1AP is a genetic modifier of the long-QT syndrome. Circulation 2009;120(17):1657–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Tomas M, Napolitano C, De Giuli L, Bloise R, Subirana I, Malovini A, et al. Polymorphisms in the NOS1AP gene modulate QT interval duration and risk of arrhythmias in the long QT syndrome. J Am Coll Cardiol 2010;55(24):2745–52. [DOI] [PubMed] [Google Scholar]
- [66].Amin AS, Giudicessi JR, Tijsen AJ, Spanjaart AM, Reckman YJ, Klemens CA, et al. Variants in the 3′, untranslated region of the KCNQ1-encoded Kv7.1 potassium channel modify disease severity in patients with type 1 long QT syndrome in an allele-specific manner. Eur Heart J 2012;33(6):714–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Refsgaard L, Holst AG, Sadjadieh G, Haunso S, Nielsen JB, Olesen MS. High prevalence of genetic variants previously associated with LQT syndrome in new exome data. Eur J Hum Genet 2012;20(8):905–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Rohatgi RK, Sugrue A, Bos JM, Cannon BC, Asirvatham SJ, Moir C, et al. Contemporary outcomes in patients with long QT syndrome. J Am Coll Cardiol 2017;70(4):453–62. [DOI] [PubMed] [Google Scholar]
- [69].Giudicessi JR, Ackerman MJ, Camilleri M. Cardiovascular safety of prokinetic agents: A focus on drug-induced arrhythmias. Neurogastroenterol Motil 2018. doi: 10.1111/nmo.13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Giudicessi JR, Ackerman MJ. Prevalence and potential genetic determinants of sensorineural deafness in KCNQ1 homozygosity and compound heterozygosity. Circ Cardiovasc Genet 2013;6(2):193–200, [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Boczek NJ, Best JM, Tester DJ, Giudicessi JR, Middha S, Evans JM, et al. Ex- ome sequencing and systems biology converge to identify novel mutations in the L-type calcium channel, CACNA1C, linked to autosomal dominant long QT syndrome. Circ Cardiovasc Genet 2013;6(3):279–89, [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Crotti L, Johnson CN, Graf E, De Ferrari GM, Cuneo BF, Ovadia M, et al. Calmodulin mutations associated with recurrent cardiac arrest in infants. Circulation 2013;127(9):1009–17, [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Reed GJ, Boczek NJ, Etheridge SP, Ackerman MJ. CALM3 mutation associated with long QT syndrome. Heart Rhythm 2015;12(2):419–22, [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Boczek NJ, Ye D, Jin F, Tester DJ, Huseby A, Bos JM, et al. Identification and functional characterization of a novel CACNA1C-mediated cardiac disorder characterized by prolonged QT intervals with hypertrophic cardiomyopathy, congenital heart defects, and sudden cardiac death. Circ Arrhythm Electrophysiol 2015;8(5):1122–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Splawski I, Timothy KW, Vincent GM, Atkinson DL, Keating MT. Molecular basis of the long-QT syndrome associated with deafness. N Engl J Med 1997;336(22):1562–7. [DOI] [PubMed] [Google Scholar]
- [76].Duggal P, Vesely MR, Wattanasirichaigoon D, Villafane J, Kaushik V, Beggs AH. Mutation of the gene for IsK associated with both Jervell and Lange-Nielsen and Romano-Ward forms of Long-QT syndrome. Circulation 1998;97(2):142–6. [DOI] [PubMed] [Google Scholar]
- [77].Dufendach KA, Giudicessi JR, Boczek NJ, Ackerman MJ. Maternal mosaicism confounds the neonatal diagnosis of type 1 Timothy syndrome. Pediatrics 2013;131(6):e1991–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Wemhoner K, Friedrich C, Stallmeyer B, Coffey AJ, Grace A, Zumhagen S, et al. Gain-of-function mutations in the calcium channel CACNA1C (Cav1.2) cause non-syndromic long-QT but not Timothy syndrome. J Mol Cell Cardiol 2015;80:186–95. [DOI] [PubMed] [Google Scholar]
- [79].Fukuyama M, Wang Q, Kato K, Ohno S, Ding WG, Toyoda F, et al. Long QT syndrome type 8: novel CACNA1C mutations causing QT prolongation and variant phenotypes. Europace 2014;16(12):1828–37. [DOI] [PubMed] [Google Scholar]
- [80].Landstrom AP, Boczek NJ, Ye D, Miyake CY, De la Uz CM, Allen HD, et al. Novel long QT syndrome-associated missense mutation, L762F, in CACNA1C-encoded L-type calcium channel imparts a slower inactivation tau and increased sustained and window current. Int J Cardiol 2016;220:290–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Bianchi L, Shen Z, Dennis AT, Priori SG, Napolitano C, Ronchetti E, et al. Cellular dysfunction of LQT5-minK mutants: abnormalities of IKs, IKr and trafficking in long QT syndrome. Hum Mol Genet 1999;8(8):1499–507. [DOI] [PubMed] [Google Scholar]
- [82].Ma L, Lin C, Teng S, Chai Y, Bahring R, Vardanyan V, et al. Characterization of a novel Long QT syndrome mutation G52R-KCNE1 in a Chinese family. Cardiovasc Res 2003;59(3):612–19. [DOI] [PubMed] [Google Scholar]
- [83].Wu DM, Lai LP, Zhang M, Wang HL, Jiang M, Liu XS, et al. Characterization of an LQT5-related mutation in KCNE1, Y81C: implications for a role of KCNE1 cytoplasmic domain in IKs channel function. Heart Rhythm 2006;3(9):1031–40. [DOI] [PubMed] [Google Scholar]
- [84].Schulze-Bahr E, Schwarz M, Hauenschild S, Wedekind H, Funke H, Haverkamp W, et al. A novel long-QT 5 gene mutation in the C-ter- minus (V109I) is associated with a mild phenotype. J Mol Med (Berl) 2001;79(9):504–9, [DOI] [PubMed] [Google Scholar]
- [85].Wu G, Ai T, Kim JJ, Mohapatra B, Xi Y, Li Z, et al. alpha-1-syntrophin mutation and the long-QT syndrome: a disease of sodium channel disruption. Circ Arrhythm Electrophysiol 2008;1(3):193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Marx SO, Kurokawa J, Reiken S, Motoike H, D’Armiento J, Marks AR, et al. Requirement of a macromolecular signaling complex for beta adrenergic receptor modulation of the KCNQ1-KCNE1 potassium channel. Science 2002;295(5554):496–9, [DOI] [PubMed] [Google Scholar]
- [87].Li Y, Chen L, Kass RS, Dessauer CW, The A-kinase anchoring protein Yotiao facilitates complex formation between adenylyl cyclase type 9 and the IKs potassium channel in heart. J Biol Chem 2012;287(35):29815–24, [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Hennessey JA, Boczek NJ, Jiang YH, Miller JD, Patrick W, Pfeiffer R, et al. A CACNA1C variant associated with reduced voltage-dependent inactivation, increased CaV1.2 channel window current, and arrhythmogenesis. PLoS One 2014;9(9):e106982, [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Song KS, Scherer PE, Tang Z, Okamoto T, Li S, Chafel M, et al. Expression of caveolin-3 in skeletal, cardiac, and smooth muscle cells. Caveolin-3 is a component of the sarcolemma and co-fractionates with dystrophin and dystrophin-associated glycoproteins. J Biol Chem 1996;271(25):15160–5. [DOI] [PubMed] [Google Scholar]
- [90].Splawski I, Shen J, Timothy KW, Lehmann MH, Priori S, Robinson JL, et al. Spectrum of mutations in long-QT syndrome genes. KVLQT1, HERG, SCN5A, KCNE1, and KCNE2. Circulation 2000;102(10):1178–85, [DOI] [PubMed] [Google Scholar]
- [91].Charpentier F, Merot J, Riochet D, Le Marec H, Escande D. Adult KCNE1-knockout mice exhibit a mild cardiac cellular phenotype. Biochem Biophys Res Commun 1998;251(3):806–10. [DOI] [PubMed] [Google Scholar]
- [92].Major P, Baczko I, Hiripi L, Odening KE, Juhasz V, Kohajda Z, et al. A novel transgenic rabbit model with reduced repolarization reserve: long QT syndrome caused by a dominant-negative mutation of the KCNE1 gene. Br J Pharmacol 2016;173(12):2046–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Cui J, Kagan A, Qin D, Mathew J, Melman YF, McDonald TV. Analysis of the cyclic nucleotide binding domain of the HERG potassium channel and interactions with KCNE2. J Biol Chem 2001;276(20):17244–51, [DOI] [PubMed] [Google Scholar]
- [94].Mazhari R, Greenstein JL, Winslow RL, Marban E, Nuss HB. Molecular interactions between two long-QT syndrome gene products, HERG and KCNE2, rationalized by in vitro and in silico analysis. Circ Res 2001;89(1):33–8. [DOI] [PubMed] [Google Scholar]
- [95].Jiang M, Zhang M, Tang DG, Clemo HF, Liu J, Holwitt D, et al. KCNE2 protein is expressed in ventricles of different species, and changes in its expression contribute to electrical remodeling in diseased hearts. Circulation 2004;109(14):1783–8. [DOI] [PubMed] [Google Scholar]
- [96].Lundquist AL, Manderfield LJ, Vanoye CG, Rogers CS, Donahue BS, Chang PA, et al. Expression of multiple KCNE genes in human heart may enable variable modulation of I(Ks). J Mol Cell Cardiol 2005;38(2):277–87. [DOI] [PubMed] [Google Scholar]
- [97].Wu DM, Jiang M, Zhang M, Liu XS, Korolkova YV, Tseng GN, KCNE2 is colocalized with KCNQ1 and KCNE1 in cardiac myocytes and may function as a negative modulator of I(Ks) current amplitude in the heart. Heart Rhythm 2006;3(12):1469–80. [DOI] [PubMed] [Google Scholar]
- [98].Roepke TK, Kontogeorgis A, Ovanez C, Xu X, Young JB, Purtell K, et al. Targeted deletion of kcne2 impairs ventricular repolarization via disruption of I(K,slow1) and I(to,f). FASEB J 2008;22(10):3648–60, [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Hu Z, Kant R, Anand M, King EC, Krogh-Madsen T, Christini DJ, et al. Kcne2 deletion creates a multisystem syndrome predisposing to sudden cardiac death. Circ Cardiovasc Genet 2014;7(1):33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Wickman K, Nemec J, Gendler SJ, Clapham DE. Abnormal heart rate regulation in GIRK4 knockout mice. Neuron 1998;20(1):103–14. [DOI] [PubMed] [Google Scholar]
- [101].Remme CA, Scicluna BP, Verkerk AO, Amin AS, van Brunschot S, Beekman L, et al. Genetically determined differences in sodium current characteristics modulate conduction disease severity in mice with cardiac sodium channelopathy. Circ Res 2009;104(11):1283–92, [DOI] [PubMed] [Google Scholar]
- [102].Maier SK, Westenbroek RE, McCormick KA, Curtis R, Scheuer T, Catter- all WA. Distinct subcellular localization of different sodium channel alpha and beta subunits in single ventricular myocytes from mouse heart. Circulation 2004;109(11):1421–7. [DOI] [PubMed] [Google Scholar]
- [103].Splawski I, Timothy KW, Tateyama M, Clancy CE, Malhotra A, Beggs AH, et al. Variant of SCN5A sodium channel implicated in risk of cardiac arrhythmia. Science 2002;297(5585):1333–6. [DOI] [PubMed] [Google Scholar]
- [104].Plant LD, Bowers PN, Liu Q, Morgan T, Zhang T, State MW, et al. A common cardiac sodium channel variant associated with sudden infant death in African Americans, SCN5A S1103Y. J Clin Invest 2006;116(2):430–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Kaab S, Crawford DC, Sinner MF, Behr ER, Kannankeril PJ, Wilde AA, et al. A large candidate gene survey identifies the KCNE1 D85N polymorphism as a possible modulator of drug-induced torsades de pointes. Circ Cardiovasc Genet 2012;5(1):91–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Nishio Y, Makiyama T, Itoh H, Sakaguchi T, Ohno S, Gong YZ, et al. D85N, a KCNE1 polymorphism, is a disease-causing gene variant in long QT syndrome. J Am Coll Cardiol 2009;54(9):812–19. [DOI] [PubMed] [Google Scholar]
- [107].Westenskow P, Splawski I, Timothy KW, Keating MT, Sanguinetti MC, Compound mutations: a common cause of severe long-QT syndrome. Circulation 2004;109(15):1834–41. [DOI] [PubMed] [Google Scholar]
- [108].Nof E, Barajas-Martinez H, Eldar M, Urrutia J, Caceres G, Rosenfeld G, ; , et al. LQT5 masquerading as LQT2: a dominant negative effect of KCNE1-D85N rare polymorphism on KCNH2 current. Europace 2011;13(10):1478–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.