Abstract
Genome rearrangements resulting in copy number variation (CNV) and loss of heterozygosity (LOH) are frequently observed during the somatic evolution of cancer and promote rapid adaptation of fungi to novel environments. In the human fungal pathogen Candida albicans, CNV and LOH confer increased virulence and antifungal drug resistance, yet the mechanisms driving these rearrangements are not completely understood. Here, we unveil an extensive array of long repeat sequences (65–6499 bp) that are associated with CNV, LOH, and chromosomal inversions. Many of these long repeat sequences are uncharacterized and encompass one or more coding sequences that are actively transcribed. Repeats associated with genome rearrangements are predominantly inverted and separated by up to ~1.6 Mb, an extraordinary distance for homology-based DNA repair/recombination in yeast. These repeat sequences are a significant source of genome plasticity across diverse strain backgrounds including clinical, environmental, and experimentally evolved isolates, and represent previously uncharacterized variation in the reference genome.
Research organism: Other
Introduction
Genome plasticity is surprisingly common in eukaryotes. DNA insertions and deletions (indels), copy number variations (CNV), and loss of heterozygosity (LOH) are frequently described during the evolution of organisms and of disease states such as cancer. In particular, the genome plasticity of fungal pathogens was recognized well before whole genome sequencing was available, including genome copy number variation (polyploidy), inter- and intra-chromosomal rearrangements, and aneuploidy (Chibana et al., 2000; Magee and Magee, 2000; Rustchenko-Bulgac, 1991; Suzuki et al., 1982). Controlled in vitro and in vivo evolution experiments in combination with whole genome sequencing have further highlighted the speed in which specific genome rearrangements provide a fitness advantage that can be selected for in these fungal pathogens (Araya et al., 2010; Croll et al., 2013; Dunham et al., 2002; Forche et al., 2011; Ford et al., 2015; Gerstein et al., 2015; Hirakawa et al., 2015; Selmecki et al., 2009; Stukenbrock et al., 2010).
Candida albicans is the most prevalent human fungal pathogen, associated with nearly half a million life-threatening infections annually, predominantly in immunocompromised individuals (Brown and Netea, 2012). C. albicans is a heterozygous diploid yeast capable of mating, yet true meiosis has not been observed. Instead, it undergoes a parasexual process that involves random chromosome loss and rare Spo11-dependent chromosome recombination events (Bennett and Johnson, 2003; Forche et al., 2008; Wang et al., 2018).
The majority of genomic diversity observed in C. albicans is attributed to asexual mitotic genome rearrangements (Forche et al., 2011; Lephart and Magee, 2006). Despite this clonal lifestyle, C. albicans isolates exhibit extensive genomic diversity in the form of de novo base substitutions, indels, ploidy variation (haploid, diploid, and polyploid), karyotypic variation due to segmental and whole chromosome aneuploidies, and allele copy number variation including LOH (Chibana et al., 2000; Forche et al., 2011; Ford et al., 2015; Hickman et al., 2013; Hirakawa et al., 2015; Magee and Magee, 2000; Rustchenko-Bulgac, 1991; Selmecki et al., 2006; Suzuki et al., 1982). Additionally, while C. albicans did not undergo an ancient whole genome duplication event like Saccharomyces cerevisiae (Butler et al., 2009; Marcet-Houben et al., 2009; Wolfe and Shields, 1997), small-scale duplication events have resulted in gene family expansions, especially in sub-telomeric regions (Anderson et al., 2012; Butler et al., 2009; Dunn et al., 2018). A comprehensive analysis of these duplication events, their evolutionary trajectories and impact on genome stability, remains largely unexplored.
Early comparative studies of the C. albicans genome identified diverse repetitive loci that contribute to genotypic and phenotypic plasticity (Braun et al., 2005; Jones et al., 2004). First, repeat analysis in C. albicans has characterized at least three major classes of long repetitive sequences: the 23 bp tandem telomeric repeat units and the 14 member telomere-associated (TLO) gene family residing in sub-telomeric regions; the Major Repeat Sequences (MRS) found, at least in part, on every C. albicans chromosome and formed by a long tandem array of ~2.1 kb RPS units flanking non-repetitive HOK and RBP-2 elements (Chibana et al., 1994; Chindamporn et al., 1998; Lephart and Magee, 2006); and the ribosomal DNA repeats (rDNA) found on ChrR, which are organized as a tandem array of up to ~200 copies of ~12 kb units (Freire-Benéitez et al., 2016; Jones et al., 2004; Rustchenko et al., 1993; Wickes et al., 1991). These long repetitive sequences can undergo both inter- and intra-locus recombination events that rapidly generate chromosome length polymorphisms, chimeric chromosomes, and telomere-telomere chromosomal fusions (Chu et al., 1992; Selmecki et al., 2006; Selmecki et al., 2010). Second, like most eukaryotes, C. albicans also encodes many ‘lone’ long terminal repeats (LTRs) and retroelements (Zorro, Tca2, Ty1/Copia) (Goodwin and Poulter, 1998; Goodwin and Poulter, 2000); however, the relative copy number of many of these genes is hypervariable between C. albicans isolates and is expanded relative to other Candida species (Butler et al., 2009; Hirakawa et al., 2015). Third, short repeat sequences (short tandem repeats and trinucleotide repeats) are significantly more frequent in protein-coding sequences of C. albicans than in S. cerevisiae and Schizosaccharomyces pombe (Braun et al., 2005; Jones et al., 2004). Fourth, expansions of multi-gene families (identified by protein alignment) were both more common and larger than the orthologous gene family size found in S. cerevisiae. These gene families often encode proteins with roles in commensalism and virulence, including the agglutinin-like sequence (ALS) family (eight genes) and other glycosylphosphatidylinositol (GPI)-anchored genes that encode large cell surface glycoproteins (five genes) (Levdansky et al., 2008; Wilkins et al., 2018). Among these gene families, recombination and/or slippage between repeat units yields extensive allelic variation, leading to functional and phenotypic diversity, similar to the FLO genes in S. cerevisiae (Hoyer et al., 1995; Kunkel, 1993; Pearson et al., 2005; Richard et al., 1999; Verstrepen et al., 2005; Zhang et al., 2003; Zhao et al., 2004). The evolution of different alleles in these repeat-containing ORFs predominantly occurs by the addition, deletion, and rearrangement of repeat units within an ORF and between different ORFs, not by the acquisition of point mutations or indels (Christiaens et al., 2012; Zhang et al., 2010). Importantly, these genomic studies focused on short repeat sequences and repeats found in protein-coding sequences. Less is known about long repeat sequences found throughout the genome, especially those encoding multiple ORFs and intergenic regions.
Over 19 years ago, Wolfe and colleagues showed that the C. albicans genome contains thousands of small chromosomal inversion events (~10 genes long) relative to S. cerevisiae. These inversions resulted in substantially different gene order between these two species (Seoighe et al., 2000). Similarly, Dujon and colleagues demonstrated that the C. albicans genome had the highest rate of genome instability due to micro- and macro-rearrangements of syntenic gene blocks, relative to 11 other hemiascomycete species (Fischer et al., 2006). The loss of synteny primarily resulted from chromosomal rearrangements, not sequence divergence of orthologous regions. A mechanism proposed for this genome instability was a higher incidence of repetitive sequences and/or a less efficient DNA repair process (Fischer et al., 2006).
The genomic diversity of C. albicans increases during in vitro and in vivo exposure to stress. For example, rates of LOH increase during exposure to elevated temperature (37°C), DNA transformation, and antifungal drugs (Bouchonville et al., 2009; Forche et al., 2011; Forche et al., 2018). LOH is also increased during in vivo models of infection (Ene et al., 2018; Forche et al., 2008; Forche et al., 2018). LOH events occur due to chromosome nondisjunction leading to whole chromosome LOH or via recombination, in which only part of the chromosome undergoes LOH. Exposure to stress also selects for isolates that have acquired adaptive mutations and genome rearrangements. For example, aneuploidy is found in ~50% of isolates resistant to the most common antifungal drug, fluconazole (FLC). The most common and only recurrent aneuploidy in different strain backgrounds is the amplification of the left arm of chromosome 5 (Chr5L), often through acquisition of a novel isochromosome structure (denoted as i(5L)), comprised of two copies of Chr5L separated by the centromere (Selmecki et al., 2006; Selmecki et al., 2008). Acquisition of i(5L) conferred FLC resistance via the amplification of two genes, ERG11 and TAC1, encoding the drug target (Erg11) and a transcriptional activator of drug efflux pumps (Tac1) (Selmecki et al., 2008; Selmecki et al., 2009). Importantly, the centromere of Chr5 contains a long inverted repeat sequence, and recombination between these repeats can form homozygous isochromosomes of both the left arm (i(5L)) and right arm of Chr5 (i(5R)) (Selmecki et al., 2006). The role of long repeat sequences in the formation of other segmental aneuploidies and other genome rearrangements has not been comprehensively addressed.
We provide evidence that long repeat sequences are involved in the formation of all observed CNV breakpoints and chromosome inversions, and many LOH breakpoints, across 33 diverse clinical and experimentally evolved isolates. Our comprehensive analysis of long repeat sequences within the C. albicans genome identified hundreds of sequences representing novel multicopy repeats, none of which include MRS, rDNA, sub-telomeric repeats, known repeat families (ALS, TLOs) or known repetitive elements (tRNAs, LTRs, retrotransposons). Long repeats that are associated with genome rearrangements (CNV, LOH, and inversions) have on average higher sequence identity than all long repeats combined. Additionally, long repeats that contain ORFs (including partial ORF sequences, single complete ORF sequences (paralogs), or multiple ORFs and intergenic sequences) are longer and associated with more genome rearrangements than long repeats that contain other genomic features (such as LTRs, retrotransposons, or tRNAs). Additionally, repeat copies involved in genome rearrangements can be located up to ~1.6 Mb apart on the same chromosome, suggesting a non-conventional, long-range mechanism for DNA double-strand break (DSB) repair and somatic genome diversification.
Results
An inverted repeat within CEN4 is associated with the formation of a novel isochromosome
To identify the mechanisms by which C. albicans isolates generate genome plasticity, we performed a comparative genomics analysis of 33 diverse clinical isolates (Supplementary file 1). This set of isolates included 11 that underwent controlled experimental evolution, where a known progenitor isolate was passaged in vitro or in vivo. Additionally, we performed comparative genomics on newly obtained clinical isolates, and clinical isolates whose genomes were published previously, including the reference isolate SC5314.
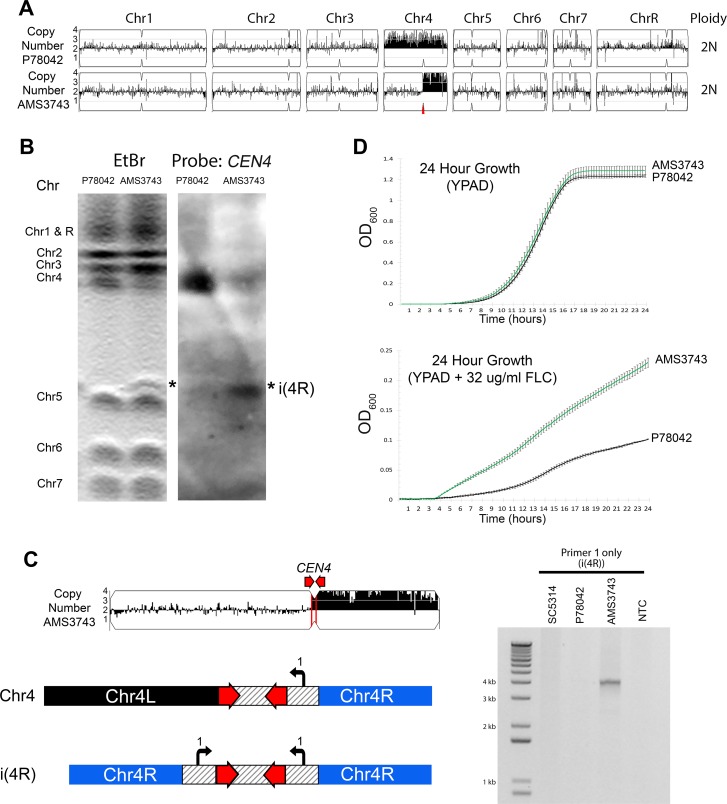
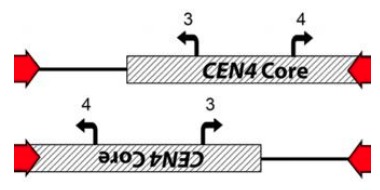
Given the significant impact of i(5L) on antifungal drug resistance, we focused first on the characterization of a novel segmental aneuploidy detected on Chr4 that arose during in vitro evolution in the presence of FLC. Initially, we passaged a FLC-sensitive clinical isolate P78042, which was trisomic for Chr4 (Hirakawa et al., 2015; Lockhart et al., 2002), in the presence of FLC (128 µg/ml) for 100 generations by serial dilution (see Materials and methods). One evolved isolate (AMS3743) was selected, based on increased fitness in FLC (see below), and the whole genome was sequenced. Read depth analysis indicated that this isolate had four copies of the right arm of Chr4 (Chr4R), but only two copies of Chr4L, and the copy number breakpoint occurred at the centromere of Chr4 (CEN4) (Figure 1A). Wildtype CEN4, like CEN5, is comprised of a CENP-A-binding core sequence (~3.1 kb) flanked by a long (524 bp) inverted repeat (Burrack et al., 2016; Ketel et al., 2009; Sanyal et al., 2004).
Figure 1. Inverted repeat at CEN4 causes a novel isochromosome leading to increased fluconazole resistance.
(A) Whole genome sequence data plotted as a log2 ratio and converted to chromosome copy number (Y-axis) and chromosome location (X-axis) using YMAP, for the progenitor clinical isolate (P78042) and an isolate obtained after 100 generations in FLC (AMS3743). The copy number breakpoint in AMS3743 occurs at CEN4 (red arrow). (B) CHEF karyotype gel stained with ethidium bromide (left panel) identifies a novel band (asterisk) above Chr5. Southern blot analysis (right panel) of the same gel using a DIG-labeled CEN4 probe identifies the full-length Chr4 homolog in P78042 and AMS3743, and the novel band in AMS3743 that is twice the size of the right arm of Chr4 in an isochromosome structure (asterisk, i(4R)). (C) PCR validation of i(4R). Schematic representation of the Chr4 homologue (top) and i(4R), where the location of a single primer sequence (Primer 1, Supplementary file 7) that flanks the CEN4 inverted repeat is indicated. PCR with Primer 1 amplified the expected product of i(4R) in AMS3743. (D) 24 hr growth curves in YPAD (top panel) and YPAD +32 µg/ml FLC (bottom panel) for P78042 (black line) and AMS3743 (green line). Average slope and standard error of the mean for three biological replicates is indicated. The average maximum slope (n = 3) of P78042 and AMS3743 in YPAD was not significantly different (0.046 and 0.046, respectively, p>0.75, t-test). The average maximum slope (n = 3) of P78042 and AMS3743 was significantly different in FLC (0.002 and 0.003, respectively, p<0.0006, t-test). OD, optical density (Figure 1—source data 1).
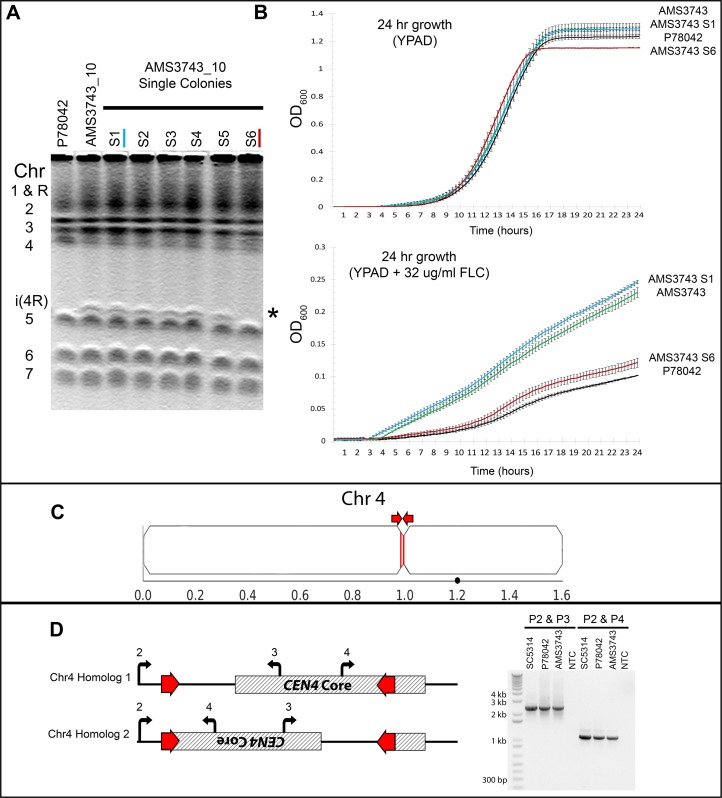
Figure 1—figure supplement 1. Long inverted repeats on Chr4 are associated with a centromere inversion and an isochromosome that confers increased fitness in FLC.
Figure 1—figure supplement 2. Sanger sequencing of CEN4 in SC5314.
To test the hypothesis that this segmental aneuploidy is an isochromosome structure, we performed CHEF karyotype analysis. Isolate AMS3743 had a novel ~1.2 Mb chromosome band that hybridized to a CEN4 probe via Southern blot (Figure 1B). This ~1.2 Mb band was twice the size of the right arm of Chr4 (~607 Kb). Consistent with an isochromosome i(4R) structure (a centromere flanked by inverted copies of Chr4R), a single primer amplified a ~4.1 kb product, from Chr4R through CEN4 and back to Chr4R in the isolate with i(4R) but did not amplify any sequence in the reference (SC5314), or progenitor (P78042) isolates (Figure 1C).
Next, we determined the impact of i(4R) on fitness in the presence and absence of FLC over a 24 hr period. In the presence of FLC, the i(4R) isolate grew significantly better than the progenitor P78042 (p<0.0006, t-test, Figure 1D). Interestingly, in the absence of FLC, the i(4R) isolate grew as well as the progenitor P78042 (Figure 1D). Furthermore, i(4R) was maintained in 12/12 populations for over ~300 generations in the absence of FLC (see Materials and methods). One of the populations, AMS3743_10, appeared to be losing i(4R) as measured by CHEF gel densitometry (see Materials and methods) and was plated for single colonies in the absence of FLC. One colony (out of six) had lost i(4R) (AMS3743_10_S6, Figure 1—figure supplement 1A). To ask if i(4R) was necessary and sufficient for the increased fitness in FLC, fitness was determined in the presence and absence of FLC. The colony that had lost i(4R) had a reduced growth rate in the presence of FLC, similar to the progenitor P78042 (Figure 1—figure supplement 1B).
Overall, these data imply that the long inverted repeat within CEN4 can generate an independent isochromosome structure comprised of two right arms of Chr4, and that i(4R) is necessary and sufficient for increased fitness in FLC. These results parallel the identification of isochromosomes associated with the long inverted repeat sequence within CEN5, which can result in the formation of i(5R) and i(5L), the latter of which confers FLC resistance (Selmecki et al., 2006; Selmecki et al., 2008).
Inverted repeat sequences are associated with inversion of centromere sequences
During our investigation of the i(4R) structure, we unveiled a surprising feature of CEN4: the CENP-A-binding core sequence of CEN4 contained two different alleles. One homologue of Chr4 contained a ~3.1 kb sequence inversion between the inverted repeat associated with CEN4. The new, inverted CEN4 sequence was detected by PCR in the reference isolate SC5314, and in the distantly related isolates P78042 and AMS3743 (Figure 1—figure supplement 1C & D). Sanger sequencing indicated that a recombination event occurred between the two arms of the inverted repeat (Figure 1—figure supplement 2). Interestingly, the CENP-A-binding core sequence of CEN4 is asymmetrically positioned on one side of the inverted repeat sequence (Figure 1—figure supplement 1D, shaded region) (Burrack et al., 2016; Sanyal et al., 2004). Therefore, this inversion caused a separation between the known CENP-A-binding core sequence of CEN4 that is located to the right and outside of the inverted repeat.
Identification of long repeat sequences throughout the C. albicans genome
Given the extensive genome rearrangements observed at the long inverted repeat associated with CEN4, we sought to characterize all long repeat sequences within the C. albicans reference genome (SC5314). All long sequence matches within SC5314 were identified by aligning the reference genome sequence to itself using the bioinformatics suite MUMmer (Kurtz et al., 2004). First, all exact sequence matches of 20 nucleotides or longer were identified, then all matches were clustered and extended to obtain a maximum-length colinear string of matches, resulting in a final list of long repeat sequences that ranged from 65 bp to 6499 bp (median 318 bp) with sequence identities of ≥80% (See Materials and methods). The genomic position and percent identity of all matched repeats was determined with MUMmer and manually verified using BLASTN and IGV (Robinson et al., 2011; Thorvaldsdóttir et al., 2013). After excluding all rDNA, MRS and sub-telomeric repeat sequences, 1974 long repeat matches were identified (Supplementary file 2). The MUMmer analysis identified five ORFs and one gene family with known, complex embedded tandem repeat sequences (PGA18, PGA55, EAP1, orf19.1725, CSA1, and the ALS gene family, herein referred to as ‘the complex tandem repeat genes’). The complexity of these repeat sequences prohibited the assignment of exact repeat copy number per genome, and they were removed from analyses when indicated. The remaining long repeat sequences cover 2.87% of the haploid reference genome (see Materials and methods).
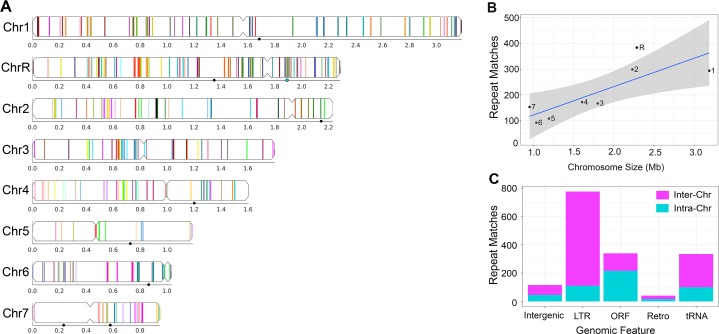
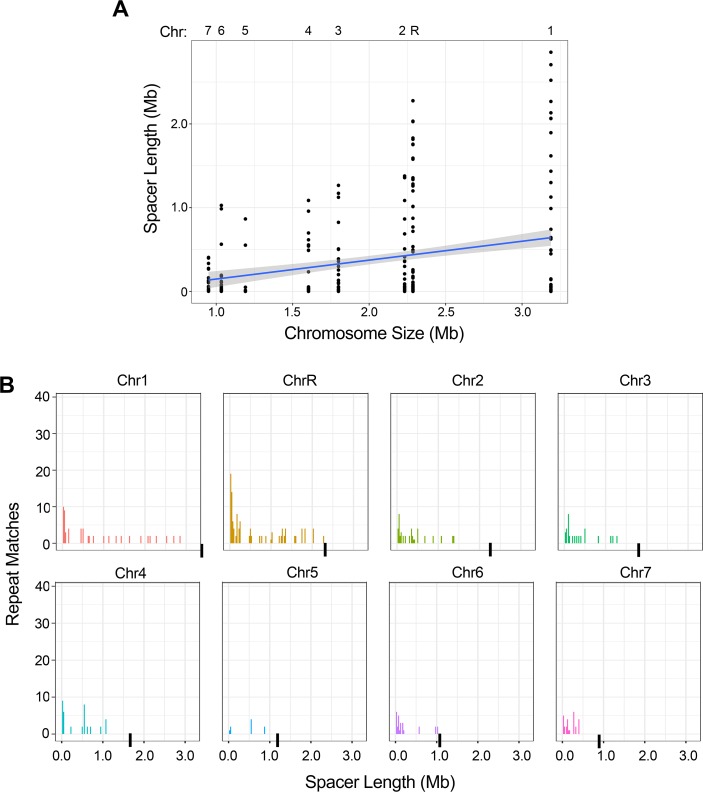
Long repeat matches occurred between sequences on the same chromosome (intra-chromosomal repeats, Figure 2A), on different chromosomes (inter-chromosomal repeats), or both. The number of all repeat matches per chromosome was correlated with chromosome size (R2 = 0.65, p<0.016, Figure 2B); however, regions of high repeat density (e.g. ChrRR near the rDNA) or low repeat density (e.g. Chr7L) were detected on some chromosome arms. This repeat density did not correlate with GC content (R2 = 0.063, p>0.32) or ORF density (R2 = 0.02, p>0.59) on any chromosome arm (Figure 2—source data 1).
Figure 2. Long repeat sequences are found across the C. albicans genome.
Detailed results for all long intra- and inter-chromosomal repeat positions, orientations, and gene features are found in Supplementary file 2. Repeats associated with the rDNA, major repeat sequences (MRS), and sub-telomeric repeats were removed prior to the analysis. (A) Representative image of the long intra-chromosomal repeat positions (colored lines – not to scale). Each repeat family is assigned a unique color within its respective chromosome. Numbers and symbols below each chromosome indicate chromosomal position (Mb), MRS position (black circles), and rDNA locus (blue circle, ChrR). (B) Number of all repeat matches (excluding the complex tandem repeat genes) on each chromosome, ordered by chromosome size (R2 = 0.65, p<0.016, gray indicates 95% confidence interval, Figure 2—source data 1). (C) The number of intra-chromosomal (Intra-Chr) and inter-chromosomal (Inter-Chr) repeat matches assigned to each genomic feature: Intergenic, LTR, ORF (excluding the complex tandem repeat genes), retrotransposon (Retro), and tRNA (Figure 2—source data 1).
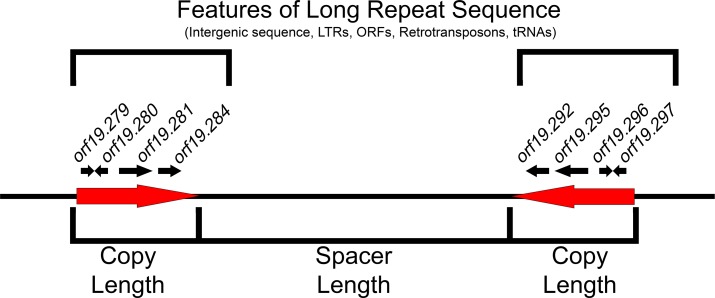
Figure 2—figure supplement 1. Features of long repeat sequences.
Figure 2—figure supplement 2. The intra-chromosomal repeats with the longest spacer length are found on the longer chromosomes.
Figure 2—figure supplement 3. Key features of long repeat sequences in C. albicans.
We next calculated the orientation and distance between matched intra-chromosomal repeat sequences (Figure 2—figure supplement 1), both important factors for reconstructing the evolutionary history of these duplication events and for analyzing the frequency and outcome of homologous recombination events that occur between repeat sequences (Lobachev et al., 1998; Ramakrishnan et al., 2018). Intra-chromosomal repeats are often generated in tandem by recombination between sister chromatids or replication slippage, and these repeats can move further away from each other by chromosomal rearrangement events (including chromosomal inversions) (Achaz et al., 2000; Reams and Roth, 2015). Indeed, intra-chromosomal repeats were predominantly tandem, although inverted and mirrored repeats also occurred (Supplementary file 2). We hypothesized that the distance between matched intra-chromosomal repeats (spacer length) would be predominantly short and that the distribution of spacer lengths on each chromosome would be similar. Strikingly, spacer length ranged from 1 bp to 2,856,212 bp (median ~82.8 kb, excluding the complex tandem repeat genes, see Materials and methods), and was correlated with chromosome size (Figure 2—figure supplement 2A, R2 = 0.066, p<0.0001). Additionally, the distribution of spacer lengths was significantly different between chromosomes (Figure 2—figure supplement 2B, p<0.035, Kruskal-Wallis with Dunn’s multiple comparison test) with the larger chromosomes (Chr1 and ChrR) containing many repeat matches that were separated by distances greater than ~1.5 Mb. The increased distance between repeat sequences likely occurred via additional large inversions, insertions or telomere-telomere recombination/fusion events.
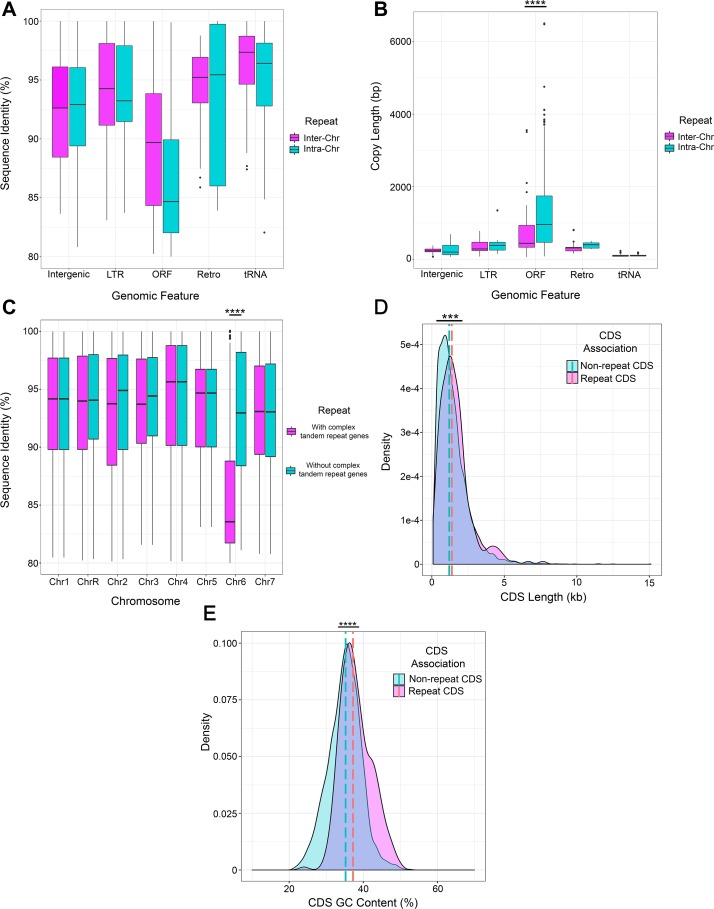
We further annotated the long repeat sequences according to the genomic features contained within each repeat (see Materials and methods). The most common long repeats contained lone long terminal repeats (LTRs) (775), followed by ORFs (339, excluding the complex tandem repeat genes), tRNAs (334), and retrotransposons (40). Repeat matches containing ORFs included partial ORF sequences (196/339, 57.8%), single complete ORF sequences (114/339, 33.6%), and multiple ORFs and intergenic sequences (29/339, 8.6%) (Supplementary file 2). Repeat matches containing complete ORFs and multiple ORFs represent paralogs and multi-gene duplication events. Additionally, there were 349 intergenic, unannotated sequences, 231 that shared high-sequence identity (>83%) with an annotated sequence found elsewhere in the genome, including known LTRs, retrotransposons, and ORFs (Supplementary file 2, ‘Unannotated Intergenic Sequence’). For example, an additional 54 LTRs were identified in the reference genome with this analysis. Interestingly, LTR matched repeat pairs were predominantly dispersed on different chromosomes (78%), while ORF matched repeat pairs were predominantly located on a single chromosome (64%, Figure 2C).
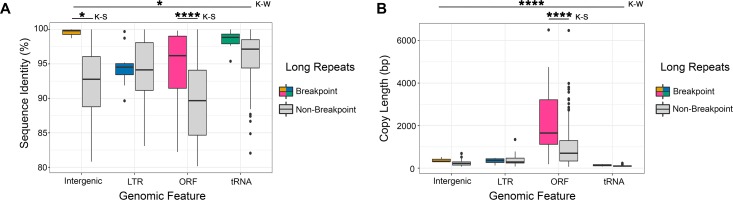
Of the matched repeat pairs, the long repeat sequences containing ORFs had the lowest median sequence identity when compared to repeats containing other features (Figure 2—figure supplement 3A, p<0.0001, Kruskal-Wallis with Dunn’s multiple comparison test). Conversely, repeats containing ORFs had significantly longer copy length than any other genomic feature (p<0.0001, Kruskal-Wallis with Dunn’s multiple comparison test) and was the only feature that had a significant increase in copy length of intra-chromosomal matches relative to inter-chromosomal matches (Figure 2—figure supplement 3B, p<0.0001, Kruskal-Wallis with Dunn’s multiple comparison test). The long repeat sequences containing ORFs were predominantly present in only two copies per genome, had pairwise coding sequences with similarly high identity, and therefore represent paralogous gene duplication events (Supplementary file 2). The origin, function, and evolutionary trajectory of these paralogs may provide insight into the evolution of fungal pathogens like C. albicans that did not undergo the ancient whole genome duplication event (Butler et al., 2009; Marcet-Houben et al., 2009; Wolfe and Shields, 1997).
The complex tandem repeat genes, for which genome copy number could not be determined, had low sequence identity and were predominantly found on Chr6 (Figure 2—figure supplement 3C). In contrast, the full-length coding sequence of all ORFs that were contained within long repeat sequences, were significantly longer (median value of 1380 bp vs 1200 bp, Figure 2—figure supplement 3D, p<0.0008, Kolmogorov-Smirnov test) and had a significantly higher GC content (median value of 37.22% vs 35.22% Figure 2—figure supplement 3E, p<0.0001, Kolmogorov-Smirnov test) than the full-length coding sequence of all ORFs not contained within long repeat sequences (genome-wide, excluding the complex tandem repeat genes, see Materials and methods). Interestingly, increased GC content was correlated with increased rates of both mitotic and meiotic recombination events in S. cerevisiae (Kiktev et al., 2018).
Identification of CNV breakpoints in isolates with segmental aneuploidies
Next, CNV breakpoints were determined across 13 additional isolates with one or more segmental aneuploidies. Six of these isolates were from in vitro evolution experiments in the presence of azole antifungal drugs (FLC or miconazole), four were from in vivo evolution experiments in a murine model of oropharyngeal candidiasis (OPC) performed in the absence of antifungal drugs, and three were human clinical isolates (Supplementary file 1). All segmental aneuploidies arose from a known euploid diploid progenitor (Abbey et al., 2014; Hirakawa et al., 2015), except two clinical isolates with unknown origin and the i(4R) isolate that arose from a trisomic progenitor, described above.
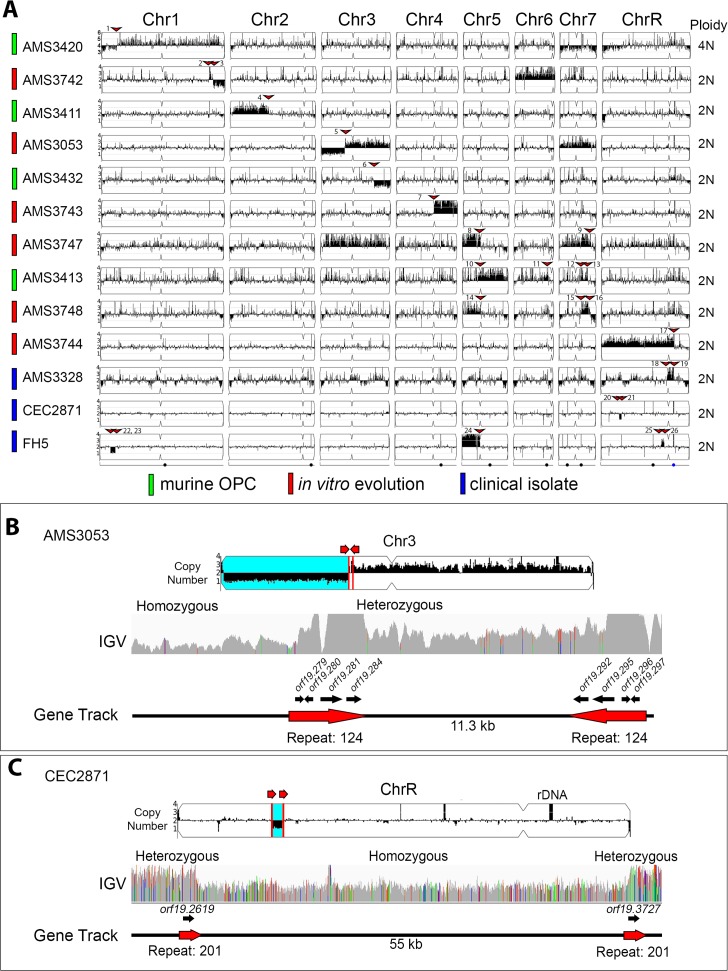
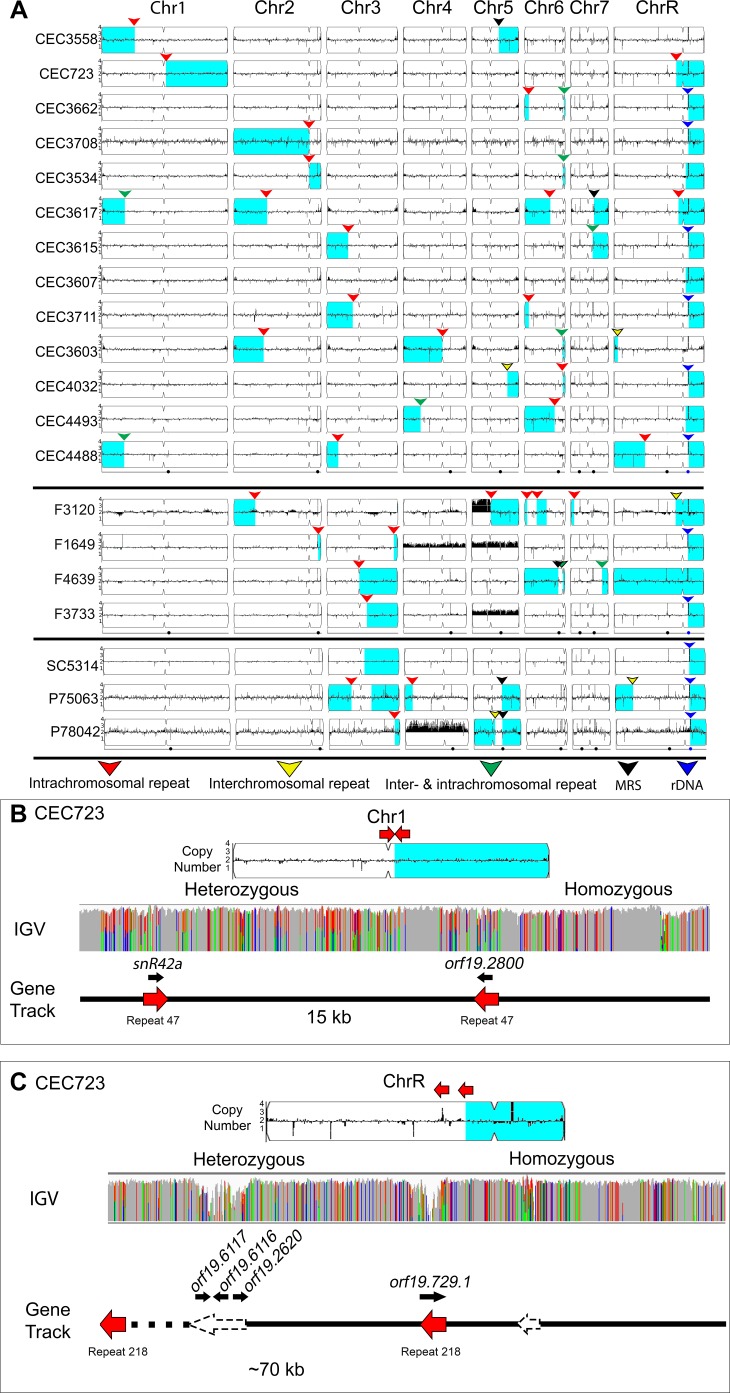
Segmental aneuploidies were initially detected by CHEF karyotype analysis and ddRAD-seq, but the coordinates of the CNV breakpoints were not known (Abbey et al., 2014; Forche et al., 2018; Mount et al., 2018; Ropars et al., 2018). The ploidy of each isolate was measured by flow cytometry and the DNA copy number of all loci was determined using whole genome sequencing (see Materials and methods). Among the 13 diverse isolates, 19 segmental aneuploidies were confirmed, with at least one segmental aneuploidy detected on each of the eight chromosomes (Figure 3A, Figure 3—figure supplement 1A–J). Segmental amplifications were more frequent (12/19, 63.2%) than segmental deletions (3/19, 15.8%). The remaining segmental aneuploidies (4/19, 21.1%) consisted of more complex rearrangements that resulted in a segmental amplification and a terminal chromosome deletion at the same breakpoint.
Figure 3. All copy number breakpoints resulting in segmental aneuploidy occur at repeat sequences.
(A) Whole genome sequence data plotted as a log2 ratio and converted to chromosome copy number (Y-axis) and chromosome location (X-axis) using YMAP. The source of each isolate is indicated in color: in vivo evolution experiments in a murine model of oropharyngeal candidiasis (OPC) (green), in vitro evolution experiments in the presence of azole antifungal drugs (red), and clinical isolates (blue). Ploidy, determined by flow cytometry, is indicated on the far right. Every copy number breakpoint occurred at a repeat sequence (red arrow), additional details are in Supplementary file 3. Location of the Major Repeat Sequences (black circle) and rDNA array (blue circle) are shown below. Example copy number breakpoints for two isolates (B–C). (B) Isolate AMS3053 underwent a complex rearrangement on Chr3L at a long inverted repeat (Repeat 124, red lines). Read depth (top panel) and allele frequency (IGV panel) data indicate the copy number breakpoint coincided with LOH (blue region) telomere proximal to the breakpoint. The inverted repeat copies (~3.2 kb, 99.5% sequence identity, separated by ~11.3 kb) each contain four complete ORFs and intergenic sequences. (C) Read depth (top panel) and allele frequency (IGV panel) data for isolate CEC2871 shows an internal chromosome deletion on ChrR with copy number breakpoints (red lines) and LOH (blue) that occur between a long tandem repeat (Repeat family 201, red arrows). The tandem repeat copies (~1.4 kb, 93.8% sequence identity, separated by ~55 kb) each contain one ORF.
Figure 3—figure supplement 1. Segmental aneuploidies occur at previously characterized and uncharacterized long repeat sequences.
All segmental aneuploidies occur at long repeat sequences
The CNV breakpoint of each segmental aneuploidy was determined using both read depth and allele ratio analysis (see Materials and methods). From the 19 segmental aneuploidies, 26 CNV breakpoints were identified because some segmental aneuploidies contained multiple breakpoints. Strikingly, every CNV breakpoint occurred within 2 kb of a long repeat sequence, ranging from 248 bp to ~4.76 kb in length. Observed breakpoints had significantly more overlap with long repeat sequences than expected given the total genome coverage of long repeat sequences (p<0.0001, two-tailed Fishers Exact Test, see Materials and methods). All but one of the repeat sequences were intra-chromosomal and separated by a distance ranging from ~3.1 kb to ~1.62 Mb (Supplementary file 3). Importantly, repeats containing ORFs were significantly more common than all other types of repeats at these breakpoints (18/26 CNV breakpoints, p<0.001, χ2 Goodness-of-fit test).
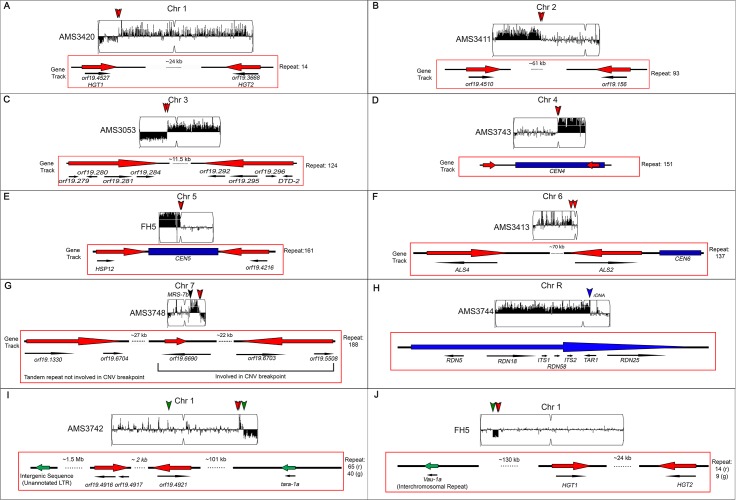
Three examples of CNV breakpoints in long repeats containing ORFs were observed in isolates AMS3053, AMS3420 and CEC2871. In both AMS3053 and AMS3420, a long inverted repeat sequence was associated with a complex segmental amplification and a terminal chromosome deletion that resulted in a long-range homozygosis event. In AMS3053, the breakpoint on Chr3L occurred within a ~1.7 kb inverted repeat sequence (>99% identity) separated by ~11.5 kb (Figure 3B). The left side of this inverted repeat contained four uncharacterized ORFs (orf19.279, orf19.280, orf19.281, orf19.284) and associated intergenic sequences, while the right side contained three uncharacterized ORFs (orf19.296, orf19.295, orf19.292) and one characterized ORF (orf19.297 DTD2) plus associated intergenic sequences. Similarly, the OPC-derived isolate AMS3420 underwent a complex segmental amplification and deletion within a ~1.6 kb inverted repeat sequence on Chr1L (91.5% identity) separated by ~26 kb, which contains the high-affinity glucose transporters HGT1 and HGT2 (Figure 3—figure supplement 1A). Long internal chromosome deletions were also observed. For example, in isolate CEC2871, a ~55 kb deletion resulted from recombination between a ~1.4 kb tandem repeat on ChrR (92.4% identity) containing ORFs of the PHO gene family (PHO112 and PHO113, Figure 3C). Proposed models for recombination events that would result in these complex segmental amplifications and deletions are described in the discussion.
Eight CNV breakpoints occurred within other long repeat sequences, including: a ~200 bp microsatellite repeat (1/26), intergenic repeats (1/26), MRS (2/26), LTRs (2/26), and the rDNA repeats (2/26) (Figure 3A, Supplementary file 3). Some segmental aneuploidies were comprised of multiple breakpoints, each associated with a different repeat family (e.g. Figure 3—figure supplement 1I & J). Interestingly, both breakpoints that occurred at the rDNA also amplified the ChrR centromere (CENR), and everything either to the telomere of the opposite chromosome arm (ChrRL) (Figure 3—figure supplement 1H), or to a microsatellite repeat sequence on ChrRL (AMS3328, Figure 3A).
In summary, all CNV breakpoints in this collection occurred at or within long repeat sequences. Inverted repeat sequences predominantly coincided with segmental amplifications and terminal chromosome deletions, while tandem repeat sequences coincided with internal chromosome deletions. Some aneuploidies were comprised of multiple breakpoints, each associated with a different repeat family. Overall, a repeat homology-associated repair mechanism appears to be driving the formation of segmental aneuploidies. Importantly, the involvement of long repeats in CNV breakpoints is independent of genetic background and environmental selection.
LOH occurs at long inter- and intra-chromosomal repeat sequences
In many of the isolates with segmental aneuploidies, the CNV also was accompanied by LOH (e.g. Figure 3B and C). To ask if long repeat sequences were associated with LOH breakpoints in the absence of detectable CNVs, we selected 20 near-euploid genomes that had at least one long-range homozygous region, but the coordinates of the LOH breakpoint were not known (Ford et al., 2015; Hirakawa et al., 2015; Ropars et al., 2018). These 20 isolates belong to nine major C. albicans clades from different origins (e.g. superficial and invasive human infections, healthy human hosts, and spoiled food) (Figure 4A, Supplementary file 1).
Figure 4. Many LOH breakpoints occur at long intra- and inter-chromosomal repeat sequences.
Whole genome sequence data plotted as a log2 ratio and converted to chromosome copy number (Y-axis) and chromosome location (X-axis) using YMAP. (A) All long-range homozygous regions (light blue) that are associated with long repeat sequences (colored arrows) are indicated for 20 diverse C. albicans isolates. LOH breakpoints and isolate information are detailed in Supplementary files 1 and 4. The type of long repeat is indicated with colored arrows: intra-chromosomal (red), inter-chromosomal (yellow), both intra- and inter-chromosomal (green), rDNA repeat (blue), and MRS (black). (B–C) Two example LOH breakpoints in isolate CEC723 that occur at long repeats (red arrows) on (B) Chr1 (repeat copy length ~1.1 kb), and (C) ChrR (repeat copy length ~3.3 kb) and continue to the right telomere of the respective chromosomes. Heterozygous and homozygous allele ratios are indicated in the IGV track. The position, orientation, and spacer length of the long repeat sequence is indicated in the gene track. ORFs (black arrows) contained within the long repeat sequences are indicated above the gene track. The LOH breakpoint on ChrR is within a repeat-dense region; additional long repeats in the region are indicated (dashed arrows).
Figure 4—figure supplement 1. Long-track homozygosis occurs on Chr3L at telomere seed sequences.
153 LOH breakpoints were identified in the 20 isolates (See Materials and methods, Supplementary file 4). 61/153 LOH breakpoints were found within 2 kb of a long repeat sequence, and, like the CNV breakpoints, these LOH breakpoints could occur on any chromosome (Figure 4A). The copy length of the repeat sequences found at LOH breakpoints ranged from 78 bp to 6499 bp (median 516 bp) with sequence identities ranging from 82.2% to 100% (median of 95.1%). Most of the repeats associated with LOH breakpoints were intra-chromosomal (46/61), in all three orientations (inverted, mirrored, and tandem), and separated by a distance ranging from 903 bp to ~1.6 Mb (median ~35.3 kb). The vast majority of long-range homozygous regions contained only one LOH breakpoint and proceeded from the breakpoint to the proximal telomere, similar to previous analyses (Ene et al., 2018; Forche et al., 2008; Forche et al., 2009; Selmecki et al., 2005). Surprisingly, four isolates had an LOH breakpoint that proceeded from one chromosome arm to the telomere on the opposite chromosome arm, causing centromere homozygosis (three events on ChrR and one event on Chr5).
One isolate, CEC723, had two long-range homozygous regions associated with intra-chromosomal repeat sequences. The first LOH breakpoint on Chr1R was associated with a ~1.1 kb mirrored repeat sequence (>99% identity) separated by ~15 kb (Figure 4B). One copy of the repeat sequence contained a snoRNA (snR42a) and the other contained an uncharacterized ORF (orf19.2800), which we predict also encodes a second copy of snR42a. The second LOH breakpoint on ChrRL was associated with a ~3.2 kb tandem repeat sequence (97.7% identity) separated by ~70 kb (Figure 4C). This breakpoint was flanked by additional long repeat sequences that were associated with CNV in other isolates, indicating that this region is a hotspot for genome rearrangements (Supplementary file 2).
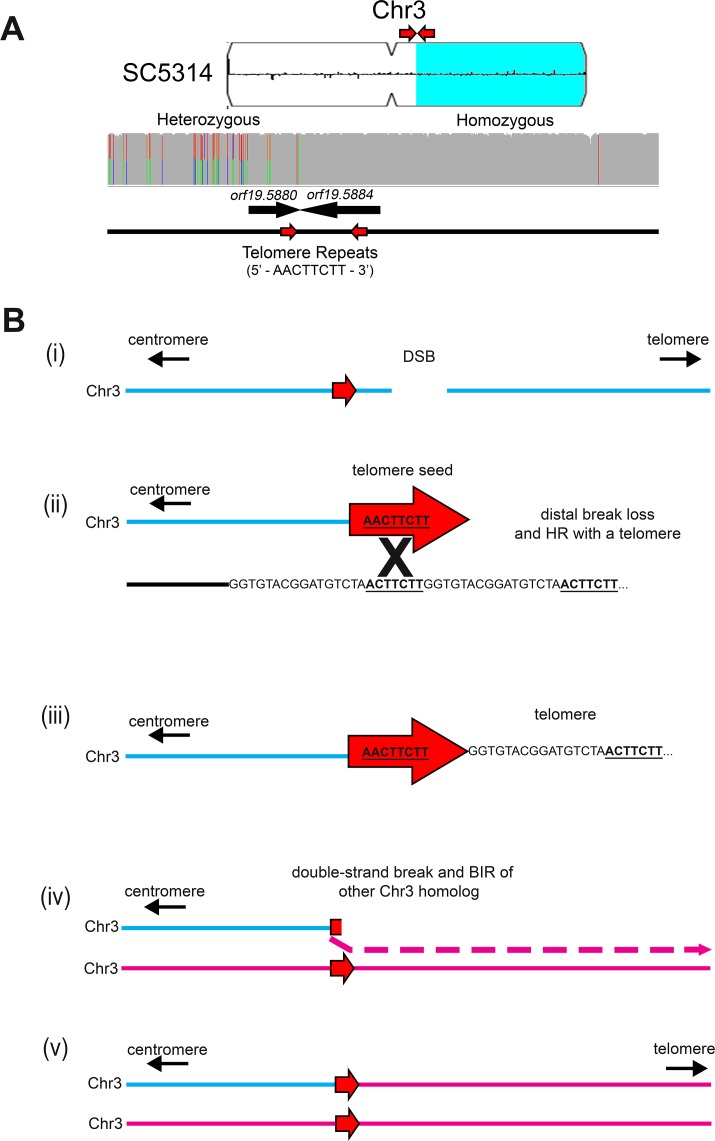
Finally, the reference isolate SC5314 contains a well-known long-range homozygous region on Chr3R. We asked if this LOH breakpoint occurred within a long repeat sequence. Remarkably, the LOH breakpoint occurred in orf19.5880 near an 8 bp sequence (AACTTCTT) identical to part of the C. albicans 23 bp telomere repeat sequence (GGTGTACGGATTGTCTAACTTCTT). Furthermore, a second copy of this same 8 bp sequence was found in an inverted orientation ~3.4 kb away in the adjacent ORF (orf19.5884). This long-range LOH event continued to the right telomere of Chr3. While LOH may have resulted from a repair template on the other homolog, an alternative model cannot be ruled out. We previously found that an LOH and CNV breakpoint that caused a segmental Chr5 truncation in the common laboratory strain BWP17 (Selmecki et al., 2005) was initiated at a 9 bp sequence (CTAACTTCT) that is almost identical to the sequence found at this breakpoint (AACTTCTT). We posit that a similar chromosome truncation, followed by reduplication of the monosomic portion of Chr3 (Figure 4—figure supplement 1A & B) may have generated the homozygosis of Chr3. These 8 bp and 9 bp telomere-like sequences occur 2160 and 249 times, respectively, within the non-telomeric portions of the C. albicans reference genome (Supplementary file 5). The presence of such a large number of potential template sequences, especially if including the telomere repeats at each chromosome end, might have driven this two-step model.
Repeat sequences cause sequence inversions and heterozygous islands
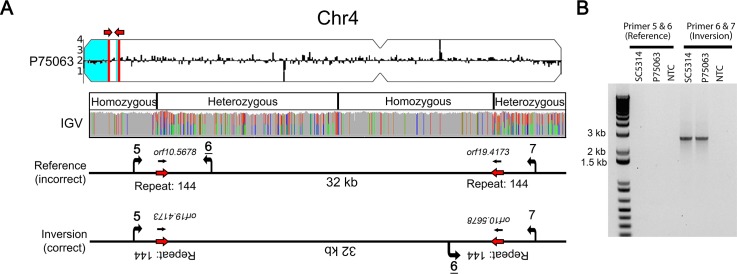
As expected, levels of heterozygosity were high within long repeat sequences due to the ability of short-read (Illumina) sequences to map to multiple positions in the genome (e.g. the heterozygous bases within repeat sequences in Figure 4B and C). Unexpectedly, between or adjacent to some long repeat sequences, heterozygous islands were observed in otherwise homozygous regions of the genome. For example, in isolate P75063, an LOH breakpoint on Chr4L was associated with a ~1.7 kb inverted repeat and resulted in a terminal homozygosis of the chromosome (Figure 5A). Adjacent to this homozygous region was an ~32 kb region that had multiple homozygous/heterozygous transitions (5’ homozygous-heterozygous-homozygous-heterozygous 3’). We hypothesized that a long sequence inversion, similar to that observed within the repeats flanking CEN4, accounted for the multiple heterozygous to homozygous transitions in this region. PCR amplification between unique sequences flanking the inverted repeat revealed a ~32 kb inversion in P75063 and SC5314 and was the only orientation that amplified by PCR; the reference orientation did not amplify, suggesting that the reference genome may be incorrect at this position (Figure 5B).
Figure 5. Long repeat sequences are associated with chromosomal inversions.
(A) Whole genome sequence read depth plotted as a log2 ratio and converted to chromosome copy number (Y-axis) and chromosome location (X-axis) using YMAP. Long-range homozygous regions (blue) on Chr4 are indicated for the isolate P75063. IGV allele ratio track indicates multiple homozygous to heterozygous transitions between a long inverted repeat (red arrows, repeat 144, copy length ~1.7 kb). Primers (5, 6, and 7, Supplementary file 7) were designed to unique sequences flanking repeat 144. (B) PCR amplification between Primers 6 and 7 identifies a ~32 kb chromosomal inversion in both the reference isolate SC5314 and P75063; the reference orientation did not amplify (Primers 5 and 6).
These two long inversions (at CEN4 and Chr4L), plus an additional seven potential sequence inversions were identified bioinformatically from a set of 21 clinical isolates (Hirakawa et al., 2015); however, none of these inversion breakpoints were characterized or validated by PCR or Sanger sequencing. We found that all potential inversions had breakpoints within long inverted repeats, and these potentially cause chromosomal inversions of ~4.1 kb to ~102.6 kb in length (median ~39.0 kb, Supplementary file 6). All but one sequence inversion (8/9) occurred within repeats containing ORFs and a high median sequence identity (98.3%). In summary, we identified examples of chromosomal inversions that occurred between long repeat sequences and provide the first molecular validation of these inversions in both the reference SC5314 and clinical isolates.
Breakpoints resulting in CNV, LOH, and inversion, occur in the longest repeat sequences with highest homology
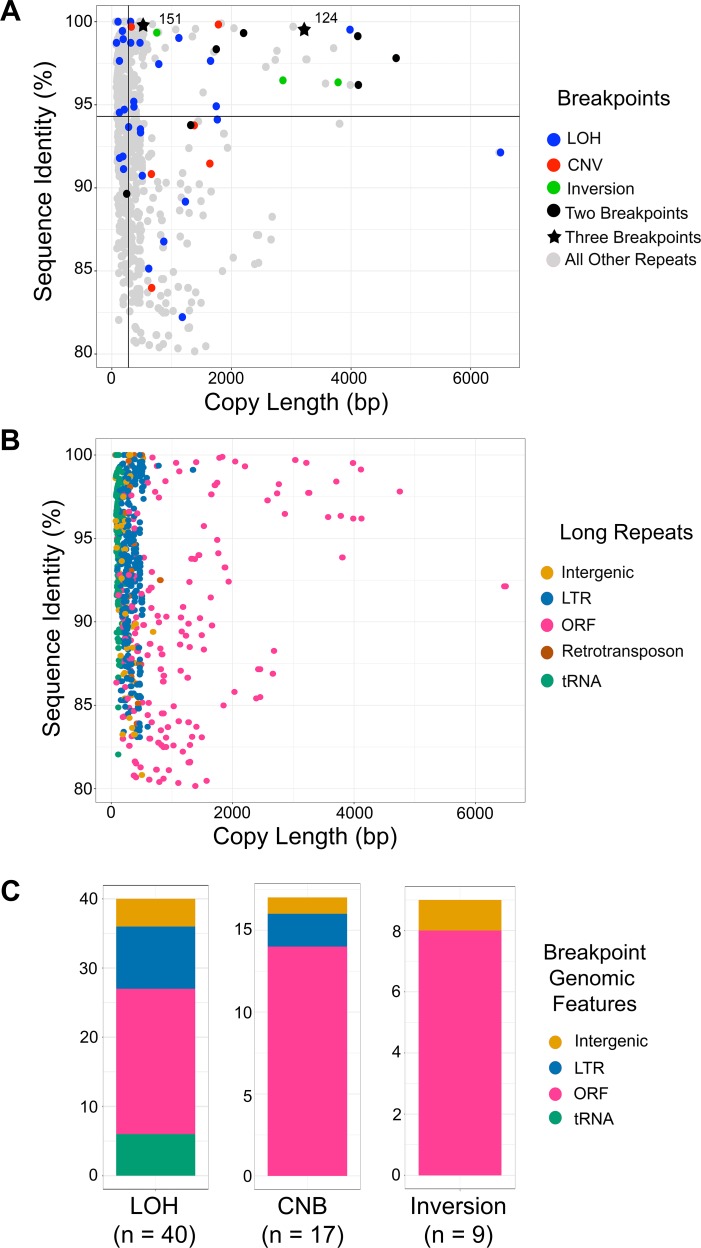
Overall, many uncharacterized long repeat sequences exist within the C. albicans genome. Repeats associated with breakpoints (CNV, LOH, and inversion) were significantly longer than all other long repeat sequences (median copy length of 785 bp vs 278 bp, p<0.0001, Kolmogorov-Smirnov test), and had a significantly higher percent sequence identity than all other long repeat sequences (median identity of 96.2% vs 94.2%, p<0.036 Kolmogorov-Smirnov test) (Figure 6A). Repeats containing ORFs were longer than repeats containing other genomic features and were the most common repeat identified at breakpoints (33/53, 62.3%, Figure 6B & C). Furthermore, repeats containing ORFs were the only genomic feature with both significantly longer copy length and significantly higher sequence identity at breakpoints than at non-breakpoints (p<0.0001 copy length, p<0.0001 sequence identity Kolmogorov-Smirnov test, Figure 6—figure supplement 1A & B). Additionally, repeat matches that contain multiple ORF sequences represent only 8.6% of all long repeats containing ORFs, yet these extra-long repeats comprise 26.8% of the observed breakpoints (Supplementary file 2). Therefore, at least under selection, genome rearrangements are occurring more often at repeats with high sequence identity, and at repeats with high sequence identity and high copy length, the latter of which includes ORFs.
Figure 6. Breakpoints associated with CNV, LOH, and inversion predominantly occur at long repeats that contain ORFs.
(A) Scatterplot of percent sequence identity and copy length of all long repeat matches in Supplementary file 2, excluding the complex tandem repeat genes. All long repeats are indicated in gray, and repeats associated with the observed breakpoints are indicated as follows: LOH (blue), CNV (red), and inversion (green). Six repeats (black circle) were associated with more than one type of breakpoint, and two repeats (black star) were associated with all three types of breakpoints. Solid black lines indicate the median repeat copy length (278 bp, vertical black line) and median percent sequence identity (94.3%, horizontal black line). Repeats associated with LOH, CNV, and inversion breakpoints have a significantly higher median copy length (p<0.0001, Kolmogorov-Smirnov test) and median sequence identity (p<0.036, Kolmogorov-Smirnov test) than all other long repeat sequences (excluding the complex tandem repeat genes, Figure 6—source data 1). (B) Scatterplot as in Figure 6A, where genomic features contained within long repeats are indicated: intergenic sequence (light brown), lone LTR (blue), ORF (pink), retrotransposon (dark brown), and tRNA (green). (C) The distribution of genomic features contained within long repeats at LOH, CNV, and inversion breakpoints. Colors indicated as in Figure 6B.
Figure 6—figure supplement 1. Breakpoint-associated repeats containing ORFs have both high sequence identity and long copy length.
Nine repeat families were associated with more than one breakpoint type (CNV, LOH, and inversion), and two of these (124 and 151) were associated with all three breakpoint types. Repeat family 124 (Figure 3B and 6A), comprised of 4 ORFs, was one of the longest repeats (~3.2 kb) and had one of the highest percent sequence identities (>99%). Repeat family 151 flanks CEN4 and was associated with the formation of the novel isochromosome i(4R), which was necessary and sufficient for increased fitness in the presence of FLC (Figure 1C and Figure 6A). Overwhelmingly, these data support that long repeat sequences found throughout the C. albicans genome are utilized to generate segmental aneuploidies, long-range LOH and sequence inversions, and that in at least one environment these rearrangements provide a significant fitness benefit to the organism.
Discussion
Genomic variation caused by CNV, LOH, and sequence inversion can drive rapid adaptation and promote tumorigenesis. Here, we examined the role of genome architecture during the formation of genetic variation in the diploid, heterozygous fungal pathogen, C. albicans. Our genome-wide analysis of 33 isolates identified long repeat sequences that had prominent roles in generating genomic diversity. These long repeats included previously uncharacterized repeat sequences, centromeric repeats, repeats found within intergenic sequences, and repeats that span multiple ORFs and intergenic sequences. Importantly, long repeat sequences were found at every CNV and sequence inversion breakpoint observed, and frequently occurred at LOH breakpoints as well. Long repeats that were associated with all breakpoints (CNV, LOH, and inversion) had on average significantly higher sequence identity compared to all repeats identified (p<0.036, Kolmogorov-Smirnov test). Furthermore, repeats containing ORFs had both significantly higher sequence identity and significantly longer copy length at breakpoints than at non-breakpoints (sequence identity p<0.0001, copy length p<0.0001 Kolmogorov-Smirnov test, Figure 6, Figure 6—figure supplement 1A and B). These results were independent of genetic background or source of isolation. Thus, long repeat sequences found across the C. albicans genome underlie the formation of significant genome variation that can increase fitness and drive adaptation.
DNA double-strand breaks are repaired using long repeat sequences found across the C. albicans genome
The genomic variants described in this study are the result of DNA double-strand breaks (DSBs) and subsequent recombination events resulting in CNVs, LOH, and sequence inversions. While the factors leading to, and the location of the initiating DSBs are unknown, the genomic variants recovered were all selected as viable, and perhaps beneficial, outcomes of the DSB repair process. DSBs are repaired by either non-homologous end-joining (NHEJ) or homologous recombination (HR). HR is thought to be a high-fidelity repair process due to the use of an intact, homologous DNA template. However, recent studies have also implicated HR in an increased rate of mutagenesis and chromosomal rearrangements (Bishop and Schiestl, 2000; Kramara et al., 2018).
We also found that the orientation of repeat copies had a major effect on the outcome of the genome rearrangements observed. Inverted repeat sequences frequently were found within 2 kb of chromosomal amplification events, while tandem repeat sequences frequently were found within 2 kb of long internal chromosomal deletions. We propose two models of HR involved in the production of genome variation observed in this study (Figure 7).
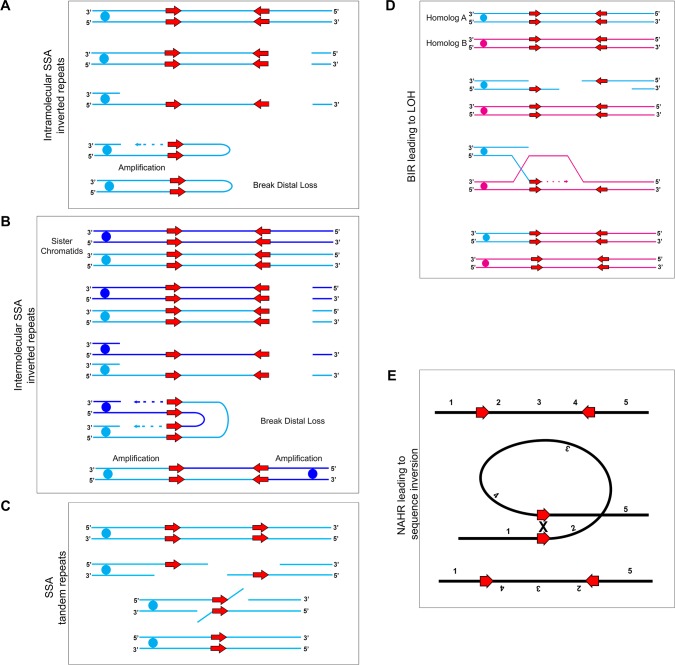
Figure 7. Mechanisms for recombination between long repeats that result in segmental amplification, deletion, LOH, and/or inversion.
(A) Intra-molecular single-strand annealing occurs after a double strand break (DSB) on a single DNA molecule undergoes 5’−3’ resection exposing two copies of an inverted repeat on the single-stranded 3’ overhang. Annealing of the two inverted repeat copies occurs followed by DNA synthesis resulting in a fold-back structure and partial chromosome truncation. (B) Inter-molecular single-strand annealing occurs when a DSB occurs on two separate DNA molecules. After 5’−3’ resection, annealing between the single-stranded inverted repeat copies of the two different DNA molecules results in the formation of a dicentric chromosome and partial chromosome truncation. (C) A single DNA molecule (blue) containing two tandem repeats (red arrows) undergoes a DSB leading to 5’−3’ resection that exposes the tandem repeats. The homologous sequences anneal and non-homologous 3’ tails are removed. The remaining gap is filled producing an intact chromosome that has undergone an internal deletion. (D) Break-Induced Replication (BIR) induces LOH between repeat sequences found on opposite homologs: Two homologs, homolog A (blue) and homolog B (magenta), contain inverted repeat sequences (red arrows). A DSB occurring on homolog A leads to strand invasion and DNA synthesis. Upon termination of synthesis of both the leading and lagging strands, all sequences to the right of the DSB are homozygous. (E) Inversion events occur due to intra-molecular recombination between inverted repeats (red arrows) flanking a unique sequence. The orientation of the reference sequence is indicated above chromosome (1-2-3-4-5). Non-Allelic Homologous Recombination (NAHR) between the inverted repeats leads to an inversion of the sequence between the repeats (1-4-3-2-5).
First, we propose that single-strand annealing (SSA) is initiated by the annealing of DNA repeats that become single-stranded after a DSB and 5’−3’ DNA resection (Figure 7A–7B) and occurs between both tandem and inverted repeat sequences (Bhargava et al., 2016; Malkova and Haber, 2012; Mehta and Haber, 2014; Ramakrishnan et al., 2018; VanHulle et al., 2007). SSA that occurs between tandem repeats leads to segmental deletion of the sequence located between the repeat sequences (Figure 7C). SSA that occurs between inverted repeats can lead to the formation of complex, often unstable dicentric and ‘fold-back’ chromosomes which then enter the breakage-fusion-bridge cycle leading to further genome instability (Aguilera and García-Muse, 2013; Croll et al., 2013; McClintock, 1939; McClintock, 1941; McClintock, 1942; VanHulle et al., 2007) (Figure 7A–7B). Evidence for dicentric chromosomes may exist in several isolates that acquired a segmental amplification of the centromere (Figure 3); however, we do not know from these data if the amplification is on the same molecule (generating a dicentric chromosome) or elsewhere in the genome.
The second HR mechanism we propose is break-induced replication (BIR) which is initiated by DSBs that have only one free end available for repair. During BIR, single-stranded DNA invades a homologous sequence followed by subsequent DNA synthesis which can copy long, chromosomal-sized DNA segments (Anand et al., 2013; Kramara et al., 2018; Malkova and Ira, 2013; Mehta and Haber, 2014). If templating and synthesis occurs on a homologous chromosome, BIR can lead to long-range homozygosis of a chromosome (Figure 7D). Processes similar to BIR have been proposed for CNV generation in a diverse set of organisms ranging from bacteria to humans (Hastings et al., 2009). These predominantly micro-homology-mediated BIR (MMBIR) events use short regions of homology to repair DSBs in a Rad51-independent manner (Hastings et al., 2009). One caveat is that the repeat sequences involved in generating genome rearrangements observed in this study are much longer than those involving MMBIR. While repair by BIR is rare in S. cerevisiae model systems, the selective benefit of the resulting genotypes generated by BIR could increase the apparent frequency with which these types of mutations are recovered in certain environments, for instance the acquisition of i(4R) in the presence of FLC (Figure 1).
C. albicans repeat copy length and spacer length
The repeat copy length associated with observed breakpoints in C. albicans are similar in copy length to transposable (Ty) elements in S. cerevisiae (~6 kb) and long interspersed nuclear elements (LINE) in the human genome (~6–7 kb), which are a major source of genome rearrangements (Chen et al., 2014; Dunham et al., 2002; Gresham et al., 2010; Higashimoto et al., 2013; Selmecki et al., 2015). Both Ty and LINE elements are high copy number repeats; LINE elements are present in thousands of copies in the human genome (Rodić and Burns, 2013). However, beyond the similarity in copy length, we rarely found high copy number repeats, like lone LTRs or retrotransposons, associated with CNV and inversion breakpoints (5.7%, Figure 6). These breakpoints predominantly occurred at repeats containing ORFs that are often present in only two copies per genome (Supplementary file 2). LOH breakpoints, on the other hand, were associated more often with LTRs (22.6%, Figure 6), which may be a result of selection or may suggest a preference for a different repair mechanism when a DSB occurs near these loci.
The repeat copy length and spacer length associated with the observed breakpoints in C. albicans are much longer than typically observed in S. cerevisiae. Segmental amplification events in S. cerevisiae are often mediated by short inverted repeat sequences, for example, 8 bp long and separated by 40 bp (Brewer et al., 2011; Lauer et al., 2018; Payen et al., 2014; Sunshine et al., 2015). The presence of a short, inverted repeat sequence within a replication fork can stimulate ligation between the leading and lagging strands, which results in replication and formation of an extra-chromosomal circle. This extra-chromosomal amplification may continue to replicate independently if it contains an origin of replication (defined as origin-dependent inverted-repeat amplification [ODIRA]; Brewer et al., 2015; Brewer et al., 2011; Payen et al., 2014). It seems unlikely that such a mechanism operates at the long distances observed between repeat sequences in C. albicans. However, it is possible that a different origin-dependent mechanism is mediating some of the rearrangements we observed (see centromere discussion below). A future challenge is to determine if/how this occurs.
The spacer length, especially between inverted repeats, has been a major focus of genome instability research. Identification and characterization of inverted repeats in S. cerevisiae has primarily focused on those repeats that are separated by very short (~80 bp) spacers (Strawbridge et al., 2010). Inverted repeats that were engineered to have variable repeat spacer lengths identified a correlation between repeat and spacer length and DSB repair. Increasing repeat copy length (from 185 bp to ~1.5 kb) and/or decreasing repeat spacer length (from ~8.5 kb to 0 bp) increased the recombination rate between repeats by up to 17,000-fold (Lobachev et al., 1998). Furthermore, spacer length alone could affect the choice of DSB repair pathway; DSB repair via inter-molecular SSA predominantly occured with a spacer length of 1 kb, while intra-molecular SSA predominantly occured with spacer length of 12 bp (Ramakrishnan et al., 2018).
Astoundingly, the C. albicans CNV and inversion breakpoints are associated with much longer repeat spacer lengths than those described in S. cerevisiae, ranging from ~3.1 kb to ~1.6 Mb (median ~30 kb) and ~3.1 kb to ~94.3 kb (median ~34.6 kb), respectively. Recombination between such long distances requires a naturally occurring, long-distance homology search. It is tempting to speculate that C. albicans may have a mechanism for long-distance resection, particular chromatin features, or a 3D-nuclear structure that facilitates recombination between inverted repeats separated by long distances.
Inverted repeat sequences directly associated with the CENP-A-binding centromere core sequences facilitate isochromosome formation
Centromeres were common breakpoints for CNV, LOH and inversion. Twelve of the 33 isolates had breakpoint events that occurred within centromeres, including those described at CEN4 and CEN5, as well as two additional centromeres that contained one copy of a long repeat sequence, CEN2 and CEN3 (Supplementary file 2). Notably, C. albicans centromeres are the earliest firing centers of DNA replication (Koren et al., 2010; Tsai et al., 2014). Therefore, errors in DNA replication may be a common source of DSBs that are repaired via HR between long repeat sequences.
Repair of a DSB within or near a centromere-associated inverted repeat can result in isochromosome formation or centromere inversion (Figure 1, Figure 1—figure supplement 1). Both of the C. albicans centromeres that are flanked by long inverted repeat sequences (CEN4 and CEN5) can form isochromosomes (Figure 1 and Selmecki et al., 2006; Selmecki et al., 2009). Exposure to the antifungal drug FLC selected for isochromosome formation at both CEN4 and CEN5. If a DSB occurs near the inverted repeat sequence, DNA synthesis via BIR will copy the entire arm of the broken chromosome, resulting in the homozygous isochromosome structures that we observed (Figure 1 and Selmecki et al., 2010; Selmecki et al., 2009). Acquisition of either isochromosome i(4R) or i(5L) was both necessary and sufficient for increased fitness in the presence of FLC (Figure 1 and Selmecki et al., 2006). Additionally, there was no fitness cost associated with either isochromosome in the absence of FLC: i(4R) was stable for ~300 generations in 12/12 populations in the absence of FLC (Figure 1—figure supplement 1). These data are in contrast to other, often whole chromosome and multiple chromosome aneuploidies that cause significant fitness defects in the absence of selection (Pavelka et al., 2010; Torres et al., 2007), but support observations that aneuploidy in general has less of a fitness cost in diploid and polyploid fungi (Hose et al., 2015; Scott et al., 2017; Selmecki et al., 2015; Tan et al., 2013).
Similarly, repair of a DSB within or near a centromere-associated inverted repeat can result in centromere inversion. Inversions are the result of intra-chromosomal non-allelic homologous recombination between inverted repeats flanking the centromere (Figure 7E). Here, we detected an inversion that occurred between inverted repeats flanking CEN4. The impact of these inversions on localization of the centromeric histone CENP-A, or of the recombination proteins Rad51 and Rad52, which are thought to recruit CENP-A, are not known. Whether or not inversion of the centromere affects chromosome stability will be important to test in future experiments.
In this study, Illumina short-read datasets were used to identify genomic features that were driving structural and allelic variation across diverse C. albicans isolates. The use of both new and previously published short-read datasets highlights the utility of this bioinformatic approach for the analysis of structural variants within this and other species. However, short-read data are unable to provide a key understanding of the molecules containing the long repeat sequences. For example, the definitive structure of chromosomal inversions, including the heterozygous CEN4 sequence, is difficult to determine with short-read data. PCR enabled rapid validation of these inversions (Figures 1 and 5); however, it required knowledge of the repeat location and unique surrounding sequences. Future long-read sequencing is needed to address the definitive structure of existing DNA molecules and potential DNA intermediates involved in recombination and resolution of CNV, LOH, and inversions.
Long repeats containing ORFs were significantly more common at breakpoints resulting in CNV, LOH and inversion than any other genomic feature
One hypothesis is that active transcription may promote DNA DSBs, due to the formation of R-loop structures (Aguilera and Gaillard, 2014; Santos-Pereira and Aguilera, 2015). Additionally, increased transcription in certain environments may increase the probability of a DNA DSB that result in genome rearrangements, as was observed at the S. cerevisiae CUP1 locus in high-copper environments (Adamo et al., 2012; Fogel et al., 1983; Hull et al., 2017; Thomas and Rothstein, 1989). Several indirect results are consistent with this hypothesis in C. albicans. First, all ORFs within a long repeat that were associated with a breakpoint were indeed actively transcribed in the reference isolate SC5314 during growth in rich medium (Bruno et al., 2010). Second, some breakpoint ORFs have increased expression in the selective environment from which the isolate with the breakpoint was obtained. For example, two different in vivo isolates, one bloodstream clinical isolate and one murine OPC-evolved isolate, have the same breakpoint on Chr1 at the inverted repeat that includes HGT1 and HGT2 (Supplementary file 2). Both HGT1 and HGT2 are induced during OPC, biofilm production and adaptation to serum (Horák, 2013; Nobile et al., 2012; Pitarch et al., 2001). Therefore, increased transcription of these repeat ORFs in vivo is a potential source of DNA damage that resulted in DSB repair.
Conclusion
In conclusion, genome rearrangements resulting in segmental aneuploidies, sequence inversions, and LOH are associated with long repeat sequence breakpoints on every chromosome. These genome rearrangements can arise rapidly, both in vitro and in vivo, and can provide an adaptive phenotype such as improved growth in antifungal drugs. Importantly, long repeat sequences are hotspots for genome variation across diverse selective environments. Indeed, several repeats were involved in all three types of genome rearrangements in different isolates. These data support the idea that the C. albicans genome is one of the most rapidly evolving genomes due to disruption of conserved syntenic sequence blocks via genome rearrangements between long repeat sequences (Fischer et al., 2006). Finally, given the frequency of long repeat sequences in the human genome, studies of C. albicans genome rearrangements can contribute to understanding the mechanisms that facilitate CNV, LOH, and inversions associated with human disease and cancer.
Materials and methods
Key resources table.
| Reagent type (species) or resource |
Designation | Source or reference |
Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Candida albicans) | SC5314 | Hirakawa et al., 2015 (doi:10.1101/gr.174623.114) | RRID:SCR_013437 | |
| Strain, strain background (C. albicans) | P78042 | Hirakawa et al., 2015 (doi:10.1101/gr.174623.114) | ||
| Strain, strain background (C. albicans) | AMS3743 | This Study | In vitro evolution of P78042 in 128 ug/ml FLC for 100 generations | |
| Strain, strain background (C. albicans) | AMS3743_10 | This Study | In vitro evolution of AMS3743 in rich medium for 300 generations | |
| Strain, strain background (C. albicans) | AMS3743_10_S6 | This Study | Single colony from AMS3743_10 | |
| Antibody | Anti-Digoxigenin-AP Fab Fragments | Roche | 11093274910 RRID:AB_2734716 | (1:5000) |
| Sequenced-based reagent | PCR Primers | This Study | Supplementary file 7 | |
| Commercial assay or kit | Illumina Nextera XT Library Prep Kit | Illumina | 105032350 | |
| Commercial assay or kit | Illumina Nextera XT Index Kit | Illumina | 105055294 | |
| Commercial assay or kit | Illumina MiSeq v2 Reagent Kit | Illumina | 15033625 | 2 × 250 cycles |
| Commercial assay or kit | Blue Pippin 1.5% agarose gel dye-free cassette | Sage Science | 250 bp - 1.5 kb DNA size range collections, Marker R2 | Target of 900 bp |
| Commercial assay or kit | Qubit dsDNA HS kit | Life Technologies | Q32854 | |
| Commercial assay or kit | PCR DIG Probe Synthesis Kit | Roche | 11636090910 | |
| Commercial assay or kit | Agilent 2100 Bioanalyzer High Sensitivity DNA Reagents | Agilent Technologies | 5067–4626 | |
| Chemical compound, drug | Fluconazole (FLC) | Alfa Aesar | J62015 | |
| Software, algorithm | MUMmer Sutie | Kurtz et al., 2004 (doi:10.1186/gb-2004-5-2-r12) | v3.0 RRID:SCR_001200 | |
| Software, algorithm | Trimmomatic | Bolger et al., 2014 (doi:10.1093/bioinformatics/btu170) | v0.33 RRID:SCR_011848 | |
| Software, algorithm | BWA |
Li, 2013
(doi:10.1093/bioinformatics/btp324) |
v0.7.12 RRID:SCR_010910 | |
| Software, algorithm | Samtools | Li et al., 2009 (doi:10.1093/bioinformatics/btp352) | v0.1.19 RRID:SCR_002105 | |
| Software, algorithm | Genome Analysis Toolkit | McKenna et al., 2010 (doi:10.1101/gr.107524.110) | v3.4–46 RRID:SCR_001876 | |
| Software, algorithm | REPuter | Kurtz et al., 2001 (doi:10.1093/nar/29.22.4633) | V1.0 https://bibiserv.cebitec.uni-bielefeld.de/reputer | |
| Software, algorithm | Yeast Analysis Mapping Pipeline | Abbey et al., 2014 (doi:10.1186/s13073-014-0100-8) | v1.0 | |
| Software, algorithm | Graphpad Prism | https://www.graphpad.com | v6.0 RRID:SCR_002798 | |
| Software, algorithm | ImageJ | https://imagej.nih.gov/ij/? | v2.0.0-rc-30/1.49 s RRID:SCR_003070 | |
| Software, algorithm | Integrative Genomics Viewer | Thorvaldsdóttir et al., 2013 (doi:10.1093/bib/bbs017) | v2.3.92 RRID:SCR_011793 | |
| Software, algorithm | R | https://www.r-project.org | v3.5.2 RRID:SCR_001905 | |
| Software, algorithm | Candida Genome Database | http://Candidagenome.org | RRID:SCR_002036 | |
| Other | Propidium Iodide | Invitrogen | P3566 | 25 ug/ml final concentration |
| Other | Ribonuclease A | MP Biomedicals | 101076 | 0.5 mg/ml final concentration |
Yeast isolates and culture conditions
All isolates used in this study are shown in Supplementary file 1. Isolates were stored at −80°C in 20% glycerol. Isolates were grown at 30°C in YPAD (yeast peptone dextrose medium; Rose W, 1990) supplemented with 40 µg ml−1 adenine and 80 µg ml−1 uridine).
In vivo evolution experiments
OPC isolates were obtained as previously described (Forche et al., 2018; Solis and Filler, 2012). Briefly, mice were orally infected with strain YJB9318 and single colony isolates were obtained from tongue tissue of mice on days 1, 2, 3, and 5 post infection and stored in 50% glycerol at −80°C for further use.
In vitro evolution experiments
Six isolates were obtained from in vitro evolution experiments in the presence of antifungal drug (Supplementary file 1). Isolate AMS3053 was obtained on 20 µg/ml Miconazole agar plates as previously described (Mount et al., 2018). Isolates AMS3742, AMS3743, AMS3747, AMS3748, and AMS3744 were obtained from liquid batch culture evolution experiments conducted in 96-well format. Progenitor isolates were plated for single colonies on YPAD and incubated for 48 hr at 30°C. Single colonies were grown to saturation in liquid YPAD at 30°C. A 1:1000 dilution was made in YPAD medium containing either 1 µg/ml or 128 µg/ml of FLC. Plates were covered with BreathEASIER tape (Electron Microscope Science) and cultured in a humidified chamber for 72 hr at 30°C. At each 72 hr time point, cells were resuspended by pipetting and transferred into fresh media via a 1:1000 dilution and cultured for another 72 hr at 30°C, for 10 consecutive passages. After the final transfer, cells were immediately collected for genomic DNA isolation and ploidy analysis by flow cytometry.
To obtain AMS3743 isolates that had lost the i(4R) (Figure 1—figure supplement 1), 12 single colonies of AMS3743 were selected on YPAD plates at 30°C after 48 hr. All 12 single colonies had i(4R) (by PCR) and were used to initiate 12 YPAD-evolved lineages, each cultured for 24 hr in 4 ml liquid YPAD at 30°C with shaking. Every 24 hr, a 1:1000 dilution was inoculated into fresh YPAD medium. Cultures were passaged for 30 days. Cells from all 12 YPAD-evolved lineages were divided into tubes for −80°C storage, genomic DNA isolation, and CHEF analysis. All 12 YPAD-evolved lineages maintained i(4R) by CHEF analysis. CHEF gel densitometry analysis (see below) identified one lineage (AMS3743_10) that had a lighter i(4R) band density relative to the rest of the genome. AMS3743_10 was plated for single colonies on a YPAD plate and incubated at 30°C for 48 hr. Six single colonies were cultured for 24 hr in 4 ml liquid YPAD at 30°C with shaking, and cells were divided into tubes for −80°C storage, genomic DNA isolation, and CHEF analysis. One of the six single colonies lost the i(4R) (AMS3743_10_S6, Figure 1—figure supplement 1).
Contour-clamped homogenous electric field (CHEF) electrophoresis
Samples were prepared as previously described (Selmecki et al., 2005). Cells were suspended in 300 µL 1.5% low-melt agarose (Bio-Rad) and digested with 1.2 mg Zymolyase (US Biological). Chromosomes were separated on a 1% Megabase agarose gel (Bio-Rad) in 0.5X TBE using a CHEF DRIII apparatus. Run conditions as follows: 60 s to 120 s switch, 6 V/cm, 120° angle for 36 hrs followed by 120 s to 300 s switch, 4.5 V/cm, 120° angle for 12 hrs.
CHEF gel densitometry
Ethidium bromide stained CHEF gels were imaged using the GelDock XR imaging system (BioRad). Images were exported as .PNG files, converted to 32-bit, and analyzed using ImageJ (v2.0.0-rc-30/1.49 s). The total lane density (gray value, area under the curve) was collected for each sample. The density associated with i(4R) was determined by drawing a box around the i(4R) density peak (box distance was from each adjacent minimums). The fraction of i(4R) relative to the entire genome was determined by normalizing the i(4R) density relative to the total lane density. The population with lowest ratio of i(4R) relative to total genome (AMS3743_10) was used for single colony analysis.
Southern hybridization
DNA from CHEF gels was transferred to BrightStar Plus nylon membrane (Invitrogen). Probing and detection of the DNA was conducted as previously described (Selmecki et al., 2005; Selmecki et al., 2008; Selmecki et al., 2009). Probes were generated by PCR incorporation of DIG-11-dUTP into target sequences following manufacturer’s instructions (Roche). Primer pairs used in probe design are listed in Supplementary file 7.
PCR
All primer sequences were designed to avoid heterozygous or SNP loci in the reference genome SC5314 and clinical isolates. Primers and primer sequences are found in Supplementary file 7. PCR conditions for i(4R) were as follows: 95°C for 3 min, followed by 32 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 5.5 min, and a final extension at 72°C for 10 min. The PCR conditions for the Chr4 inversion (Figure 5) were the same as above, except the annealing temperature was 53°C and the extension time was 3.25 min.
Flow cytometry
Cells were prepared as previously described (Todd et al., 2018). Briefly, cells were grown to a density of 1 × 107 in liquid medium and gently spun down (500 x g) for 3 min. The supernatant was removed and cells were fixed with 70% (v/v) ethanol for at least 1 hr at room temperature. Cells were then washed twice with 50 mM sodium citrate and sonicated (Biorupter Fisher Science) for 10–15 s at 30% power to separate the cells. Following sonication, cells were centrifuged and resuspended with 50 mM sodium citrate and incubated for at least 3 hr at 37°C in 0.5 mg ml−1 RNase A (MP Biomedicals) in 50 mM sodium citrate (Fisher Scientific). Cells were stained with 25 µg ml−1 propidium iodide (Invitrogen) overnight in the dark at 37°C. Cells were sonicated for 5–10 s at 15% power, and 30,000 cells were analyzed on a ZE5 cell analyzer (BioRad). Data were analyzed in FlowJo (https://www.flowjo.com/solutions/flowjo/downloads) (v10.4.1).
Growth curve analysis
Growth curves were determined using a BioTek Epoch plate reader. Culture medium included YPAD or YPAD +32 µg/ml FLC (Alfa Aesar) Approximately 5 × 103 cells were inoculated into 200 µl culture medium in a clear, flat bottomed 96-well plate (Thermo Scientific). The plate was incubated at 30°C with double orbital shaking at 256 rpm, and the OD600 was measured every 15 min. Data were collected with Gen5 Software (BioTek) and exported to Microsoft Excel for downstream analysis. All growth curves were conducted in individual biological triplicate on separate days.
Illumina whole genome sequencing
Genomic DNA was isolated with phenol chloroform as described previously (Selmecki et al., 2006). Libraries were prepared using the NexteraXT DNA Sample Preparation Kit following the manufacturer’s instructions (Illumina). DNA fragments between 600 and 1200 bp were selected for sequencing using a Blue Pippin 1.5% agarose gel dye-free cassette (Sage Science). Library fragments were analyzed with a Bioanalyzer High Sensitivity DNA Chip (Agilent Technologies) and Qubit High Sensitivity dsDNA (Life Technologies). Libraries were sequenced using paired-end, 2 × 250 reads on an Illumina MiSeq (Creighton University). Adaptor sequences and low-quality reads were trimmed using Trimmomatic (v0.33 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36 TOPHRED33) (Bolger et al., 2014). Reads were mapped to the Candida albicans reference genome (A21-s02-m09-r08) obtained 7 of October 2015 from the Candida Genome Database website: http://www.candidagenome.org/download/sequence/C_albicans_SC5314/Assembly21/archive/ C_albicans_SC5314_version_A21-s02-m09-r08_chromosomes.fasta.gz). The reads were mapped using the Burrows-Wheeler Aligner MEM algorithm using default parameters (BWA v0.7.12) (Li, 2013). Duplicate PCR amplicons were removed using Samtools (v0.1.19) (Li et al., 2009), and reads were realigned around possible indels using Genome Analysis Toolkit’s RealignerTargetCreator and IndelRealigner (-model USE_READS -targetIntervals) (v3.4–46) (McKenna et al., 2010). All WGS data have been deposited in the National Center for Biotechnology Information Sequence Read Archive database as PRJNA510147. Sequence data obtained from published datasets are noted in Supplementary file 1.
Identification of aneuploidy and copy number breakpoints
Preliminary identification of chromosomes containing CNVs was conducted using Illumina whole genome sequence data and the Yeast Mapping Analysis Pipeline (YMAP v1.0). Fastq files were uploaded to YMAP and read depth was plotted as a function of chromosome location using the reference genome Candida albicans (A21-s02-mo8-r09), with correction for chromosome end bias and GC content (Abbey et al., 2014). The average normalized genome coverage was determined for 45.5 kb non-overlapping windows across each chromosome using the YMAP GBrowse CNV track. The largest absolute difference between the average normalized genome coverage of two consecutive 45.5 kb windows was identified. To further refine CNV breakpoints, fastq files were aligned to the reference genome as above (Illumina Whole Genome Sequencing), read depth was calculated for every base pair in the nuclear genome using Samtools (samtools depth -aa) (v0.1.19), and normalized by read depth of the total nuclear genome using R (v3.5.2). The two consecutive 45.5 kb windows were further sub-divided into 5 kb windows. The average normalized read depth was determined for these 5 kb windows and a rolling mean of every two consecutive 5 kb windows was determined. CNV breakpoint boundaries were identified when 75% of four consecutive means had an average normalized read depth that deviated from the average normalized nuclear genome read depth by more than 25% in tetraploids or 50% in diploids (Ford et al., 2015). Boundaries were confirmed by visual inspection in Integrative Genomics Viewer (IGV v2.3.92) (Thorvaldsdóttir et al., 2013). CNV breakpoints were then determined using visual inspection of total read depth and allele ratio analysis (when the breakpoint was surrounded by heterozygous sequence) within unique, non-repeat sequences. CNV breakpoint positions were compared to Supplementary file 2 and breakpoints were assigned a repeat name if they fell within 2 kb of a long repeat sequence.
Enrichment of CNV breakpoints at long repeat sequences
Enrichment analysis of CNV breakpoints was conducted using a two-tailed Fisher’s Exact Test in Bedtools (Bedtools v2.28.0) with default parameters (Quinlan and Hall, 2010). Briefly, two bed files were generated with 1) the start and stop positions of all long repeat sequences and 2) the start and stop positions of all long repeat sequences located within 2 kb of a CNV breakpoint (Supplementary file 2, excluding the complex tandem repeat genes). The overlap of observed breakpoints and long repeat sequences was compared to the expected overlap between CNV breakpoints and long repeat sequences, given the total genome coverage of long repeat sequences. The minimum overlap required was a single base pair between a CNV breakpoint and repeat sequence.
Identification of long-range homozygosity breakpoints
Illumina whole genome sequence data were analyzed using YMAP (v1.0) and IGV (v2.3.92). First, fastq files were uploaded to YMAP and the density of heterozygous SNPs was determined for non-overlapping 5 kb windows and plotted by chromosomal position in standard SNP/LOH view (default parameters, baseline ploidy was 2N for all isolates except AMS3420, which was 4N). Approximate positions of all long-range homozygous and heterozygous transitions were determined within 20–25 kb. To further refine LOH breakpoints, fastq files were aligned to the reference genome as above (Illumina Whole Genome Sequencing) and visualized in IGV. All heterozygous to homozygous (and vice versa) transitions were recorded when four or more consecutive loci were heterozygous and transitioned to four or more homozygous loci (and vice versa). The minimum distance covered by the four or more consecutive loci was greater than 300 bp and all four of the loci were located within unique, non-repeat sequences. Additionally, all heterozygous loci utilized for breakpoint analysis had an alternate allele frequency greater than or equal to 20%, read depth greater than 10 reads, and both forward and reverse strands that supported the alternate allele (Selmecki et al., 2015). The breakpoints of these long-range homozygous tracks (‘LOH breakpoints’) were recorded as the last heterozygous locus and the first homozygous locus of the heterozygous > homozygous transition, and vice versa for the homozygous > heterozygous transition. Long-range LOH breakpoints were then compared to Supplementary file 2 and were assigned a repeat number if they fell within 2 kb of a long repeat sequence (Supplementary file 4).
Identification of inversion breakpoints
Additional positions of predicted chromosomal inversions were obtained from Hirakawa et al. (2015), Table S13. Coordinates corresponding to potential inversions were obtained using BreakDancer or NUCmer (Hirakawa et al., 2015). The distance between the BreakDancer or NUCmer coordinates (start and stop) and the nearest long repeat sequence was determined. If a long repeat sequence occurred within 2 kb of either BreakDancer or NUCmer coordinates, the repeat number and family were recorded. Disagreement between BreakDancer and NUCmer coordinates that coincided with breakpoints in different repeat families (representing more complex chromosome rearrangements or inversions) were removed from the analysis. Additionally, all NUCmer or Breakdancer positions that occurred within ALS gene family repeats were removed from the analysis because the BreakDancer and NUCmer coordinates did not support a consistent length of sequence inversion (likely due to mapping errors within and between ALS repeats). The long repeat sequences identified at these potential inversion breakpoints, including those shared across different isolates, are summarized in Supplementary file 6.
Microsatellite repeat identification
Short repetitive sequences found at either copy number breakpoints or allele ratio breakpoints were analyzed using REPuter (Kurtz et al., 2001) with a minimum repeat length of 8 bp. Analysis was conducted using the forward, reverse, complement, and palindromic match direction.
Identification of long repeat sequences
Repeat sequences within the C. albicans genome were identified using the MUMmer suite (v3.0) (Kurtz et al., 2004). Whole genome sequence alignment with NUCmer (nucmer --maxmatch --nosimplify) identified all maximum-length matches with 100% sequence identity (minimum match length of 20 bp) within the Candida albicans SC5314 reference genome (A21-s02-m09-r08, obtained 7 of October 2015 from the Candida Genome Database (CGD): http://www.candidagenome.org/download/sequence/C_albicans_SC5314/Assembly21/archive/ C_albicans_SC5314_version_A21-s02-m09-r08_chromosomes.fasta.gz). All maximum length matches were identified, regardless of their uniqueness (meaning all matches in the genome were identified). Then, all sequence matches were clustered and extended to obtain a maximum-length colinear string of matches if they were separated by no more than 90 nucleotides (NUCmer default parameters). Three repeat matches shared less than 80% sequence identity, therefore an 80% cutoff was used for the final long repeat analysis (Supplementary file 2), similar to previous studies (Achaz et al., 2000; Warren et al., 2014). All sequences that self-aligned to the same genomic position were removed.
Repeat matches were annotated using the reference genome feature file (C_albicans_SC5314_version_A21-s02-m09-r08_Chromosomal_feature file) and repeat tracks obtained from CGD (Skrzypek et al., 2017). To highlight uncharacterized long repeat sequences, repeats associated with the three major classes of repetitive DNA in C. albicans were removed, including the rDNA locus, MRS sequences (RPS, HOK, and RB2), telomere-proximal regions, as well as ambiguous sequences (containing poly-N nucleotides). These regions are highly variable and difficult to analyze with short-read sequencing techniques (Chibana et al., 2000; Chibana et al., 1994; Chindamporn et al., 1998; Goodwin and Poulter, 2000; Hoyer and Cota, 2016; Hoyer et al., 1995; Levdansky et al., 2008). Telomere-proximal regions were determined as the region from each chromosome end to the first confirmed, non-repetitive-genome feature, similar to previous studies (Ene et al., 2018; Hirakawa et al., 2015): Chr1: 1–10000, Chr1:3181000–3188548, Chr2: 1–5000, Chr2: 2228650–2232035, Chr3: 1–15000, Chr3: 1787000–1799406, Chr4: 1–2700, Chr4: 1597200–1603443, Chr5: 1–3800, Chr5: 1183000–1190928, Chr6: 1–3000, Chr6: 1031500–1033530, Chr7: 1–75, Chr7: 942300–949616, ChrR: 1–4500, ChrR: 2286355–2286389. Telomere-associated genes, including TLO genes, that were not positioned in these telomere-proximal regions were maintained.
All long repeat sequences were verified using BLAST and IGV. Repeat copies that were on the same chromosome were defined as either tandem, mirrored, or inverted using the repeat start and end positions obtained from NUCmer and manually inspected in IGV. Tandem repeat sequences are in the same orientation on the same strand, mirrored repeat sequences are in opposite orientations on the same strand, and inverted repeat sequences are in opposite orientations on the opposite strand. Spacer length was obtained by calculating the shortest distance between repeat matches.
After the post-alignment annotations and filtration, repeats were combined into repeat families if they shared an identical match. For example, if repetitive sequence A was matched with sequence B, sequences A and B were combined into one family. In some instances, a sequence matched with more than one sequence (e.g. A matched with B and C). In these cases, all matched sequences were combined into one family. In total, 230 repeat families were identified with sequence identities of ≥80% (median value of 92.9%) between all copies of the repeat within a family. Of these 230 families, 68 included more than two copies per genome (Supplementary file 2).
The fraction of the genome covered by long repeat sequences was determined by multiplying the average copy length of each repeat family by the number of copies of that repeat family found throughout the genome (excluding the complex tandem repeat genes). The sum of the average copy length of all repeat families (409129 bp) was then divided by the length of the haploid Candida albicans SC5314 reference genome (excluding the mt-DNA, 14280189 bp) to determine that 2.87% of the genome is covered by long repeat sequences (Figure 2—source data 1).
Annotation of repeat sequences
The long repeat sequences were annotated according to the genomic features contained within each matched repeat sequence using the C. albicans genome feature file described above. The genomic features included were: lone long terminal repeats (LTRs) lacking ORFs, retrotransposons, tRNAs, ORFs, and intergenic sequences. Repeat matches containing ORFs included partial ORF sequences, single complete ORF sequences, and multiple ORFs and intergenic sequences. In cases where one repeat copy contained a genome feature, but the other repeat copy contained an intergenic sequence (no genome feature), this latter repeat was flagged as ‘Unannotated Intergenic Sequence’ and both repeat copies were assigned the feature found at the annotated repeat copy (Supplementary file 2). All unannotated sequences were verified in both V21 and V22 of the C. albicans reference genome (Skrzypek et al., 2017).
Of the known LTRs present within the C. albicans genome, only five were not detected in the MUMmer analysis. Analysis of the five undetected LTRs using BLASTN revealed that they lacked an exact match of 20 nucleotides required to establish a matched repeat pair.
All full-length ORF coding sequences within the C. albicans reference genome (C_albicans_SC5314_version_A21-s02-m09-r08_chromosomes.fasta.gz) were analyzed for length and GC content using EMBOSS Infoseq (http://imed.med.ucm.es/cgi-bin/emboss.pl?_action=input&_app=infoseq). All full-length ORF coding sequences were divided into coding sequences that were contained within long repeat sequences or coding sequences that were not contained within long repeat sequences (excluding the complex tandem repeat genes, Supplementary file 2, Figure 2—figure supplement 3D and E). If a long repeat sequence contained a partial ORF sequence, the full-length coding sequence was used in the analysis. Similarly, if a long repeat sequence contained multiple ORF sequences, the full-length coding sequence of each ORF were included in the analysis.
Exclusion of complex tandem repeat genes
Five ORFs and one gene family with known, complex embedded tandem repeats were confirmed by NUCmer (PGA18, PGA55, EAP1, Adhesin-like orf19.1725, CSA1, and the ALS gene family comprised of eight ORFs, Supplementary file 2) (Levdansky et al., 2008; Wilkins et al., 2018). Assignment of a genome copy count was not possible for these tandem repeat genes due to the extreme complexity of matched repeat sequences. For this reason, all repeat copy counts and analysis using copy counts exclude the complex tandem repeat genes listed above and are indicated throughout the text (Supplementary file 2).
Statistical analyses
For this study, biological replicates are defined as a single, independent culture derived from a frozen −80°C glycerol stock. Data were analyzed using GraphPad Prism v6 and made into graphical representations using RSudio v1.1.463. All p-values below 0.05 were considered significant.
Acknowledgements
We thank all members of the Selmecki laboratory, especially Curtis Focht, Alison Guyer, Robert Thomas, and Annette Beach for technical assistance. We thank Dr. Robin Dowell, Dr. Mary Ann Allen, and Dr. Hung-Ji Tsai for feedback on the manuscript and helpful discussions. Support for this research was provided by LB692 NE Tobacco Settlement Biomedical Research Development New Initiative Grant (to AS), NE Established Program to Stimulate Competitive Research (EPSCoR) First Award (to AS), NE Department of Health and Human Services (LB506-2017-55) award (to AS), CURAS Faculty Research Fund Award (to AS), and NIH-NCRR COBRE grant P20RR018788 sub-award (to AS). AF was supported by NIH grant R15 AI090633. The sequencing datasets generated during this study are available in the Sequence Read Archive repository under project PRJNA510147.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Anna Selmecki, Email: annaselmecki@creighton.edu.
Kevin J Verstrepen, VIB-KU Leuven Center for Microbiology, Belgium.
Detlef Weigel, Max Planck Institute for Developmental Biology, Germany.
Funding Information
This paper was supported by the following grants:
Nebraska LB692 New Initiatives Grants LB692 NE Tobacco Settlement Biomedical Research Development New Initiative Grant to Anna Selmecki.
Nebraska's Established Program to Stimulate Competitive Research EPSCoR First Award to Anna Selmecki.
Nebraska Department of Health and Human Services LB506-2017-55 to Anna Selmecki.
Creighton University CURAS Faculty Faculty Research Fund to Anna Selmecki.
National Center for Research Resources P20RR018788 sub award to Anna Selmecki.
National Institutes of Health R15 AI090633 to Anja Forche.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Data curation, Formal analysis, Supervision, Funding acquisition, Validation, Investigation, Visualization, Methodology, Writing—original draft, Writing—review and editing.
Data curation, Formal analysis, Validation, Investigation, Visualization, Methodology.
Resources, Data curation, Funding acquisition, Validation, Writing—original draft, Writing—review and editing.
Conceptualization, Resources, Data curation, Formal analysis, Supervision, Funding acquisition, Validation, Investigation, Methodology, Writing—original draft, Writing—review and editing.
Additional files
Data availability
All data generated and analyzed during this study are included in the manuscript and supporting files. Source data files have a been provided for Figure 1, Figure 1—figure supplement 1, Figure 2, Figure 2—figure supplement 2, Figure 2—figure supplement 3, Figure 6, and Figure 6—figure supplement 1. All genomic data are deposited in SRA under accession PRJNA510147.
The following dataset was generated:
Todd RT, Forche A, Selmecki A. 2019. Segmental Aneuploidies in Candida albicans. NCBI SRA. PRJNA510147
The following previously published datasets were used:
Hirakawa MP, Martinez DA, Sakthikumar S, Anderson MZ, Berlin A, Gujja S, Zeng Q, Zisson E, Wang JM, Greenberg JM, Berman J, Bennett RJ, Cuomo CA. 2013. Candida albians Umbrella Comparative genomics project. NCBI SRA. PRJNA193498
Ford CB, Funt JM, Abbey D, Issi L, Guiducci C, Martinez DA, Delorey T, Li BY, White TC, Cuomo C, Rao R, Berman J, Thompson D, Regev A. 2014. The evolution of gradual acquisition of drug reisstance in clinical isolates of Candida albicans. NCBI SRA. PRJNA257929
Ropars J, Maufrais C, Diogo D, Marcet-Houben M, Perin A, Sertour N, Mosca K, Permal E, Laval G, Bouchier C, Ma L, Schwarts K, Voelz K, May RC, Poulain J, Battail C, Wincker P, Borman AM, Chowdhary A, Fan S, Kim SH, Le Pape P, Romeo O, Shin JH, Gabaldon T, Sherlock G, Bougnoux M, d'Enfert C. 2018. Whole Genome sequencing of 182 isolates of the fungal pathogen of humans Candida albicans. NCBI SRA. PRJNA432884
References
- Abbey DA, Funt J, Lurie-Weinberger MN, Thompson DA, Regev A, Myers CL, Berman J. YMAP: a pipeline for visualization of copy number variation and loss of heterozygosity in eukaryotic pathogens. Genome Medicine. 2014;6:100. doi: 10.1186/s13073-014-0100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achaz G, Coissac E, Viari A, Netter P. Analysis of intrachromosomal duplications in yeast saccharomyces cerevisiae: a possible model for their origin. Molecular Biology and Evolution. 2000;17:1268–1275. doi: 10.1093/oxfordjournals.molbev.a026410. [DOI] [PubMed] [Google Scholar]
- Adamo GM, Lotti M, Tamás MJ, Brocca S. Amplification of the CUP1 gene is associated with evolution of copper tolerance in saccharomyces cerevisiae. Microbiology. 2012;158:2325–2335. doi: 10.1099/mic.0.058024-0. [DOI] [PubMed] [Google Scholar]
- Aguilera A, Gaillard H. Transcription and recombination: when RNA meets DNA. Cold Spring Harbor Perspectives in Biology. 2014;6:a016543. doi: 10.1101/cshperspect.a016543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A, García-Muse T. Causes of genome instability. Annual Review of Genetics. 2013;47:1–32. doi: 10.1146/annurev-genet-111212-133232. [DOI] [PubMed] [Google Scholar]
- Anand RP, Lovett ST, Haber JE. Break-induced DNA replication. Cold Spring Harbor Perspectives in Biology. 2013;5:a010397. doi: 10.1101/cshperspect.a010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MZ, Baller JA, Dulmage K, Wigen L, Berman J. The three clades of the telomere-associated TLO gene family of Candida Albicans have different splicing, localization, and expression features. Eukaryotic Cell. 2012;11:1268–1275. doi: 10.1128/EC.00230-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya CL, Payen C, Dunham MJ, Fields S. Whole-genome sequencing of a laboratory-evolved yeast strain. BMC Genomics. 2010;11:88. doi: 10.1186/1471-2164-11-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ, Johnson AD. Completion of a parasexual cycle in Candida Albicans by induced chromosome loss in tetraploid strains. The EMBO Journal. 2003;22:2505–2515. doi: 10.1093/emboj/cdg235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava R, Onyango DO, Stark JM. Regulation of Single-Strand annealing and its role in genome maintenance. Trends in Genetics. 2016;32:566–575. doi: 10.1016/j.tig.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop AJR, Schiestl RH. Homologous recombination as a mechanism for genome rearrangements: environmental and genetic effects. Human Molecular Genetics. 2000;9:2427–2434. doi: 10.1093/hmg/9.16.2427. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchonville K, Forche A, Tang KES, Selmecki A, Berman J. Aneuploid Chromosomes Are Highly Unstable during DNA Transformation of Candida albicans. Eukaryotic Cell. 2009;8:1554–1566. doi: 10.1128/EC.00209-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun BR, van het Hoog M, d'Enfert C, Martchenko M, Dungan J, Kuo A, Inglis DO, Uhl MA, Hogues H, Berriman M, Lorenz M, Levitin A, Oberholzer U, Bachewich C, Harcus D, Marcil A, Dignard D, Iouk T, Zito R, Frangeul L, Tekaia F, Rutherford K, Wang E, Munro CA, Bates S, Gow NA, Hoyer LL, Köhler G, Morschhäuser J, Newport G, Znaidi S, Raymond M, Turcotte B, Sherlock G, Costanzo M, Ihmels J, Berman J, Sanglard D, Agabian N, Mitchell AP, Johnson AD, Whiteway M, Nantel A. A Human-Curated annotation of the Candida albicans genome. PLOS Genetics. 2005;1:e1–e57. doi: 10.1371/journal.pgen.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BJ, Payen C, Raghuraman MK, Dunham MJ. Origin-dependent inverted-repeat amplification: a replication-based model for generating palindromic amplicons. PLOS Genetics. 2011;7:e1002016. doi: 10.1371/journal.pgen.1002016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BJ, Payen C, Di Rienzi SC, Higgins MM, Ong G, Dunham MJ, Raghuraman MK. Origin-Dependent Inverted-Repeat Amplification: Tests of a Model for Inverted DNA Amplification. PLOS Genetics. 2015;11:e1005699. doi: 10.1371/journal.pgen.1005699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GD, Netea MG. Exciting Developments in the Immunology of Fungal Infections. Cell Host & Microbe. 2012;11:422–424. doi: 10.1016/j.chom.2012.04.010. [DOI] [PubMed] [Google Scholar]
- Bruno VM, Wang Z, Marjani SL, Euskirchen GM, Martin J, Sherlock G, Snyder M. Comprehensive annotation of the transcriptome of the human fungal pathogen Candida albicans using RNA-seq. Genome Research. 2010;20:1451–1458. doi: 10.1101/gr.109553.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrack LS, Hutton HF, Matter KJ, Clancey SA, Liachko I, Plemmons AE, Saha A, Power EA, Turman B, Thevandavakkam MA, Ay F, Dunham MJ, Berman J. Neocentromeres Provide Chromosome Segregation Accuracy and Centromere Clustering to Multiple Loci along a Candida albicans Chromosome. PLOS Genetics. 2016;12:e1006317. doi: 10.1371/journal.pgen.1006317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G, Rasmussen MD, Lin MF, Santos MAS, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, Agrafioti I, Arnaud MB, Bates S, Brown AJP, Brunke S, Costanzo MC, Fitzpatrick DA, de Groot PWJ, Harris D, Hoyer LL, Hube B, Klis FM, Kodira C, Lennard N, Logue ME, Martin R, Neiman AM, Nikolaou E, Quail MA, Quinn J, Santos MC, Schmitzberger FF, Sherlock G, Shah P, Silverstein KAT, Skrzypek MS, Soll D, Staggs R, Stansfield I, Stumpf MPH, Sudbery PE, Srikantha T, Zeng Q, Berman J, Berriman M, Heitman J, Gow NAR, Lorenz MC, Birren BW, Kellis M, Cuomo CA. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhou W, Zhang L, Zhang F. Genome Architecture and Its Roles in Human Copy Number Variation. Genomics & Informatics. 2014;12:136–144. doi: 10.5808/GI.2014.12.4.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibana H, Iwaguchi S, Homma M, Chindamporn A, Nakagawa Y, Tanaka K. Diversity of tandemly repetitive sequences due to short periodic repetitions in the chromosomes of Candida albicans. Journal of Bacteriology. 1994;176:3851–3858. doi: 10.1128/jb.176.13.3851-3858.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibana H, Beckerman JL, Magee PT. Fine-Resolution Physical Mapping of Genomic Diversity in Candida albicans. Genome Research. 2000;10:1865–1877. doi: 10.1101/gr.148600. [DOI] [PubMed] [Google Scholar]
- Chindamporn A, Nakagawa Y, Mizuguchi I, Chibana H, Doi M, Tanaka K. Repetitive sequences (RPSs) in the chromosomes of Candida albicans are sandwiched between two novel stretches, HOK and RB2, common to each chromosome. Microbiology. 1998;144:849–857. doi: 10.1099/00221287-144-4-849. [DOI] [PubMed] [Google Scholar]
- Christiaens JF, Van Mulders SE, Duitama J, Brown CA, Ghequire MG, De Meester L, Michiels J, Wenseleers T, Voordeckers K, Verstrepen KJ. Functional divergence of gene duplicates through ectopic recombination. EMBO reports. 2012;13:1145–1151. doi: 10.1038/embor.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu WS, Rikkerink EH, Magee PT. Genetics of the white-opaque transition in Candida albicans: demonstration of switching recessivity and mapping of switching genes. Journal of Bacteriology. 1992;174:2951–2957. doi: 10.1128/jb.174.9.2951-2957.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croll D, Zala M, McDonald BA. Breakage-fusion-bridge cycles and large insertions contribute to the rapid evolution of accessory chromosomes in a fungal pathogen. PLOS Genetics. 2013;9:e1003567. doi: 10.1371/journal.pgen.1003567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham MJ, Badrane H, Ferea T, Adams J, Brown PO, Rosenzweig F, Botstein D. Characteristic genome rearrangements in experimental evolution of saccharomyces cerevisiae. PNAS. 2002;99:16144–16149. doi: 10.1073/pnas.242624799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn MJ, Kinney GM, Washington PM, Berman J, Anderson MZ. Functional diversification accompanies gene family expansion of MED2 homologs in candida albicans. PLOS Genetics. 2018;14:e1007326. doi: 10.1371/journal.pgen.1007326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ene IV, Farrer RA, Hirakawa MP, Agwamba K, Cuomo CA, Bennett RJ. Global analysis of mutations driving microevolution of a heterozygous diploid fungal pathogen. PNAS. 2018;115:E8688–E8697. doi: 10.1073/pnas.1806002115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer G, Rocha EP, Brunet F, Vergassola M, Dujon B. Highly variable rates of genome rearrangements between hemiascomycetous yeast lineages. PLOS Genetics. 2006;2:e32. doi: 10.1371/journal.pgen.0020032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel S, Welch JW, Cathala G, Karin M. Gene amplification in yeast: cup1 copy number regulates copper resistance. Current Genetics. 1983;7:347–355. doi: 10.1007/BF00445874. [DOI] [PubMed] [Google Scholar]
- Forche A, Alby K, Schaefer D, Johnson AD, Berman J, Bennett RJ. The parasexual cycle in Candida Albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLOS Biology. 2008;6:e110. doi: 10.1371/journal.pbio.0060110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A, Magee PT, Selmecki A, Berman J, May G. Evolution in Candida Albicans populations during a single passage through a mouse host. Genetics. 2009;182:799–811. doi: 10.1534/genetics.109.103325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A, Abbey D, Pisithkul T, Weinzierl MA, Ringstrom T, Bruck D, Petersen K, Berman J. Stress alters rates and types of loss of heterozygosity in candida albicans. mBio. 2011;2:e00129-11. doi: 10.1128/mBio.00129-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A, Cromie G, Gerstein AC, Solis NV, Pisithkul T, Srifa W, Jeffery E, Abbey D, Filler SG, Dudley AM, Berman J. Rapid phenotypic and genotypic diversification after exposure to the oral host niche in Candida Albicans. Genetics. 2018;209:725–741. doi: 10.1534/genetics.118.301019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford CB, Funt JM, Abbey D, Issi L, Guiducci C, Martinez DA, Delorey T, Li BY, White TC, Cuomo C, Rao RP, Berman J, Thompson DA, Regev A. The evolution of drug resistance in clinical isolates of candida albicans. eLife. 2015;4:e00662. doi: 10.7554/eLife.00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire-Benéitez V, Price RJ, Tarrant D, Berman J, Buscaino A. Candida Albicans repetitive elements display epigenetic diversity and plasticity. Scientific Reports. 2016;6:22989. doi: 10.1038/srep22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein AC, Fu MS, Mukaremera L, Li Z, Ormerod KL, Fraser JA, Berman J, Nielsen K. Polyploid titan cells produce haploid and aneuploid progeny to promote stress adaptation. mBio. 2015;6:e01340-15. doi: 10.1128/mBio.01340-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin TJ, Poulter RT. The CARE-2 and rel-2 repetitive elements of Candida Albicans contain LTR fragments of a new retrotransposon. Gene. 1998;218:85–93. doi: 10.1016/S0378-1119(98)00362-X. [DOI] [PubMed] [Google Scholar]
- Goodwin TJ, Poulter RT. Multiple LTR-retrotransposon families in the asexual yeast candida albicans. Genome Research. 2000;10:174–191. doi: 10.1101/gr.10.2.174. [DOI] [PubMed] [Google Scholar]
- Gresham D, Usaite R, Germann SM, Lisby M, Botstein D, Regenberg B. Adaptation to diverse nitrogen-limited environments by deletion or extrachromosomal element formation of the GAP1 locus. PNAS. 2010;107:18551–18556. doi: 10.1073/pnas.1014023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLOS Genetics. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman MA, Zeng G, Forche A, Hirakawa MP, Abbey D, Harrison BD, Wang YM, Su CH, Bennett RJ, Wang Y, Berman J. The 'obligate diploid' Candida albicans forms mating-competent haploids. Nature. 2013;494:55–59. doi: 10.1038/nature11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashimoto K, Maeda T, Okada J, Ohtsuka Y, Sasaki K, Hirose A, Nomiyama M, Takayanagi T, Fukuzawa R, Yatsuki H, Koide K, Nishioka K, Joh K, Watanabe Y, Yoshiura K, Soejima H. Homozygous deletion of DIS3L2 exon 9 due to non-allelic homologous recombination between LINE-1s in a japanese patient with Perlman syndrome. European Journal of Human Genetics. 2013;21:1316–1319. doi: 10.1038/ejhg.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa MP, Martinez DA, Sakthikumar S, Anderson MZ, Berlin A, Gujja S, Zeng Q, Zisson E, Wang JM, Greenberg JM, Berman J, Bennett RJ, Cuomo CA. Genetic and phenotypic intra-species variation in candida albicans. Genome Research. 2015;25:413–425. doi: 10.1101/gr.174623.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horák J. Regulations of sugar transporters: insights from yeast. Current Genetics. 2013;59:1–31. doi: 10.1007/s00294-013-0388-8. [DOI] [PubMed] [Google Scholar]
- Hose J, Yong CM, Sardi M, Wang Z, Newton MA, Gasch AP. Dosage compensation can buffer copy-number variation in wild yeast. eLife. 2015;4:e05462. doi: 10.7554/eLife.05462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer LL, Scherer S, Shatzman AR, Livi GP. Candida Albicans ALS1: domains related to a saccharomyces cerevisiae sexual agglutinin separated by a repeating motif. Molecular Microbiology. 1995;15:39–54. doi: 10.1111/j.1365-2958.1995.tb02219.x. [DOI] [PubMed] [Google Scholar]
- Hoyer LL, Cota E. Candida Albicans Agglutinin-Like sequence (Als) Family vignettes: a review of als protein structure and function. Frontiers in Microbiology. 2016;7:280. doi: 10.3389/fmicb.2016.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull RM, Cruz C, Jack CV, Houseley J. Environmental change drives accelerated adaptation through stimulated copy number variation. PLOS Biology. 2017;15:e2001333. doi: 10.1371/journal.pbio.2001333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T, Federspiel NA, Chibana H, Dungan J, Kalman S, Magee BB, Newport G, Thorstenson YR, Agabian N, Magee PT, Davis RW, Scherer S. The diploid genome sequence of candida albicans. PNAS. 2004;101:7329–7334. doi: 10.1073/pnas.0401648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketel C, Wang HS, McClellan M, Bouchonville K, Selmecki A, Lahav T, Gerami-Nejad M, Berman J. Neocentromeres form efficiently at multiple possible loci in Candida Albicans. PLOS Genetics. 2009;5:e1000400. doi: 10.1371/journal.pgen.1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiktev DA, Sheng Z, Lobachev KS, Petes TD. GC content elevates mutation and recombination rates in the yeast saccharomyces cerevisiae. PNAS. 2018;115:E7109–E7118. doi: 10.1073/pnas.1807334115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren A, Tsai HJ, Tirosh I, Burrack LS, Barkai N, Berman J. Epigenetically-inherited centromere and neocentromere DNA replicates earliest in S-phase. PLOS Genetics. 2010;6:e1001068. doi: 10.1371/journal.pgen.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramara J, Osia B, Malkova A. Break-Induced replication: the where, the why, and the how. Trends in Genetics. 2018;34:518–531. doi: 10.1016/j.tig.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA. Nucleotide repeats. Slippery DNA and diseases. Nature. 1993;365:207–208. doi: 10.1038/365207a0. [DOI] [PubMed] [Google Scholar]
- Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Research. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biology. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer S, Avecilla G, Spealman P, Sethia G, Brandt N, Levy SF, Gresham D. Single-cell copy number variant detection reveals the dynamics and diversity of adaptation. PLOS Biology. 2018;16:e3000069. doi: 10.1371/journal.pbio.3000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lephart PR, Magee PT. Effect of the major repeat sequence on mitotic recombination in Candida Albicans. Genetics. 2006;174:1737–1744. doi: 10.1534/genetics.106.063271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levdansky E, Sharon H, Osherov N. Coding fungal tandem repeats as generators of fungal diversity. Fungal Biology Reviews. 2008;22:85–96. doi: 10.1016/j.fbr.2008.08.001. [DOI] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The sequence alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Aligning sequence reads clone sequence and assembly contigs with BWA-MEM. arXiv. 2013 https://arxiv.org/abs/1303.3997
- Lobachev KS, Shor BM, Tran HT, Taylor W, Keen JD, Resnick MA, Gordenin DA. Factors affecting inverted repeat stimulation of recombination and deletion in saccharomyces cerevisiae. Genetics. 1998;148:1507–1524. doi: 10.1093/genetics/148.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart SR, Pujol C, Daniels KJ, Miller MG, Johnson AD, Pfaller MA, Soll DR. In Candida Albicans, white-opaque switchers are homozygous for mating type. Genetics. 2002;162:737–745. doi: 10.3410/f.1004386.154857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee BB, Magee PT. Induction of mating in Candida Albicans by construction of MTLa and MTLalpha strains. Science. 2000;289:310–313. doi: 10.1126/science.289.5477.310. [DOI] [PubMed] [Google Scholar]
- Malkova A, Haber JE. Mutations arising during repair of chromosome breaks. Annual Review of Genetics. 2012;46:455–473. doi: 10.1146/annurev-genet-110711-155547. [DOI] [PubMed] [Google Scholar]
- Malkova A, Ira G. Break-induced replication: functions and molecular mechanism. Current Opinion in Genetics & Development. 2013;23:271–279. doi: 10.1016/j.gde.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcet-Houben M, Marceddu G, Gabaldón T. Phylogenomics of the oxidative phosphorylation in fungi reveals extensive gene duplication followed by functional divergence. BMC Evolutionary Biology. 2009;9:295. doi: 10.1186/1471-2148-9-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The behavior in successive nuclear divisions of a chromosome broken at meiosis. PNAS. 1939;25:405–416. doi: 10.1073/pnas.25.8.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The stability of broken ends of chromosomes in zea mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The fusion of broken ends of chromosomes following nuclear fusion. PNAS. 1942;28:458–463. doi: 10.1073/pnas.28.11.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A, Haber JE. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harbor Perspectives in Biology. 2014;6:a016428. doi: 10.1101/cshperspect.a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount HO, Revie NM, Todd RT, Anstett K, Collins C, Costanzo M, Boone C, Robbins N, Selmecki A, Cowen LE. Global analysis of genetic circuitry and adaptive mechanisms enabling resistance to the azole antifungal drugs. PLOS Genetics. 2018;14:e1007319. doi: 10.1371/journal.pgen.1007319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, Tuch BB, Andes DR, Johnson AD. A recently evolved transcriptional network controls biofilm development in candida albicans. Cell. 2012;148:126–138. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–325. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payen C, Di Rienzi SC, Ong GT, Pogachar JL, Sanchez JC, Sunshine AB, Raghuraman MK, Brewer BJ, Dunham MJ. The Dynamics of Diverse Segmental Amplifications in Populations of Saccharomyces cerevisiae Adapting to Strong Selection. G3: Genes|Genomes|Genetics. 2014;4:399–409. doi: 10.1534/g3.113.009365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nature Reviews Genetics. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- Pitarch A, Díez-Orejas R, Molero G, Pardo M, Sánchez M, Gil C, Nombela C. Analysis of the serologic response to systemic candida albicans infection in a murine model. Proteomics. 2001;1:550–559. doi: 10.1002/1615-9861(200104)1:4<550::AID-PROT550>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan S, Kockler Z, Evans R, Downing BD, Malkova A. Single-strand annealing between inverted DNA repeats: pathway choice, participating proteins, and genome destabilizing consequences. PLOS Genetics. 2018;14:e1007543. doi: 10.1371/journal.pgen.1007543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reams AB, Roth JR. Mechanisms of gene duplication and amplification. Cold Spring Harbor Perspectives in Biology. 2015;7:a016592. doi: 10.1101/cshperspect.a016592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard GF, Dujon B, Haber JE. Double-strand break repair can lead to high frequencies of deletions within short CAG/CTG trinucleotide repeats. Molecular and General Genetics MGG. 1999;261:871–882. doi: 10.1007/s004380050031. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nature Biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodić N, Burns KH. Long interspersed element-1 (LINE-1): passenger or driver in human neoplasms? PLOS Genetics. 2013;9:e1003402. doi: 10.1371/journal.pgen.1003402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropars J, Maufrais C, Diogo D, Marcet-Houben M, Perin A, Sertour N, Mosca K, Permal E, Laval G, Bouchier C, Ma L, Schwartz K, Voelz K, May RC, Poulain J, Battail C, Wincker P, Borman AM, Chowdhary A, Fan S, Kim SH, Le Pape P, Romeo O, Shin JH, Gabaldon T, Sherlock G, Bougnoux ME, d'Enfert C. Gene flow contributes to diversification of the major fungal pathogen candida albicans. Nature Communications. 2018;9:2253. doi: 10.1038/s41467-018-04787-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose W H. Methods in Yeast Genetics. COld Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Rustchenko EP, Curran TM, Sherman F. Variations in the number of ribosomal DNA units in morphological mutants and normal strains of Candida Albicans and in normal strains of saccharomyces cerevisiae. Journal of Bacteriology. 1993;175:7189–7199. doi: 10.1128/jb.175.22.7189-7199.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustchenko-Bulgac EP. Variations of Candida Albicans electrophoretic karyotypes. Journal of Bacteriology. 1991;173:6586–6596. doi: 10.1128/jb.173.20.6586-6596.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Pereira JM, Aguilera A. R loops: new modulators of genome dynamics and function. Nature Reviews Genetics. 2015;16:583–597. doi: 10.1038/nrg3961. [DOI] [PubMed] [Google Scholar]
- Sanyal K, Baum M, Carbon J. Centromeric DNA sequences in the pathogenic yeast candida albicans are all different and unique. PNAS. 2004;101:11374–11379. doi: 10.1073/pnas.0404318101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott AL, Richmond PA, Dowell RD, Selmecki AM. The influence of polyploidy on the evolution of yeast grown in a Sub-Optimal carbon source. Molecular Biology and Evolution. 2017;34:2690–2703. doi: 10.1093/molbev/msx205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki A, Bergmann S, Berman J. Comparative genome hybridization reveals widespread aneuploidy in Candida Albicans laboratory strains. Molecular Microbiology. 2005;55:1553–1565. doi: 10.1111/j.1365-2958.2005.04492.x. [DOI] [PubMed] [Google Scholar]
- Selmecki A, Forche A, Berman J. Aneuploidy and isochromosome formation in drug-resistant candida albicans. Science. 2006;313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki A, Gerami-Nejad M, Paulson C, Forche A, Berman J. An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Molecular Microbiology. 2008;68:624–641. doi: 10.1111/j.1365-2958.2008.06176.x. [DOI] [PubMed] [Google Scholar]
- Selmecki AM, Dulmage K, Cowen LE, Anderson JB, Berman J. Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLOS Genetics. 2009;5:e1000705. doi: 10.1371/journal.pgen.1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki A, Forche A, Berman J. Genomic plasticity of the human fungal pathogen candida albicans. Eukaryotic Cell. 2010;9:991–1008. doi: 10.1128/EC.00060-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki AM, Maruvka YE, Richmond PA, Guillet M, Shoresh N, Sorenson AL, De S, Kishony R, Michor F, Dowell R, Pellman D. Polyploidy can drive rapid adaptation in yeast. Nature. 2015;519:349–352. doi: 10.1038/nature14187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoighe C, Federspiel N, Jones T, Hansen N, Bivolarovic V, Surzycki R, Tamse R, Komp C, Huizar L, Davis RW, Scherer S, Tait E, Shaw DJ, Harris D, Murphy L, Oliver K, Taylor K, Rajandream MA, Barrell BG, Wolfe KH. Prevalence of small inversions in yeast gene order evolution. PNAS. 2000;97:14433–14437. doi: 10.1073/pnas.240462997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrzypek MS, Binkley J, Binkley G, Miyasato SR, Simison M, Sherlock G. The candida genome database (CGD): incorporation of assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Research. 2017;45:D592–D596. doi: 10.1093/nar/gkw924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis NV, Filler SG. Mouse model of oropharyngeal candidiasis. Nature Protocols. 2012;7:637–642. doi: 10.1038/nprot.2012.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawbridge EM, Benson G, Gelfand Y, Benham CJ. The distribution of inverted repeat sequences in the saccharomyces cerevisiae genome. Current Genetics. 2010;56:321–340. doi: 10.1007/s00294-010-0302-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenbrock EH, Jørgensen FG, Zala M, Hansen TT, McDonald BA, Schierup MH. Whole-genome and chromosome evolution associated with host adaptation and speciation of the wheat pathogen mycosphaerella graminicola. PLOS Genetics. 2010;6:e1001189. doi: 10.1371/journal.pgen.1001189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunshine AB, Payen C, Ong GT, Liachko I, Tan KM, Dunham MJ. The fitness consequences of aneuploidy are driven by condition-dependent gene effects. PLOS Biology. 2015;13:e1002155. doi: 10.1371/journal.pbio.1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Nishibayashi S, Kuroiwa T, Kanbe T, Tanaka K. Variance of ploidy in Candida Albicans. Journal of Bacteriology. 1982;152:893–896. doi: 10.1128/jb.152.2.893-896.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z, Hays M, Cromie GA, Jeffery EW, Scott AC, Ahyong V, Sirr A, Skupin A, Dudley AM. Aneuploidy underlies a multicellular phenotypic switch. PNAS. 2013;110:12367–12372. doi: 10.1073/pnas.1301047110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in Bioinformatics. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd RT, Braverman AL, Selmecki A. Flow Cytometry Analysis of Fungal Ploidy. Current Protocols in Microbiology. 2018;50:e58. doi: 10.1002/cpmc.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- Tsai HJ, Baller JA, Liachko I, Koren A, Burrack LS, Hickman MA, Thevandavakkam MA, Rusche LN, Berman J. Origin replication complex binding, nucleosome depletion patterns, and a primary sequence motif can predict origins of replication in a genome with epigenetic centromeres. mBio. 2014;5:e01703–e01714. doi: 10.1128/mBio.01703-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanHulle K, Lemoine FJ, Narayanan V, Downing B, Hull K, McCullough C, Bellinger M, Lobachev K, Petes TD, Malkova A. Inverted DNA repeats channel repair of distant double-strand breaks into chromatid fusions and chromosomal rearrangements. Molecular and Cellular Biology. 2007;27:2601–2614. doi: 10.1128/MCB.01740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen KJ, Jansen A, Lewitter F, Fink GR. Intragenic tandem repeats generate functional variability. Nature Genetics. 2005;37:986–990. doi: 10.1038/ng1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JM, Bennett RJ, Anderson MZ. The genome of the human pathogen Candida albicans is shaped by mutation and cryptic sexual recombination. mBio. 2018;9:e01205-18. doi: 10.1128/mBio.01205-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren IA, Ciborowski KL, Casadei E, Hazlerigg DG, Martin S, Jordan WC, Sumner S. Extensive local gene duplication and functional divergence among paralogs in Atlantic salmon. Genome Biology and Evolution. 2014;6:1790–1805. doi: 10.1093/gbe/evu131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickes B, Staudinger J, Magee BB, Kwon-Chung KJ, Magee PT, Scherer S. Physical and genetic mapping of Candida Albicans: several genes previously assigned to chromosome 1 map to chromosome R, the rDNA-containing linkage group. Infection and Immunity. 1991;59:2480–2484. doi: 10.1128/iai.59.7.2480-2484.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins M, Zhang N, Schmid J. Biological roles of Protein-Coding tandem repeats in the yeast Candida Albicans. Journal of Fungi. 2018;4:78. doi: 10.3390/jof4030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- Zhang N, Harrex AL, Holland BR, Fenton LE, Cannon RD, Schmid J. Sixty alleles of the ALS7 open reading frame in Candida Albicans: als7 is a hypermutable contingency locus. Genome Research. 2003;13:2005–2017. doi: 10.1101/gr.1024903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Cannon RD, Holland BR, Patchett ML, Schmid J. Impact of genetic background on allele selection in a highly mutable candida albicans gene, PNG2. PLOS ONE. 2010;5:e9614. doi: 10.1371/journal.pone.0009614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Oh SH, Cheng G, Green CB, Nuessen JA, Yeater K, Leng RP, Brown AJ, Hoyer LL. ALS3 and ALS8 represent a single locus that encodes a candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiology. 2004;150:2415–2428. doi: 10.1099/mic.0.26943-0. [DOI] [PubMed] [Google Scholar]