Abstract
BACKGROUND:
Malignant peripheral nerve sheath tumors (MPNSTs) are aggressive sarcomas that arise at an estimated frequency of 8% to 13% in individuals with neurofibromatosis type 1 (NF1). Compared with their sporadic counterparts, NF1-associated MPNSTs (NF1-MPNSTs) develop in young adults, frequently recur (approximately 50% of cases), and carry a dismal prognosis. As such, most individuals affected with NF1-MPNSTs die within 5 years of diagnosis, despite surgical resection combined with radiotherapy and chemotherapy.
METHODS:
Clinical genomic profiling was performed using 1000 ng of DNA from 7 cases of NF1-MPNST, and bioinformatic analyses were conducted to identify genes with actionable mutations.
RESULTS:
A total of 3 women and 4 men with NF1-MPNST were identified (median age, 38 years). Nonsynonymous mutations were discovered in 4 genes (neurofibromatosis type 1 [NF1], ROS proto-oncogene 1 [ROS1], tumor protein p53 [TP53], and tyrosine kinase 2 [TYK2]), which in addition were mutated in other MPNST cases in this sample set. Consistent with their occurrence in individuals with NF1, all tumors had at least 1 mutation in the NF1 gene. Whereas TP53 gene mutations are frequently observed in other cancers, ROS1 mutations are common in melanoma (15%−35%), another neural crest-derived malignancy. In contrast, TYK2 mutations are uncommon in other malignancies (<7%). In the current series, recurrent TYK2 mutations were identified in 2 cases of NF1-MPNST (30% of cases), whereas TYK2 protein overexpression was observed in 60% of MPNST cases using an independently generated tissue microarray, regardless of NF1 status.
CONCLUSIONS:
Clinical genomic analysis of the current series of NF1-MPNST cases found that TYK2 is a new gene mutated in MPNST. Future work will focus on examining the utility of TYK2 expression as a biomarker and therapeutic target for these cancers
Keywords: cancer predisposition, neurofibromatosis, ROS proto-oncogene 1 (ROS1), sarcoma, tyrosine kinase 2 (TYK2)
INTRODUCTION
Neurofibromatosis type 1 (NF1) is one of the most common inherited tumor predisposition syndromes, affecting 1 in 2500 individuals worldwide.1 As such, individuals with NF1 begin life with 1 mutated (germline) copy and 1 functional copy of the NF1 gene in every cell in their body.2 The presence of this germline mutation significantly increases their risk of developing cancer because tumorigenesis requires only somatic loss of the remaining functional NF1 gene. In this regard, individuals with NF1 are prone to the development of central and peripheral nervous system malignancies, including malignant peripheral nerve sheath tumors (MPNSTs).
MPNSTs are aggressive soft tissue sarcomas3 that arise in individuals with NF1 at an estimated frequency of 8% to 13%. In contrast to sporadic MPNSTs, which arise in adults during their 60s and 70s, NF1-associated MPNSTs (NF1-MPNSTs) tend to occur in young patients, usually in their 20s and 30s.4 These cancers are composed of neoplastic Schwann cells, and in the setting of NF1, most often arise from a benign precursor lesion, termed a plexiform neurofibroma. When MPNSTs are diagnosed, they initially are treated with surgery and radiotherapy, and in some situations, chemotherapy is added. However, despite aggressive initial management with multimodality therapy, approximately 50% of affected individuals will experience cancer recurrence or metastatic disease, and the majority of patients die within 5 years of their diagnosis.5–7 Moreover, metastatic tumors are particularly difficult to treat because they inevitably become resistant to chemotherapy. Given the poor prognosis of these tumors, it is essential that better strategies be developed to detect and treat these aggressive malignancies.
To identify improved treatments for patients with NF1-MPNSTs, several groups have used genetically engineered mouse models of these cancers, resulting in the discovery of new biologically targeted therapies.8–14 Based on these basic discovery efforts, mechanistic target of rapamycin (mTOR), mitogen-activated kinase kinase (MEK), and heat shock protein 90 (HSP90) inhibitors have all entered the clinical trials pipeline as promising targeted therapies for NF1 tumors (ClinicalTrials.gov Identifier NCT01661283, NCT01885195, and NCT020088770). As a complementary approach to mouse model-driven discovery,15–17 in the current study, we leveraged clinical-grade genomic profiling on MPNSTs from 7 subjects with NF1 to identify new potentially actionable gene mutations.
MATERIALS AND METHODS
Patients
Tumors were obtained from individuals diagnosed with NF1 according to established criteria18 and treated at the Washington University/St. Louis Children’s Hospital Neurofibromatosis Clinical Program. This study was performed under active Human Studies Protocols approved by the Institutional Review Boards at each respective institution in accordance with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. Tumor DNA samples were obtained from formalin-fixed paraffin-embedded (FFPE) blocks or frozen tissue (when available) obtained at the time of surgical resection or biopsy.
Laser Capture Microdissection
Images of hematoxylin and eosin-stained slides were created on an Aperio ScanScope XT (Leica Biosystems, Buffalo Grove, Ill). The Aurora mScope viewer (Aurora Interactive Ltd, Montreal, Quebec, Canada) was used to view the images and obtain images at × 20 magnification. A pathologist examined a digitized hematoxylin and eosin-stained slide to determine and outline tumor nests, directing the laser capture microdissection (LCM) to areas of high tumor cell content on the adjacent FFPE sections. A total of 5 to 10 sections (10-μm thick) from each FFPE block were cut on a microtome (Leica RM 2255; Leica Biosystems), air-dried for ≥ 2 hours, and placed at 60°Cfor 30 minutes. The sections were stored at 4°C until use or were immediately stained with a rapid hematoxylin and eosin ethanol-based staining protocol. Within 10 minutes of air-drying, the slide was placed on the LCM stage of the Arcturus PixCell instrument (Thermo Fisher Scientific, Waltham, Mass) for microdissection. The desired cells were microdissected into the cap of a 500-μL, safe-lock tube filled with 50 μL of ALT buffer (Qiagen, Valencia, Calif). This procedure was repeated on the next slide until a total of 2000 to 5000 cells of interest had been captured for each case.
DNA Extraction
LCM-derived DNA samples were isolated from 2000 to 5000 cells using a QIAamp DNA Micro Kit (Qiagen category 56304; Qiagen) after RNase A treatment, according to the manufacturer’s instructions. DNA was quantified using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific).
Sequencing
For these studies, we employed next-generation sequencing using a protocol similar to Clinical Laboratory Improvement Amendments-approved mutation testing available for patients with cancer treated at the study institution (Genomics and Pathology Services; https://gps.wustl.edu/patient care/). The list of sequenced genes in this set are listed in Supporting Information Table 1. In brief, genomic DNA was sonicated to an average size of approximately 200 base pairs. The fragments were blunt ended, “A” tailed, and ligated to Illumina sequencing adapters (Illumina Inc, San Diego, Calif). The ligated fragments were amplified for 7 polymerase chain reaction cycles. Exome targets were enriched with Agilent Custom SureSelect reagents (Agilent Technologies, Santa Clara, Calif) according to the manufacturer’s protocols. Enriched fragments were amplified for 14 cycles with primers that incorporate a unique indexing sequence tag. The resulting library fragments were sequenced from each end (paired-end reads) for 101 bases using an Illumina HiSeq-2500 instrument (Illumina Inc).
Analysis
Sequence reads were mapped to the human reference genome (GRCh37/hg19) using the Novoalign tool (www.novocraft.com/products/novoalign/).19 Marking and removing polymerase chain reaction duplicates were performed using the PICARD tool (http://broadinstitute.github.io/picard/). Variants were determined using the GATK Unified Genotyper (https://www.broadinstitute.org/gatk/).20 Variant filtration and selection was performed with the GATK Filter Variants tool. Variants with low depth, strand bias, etc, were discarded. Filters used include: filter out snps if depth <50 or phredFisher StrandBias > 100 and filter out indels if readDepth (DP) < 50, phredFisherStrandBias > 100, or Homo-polymerRun > 7. Filtered data was annotated using Annovar (hg19 build) (http://annovar.openbioinformatics.org/en/latest/).21 Filtered variants initially were classified into 1 of 4 levels using the bioinformatics pipeline. The classification scheme is described in Table 1. The driver genes used for level 1 significance were tumor protein 53 (TP53), NF1, cyclin-dependent kinase inhibitor 2A (CDKN2A), epidermal growth factor receptor (EGFR), platelet-derived growth factor (PDGF), Erb-B2 receptor tyrosine kinase 2 (ERBB2), KIT, MET, platelet-derived growth factor receptor (PDGFR), phosphatase and tensin homolog (PTEN), Janus kinase 2 (JAK2), and patched-1 protein (PTCH1). Variants were classified into categories 1, 2, 3, and 4. Variants were then manually reviewed, and recurrent variants occurring in exonic sequences were identified.
TABLE 1.
Variant Classification Scheme
| Variant Level | Significance | Details |
|---|---|---|
| 1 | Known driver for MPNSTs | Variants reported in various publications to be associated with MPNST |
| 2 | Reported in cancer or other diseases | Previously reported as a somatic variant in COSMIC or reported as clinically significant (pathogenic) in ClinVar archive |
| 3 | Common variants | Synonymous/intergenic variants or variants with 1000 genome maf > 0.05 or ESP6500maf>0.05 |
| 4 | Unknown | Novel variant of unknown significance |
Abbreviation: COSMIC, Catalogue Of Somatic Mutations In Cancer; MPNST, malignant peripheral nerve sheath tumor; maf, minor allele frequency.
Immunohistochemistry and Tissue
Microarray Generation
Immunohistochemical staining was performed using a rabbit polyclonal antibody to neurofibromin (1:500 dilution; Santa Cruz Biotechnology, Dallas, Tex) or a rabbit polyclonal antibody to TYK2 antibody (ab39550, dilution 1:1000; Abcam, Cambridge, Mass;) with citrate antigen retrieval. Immunohistochemistry for histone 3 lysine trimethylation (H3K27 me3) was performed using rabbit monoclonal antibodies (C36B11, 1:50 dilution; Cell Signaling Technology, Danvers, Mass) and Trisethylenediaminetetraacetic acid (EDTA) antigen retrieval. Previously generated tissue microarray blocks contained at least 2 cores of tissue (2 mm in diameter) from the most representative areas of the tumor, as well as from normal peripheral nerve.22 Breast carcinoma was used as a positive control for TYK2 staining. Images for TYK2 and neurofibromin immunohistochemistry were acquired at × 200 magnification using a Nikon Eclipse E600 microscope (Nikon USA, Melville, NY) equipped with an optical camera (Leica EC3; Leica Biosystems) and Leica Application suite image analysis software (Version 2.1.0; Leica Biosystems). Tumors with strong immunostaining in >80% of the cells were scored as positive, whereas those with weak or no immunostaining were deemed negative. H3K27 me3 immunoreactivity was evaluated by one of the authors (M.P.) who was blinded to the other molecular features of the cases. Tumors with nuclear staining in <5% of the tumor cells, in the presence of an internal positive control (eg, endothelial cells), were scored as having a loss of H3K27 me3.
Survival Analysis
Clinical data from the MPNST cases from the study institutions were accessed by electronic medical records. Patients were censored by date of death (Social Security Death Index) or date of last follow-up. Overall survival data was generated by Kaplan-Meier analysis and the log-rank test using GraphPad Prism statistical software (version 5.03; GraphPad Software, San Diego, Calif).
RESULTS
We collected clinical data on 7 adults with NF1-associated MPNSTs: 3 women and 4 men (Table 2). The average age of the subjects at the time of diagnosis of MPNST was 39 years (median, 38 years; range, 20–50 years), which is consistent with previous studies reporting a lower age of onset for NF1-MPNSTs.22,23 One subject was found to have metastatic disease at the time of diagnosis whereas the other individuals presented with localized disease. At the time of the study, 2 subjects died, 4 were being actively followed at Barnes-Jewish Hospital/Washington University, and 1 individual was lost to follow-up. All DNA that was sequenced was isolated from the primary tumors by microdissection to ensure that samples contained pure tumor. We specifically selected a sequencing platform that is similar to the Clinical Laboratory Improvement Amendments-approved service at the study institution to identify actionable mutations in cancer relevant to the continued application of this discovery modality for future precision oncology treatments.
TABLE 2.
Clinical Characteristics of NF1-MPNSTs
| Subject ID | Age at Diagnosis, Years | Sex | Tumor Location | Tumor Size/Grade | Surgical Margin Status | Disease Status | Metastasis | Time From Diagnosis to Metastasis, Months | Adjuvant Treatment | OS, Months |
|---|---|---|---|---|---|---|---|---|---|---|
| 1111 | 20 | Woman | Left thigh | 7.5 cm/high | Positive | Deceased | Lung | 9 | XRT | 31 |
| 2222 | 37 | Man | Left pelvis | 8.8 cm/high | Positive | NED | No | NA | XRTand chemo | 39 |
| 1405601 | 38 | Man | Right thigh | 9 cm/high | NA | Deceased | Lung, bone | At diagnosis | NA | 19 |
| 1405593 | 34 | Woman | Chest wall | 20 cm/low | Negative | NED | No | NA | XRT | 49 |
| 1405577 | 50 | Man | Sacral | 13 cm/high | Positive | Recurred | No | NA | XRTand chemo | 44 |
| 1405576 | 44 | Man | Abdominal wall | 13 cm/high | Positive | NED | No | NA | XRTand chemo | 45 |
| 1405575 | 48 | Woman | Buttocks | 6 cm/low | Positive | unknown | NA | NA | NA | Lost to follow-up |
Abbreviations: chemo, chemotherapy; ID, identification; NA, not applicable; NED, no evidence of disease; NF1-MPNST, neurofibromatosis type 1-associated malignant peripheral nerve sheath tumors; OS, overall survival; XRT, radiotherapy.
On average, there were 67 mutations per tumor categorized as variants in: 1) genes associated with MPNST; 2) genes previously reported to be mutated in other cancers; or 3) mutations for which the clinical significance is currently unknown (Table 3). Mutations were identified in 4 genes previously implicated in MPNST pathogenesis in both human studies and genetically engineered mouse models (NF1, TP53, EGFR, and PDGFR-α [PDGFRA]).24–28 Variants also were identified in other genes potentially involved in MPNST pathogenesis (GNAQ, SLCO1B1, LAMA2, SLC34A2, PTCH1, RB1, KDR, CYP2A6, MYC, FLT4, and PSMD2).17,29–34 In addition, there were variants identified in genes not previously implicated in MPNST pathogenesis (TYK2, CYP2D6, ABCB1, CSF1R, MAP3K1, JAK3, GNA11, FLT3, ROS1, APC, HTR2B, DPYD, ATRX, EV12A, CYP2C19, ALK, GNAS, CYP2B6, BRCA1, NOTCH1, DDR2, RAF1, SMARCB1, FLT1, smoothened, frizzled class receptor [SMO], ESR1, ERBB4, CREBBBP, and ABL1). Mutations in many of these genes have been identified in other malignancies.
TABLE 3.
Genes With Exonic Mutations
| NF1 | GNAQ | TYK2 |
| TP53 | SLC01B1 | CYP2D6 |
| EGFR | LAMA2 | ABCB1 |
| PDGFRA | SLC34A2 | CSF1R |
| PTCH1 | MAP3K1 | |
| RB1 | JAK3 | |
| KDR | GNA11 | |
| CYP2A6 | FLT3 | |
| MYC | R0S1 | |
| FLT4 | APC | |
| PSMD2 | HTR2B | |
| DPYD | ||
| ATRX | ||
| EV12A | ||
| CYP2C19 | ||
| ALK | ||
| GNAS | ||
| CYP2B6 | ||
| BRCA1 | ||
| N0TCH1 | ||
| DDR2 | ||
| RAF1 | ||
| SMARCB1 | ||
| FLT1 | ||
| SMO | ||
| ESR1 | ||
| ERBB4 | ||
| CREBBP | ||
| ABL1 | ||
Abbreviations: ABCB1, ATP-binding cassette subfamily B member 1; ABL1, Abelson murine leukemia viral oncogene homolog 1; ALK, anaplastic lymphoma kinase; APC, adenomatous polyposis coli; ATRX, alpha thalassemia/mental retardation syndrome X-linked; CREBBP, CREB-binding protein; CSF1R, colony-stimulating factor 1 receptor; CYP2A6, cytochrome P450 2A6; CYP2B6, cytochrome P450 family 2 subfamily B member 6; CYP2C19, cytochrome P450 family 2 subfamily C member 19; CYP2D6, cytochrome P450 family 2 subfamily D member 6; DDR2, discoidin domain receptor tyrosine kinase 2; DPYD, dihydropyrimidine dehydrogenase; EGFR, epidermal growth factor receptor; ERBB4, Erb-B2 receptor tyrosine kinase 4; ESR1, estrogen receptor 1; EV12A, ectropic viral integration site 2A; FLT1, Fms-related tyrosine kinase 1; FLT3, Fms-like tyrosine kinase 3; FLT4, Fms-related tyrosine kinase 4; GNA11, G protein subunit a 11; GNAQ, G protein subunit a Q; HTR2B, 5-hydroxytryptamine receptor 2B; JAK3, Janus kinase 3; KDR, kinase insert domain receptor; LAMA2, laminin subunit a 2; MAP3K1, mitogen-activated protein kinase kinase kinase 1; NF1, neurofibromatosis type 1; PDGFRA, platelet-derived growth factor receptor a; PSMD2, proteasome 26S subunit, non-ATPase 2; PTCH1, patched-1 protein; RB1, retinoblastoma 1; ROS1, ROS proto-oncogene 1; SLC34A2, solute carrier family 34 member 2; SLCO1B1, solute carrier organic anion transporter family member 1B1; SMARCB1, SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily B, member 1; SMO, smoothened, frizzled class receptor; TP53, tumor protein p53; TYK2, tyrosine kinase 2.
a Gray shading indicates strong murine and human data, green shading indicates published molecular/sequencing data, and pink shading indicates new variants of unknown significance.
After manual review of our sequencing data, 4 of these genes were found to be recurrently mutated in other NF1-MPNSTs in the current series, and harbored mutations within potential functional domains (NF1, TP53, ROS1, and TYK2) (Table 4). Consistent with these tumors arising in adults with NF1, all tumors harbored at least 1 mutation predicted to be pathogenic in the NF1 gene and 86% of tumors (6 of 7 tumors) exhibited loss of NF1 protein (neurofibromin) expression by immunohistochemistry (see Supporting Information Fig. 1). It is interesting to note that the 1 tumor that retained neurofibromin expression contained only 1 mutation predicted to be pathogenic whereas the other 6 tumors contained 2 mutations predicted to be pathogenic. Although ROS1 mutations are reported to occur in 15% to 35% of melanomas, TP53 mutations are commonly reported in other sarcomas, malignant gliomas (glioblastoma, glioblastoma multiforme), and melanoma (Table 5).29,35
TABLE 4.
Genes With Recurrent Mutations Located in Predicted Functional Protein Domainsa
| Tumor | |||||||
|---|---|---|---|---|---|---|---|
| Gene | 1405601 | 1405593 | 1405577 | 1405576 | 1405575 | 1111 | 2222 |
| NF1 | |||||||
| ROS1 | |||||||
| TP53 | |||||||
| TYK2 | |||||||
Abbreviations: NF1, neurofibromatosis type 1; ROS1, ROS proto-oncogene 1; TP53, tumor protein p53; TYK2, tyrosine kinase 2.
Black boxes indicate mutation, whereas the white boxes denote the absence of a mutation.
TABLE 5.
Genes Mutated in Other Cancers
| Gene | Sarcoma | GBM | Melanoma | Pheochromocytoma |
|---|---|---|---|---|
| NF1 | 3.4%−8.8% | 6.1%−17.6% | 12%−45% | 8.7% |
| ROS1 | 5% | 1 %−2.2% | 15.4%−35% | 0.6% |
| TP53 | 13.5%−45% | 29.7%−35.2% | 12.1%−25% | 0.6% |
| TYK2 | 1.9%−6.7% | 2.2% | 3.2%−5% | 0 |
Abbreviations: GBM, glioblastoma multiforme; NF1, neurofibromatosis type 1; ROS1, ROS proto-oncogene 1; TP53, tumor protein p53; TYK2, tyrosine kinase 2.
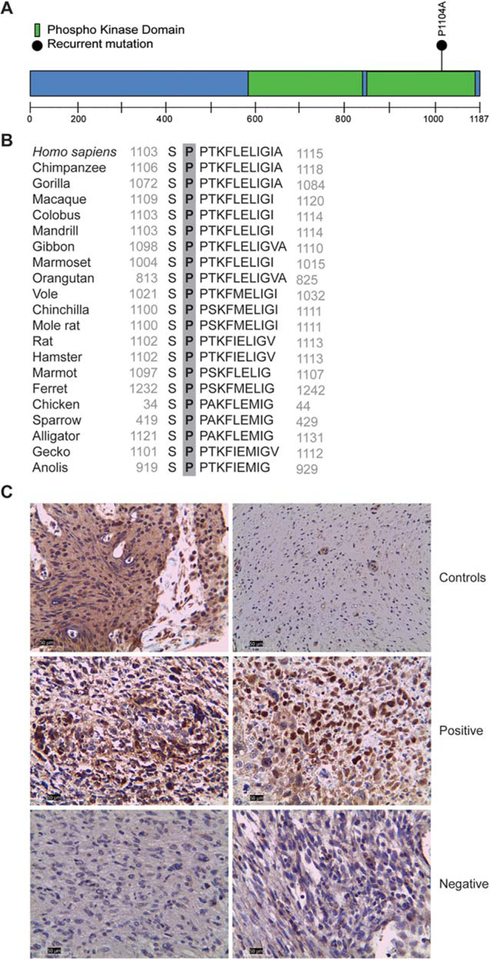
It is important to note that the specific mutation identified in the TYK2 gene (Pro1104Ala) (Fig. 1A) previously has been identified in one case of diffuse large B-cell lymphoma,29,35 and positive TYK2 immunoreactivity and increased signaling has been demonstrated in other cancers.36 Moreover, this specific mutation occurs within a highly conserved domain (Fig. 1B), in which it has been associated with autoimmune disease.37,38 In addition, this mutation was predicted to be pathogenic using several programs, including SIFT (Sorting Intolerant from Tolerant),39 PolyPhen2,40 and Mutation Assessor.41 Furthermore, this mutation has been predicted to be oncogenic, based on a previously published computational analysis.42 In light of all these observations, we performed immunohistochemistry to determine whether there was deregulated TYK2 expression in the original 7 cases of NF1-MPNST. Although strong immunoreactivity was detected in both NF1-MPNST cases with the TYK2-Pro1104Ala mutation, little or no immunoreactivity was observed in NF1-MPNST cases lacking this mutation (Fig. 1C).
Figure 1.
Tyrosine kinase 2 (TYK2) mutation and protein expression in malignant peripheral nerve sheath tumors (MPNSTs). (A) Schematic representation of the predicted TYK2 protein sequence. The location of the exonic mutations is denoted above the protein schematic (amino acid residue).(B) The proline (P) residue at position 1104 is highly conserved in vertebrates. (C) TYK2 immunoreactivity was observed in TYK2-mutant, but not in TYK2-wild-type, neurofibromatosis type 1-associated MPNSTs. Breast carcinoma was included as a positive control, whereas normal sural nerve served as a negative control for TYK2 immunostaining.
Because 30% of the original 7 cases of NF1-MPNSTs harbored this TYK2 mutation and exhibited strong TYK2 protein expression that correlated with mutation status, we next examined TYK2 protein expression in a larger set of MPNSTs. Using an independently generated tissue microarray containing 28 MPNST cases (both sporadic and those associated with NF1), 1 neurofibroma, and 1 schwannoma, we observed strong immunopositivity in 17 MPNST cases (approximately 60%: 55% of sporadic cases [6 of 11 cases] and 65% of NF1-associated cases [11 of 17 cases]). In contrast, neither of the benign tumors (neurofibroma and schwannoma) were found to be immunopositive for TYK2 (data not shown), similar to normal peripheral nerve. There was no significant difference noted with regard to overall patient survival based on TYK2 expression (P = .4893).
DISCUSSION
MPNSTs are aggressive sarcomas with limited treatment options and poor overall survival given the high rates of recurrence and metastatic disease. This is particularly true for MPNSTs arising within the context of NF1, in which individuals present at an earlier age than their counterparts with sporadic disease and thus often die at a young age, often in their 20s and 30s.43,44 For this reason, it is critical to identify more effective therapies for this subgroup of individuals with MPNST. Based on the emergence of molecularly targeted therapies, we sought to apply a clinically applicable genomic profiling platform to discover potentially targetable mutations in cases of NF1-MPNST. The current study raises 3 important points.
First, NF1 gene mutations were identified in all the subjects analyzed, which is consistent with their diagnosis of NF1.45,46 There was no specific clustering of mutations and all were predicted to result in impaired neurofibromin function (ie, frameshift, deletion, and nonsense mutations). Consistent with biallelic NF1 gene inactivation in NF1-MPNSTs, immunohistochemistry revealed loss of neurofibromin expression in approximately 86% of these tumors. It is interesting to note that the 1 tumor that retained neurofibromin expression harbored only a single mutation predicted to be pathogenic, whereas the other 6 tumors contained 2 NF1 gene mutations predicted to be pathogenic.
Second, we identified genes previously implicated in MPNST pathogenesis. In this regard, approximately 30% of subjects (2 of 7 individuals) harbored a mutation in the TP53 gene. These mutations were located in the DNA-binding domain, and would be predicted to result in loss of p53 function.47 Although EGFR26 and PDGFRA48 mutations were identified in nonfunctional domains, to the best of our knowledge, the significance of these variants is unknown. In addition, we found nucleotide var iants in several NOTCH signaling pathway genes (PTCH1, NOTCH1, and SMO); however, these mutations were not located in functional domains and their biological significance is unclear. Although our clinical sequencing platform examines many of the known genes involved in MPNST pathogenesis (including TP53, NF1, CDKN2A, EGFR, PDGF, ERBB2, KIT, MET, PDGFR, PTEN, JAK2, and PTCH1), there are some important genes not included on this platform, such as polycomb repressive complex 2 (PRC2)/polycomb repressive complex 2 subunit (SUZ12), which is deregulated in as many as 70% of MPNSTs.9,49 As a surrogate for PRC2/SUZ12 loss, we performed immunohistochemistry to examine H3K27 me3, a known downstream target of SUZ12.50 Consistent with PRC2/SUZ12 involvement, approximately 60% of the NF1-MPNSTs (20 of 35 NF1-MPNSTs) demonstrated loss of H3K27 me3 (data not shown).
Third, we identified 2 genes with mutations (ROS1 and TYK2) in 2 of the 7 NF1-MPNST cases in the current series. ROS1 is a receptor tyrosine kinase that is rearranged in approximately 1% of lung cancer cases.51–53 The 2NF1-MPNST cases in the current study harbored single-nucleotide variants, which resulted in nonsynonymous mutations within the predicted fibronectin type domain of ROS1, rather than the kinase domain in which drugs such as crizotinib act.51 As such, the significance of this specific mutation to our knowledge is unknown, and is not likely to result in a protein targetable by currently available inhibitors. Further analysis of this mutation in the biology of MPNST will be required.
In contrast, TYK2 was mutated or its protein product overexpressed in approximately one-half of MPNST cases examined in the current study, which is significantly greater than that observed in other sarcomas, malignant gliomas, melanomas, or pheochromocytomas (<7% of all tumors). Moreover, the particular recurrent mutation (Pro1104Ala) was located within a highly evolutionarily conserved Jak1 homology domain (KH2; α helical region). TYK2 is a kinase molecule that associates with the cytoplasmic domain of cytokine receptors54 to activate STAT signaling and promote cancer cell survival by upregulating the BCL2 pro-survival gene.55 Relevant to cancer therapeutics, TYK2 protein stability is in part mediated by HSP90, the molecular chaperone protein, such that HSP90 targeting using small molecule inhibitors results in the rapid degradation of TYK2, reduced BCL2 expression, and apoptosis in patients with T-cell leukemia.56
The discovery of tumors with TYK2 immunoreactivity may serve as an indirect biomarker with which to identify those specific individuals whose MPNSTs are more likely to respond to agents such as ganetespib in combination with radiotherapy and targeted or conventional chemotherapy. In this regard, HSP90 inhibitors currently are being examined in clinical trials for individuals with NF1-MPNST (ClinicalTrials.gov Identifier NCT02008877).57 In addition, the established role of TYK2 in regulating cell survival in cancer raises the intriguing possibility that small molecular inhibitors that target HSP90 will increase TYK2/BCL2-mediated MPNST cell death and result in more effective clinical outcomes. Future mechanistic studies will be required to determine whether TYK2 expression predicts patient response to HSP90 inhibitors.
Supplementary Material
FUNDING SUPPORT
Funded in part by generous gifts from Schnuck Markets Inc (to David H. Gutmann) and the St. Louis Men’s Group Against Cancer (to David H. Gutmann).
Footnotes
CONFLICT OF INTEREST DISCLOSURES
Angela C. Hirbe is supported by the T32 Hematology Training Grant (HL007088) and a SARC Career Development Grant.
REFERENCES
- 1.Huson SM, Harper PS, Compston DA. Von Recklinghausen neurofibromatosis. A clinical and population study in south-east Wales. Brain. 1988;111(pt 6):1355–1381. [DOI] [PubMed] [Google Scholar]
- 2.Ferner RE, Huson SM, Thomas N, et al. Guidelines for the diagnosis and management of individuals with neurofibromatosis 1. J Med Genet. 2007;44:81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferner RE, Gutmann DH. International consensus statement on malignant peripheral nerve sheath tumors in neurofibromatosis. Cancer Res. 2002;62:1573–1577. [PubMed] [Google Scholar]
- 4.Evans DG, Baser ME, McGaughran J, Sharif S, Howard E, MoranA. Malignant peripheral nerve sheath tumours in neurofibromatosis1. J Med Genet. 2002;39:311–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hruban RH, Shiu MH, Senie RT, Woodruff JM. Malignant peripheral nerve sheath tumors of the buttock and lower extremity. A study of 43 cases. Cancer. 1990;66:1253–1265. [DOI] [PubMed] [Google Scholar]
- 6.Kourea HP, Bilsky MH, Leung DH, Lewis JJ, Woodruff JM. Subdiaphragmatic and intrathoracic paraspinal malignant peripheral nerve sheath tumors: a clinicopathologic study of 25 patients and 26 tumors. Cancer. 1998;82:2191–2203. [DOI] [PubMed] [Google Scholar]
- 7.Wong WW, Hirose T, Scheithauer BW, Schild SE, Gunderson LL. Malignant peripheral nerve sheath tumor: analysis of treatment outcome. Int J Radiat Oncol Biol Phys. 1998;42:351–360. [DOI] [PubMed] [Google Scholar]
- 8.Bhola P, Banerjee S, Mukherjee J, et al. Preclinical in vivo evaluation of rapamycin in human malignant peripheral nerve sheath ex-plant xenograft. Int J Cancer. 2010;126:563–571. [DOI] [PubMed] [Google Scholar]
- 9.De Raedt T, Beert E, Pasmant E, et al. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature. 2014;514:247–251. [DOI] [PubMed] [Google Scholar]
- 10.De Raedt T, Walton Z, Yecies JL, et al. Exploiting cancer cell vulnerabilities to develop a combination therapy for ras-driven tumors. Cancer Cell. 2011;20:400–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johannessen CM, Johnson BW, Williams SM, et al. TORC1 is essential for NF1-associated malignancies. Curr Biol. 2008;18:56–62. [DOI] [PubMed] [Google Scholar]
- 12.Johansson G, Mahller YY, Collins MH, et al. Effective in vivo targeting of the mammalian target of rapamycin pathway in malignant peripheral nerve sheath tumors. Mol Cancer Ther. 2008;7:1237–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lock R, Ingraham R, Maertens O, et al. Cotargeting MNK and MEK kinases induces the regression of NF1-mutant cancers. J Clin Invest. 2016;126:2181–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watson AL, Anderson LK, Greeley AD, et al. Co-targeting the MAPK and PI3K/AKT/mTOR pathways in two genetically engineered mouse models of schwann cell tumors reduces tumor grade and multiplicity. Oncotarget. 2014;5:1502–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu J, Keng VW, Patmore DM, et al. Insertional mutagenesis identifies a STAT3/Arid1b/beta-catenin pathway driving neurofibroma initiation. Cell Rep. 2016;14:1979–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watson AL, Rahrmann EP, Moriarity BS, et al. Canonical Wnt/beta-catenin signaling drives human schwann cell transformation, progression, and tumor maintenance. Cancer Discov. 2013;3:674–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rahrmann EP, Watson AL, Keng VW, et al. Forward genetic screen for malignant peripheral nerve sheath tumor formation identifies new genes and pathways driving tumorigenesis. Nat Genet. 2013;45: 756–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Institutes of Health Consensus Development Conference Statement: neurofibromatosis. Bethesda, Md., USA, July 13–15, 1987. Neurofibromatosis; 1988;1:172–178. [PubMed] [Google Scholar]
- 19.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008; 18:1851–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Tool-kit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirbe AC, Pekmezci M, Dahiya S, et al. BRAFV600E mutation in sporadic and neurofibromatosis type 1-related malignant peripheral nerve sheath tumors. Neuro Oncol. 2014;16:466–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ducatman BS, Scheithauer BW, Piepgras DG, Reiman HM, Ilstrup DM. Malignant peripheral nerve sheath tumors. A clinicopathologic study of 120 cases. Cancer. 1986;57:2006–2021. [DOI] [PubMed] [Google Scholar]
- 24.Brosius SN, Turk AN, Byer SJ, et al. Neuregulin-1 overexpression and Trp53 haploinsufficiency cooperatively promote de novo malignant peripheral nerve sheath tumor pathogenesis. Acta Neuropathol. 2014;127:573–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Legius E, Dierick H, Wu R, et al. TP53 mutations are frequent in malignant NF1 tumors. Genes Chromosomes Cancer. 1994;10:250–255. [DOI] [PubMed] [Google Scholar]
- 26.Wu J, Patmore DM, Jousma E, et al. EGFR-STAT3 signaling promotes formation of malignant peripheral nerve sheath tumors. Onco-gene. 2014;33:173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cichowski K, Shih TS, Schmitt E, et al. Mouse models of tumor development in neurofibromatosis type 1. Science. 1999;286:2172–2176. [DOI] [PubMed] [Google Scholar]
- 28.Vogel KS, Klesse LJ, Velasco-Miguel S, Meyers K, Rushing EJ, Parada LF. Mouse tumor model for neurofibromatosis type 1. Science. 1999;286:2176–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang Y, Elahi A, Denley RC, Rao PH, Brennan MF, Jhanwar SC. Molecular characterization of permanent cell lines from primary, metastatic and recurrent malignant peripheral nerve sheath tumors (MPNST) with underlying neurofibromatosis-1. Anticancer Res. 2009;29:1255–1262. [PubMed] [Google Scholar]
- 31.Mantripragada KK, Spurlock G, Kluwe L, et al. High-resolution DNA copy number profiling of malignant peripheral nerve sheath tumors using targeted microarray-based comparative genomic hybridization. Clin Cancer Res. 2008;14:1015–1024. [DOI] [PubMed] [Google Scholar]
- 32.Reuss DE, Mucha J, Hagenlocher C, et al. Sensitivity of malignant peripheral nerve sheath tumor cells to TRAIL is augmented by loss of NF1 through modulation of MYC/MAD and is potentiated by curcumin through induction of ROS. PLoS One. 2013;8:e57152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McPherson JR, Ong CK, Ng CC, et al. Whole-exome sequencing of breast cancer, malignant peripheral nerve sheath tumor and neurofibroma from a patient with neurofibromatosis type 1. Cancer Med. 2015;4:1871–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spurlock G, Knight SJ, Thomas N, Kiehl TR, Guha A, UpadhyayaM. Molecular evolution of a neurofibroma to malignant peripheral nerve sheath tumor (MPNST) in an NF1 patient: correlation between histopathological, clinical and molecular findings. J Cancer Res Clin Oncol. 2010;136:1869–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ide H, Nakagawa T, Terado Y, Kamiyama Y, Muto S, Horie S. Tyk2 expression and its signaling enhances the invasiveness of prostate cancer cells. Biochem Biophys Res Commun. 2008;369:292–296. [DOI] [PubMed] [Google Scholar]
- 37.Eyre S, Bowes J, Diogo D, et al. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat Genet. 2012;44:1336–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Z, Gakovic M, Ragimbeau J, et al. Two rare disease-associated Tyk2 variants are catalytically impaired but signaling competent. J Immunol. 2013;190:2335–2344. [DOI] [PubMed] [Google Scholar]
- 39.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. [DOI] [PubMed] [Google Scholar]
- 40.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaminker JS, Zhang Y, Waugh A, et al. Distinguishing cancer-associated missense mutations from common polymorphisms. Cancer Res. 2007;67:465–473. [DOI] [PubMed] [Google Scholar]
- 43.Le Guellec S, Decouvelaere AV, Filleron T, et al. Malignant peripheral nerve sheath tumor is a challenging diagnosis: a systematic pa thology review, immunohistochemistry, and molecular analysis in 160 patients from the French Sarcoma Group Database. Am J Surg Pathol. 2016;40:896–908. [DOI] [PubMed] [Google Scholar]
- 44.Kolberg M, Holand M, Agesen TH, et al. Survival meta-analyses for > 1800 malignant peripheral nerve sheath tumor patients with and without neurofibromatosis type 1. Neuro Oncol. 2013;15: 135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perry A, Roth KA, Banerjee R, Fuller CE, Gutmann DH. NF1 deletions in S-100 protein-positive and negative cells of sporadic and neurofibromatosis 1 (NF1)-associated plexiform neurofibromas and malignant peripheral nerve sheath tumors. Am J Pathol. 2001;159: 57–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guha A, Lau N, Huvar I, et al. Ras-GTP levels are elevated in human NF1 peripheral nerve tumors. Oncogene. 1996;12:507–513. [PubMed] [Google Scholar]
- 47.Kato S, Han SY, Liu W, et al. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci U S A. 2003;100:8424–8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zietsch J, Ziegenhagen N, Heppner FL, Reuss D, von Deimling A, Holtkamp N. The 4q12 amplicon in malignant peripheral nerve sheath tumors: consequences on gene expression and implications for sunitinib treatment. PLoS One. 2010;5:e11858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang M, Wang Y, Jones S, et al. Somatic mutations of SUZ12 in malignant peripheral nerve sheath tumors. Nat Genet. 2014;46: 1170–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gall Troselj K, Novak Kujundzic R, Ugarkovic D. Polycomb repressive complex’s evolutionary conserved function: the role of EZH2 status and cellular background. Clin Epigenetics. 2016;8:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jorge SE, Schulman S, Freed JA, et al. Responses to the multitargeted MET/ALK/ROS1 inhibitor crizotinib and co-occurring mutations in lung adenocarcinomas with MET amplification or MET exon 14 skipping mutation. Lung Cancer. 2015;90:369–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuang BH, Zhang MQ, Xu LH, et al. Proline-rich tyrosine kinase 2 and its phosphorylated form pY881 are novel prognostic markers for non-small-cell lung cancer progression and patients’ overall survival. Br J Cancer. 2013;109:1252–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muller S, Chen Y, Ginter T, et al. SIAH2 antagonizes TYK2-STAT3 signaling in lung carcinoma cells. Oncotarget. 2014;5:3184–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan H, Piazza F, Krishnan K, Pine R, Krolewski JJ. Definition of the interferon-alpha receptor-binding domain on the TYK2 kinase. J Biol Chem. 1998;273:4046–4051. [DOI] [PubMed] [Google Scholar]
- 55.Sanda T, Tyner JW, Gutierrez A, et al. TYK2-STAT1-BCL2 pathway dependence in T-cell acute lymphoblastic leukemia. Cancer Discov. 2013;3:564–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taipale M, Krykbaeva I, Koeva M, et al. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell. 2012;150:987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Akahane K, Sanda T, Mansour MR, et al. HSP90 inhibition leads to degradation of the TYK2 kinase and apoptotic cell death in T-cell acute lymphoblastic leukemia. Leukemia. 2016;30:219–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.