Abstract
Basal cell carcinomas (BCCs) rely on Hedgehog (HH) pathway growth signal amplification by the microtubule-based organelle, the primary cilium. Despite naive tumors responsiveness to Smoothened inhibitors (Smoi), resistance in advanced tumors remains frequent. While the resistant BCCs usually maintain HH pathway activation, squamous cell carcinomas with Ras/MAPK pathway activation also arise, with the molecular basis of tumor type and pathway selection still obscure. Here we identify the primary cilium as a critical determinant controlling tumor pathway switching. Strikingly, Smoi-resistant BCCs possess an increased mutational load in ciliome genes, resulting in reduced primary cilia and HH pathway activation compared to naive or Gorlin patient BCCs. Gene set enrichment analysis of resistant BCCs with a low HH pathway signature reveals increased Ras/MAPK pathway activation. Tissue analysis confirms an inverse relationship between primary cilia presence and Ras/MAPK activation, and primary cilia removal in BCCs potentiates Ras/MAPK pathway activation. Moreover, activating Ras in HH-responsive cell lines confers resistance to both canonical (vismodegib) and non-canonical (aPKC and MRTF inhibitors) HH pathway inhibitors, while conferring sensitivity to MAPK inhibitors. Our results provide insights into BCC treatment and identify the primary cilium as an important lineage gatekeeper, preventing HH to Ras/MAPK pathway switching.
Introduction
Basal cell carcinoma (BCC) is the most common skin cancer, affecting more than 50% of Caucasians during their lifetime (Kasper et al., 2012). BCCs depend on a deregulated Hedgehog (HH) signaling pathway, leading to Smoothened (Smo) de-repression and the activation of GLI transcription factors (Epstein, 2008, Rubin and de Sauvage, 2006). Facilitating high pathway output is the primary cilium, a microtubule-based organelle where the core components of the HH pathway co-localize upon signal transduction (Bangs and Anderson, 2017, Pak and Segal, 2016, Wu et al., 2017). Intense study over the past decade demonstrates the necessity of the primary cilium for HH pathway activity, as human ciliopathies phenocopy HH mutations.
While Smo inhibitors (Smoi) potently prevent naïve BCC growth (Migden et al., 2015, Rubin and de Sauvage, 2006), resistance emerges rapidly with early and advanced sporadic BCC demonstrating 40 to 60% resistance respectively (Axelson et al., 2013, Chang and Oro, 2012, Sekulic et al., 2012, Tang et al., 2012). While sensitive BCC harbor reduced HH pathway activation under Smoi (Migden et al., 2015), detailed genomic interrogation of Smoi resistant BCCs has identified both genetic alterations in Smo and downstream compensatory mechanisms that maintain BCC growth through HH pathway activation. These include point mutations in Smoi binding-pocket or Smo activating mutations (Atwood et al., 2015, Sharpe et al., 2015, Yauch et al., 2009), amplification of Gli2 or the HH target gene Cyclin D1 (Buonamici et al., 2010, Dijkgraaf et al., 2011), and non-canonical mechanisms to bolster HH pathway output through enhanced PI3K, aPKC signaling or G-actin-mediated activation of the MRTF/SRF complex (Atwood et al., 2013, Buonamici et al., 2010, Whitson et al., 2018). These results demonstrate the diverse mechanisms used to maintain GLI activity in resistant BCCs and suggest the existence of other as yet undiscovered resistance pathways.
An emerging resistance route to Smoi resistance occurs through the switching of BCC to squamous cell carcinoma (SCC) (Otsuka et al., 2015, Ransohoff et al., 2015, Saintes et al., 2015, Zhao et al., 2015). Primarily seen in patients with sporadic, not syndromic BCCs, multiple independent studies have found that the SCC arising during Smoi treatment shares high sequence similarity to the naïve BCC, supporting the idea that these SCC emerge directly from the naïve BCC (Ransohoff et al., 2015, Zhao et al., 2015). Rather than HH dependence for cell growth, SCCs depend instead on the Ras/MAP kinase (MAPK) pathway activation and loss of Notch signaling for proliferation and blocking of differentiation respectively (Lee et al., 2014, Lefort et al., 2007). While previous work in transgenic mice illustrates how Ras pathway blockade in the context of SCC development leads to hair follicle-lineage sebaceous adenomas with increased HH signaling (Gerdes et al., 2006), the factors determining BCC-to-SCC pathway switching in human tumor setting remain mysterious.
In this study, we find increased levels of mutations in genes encoding the ciliome as well as reduced primary cilia in human resistant BCC. Importantly, loss of cilia correlates with lower HH and higher Ras/MAPK pathway activation. Consistently, both pharmacological and genetic depletions of primary cilia inhibit HH pathway, while potentiating Ras/MAPK pathway activation. Using GLI-dependent cell lines, we show that Ras/MAPK pathway activation confers resistance to both canonical and non-canonical HH pathway inhibitors, while conferring sensitivity to MEK inhibitors, suggesting a switch from a GLI-dependent to a GLI-independent cell state. Altogether, these results identify primary cilia as gatekeepers between HH and Ras/MAPK signaling pathways in BCC. They support the switch to Ras/MAPK pathway as a potential mechanism of resistance to both canonical and non-canonical HH pathway inhibitors.
Results
Human resistant BCCs harbor reduced primary cilia.
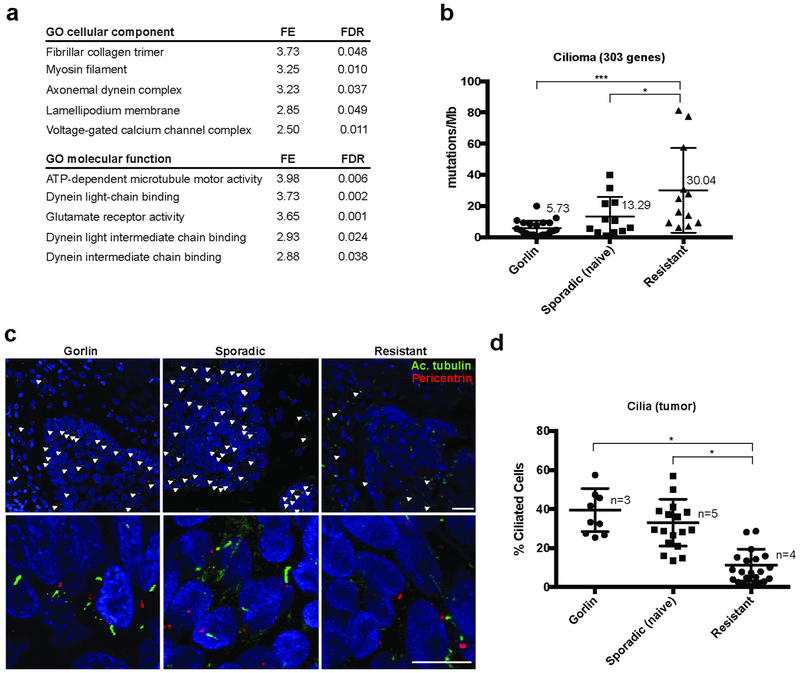
We have previously observed that the rate of resistance to Smoi differed dramatically between different patient populations, with syndromic Gorlin’s syndrome patients demonstrating significantly less resistance despite hundreds of tumors (Tang et al., 2012). To gain insight into the basis of this resistance, we submitted our previously identified list of genes commonly mutated in resistant BCCs (Atwood et al., 2015) (GEO accession number GSE58377; Suppl. Table 1) to gene ontology analysis. Remarkably, analyses for both cellular component and molecular function highlighted constitutive and functional components of the primary cilium (Figure 1a). To quantify mutations in ciliary genes, we reported the mutations in the coding sequences of the 303 ciliary genes comprised in the SYSCILIA list (van Dam et al., 2013) for Gorlin patients, sporadic naïve and vismodegib-resistant BCCs (National Institutes of Health Sequence Read Archive SAMN07507265 – SAMN07507288, GEO accession number GSE58377; Suppl. Fig. S1a). Importantly, the mutation rates found in the ciliome (5.73 and 13.29 mut/Mb for Gorlin patient and sporadic BCCs respectively) were significantly lower than the genome-wide mutation rates previously reported in BCCs (21 and 65 mut/Mb respectively) (Bonilla et al., 2016), indicating that the ciliome is conserved during the early stages of BCC progression. This is consistent with the prominent role of cilia for HH pathway activation and maintenance. However, ciliome mutations were found in significantly higher number in resistant compared to sporadic or Gorlin patients BCC (Fig. 1b), including when controlling for the large gene size of ciliary protein-encoding genes.
Figure 1. Human resistant BCC harbor reduced primary cilia.
(a) Top-lists of GO cellular components and molecular functions associated with genes commonly mutated in resistant BCCs.
(b) Quantification of the mutations found in the ciliome of Gorlin patients, sporadic naïve and resistant human BCCs using whole exome sequencing. Numbers indicate the mean values for each category.
(c) Acetylated tubulin (cilia shaft), pericentrin (cilia body) and DAPI immunostainings of Gorlin patient, sporadic naïve and resistant human BCCs. White arrowheads indicate cilia.
(d) Quantification of cilia in the tumor compartment shown in (c). Dots represent microscope fields, distributed across n>3 tumors.
FE stands for fold enrichment. FDR stands for false-discovery rate. Scale bars indicate 25μm. Horizontal bars and error bars in (b) and (d) represent the mean ± SD. *p < 0.05, ***p < 0.001.
To confirm that these sequence alterations in ciliome components were reflected in altered ciliogenesis, we compared Gorlin patients, sporadic naïve and resistant BCCs for cilia by immunostaining. Consistent with the sequencing data, we found that resistant BCCs harbored significantly reduced cilia number (Fig. 1c). To question the potential contribution of Smoi in reduced ciliogenesis (Otsuka et al., 2015), we compared primary cilia in both the tumor and stromal compartments. Importantly, cilia were lost specifically in the tumor epithelium, and not in the stromal compartment (Fig. 1d and Suppl. Fig. S1b). Moreover, Gorlin patients BCC treated with Smoi harbored conserved cilia relatively to resistant BCC, despite efficient HH pathway inhibition, evaluated by nuclear Gli1 staining intensity (Suppl. Fig. S1c and S1d). Finally, as ciliogenesis may be affected by a higher proliferation rate, we compared proliferation rates of sporadic naïve to resistant BCC using Ki67 immunostaining and failed to detect differences (Suppl. Fig. S1e). Altogether, these observations suggest a major contribution of the genetic mutations rather than the Smoi therapy or the proliferation status in the reduced ciliogenesis observed in resistant BCCs.
Loss of primary cilia correlates with HH pathway inactivation and Ras/MAPK pathway activation in a subset of resistant BCCs.
The observation of reduced cilia questions previous demonstration that BCCs uniformly depend on the HH pathway for growth; indeed, all Smoi resistant BCCs our group and others have characterized maintain or upregulate HH pathway target genes (Atwood et al., 2015, Sharpe et al., 2015). This led us to investigate alternative signaling mechanisms upon which BCCs with altered cilia depend for growth, a phenomenon well-characterized in many tumor types as pathway switching (Chandarlapaty, 2012, Chandarlapaty et al., 2011, Serra et al., 2011). Our group and others previously reported the emergence of SCCs from vismodegib-treated BCCs (Otsuka et al., 2015, Ransohoff et al., 2015, Zhao et al., 2015). As Ras/MAPK pathway is a main driver of SCC progression (Lee et al., 2014, Lefort et al., 2007, Pickering et al., 2014), and was shown to promote resistance to sonidegib, a Smoi, in an experimental model of medulloblastoma (Zhao et al., 2015), we hypothesized that BCC may transition from HH to Ras/MAPK pathway upon Smoi therapy.
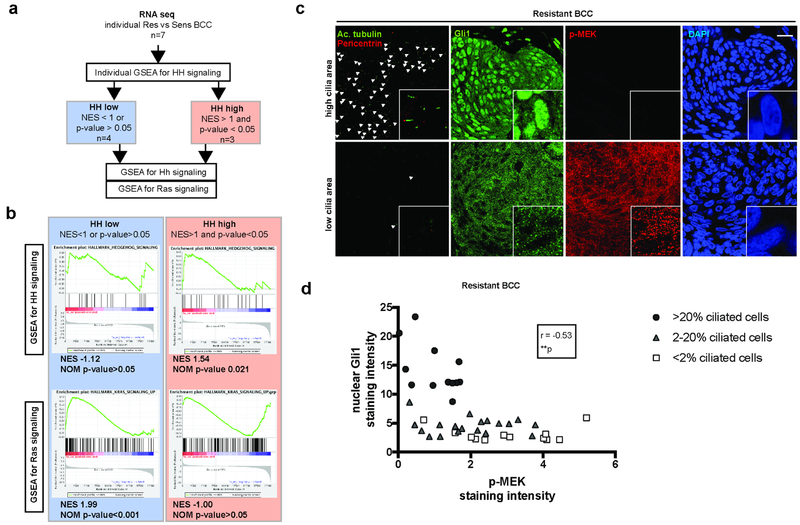
To determine whether Ras pathway was activated in resistant BCC, we used previously obtained RNA sequencing data (GEO accession number GSE58377) (Atwood et al., 2015). As Ras pathway is activated during normal skin differentiation, we decided to compare transcriptomes of resistant to sporadic naïve BCC samples (rather than normal skin) obtained from the same patient. In this setting, we identified 7 matched pairs. When the RNA sequencing data were analyzed together, GSEA identified HH pathway (as previously reported in (Atwood et al., 2015, Sharpe et al., 2015)), but failed to identify Ras pathway activation in resistant BCCs (Suppl. Fig. S2a). However, we may reasonably expect the activation of the Ras pathway to occur as an alternative rather than an additional mechanism to HH pathway activation in resistant BCC. Therefore, we distinguished “HH high” paired samples defined by a significant enrichment for HH pathway (NES>1 and p-value<0.05) from “HH low” paired samples (NES<1 or p-value>0.05) (Fig. 2a). When analyzed separately, we confirmed the absence of Ras pathway enrichment in the “HH high” group, whereas Ras pathway was significantly enriched in the “HH low” group (Fig. 2b and Suppl. Fig. S2b).
Figure 2. Loss of primary cilia goes with HH pathway inactivation and Ras/MAPK pathway activation in a subset of resistant BCCs.
(a) Algorithm for the analysis of RNA-sequencing obtained from vismodegib-sensitive and - resistant human BCCs.
(b) GSEA for HH and Ras pathways activation in “HH-high” versus “HH-low” human resistant BCCs. NES stands for normalized enrichment score.
(c) Adjacent immunostainings for cilia (acetylated tubulin and pericentrin), Gli1 (as a readout for HH pathway activation) and p-MEK (as readout for Ras/MAPK pathway activation) in resistant human BCCs. White arrowheads indicate cilia. Higher magnifications are shown in framed pictures.
(d) Correlation between nuclear Gli1 and p-MEK relative intensity in resistant human BCCs, annotated for cilia density. Dots represent microscope fields, distributed across 4 different resistant BCC tumors.
Scale bars indicate 25μm. **p < 0.01. r stands for Spearman correlation factor.
To further document their reciprocal relationship, we quantified cilia, nuclear Gli1 (as readout for HH pathway activation, Suppl. Fig. S1d) and p-MEK (as readout for Ras/MAPK pathway activation, Suppl. Fig. S1d) in various areas across resistant BCCs on immediate adjacent sections. Overall, cilia, nuclear Gli1 and p-MEK immunostainings revealed intra- and inter-tumoral heterogeneity. Interestingly, nuclear Gli1 and p-MEK immunostainings revealed a non-continuous inverse correlation identifying both Gli1high/p-MEKlow and Gli1low/p-MEKhigh areas (n= 43 sections examined, Fig. 2c and 2d; Suppl. Fig. S2c). Notably, cilia were mostly conserved in the Gli1high/p-MEKlow areas, while mostly lost in the Gli1low/p-MEKhigh areas (Fig. 2c and 2d). As expected, primary cilia showed a significant positive correlation with HH pathway activation (Spearman correlation factors of 0.80, Suppl. Fig. S2d), while a significant negative correlation with Ras/MAPK pathway activation (Spearman correlation factors of −0.61, Suppl. Fig. S2e). Altogether, our results identify an antagonistic relationship between cilia/HH pathway and Ras/MAPK pathway in a subset of resistant BCCs.
Loss of primary cilia potentiates Ras/MAPK pathway activation in resistant BCC cell lines.
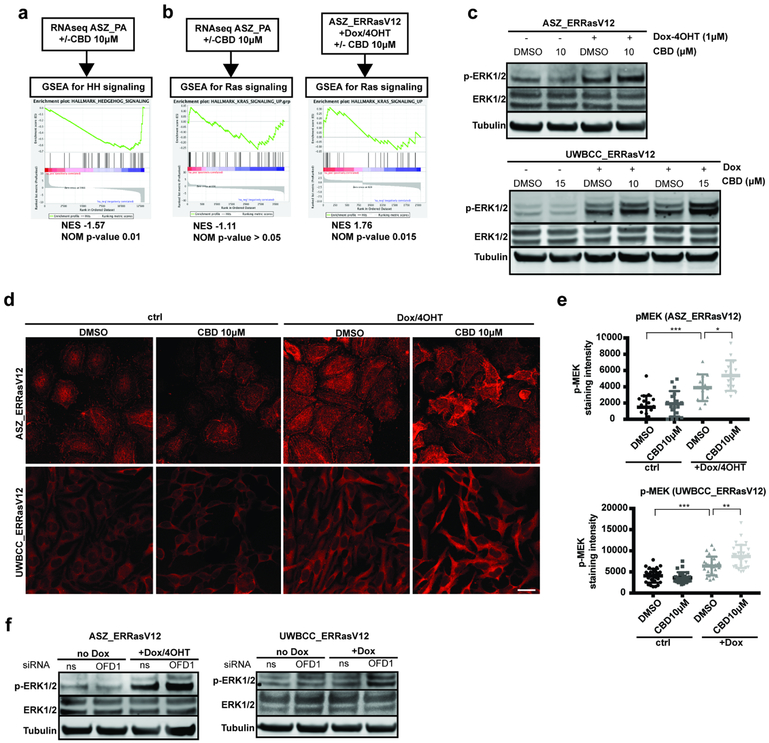
Our sequencing and histological results suggest a strong reciprocal relationship between the presence of normal cilia/HH pathway activation and Ras/MAPK pathway activation. To confirm the cilia-dependency of the HH pathway in vitro, we used the GLI-dependent murine (ASZ) and human (UWBCC) BCC cell lines. First we mimicked loss of cilia using ciliobrevin D (CBD), a cytoplasmic dynein inhibitor known to impede cilia formation (Fu et al., 2014). Both ASZ and UWBCC cell lines showed a dose-dependent reduction in both the fraction of ciliated cells and the cilia length upon CBD treatment (Suppl. Fig. S3a-c). To determine HH pathway response upon CBD treatment, we performed RNA sequencing on ASZ upon DMSO (control) or CBD (10μM) (GEO accession number GSE120954). GSEA revealed a significant negative enrichment for HH pathway upon CBD (Fig. 3a), demonstrating HH pathway inactivation upon loss of cilia. This was confirmed in both ASZ and UWBCC cell lines showing a dose-dependent reduction in Gli1 mRNA in response to CBD treatment (Suppl. Fig. S3d and S3e). To confirm these results, we tested a complementary genetic approach, using mRNA silencing of Oral-facial-digital syndrome 1 (OFD1), a gene essential for primary cilia formation (Ferrante et al., 2006). OFD1_siRNA efficiently silenced OFD1 mRNA (Suppl. Fig. S3f and S3g), prevented cilia formation (Suppl. Fig. S3h-j) and inhibited HH pathway activation in both ASZ and UWBCC cells (Suppl. Fig. S3k), confirming the cilia-dependency of the HH pathway in GLI-dependent BCC cell lines.
Figure 3. Loss of primary cilia potentiates Ras/MAPK pathway activation.
(a) GSEA for HH pathway activation in RNA-sequencing datas obtained from ASZ_PA upon DMSO (control) versus CBD (10μM) treatment.
(b) GSEA for Ras pathway activation in RNA-sequencing datas obtained from ASZ_PA or ASZ_ERRasV12/Dox-4OHT upon DMSO (control) versus CBD (10μM) treatment.
(c) Western blot for p-ERK1/2 (as readout for Ras/MAPK pathway activation), total ERK1/2 and β tubulin (loading control) in ASZ_ERRasV12 and UWBCC_ERRasV12 upon doxycycline/4OHT and DMSO (control) versus CBD treatment.
(d) p-MEK immunostaining (as readout for Ras/MAPK pathway activation) in ASZ_ERRasV12 and UWBCC_ERRasV12 upon doxycycline/4OHT and DMSO (control) versus CBD treatment.
(e) Quantification of p-MEK immunostaining in ASZ_ERRasV12 and UWBCC_ERRasV12 upon doxycycline/4OHT and DMSO (control) versus CBD treatment.
(f) Western blot for p-ERK1/2, total ERK1/2 and β tubulin (loading control) in ASZ_ERRasV12 and UWBCC_ERRasV12 upon doxycycline/4OHT after transfection with ns_siRNA versus OFD1_siRNA.
Scale bars indicate 25μm. Horizontal and error bars in (e) represent the mean ± SD. *p < 0.05, **p < 0.01 ***p < 0.001.
We then wondered whether loss of cilia may affect Ras/MAPK pathway activation as well. Therefore, we first used the RNA-sequencing data obtained from ASZ cells lines upon DMSO (control) or CBD (10μM) (GEO accession number GSE120954). GSEA for Ras pathway activation did not reveal any significant enrichment upon CBD treatment (Fig. 3b), indicating that loss of cilia fails to elicit Ras pathway activation by itself. We thus hypothesized that loss of cilia sustains pathway activity once activated. Therefore, we subcloned the previously reported 4-hydroxy-tamoxifen (4OHT) -dependent constitutive RasV12 (Dajee et al., 2002) under the doxycycline (DOX)–inducible promoter in the piggyBac transposon vector. Stable transfections were performed in both the Smoi-sensitive 3T3 cell line and Smoi-resistant ASZ and UWBCC BCC cell lines. As shown by western blot for p-ERK1/2 and p-MEK immunostaining (as readout for Ras/MAPK pathway activation), all three obtained cell lines showed inducible Ras pathway activation upon doxycycline and 4OHT stimulation (Suppl. Fig. S4a-d). Importantly, similar levels of Ras/MAPK induction compared to human SCC cell line SCC-13 (Suppl. Fig. S4e and S4f), confirming physiological activation of the Ras/MAPK pathway. To test the effect of the loss of cilia on activated Ras/MAPK pathway, we performed RNA-sequencing on ASZ_ERRasV12/Dox-4OHT upon either DMSO (control) or CBD (10μM) (GEO accession number GSE120954). In this setting, GSEA revealed a significant positive enrichment for Ras signaling in CBD-treated cells (Fig. 3b), suggesting that loss of cilia potentiates Ras pathway activation. This result was confirmed by Western blot for p-ERK1/2. Whereas CBD treatment did not affect ERK1/2 phosphorylation on ASZ_ERRasV12 or UWBCC_ERRasV12, it did enhance ERK1/2 phosphorylation upon concomitant Ras pathway activation through Dox/4OHT stimulation (Fig. 3c and Suppl. Fig. S5a). Similar observations were done using p-MEK immunostaining (Fig. 3d and 3e). Notably, the higher individual values (at the cellular level) of MEK phosphorylation observed in both ASZ_ERRasV12/Dox-4OHT and UWBCCERRasV12/Dox upon CBD treatment compared to control (DMSO) suggest a potentiation of Ras pathway activation rather than a selection process. As an alternative approach to pharmacological depletion of primary cilia, we measured p-ERK1/2 level by Western blot in ASZ_ERRasV12 and UWBCC_ERRasV12 previously treated with either non-silencing or OFD1 siRNAs. OFD1_siRNA efficiently potentiated Ras/MAPK pathway activation induced upon Dox-4OHT stimulation, in both ASZ and UWBCC cell lines (Fig. 3f and Suppl. Fig. S5b), confirming the role of primary cilia in antagonizing Ras/MAPK pathway activity.
HH to Ras/MAPK pathway switching confers resistance to canonical and non-canonical HH pathway inhibitors and sensitivity to MEK inhibitor UO126 in vitro.
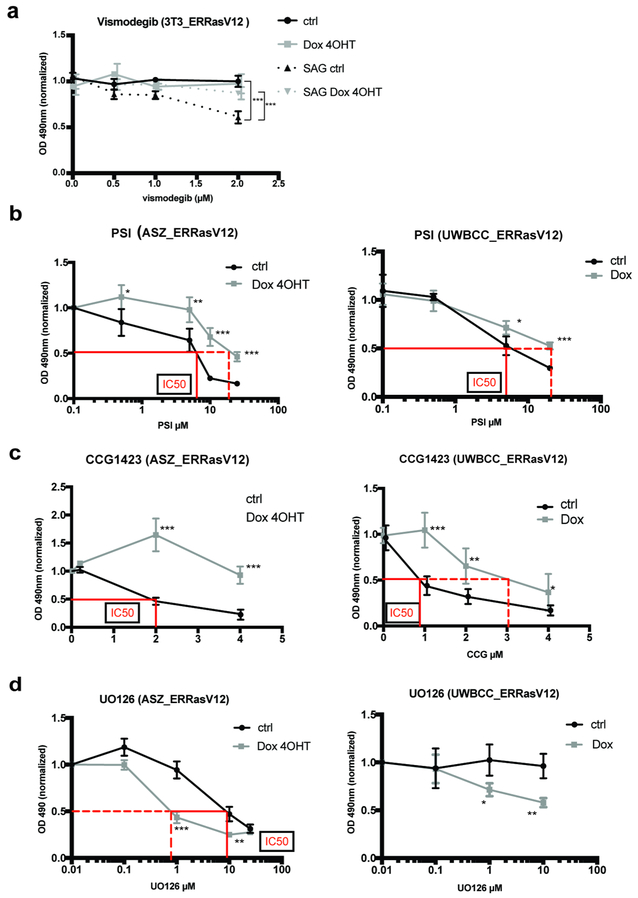
Finally, we interrogated the extent pathway switching affects BCC sensitivity to targeted therapy. We first tested the effect of Ras pathway activation on sensitivity to Smoi, using the 3T3_ERRasv12, that shows physiological sensitivity to vismodegib upon stimulation with Smoothened agonist (SAG). Interestingly, SAG-induced sensitivity to vismodegib was lost upon concomitant activation of the Ras pathway (Fig. 4a). To determine whether this effect was Smoi-specific, we then induced Ras pathway activation under HH pathway inhibitors acting downstream of Smo inhibition, in both ASZ and UWBCC BCC cell lines. Similarly, Ras pathway activation in ASZ_ERRasV12 and UWBCC_ERRasV12 conferred resistance to both PSI (an aPKC inhibitor, acting downstream of Smo (Mirza et al., 2017)) and CCG1423 (a MRTF inhibitor, implicated in non-canonical HH pathway activation (Whitson et al., 2018)), as illustrated by the right shift of the IC50 values (Fig. 4b and 4c). Although non-canonical HH inhibitors have not been tested in clinical practice yet, these results suggest that HH to Ras pathway switching may confer a distinct cell state from that driven by canonical or non-canonical HH activation. To test this idea, we investigated whether resistance to HH inhibitors might be overcome by Ras/MAPK inhibition. Indeed, both ASZ_ERRasV12 and UWBCC_ERRasV12 cell lines were sensitive to MEK inhibitor UO126 upon Ras pathway activation, as illustrated by the left shift of the IC50 values (Fig. 4d).
Figure 4. HH to Ras pathway switching confers resistance to canonical and non-canonical HH pathway inhibitors and sensitivity to MEK inhibitor UO126 in vitro.
(a) Vismodegib sensitivity assessed by MTT assay in 3T3_ERRasV12 upon stimulation with Smoothened agonist (SAG, 100nM) and Doxycycline/4OHT.
(b) PSI (aPKC inhibitor) sensitivity assessed by MTT assay in ASZ_ERRasV12 and UWBCC_ERRasV12 upon Doxycycline/4OHT.
(c) CCG1423 (MRTF inhibitor) sensitivity assessed by MTT assay in ASZ_ERRasV12 or UWBCC_ERRasV12 upon Doxycycline/4OHT.
(d) UO126 (MEK inhibitor) sensitivity assessed by MTT assay in ASZ_ERRasV12 or UWBCC_ERRasV12 upon Doxycycline/4OHT.
When reached, IC50 (half-maximal inhibitory concentration) are indicated in red lines, both in the absence (full line) or presence (dashed line) of Doxycycline/4OHT. Dots and error bars represent the mean ± SD for n ≥ 6 per group. *p < 0.05, **p < 0.01 ***p < 0.001.
Altogether, these results identify HH to Ras/MAPK pathway switching as a mechanism for BCC resistance distinct from both canonical and non-canonical HH pathway activation and MAPK inhibition as a promising therapy to prevent BCC escape from HH inhibition. Importantly, they identify the primary cilia as key regulators of this switch.
Discussion
Here we show the correlation of resistance with loss of the primary cilium and the antagonistic relationship between the primary cilium and Ras/MAPK activity, thereby providing insight into BCC pathway switching and resistance to Smoi.
Our work provides an important contrast to previous mechanisms as we find an accumulation of mutations in genes encoding components of the primary cilia, reduced numbers of cells with primary cilium, reduced HH pathway output, and activation of Ras/MAPK signaling. The cilium is composed of over 300 proteins that must work together for proper HH signaling (van Dam et al., 2013). The frequency of recurrent coding region variants that include nonsense and small deletions argues for the existence of mutations of more than one ciliary protein in each tumor cell and a reduction in ciliary function. However, because of the observed complex ciliary genotypes, we can only speculate as to the precise phenotype in each cell, especially given the known functional variability of each genotype in distinct biological contexts (Arnaiz et al., 2014). This complexity likely explains the observed sporadic loss of full-length mature cilia despite the frequency of ciliary mutations.
Elevated mutation frequencies of ciliome genes have been detected in other advanced cancers, including breast carcinoma (Menzl et al., 2014), melanoma (Zingg et al., 2018), ovarian cancer (Egeberg et al., 2012) and thyroid cancer (Lee et al., 2018, Menzl et al., 2014), suggesting that the cilia may function to enforce particular lineages or cell states outside the skin. Recently, Zhao et al. identified mutations in ciliary genes conferring resistance to sonidegib in medulloblastoma (Zhao et al., 2017). In their model, HH pathway reduction upon loss of cilia induces tumor cells to go into a “persister” state, where low proliferation rate reduces sensitivity to Smoi, while allowing for further HH pathway activating genetic alterations to accumulate. Here we show that loss of cilia, in addition to reducing HH pathway, appears permissive for the dominant oncogenic Ras/MAPK pathway, involved in both skin differentiation (Dajee et al., 2002, Dlugosz et al., 1994, Lin and Lowe, 2001, Mainiero et al., 1997) and SCC carcinogenesis (Dajee et al., 2003, To et al., 2005, Woodworth et al., 2004). Our data indicate that Ras pathway activation constitutes a distinct state, as it confers resistance to both canonical and non-canonical HH inhibitors. We may thus reasonably expect Ras activation to appear upon and confer resistance to newly approved HHI (as sonidegib (Danial et al., 2016, Migden et al., 2015)) or pre-clinical HH inhibitors currently in development. Here, as a proof-of-principle for the use of MAPK inhibitors for the treatment of BCC, we show that Ras activation in BCC cell lines actually confers sensitivity to p-MEK inhibitor UO126. However, given the existence of rapid resistance or limited efficiency of Smo and Ras/MAPK inhibitors in clinical trials of patients with non-syndromic tumors (Axelson et al., 2013, Santarpia et al., 2012, Sekulic et al., 2012), an alternative strategy may consist in targeting the crosstalk between cilia and Ras/MAPK pathway, thereby potentially preventing pathway switching and reducing the frequency of resistance. Our data did not identify the mechanism of Ras/MAPK regulation by primary cilia. Interestingly, Abdul-Majeed, S. et al. reported that, in polycystic kidney disease, reduced intracellular Ca2+ resulting from reduced ciliogenesis decreases phosphodiesterase 1-mediated conversion of cAMP to AMP, inducing downstream MAPK targets activation (Abdul-Majeed and Nauli, 2011). Alternatively, Ras pathway activation was reported to induce primary cilia disassembly and reduce HH pathway activation through the induction of AurorA kinase and Dyrk1b respectively (Furukawa et al., 2006, Kobayashi et al., 2017, Lauth et al., 2010, Seeley et al., 2009), suggesting that restoring ciliogenesis may restore sensitivity to HH inhibition. These mechanisms however need to be further validated experimentally in BCC. Therefore, experimental models fully recapitulating the BCC-to-SCC transition will be required to help deciphering the implication of primary cilia both dynamically and mechanistically in this intriguing process.
In conclusion, this work identifies the pathway switching from HH to Ras/MAPK as a further mechanism of resistance in BCCs treated with Smoi, emphasizing the need for combined and tailored targeted therapies as BCCs display various ways to escape Smo inhibition. Importantly, we bring experimental evidence for the role of primary cilia as pivotal regulators of the antagonistic HH and Ras/MAPK pathways, thereby opening perspectives for the treatment of advanced resistant BCC.
Material and Methods
Human samples.
Study on human paraffin samples was performed with approval from the Stanford University Institutional Review Board, protocol #18325 (Stanford, CA), with a waiver of written, informed consent from participants for the paraffin blocks. Histological diagnoses of Gorlin patients, sporadic naive and resistant BCC samples used in this study were previously confirmed in (Chiang et al., 2018) and (Atwood et al., 2015). Histological diagnoses of BCC and SCC samples used for Gli1 and and p-MEK immunostainings were confirmed by an independent dermatopathologist.
Whole exome sequencing and analysis.
Gene ontology analysis was performed with Panther 13.1 using GO annotations database (released 2018-02-02) on the list of previously identified genes specifically mutated in resistant BCC compared to normal skin (GEO accession number GSE58377) (Atwood et al., 2015). Ciliome mutations were quantified from whole-exome sequencing data from 21 Gorlin patients, 12 sporadic, 12 resistant BCCs and normal skin pairs previously obtained as described in (Chiang et al., 2018) and (Atwood et al., 2015) and deposited under National Institutes of Health Sequence Read Archive SAMN07507265 – SAMN07507288 and GEO GSE58377 accession numbers respectively. Custom scripts were written to count the total number of mutations in a specified set of “cilia genes” known to be important in cilia function (ciliome) (van Dam et al., 2013) and to calculate the lengths of these genes. Ciliome mutations were quantified as the number of mutations per Mb of the whole ciliome (1.303754 Mb).
Cell lines and cell culture.
ASZ001 cells (mouse BCC cell line) were grown in M154CF (ThermoFisher Scientific) supplemented with 2% chelated FBS, 1% P/S and CaCl2 0.05mM (So et al., 2006). UWBCC cells (human BCC cell line) were grown in EpilifeCF (ThermoFisher Scientific) supplemented with 7% chelated FBS, 1% P/S and CaCl2 0.05mM (Noubissi et al., 2014). NIH-3T3 were grown in DMEM (ThermoFisher Scientific) supplemented with 10% FBS and 1% P/S. SCC-13 SCC cells (human SCC cell line) were generously provided by Dr. C. Lee (Stanford, CA) and grown in KSFM (ThermoFisher Scientific) supplemented with 1% P/S, 0.25μg/ml bovine pituitary extract (BPE), 0.2ng/ml EGF and 0.3mM CaCl2 (ThermoFisher Scientific). Experiments were carried out in serum-starved conditions.
RNA sequencing and analysis.
RNA samples were obtained from adherent cells using RNeasy kit from Qiagen according to manufacturer’s instructions. Library preparation, sequencing, and mapping were performed as described previously (Atwood et al., 2015). Alignment was performed using TopHat with mm9 as a reference genome. The DEseq R package was used to generate a preranked list of genes differentially expressed in ASZ +/− CBD or ASZERRasV12/Dox-4OHT +/− CBD before submission to GSEA2.2.4 software for HH or Ras pathway enrichment. Mouse RNA-seq data generated in this manuscript were submitted to GEO (GSE120954). An additional GSEA was carried out using RNA-sequencing data from published human resistant and sensitive BCCs (GEO accession number 58377(Atwood et al., 2015)). For gene expression, resistant BCCs were compared to sensitive BCCs obtained from the same patient. Genes differentially expressed were preranked according to log2FC before submission to GSEA2.2.4 software for HH or Ras pathway enrichment.
MTT assays.
For MTT assays, Ras pathway activation was obtained using medium containing Doxycycline and 4-OHT. After 24 hours, the medium was replaced with medium containing DMSO or the tested compound and incubated for 48 hours. Cell viability was measured using CellTiter 96 Aqueous One Solution (Promega, Madison, WI) and normalized to control (DMSO-treated cells). Each condition was tested in 6-plicate. For each drug and cell line, IC50 was determined as the drug concentration giving the half-maximal response compared to the control (DMSO-treated) conditions. Ciliobrevin (CBD, Sigma-Aldrich), Vismodegib (GDC-0449, Selleckchem,Houston, TX), CCG1423 (Cayman Chemicals, Ann Harbor, MI) and PSI (2549, Tocris, Minneapolis, MN) were suspended in DMSO.
Statistical analyses.
Statistical comparisons were performed by a two-tailed Student’s t test or one-way ANOVA with Tukey’s post-test for multiple comparisons. Statistical analyses were performed using GraphPad Prism (La Jolla, CA). The data were considered to be significantly different when p < 0.05.
A more complete and detailed description of the methods is included in Supplementary Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by an SNSF fellowship (to FK), a Fondation Rene Touraine fellowship (to FK), F32 CA200108-01 NIH grant (to NH) and NIH R01 ARO46786 and ARO54780 (to AEO). The authors declare no competing financial interests.
Footnotes
Conflict of interest statement
The authors state no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdul-Majeed S, Nauli SM. Calcium-mediated mechanisms of cystic expansion. Biochim Biophys Acta 2011;1812(10):1281–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaiz O, Cohen J, Tassin AM, Koll F. Remodeling Cildb, a popular database for cilia and links for ciliopathies. Cilia 2014;3:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood SX, Li M, Lee A, Tang JY, Oro AE. GLI activation by atypical protein kinase C iota/lambda regulates the growth of basal cell carcinomas. Nature 2013;494(7438):484–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood SX, Sarin KY, Whitson RJ, Li JR, Kim G, Rezaee M, et al. Smoothened variants explain the majority of drug resistance in basal cell carcinoma. Cancer Cell 2015;27(3):342–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelson M, Liu K, Jiang X, He K, Wang J, Zhao H, et al. U.S. Food and Drug Administration approval: vismodegib for recurrent, locally advanced, or metastatic basal cell carcinoma. Clin Cancer Res 2013;19(9):2289–93. [DOI] [PubMed] [Google Scholar]
- Bangs F, Anderson KV. Primary Cilia and Mammalian Hedgehog Signaling. Cold Spring Harb Perspect Biol 2017;9(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla X, Parmentier L, King B, Bezrukov F, Kaya G, Zoete V, et al. Genomic analysis identifies new drivers and progression pathways in skin basal cell carcinoma. Nat Genet 2016;48(4):398–406. [DOI] [PubMed] [Google Scholar]
- Buonamici S, Williams J, Morrissey M, Wang A, Guo R, Vattay A, et al. Interfering with resistance to smoothened antagonists by inhibition of the PI3K pathway in medulloblastoma. Sci Transl Med 2010;2(51):51ra70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandarlapaty S Negative feedback and adaptive resistance to the targeted therapy of cancer. Cancer Discov 2012;2(4):311–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell 2011;19(1):58–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AL, Oro AE. Initial assessment of tumor regrowth after vismodegib in advanced Basal cell carcinoma. Arch Dermatol 2012;148(11):1324–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang A, Jaju PD, Batra P, Rezaee M, Epstein EH Jr., Tang JY, et al. Genomic Stability in Syndromic Basal Cell Carcinoma. J Invest Dermatol 2018;138(5):1044–51. [DOI] [PubMed] [Google Scholar]
- Dajee M, Lazarov M, Zhang JY, Cai T, Green CL, Russell AJ, et al. NF-kappaB blockade and oncogenic Ras trigger invasive human epidermal neoplasia. Nature 2003;421(6923):639–43. [DOI] [PubMed] [Google Scholar]
- Dajee M, Tarutani M, Deng H, Cai T, Khavari PA. Epidermal Ras blockade demonstrates spatially localized Ras promotion of proliferation and inhibition of differentiation. Oncogene 2002;21(10):1527–38. [DOI] [PubMed] [Google Scholar]
- Danial C, Sarin KY, Oro AE, Chang AL. An Investigator-Initiated Open-Label Trial of Sonidegib in Advanced Basal Cell Carcinoma Patients Resistant to Vismodegib. Clin Cancer Res 2016;22(6):1325–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkgraaf GJ, Alicke B, Weinmann L, Januario T, West K, Modrusan Z, et al. Small molecule inhibition of GDC-0449 refractory smoothened mutants and downstream mechanisms of drug resistance. Cancer Res 2011;71(2):435–44. [DOI] [PubMed] [Google Scholar]
- Dlugosz AA, Cheng C, Williams EK, Dharia AG, Denning MF, Yuspa SH. Alterations in murine keratinocyte differentiation induced by activated rasHa genes are mediated by protein kinase C-alpha. Cancer Res 1994;54(24):6413–20. [PubMed] [Google Scholar]
- Egeberg DL, Lethan M, Manguso R, Schneider L, Awan A, Jorgensen TS, et al. Primary cilia and aberrant cell signaling in epithelial ovarian cancer. Cilia 2012;1(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein EH. Basal cell carcinomas: attack of the hedgehog. Nat Rev Cancer 2008;8(10):743–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante MI, Zullo A, Barra A, Bimonte S, Messaddeq N, Studer M, et al. Oral-facial-digital type I protein is required for primary cilia formation and left-right axis specification. Nat Genet 2006;38(1):112–7. [DOI] [PubMed] [Google Scholar]
- Fu W, Asp P, Canter B, Dynlacht BD. Primary cilia control hedgehog signaling during muscle differentiation and are deregulated in rhabdomyosarcoma. Proc Natl Acad Sci U S A 2014;111(25):9151–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T, Kanai N, Shiwaku HO, Soga N, Uehara A, Horii A. AURKA is one of the downstream targets of MAPK1/ERK2 in pancreatic cancer. Oncogene 2006;25(35):4831–9. [DOI] [PubMed] [Google Scholar]
- Gerdes MJ, Myakishev M, Frost NA, Rishi V, Moitra J, Acharya A, et al. Activator protein-1 activity regulates epithelial tumor cell identity. Cancer Res 2006;66(15):7578–88. [DOI] [PubMed] [Google Scholar]
- Kasper M, Jaks V, Hohl D, Toftgard R. Basal cell carcinoma - molecular biology and potential new therapies. J Clin Invest 2012;122(2):455–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Nakazono K, Tokuda M, Mashima Y, Dynlacht BD, Itoh H. HDAC2 promotes loss of primary cilia in pancreatic ductal adenocarcinoma. EMBO Rep 2017;18(2):334–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauth M, Bergstrom A, Shimokawa T, Tostar U, Jin Q, Fendrich V, et al. DYRK1B-dependent autocrine-to-paracrine shift of Hedgehog signaling by mutant RAS. Nat Struct Mol Biol 2010;17(6):718–25. [DOI] [PubMed] [Google Scholar]
- Lee CS, Bhaduri A, Mah A, Johnson WL, Ungewickell A, Aros CJ, et al. Recurrent point mutations in the kinetochore gene KNSTRN in cutaneous squamous cell carcinoma. Nat Genet 2014;46(10):1060–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Yi S, Won M, Song YS, Yi HS, Park YJ, et al. Loss-of-function of IFT88 determines metabolic phenotypes in thyroid cancer. Oncogene 2018. [DOI] [PubMed] [Google Scholar]
- Lefort K, Mandinova A, Ostano P, Kolev V, Calpini V, Kolfschoten I, et al. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKalpha kinases. Genes Dev 2007;21(5):562–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AW, Lowe SW. Oncogenic ras activates the ARF-p53 pathway to suppress epithelial cell transformation. Proc Natl Acad Sci U S A 2001;98(9):5025–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainiero F, Murgia C, Wary KK, Curatola AM, Pepe A, Blumemberg M, et al. The coupling of alpha6beta4 integrin to Ras-MAP kinase pathways mediated by Shc controls keratinocyte proliferation. EMBO J 1997;16(9):2365–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzl I, Lebeau L, Pandey R, Hassounah NB, Li FW, Nagle R, et al. Loss of primary cilia occurs early in breast cancer development. Cilia 2014;3:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migden MR, Guminski A, Gutzmer R, Dirix L, Lewis KD, Combemale P, et al. Treatment with two different doses of sonidegib in patients with locally advanced or metastatic basal cell carcinoma (BOLT): a multicentre, randomised, double-blind phase 2 trial. Lancet Oncol 2015;16(6):716–28. [DOI] [PubMed] [Google Scholar]
- Mirza AN, Fry MA, Urman NM, Atwood SX, Roffey J, Ott GR, et al. Combined inhibition of atypical PKC and histone deacetylase 1 is cooperative in basal cell carcinoma treatment. JCI Insight 2017;2(21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noubissi FK, Kim T, Kawahara TN, Aughenbaugh WD, Berg E, Longley BJ, et al. Role of CRD-BP in the growth of human basal cell carcinoma cells. J Invest Dermatol 2014;134(6):1718–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka A, Dreier J, Cheng PF, Nageli M, Lehmann H, Felderer L, et al. Hedgehog pathway inhibitors promote adaptive immune responses in basal cell carcinoma. Clin Cancer Res 2015;21(6):1289–97. [DOI] [PubMed] [Google Scholar]
- Pak E, Segal RA. Hedgehog Signal Transduction: Key Players, Oncogenic Drivers, and Cancer Therapy. Dev Cell 2016;38(4):333–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering CR, Zhou JH, Lee JJ, Drummond JA, Peng SA, Saade RE, et al. Mutational landscape of aggressive cutaneous squamous cell carcinoma. Clin Cancer Res 2014;20(24):6582–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransohoff KJ, Tang JY, Sarin KY. Squamous Change in Basal-Cell Carcinoma with Drug Resistance. N Engl J Med 2015;373(11):1079–82. [DOI] [PubMed] [Google Scholar]
- Rubin LL, de Sauvage FJ. Targeting the Hedgehog pathway in cancer. Nat Rev Drug Discov 2006;5(12):1026–33. [DOI] [PubMed] [Google Scholar]
- Saintes C, Saint-Jean M, Brocard A, Peuvrel L, Renaut JJ, Khammari A, et al. Development of squamous cell carcinoma into basal cell carcinoma under treatment with Vismodegib. J Eur Acad Dermatol Venereol 2015;29(5):1006–9. [DOI] [PubMed] [Google Scholar]
- Santarpia L, Lippman SM, El-Naggar AK. Targeting the MAPK-RAS-RAF signaling pathway in cancer therapy. Expert Opin Ther Targets 2012;16(1):103–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley ES, Carriere C, Goetze T, Longnecker DS, Korc M. Pancreatic cancer and precursor pancreatic intraepithelial neoplasia lesions are devoid of primary cilia. Cancer Res 2009;69(2):422–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulic A, Migden MR, Oro AE, Dirix L, Lewis KD, Hainsworth JD, et al. Efficacy and safety of vismodegib in advanced basal-cell carcinoma. N Engl J Med 2012;366(23):2171–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra V, Scaltriti M, Prudkin L, Eichhorn PJ, Ibrahim YH, Chandarlapaty S, et al. PI3K inhibition results in enhanced HER signaling and acquired ERK dependency in HER2-overexpressing breast cancer. Oncogene 2011;30(22):2547–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe HJ, Pau G, Dijkgraaf GJ, Basset-Seguin N, Modrusan Z, Januario T, et al. Genomic analysis of smoothened inhibitor resistance in basal cell carcinoma. Cancer Cell 2015;27(3):327–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So PL, Langston AW, Daniallinia N, Hebert JL, Fujimoto MA, Khaimskiy Y, et al. Longterm establishment, characterization and manipulation of cell lines from mouse basal cell carcinoma tumors. Exp Dermatol 2006;15(9):742–50. [DOI] [PubMed] [Google Scholar]
- Tang JY, Mackay-Wiggan JM, Aszterbaum M, Yauch RL, Lindgren J, Chang K, et al. Inhibiting the hedgehog pathway in patients with the basal-cell nevus syndrome. N Engl J Med 2012;366(23):2180–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To MD, Perez-Losada J, Mao JH, Balmain A. Crosstalk between Pten and Ras signaling pathways in tumor development. Cell Cycle 2005;4(9):1185–8. [DOI] [PubMed] [Google Scholar]
- van Dam TJ, Wheway G, Slaats GG, Group SS, Huynen MA, Giles RH. The SYSCILIA gold standard (SCGSv1) of known ciliary components and its applications within a systems biology consortium. Cilia 2013;2(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitson RJ, Lee A, Urman NM, Mirza A, Yao CY, Brown AS, et al. Noncanonical hedgehog pathway activation through SRF-MKL1 promotes drug resistance in basal cell carcinomas. Nat Med 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodworth CD, Michael E, Smith L, Vijayachandra K, Glick A, Hennings H, et al. Strain-dependent differences in malignant conversion of mouse skin tumors is an inherent property of the epidermal keratinocyte. Carcinogenesis 2004;25(9):1771–8. [DOI] [PubMed] [Google Scholar]
- Wu F, Zhang Y, Sun B, McMahon AP, Wang Y. Hedgehog Signaling: From Basic Biology to Cancer Therapy. Cell Chem Biol 2017;24(3):252–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch RL, Dijkgraaf GJ, Alicke B, Januario T, Ahn CP, Holcomb T, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science 2009;326(5952):572–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Pak E, Ornell KJ, Pazyra-Murphy MF, MacKenzie EL, Chadwick EJ, et al. A Transposon Screen Identifies Loss of Primary Cilia as a Mechanism of Resistance to SMO Inhibitors. Cancer Discov 2017;7(12):1436–49. [DOI] [PubMed] [Google Scholar]
- Zhao X, Ponomaryov T, Ornell KJ, Zhou P, Dabral SK, Pak E, et al. RAS/MAPK Activation Drives Resistance to Smo Inhibition, Metastasis, and Tumor Evolution in Shh Pathway-Dependent Tumors. Cancer Res 2015;75(17):3623–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingg D, Debbache J, Pena-Hernandez R, Antunes AT, Schaefer SM, Cheng PF, et al. EZH2-Mediated Primary Cilium Deconstruction Drives Metastatic Melanoma Formation. Cancer Cell 2018;34(1):69–84 e14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.