Abstract
Coordination of iron acquisition and heme synthesis is required for effective erythropoiesis. The small teleost zebrafish (Danio rerio) is an ideal vertebrate animal model to replicate various aspects of human physiology and provides an efficient and cost-effective way to model human pathophysiology. Importantly, zebrafish erythropoiesis largely resembles mammalian erythropoiesis. Gene discovery by large-scale forward mutagenesis screening has identified key components in heme and iron metabolism. Reverse genetic screens using morpholino-knockdown and CRISPR/Cas9 have further accelerated gene functional studies, taking advantages of genetic tractability of zebrafish embryos. Ultimately, the ex utero development of zebrafish embryos combined with their transparency and developmental plasticity could provide a deep understanding of the role of iron and heme metabolism during early vertebrate embryonic development.
Keywords: zebrafish, porphyria, heme transport, iron metabolism, erythropoiesis
1. Introduction
The teleost fish, Danio rerio, also known as zebrafish, has gained greater attention as a model organism for studying vertebrate development owing to its advantage in developmental biology. Adult zebrafish are relatively small and one breeding pair can prod uce 100–200 progenies per spawning each week, which allows easy maintenance of animal strains with small space and ensures production of numerous embryos for experiments (genetic screening work and building transgenic lines). The external fertilization and rapid development ex utero, combined with the property of transparency, allow direct visualization and manipulation of developmental processes in early embryos, which are not possible in mice. The techniques for transgenesis and gene manipulation are well-developed, rendering zebrafish as a genetically-tractable vertebrate animal model [1].
2. Zebrafish erythropoiesis resembles those of higher vertebrates
Zebrafish has a similar hematopoietic program compared to higher vertebrates, spanning from developmental waves of hematopoiesis to conservation of producing comparable blood cell components. Two successive waves of hematopoiesis, primitive and definitive, are also found in the zebrafish [2, 3]. Additionally, crucial regulators have been isolated as orthologs of many essential mammalian hematopoietic regulators and functional conservation of these factors in zebrafish is validated by morpholino-mediated knockdown or identified mutants [4]. These conserved properties of zebrafish hematopoiesis make it a good model for hematopoietic study and further applicable to mammals.
Primitive erythropoiesis in zebrafish occurs in the intermediate cell mass (ICM) of developing embryos, which is functionally equivalent to extra-embryonic yolk sac blood island region in mammals [5]. Primitive erythroid lineage arises from ICM which is previously developed from posterior lateral mesoderm (PLM) region. Primitive erythropoiesis in zebrafish begins at 4-somite stage with the onset expression of erythroid-specific transcription factor GATA1, a zinc-finger transcription factor for early primitive erythroid progenitors differentiation [6]. The circulating primitive erythroblasts further proliferate and differentiate into mature erythrocytes through a series of morphological alterations, with elongated nucleus around 4 dpf (days post fertilization), and functionally survive to 10 dpf in peripheral blood stream [7]. Mature erythrocytes display unique morphological characteristics, distinguishable from adult counterparts by less cytoplasm and remaining nucleus.
Consistent with hematopoietic programs in mammals, definitive hematopoiesis in zebrafish marks its distinctive wave by generating all blood cell lineages throughout the whole lifespan. The initiation site of zebrafish definitive hematopoiesis has been identified by expression patterns of mammalian definitive hematopoietic regulator orthologues, C-MYB and RUNX1. These two transcription factors dictate the formation of definitive HSPCs (hematopoietic stem/progenitor cells) [8]. In zebrafish, initial expression of runx1 and c-myb is located in the VDA (ventral wall of dorsal aorta) from 26–48 hpf (hours post fertilization), suggesting that definitive HSPCs are generated in the VDA. The initiation of definitive hematopoiesis in VDA happens as early as 36 hpf [9]. At around 4–5 dpf, hematopoietic cells are identifiable in pronephros/kidney and thereafter by 13 dpf. The fish kidney functions as a counterpart of the mammalian bone marrow to sustain definitive hematopoiesis throughout its lifespan [10]. Posterior blood island (PBI), also called caudal hematopoietic tissue (CHT) and located between the caudal artery and the caudal vein at the end of the tail, is recognized as a fetal hematopoietic organ in zebrafish and an intermediate hematopoietic site covering the period between VDA and kidney as a counterpart to fetal liver in mammals [11, 12]. Definitive erythrocytes are postulated to populate the circulation at around 5 dpf as RBCs emerge at this stage in bloodless mutants with defects in producing primitive erythrocytes [13]. Transfusion experiments reveal that primitive erythrocytes are the major circulating erythroid components for the first 5 days and thereafter are gradually replaced by presumptive definitive erythrocytes [7]. Like mammals, these definitive erythrocytes switch to express adult form of globins and could be discriminated morphologically from primitive counterparts [14, 15].
The kidney marrow (head kidney) is an adult hematopoietic organ in zebrafish which is functionally equivalent to BM in mammals. Based on forward scatter (cell size) and side scatter (granularity), hematopoietic cells in the kidney marrow could be fractioned into four populations: immature precursors of all lineages, lymphoid cells, mature erythroid cells and myelo-monocytic cells (neutrophils, monocytes, macrophages, and eosinophils) [16]. Lethal irradiation followed by transplantation rescue in zebrafish reveals presence of definitive HSCs in the kidney marrow maintaining lifelong hematopoiesis program [17].
Spleen is postulated to be another hematopoietic tissue in the adult zebrafish, although clear evidence is lacking to define its hematopoietic activity. Compared to the kidney marrow, zebrafish spleen is not well-characterized. It was proposed that zebrafish spleen may function as a reservoir for RBCs where erythrocytes are stored and destroyed [18]. Splenic macrophages can be found in the red pulp and contain phagosomes with erythrocytes and other cellular debris, indicating the possibility of active EP in the zebrafish spleen [19, 20].
3. Iron acquisition and heme biosynthesis during erythropoiesis
The need for iron for heme synthesis during erythropoiesis mandates an efficient pathway to uptake extracellular iron (Fig.1). Erythroid cells rely on a high affinity system composed of transferrin (TF) and transferrin receptor (TFR). One molecule of tra nsferrin can bind to two ferric iron atoms with an association coefficient of 10−20 M at physiological pH [21]. Transferrin receptor 1 (TFR1, also known as CD71) tightly binds to TF, permitting developing erythroid cells to uptake iron efficiently from circulation. The complex of iron-bound TF and TFR1 is internalized by receptor-mediated endocytosis and the ferric iron is then released from TF as the endosomes acidify. The released ferric iron is reduced to ferrous by STEAP3 (six-transmembrane epithelial antigen of prostate 3 reductase) [22]. The ferrous iron is transported out of the endosome by DMT1 (divalent metal transporter 1, also known as NRAMP2 and SLC11A2) [23]. The apo-TF/TFR1 complex is then recycled back to the cell surface, where apo-TF dissociates from the TFR1 and re-enters the circulation. The holo-TF/TFR1 and apo-TF/TFR1 cycle ensures optimal iron uptake from the circulation for hemoglobin production.
Fig.1. Hene synthesis and iron metabolism in erythroid cells.
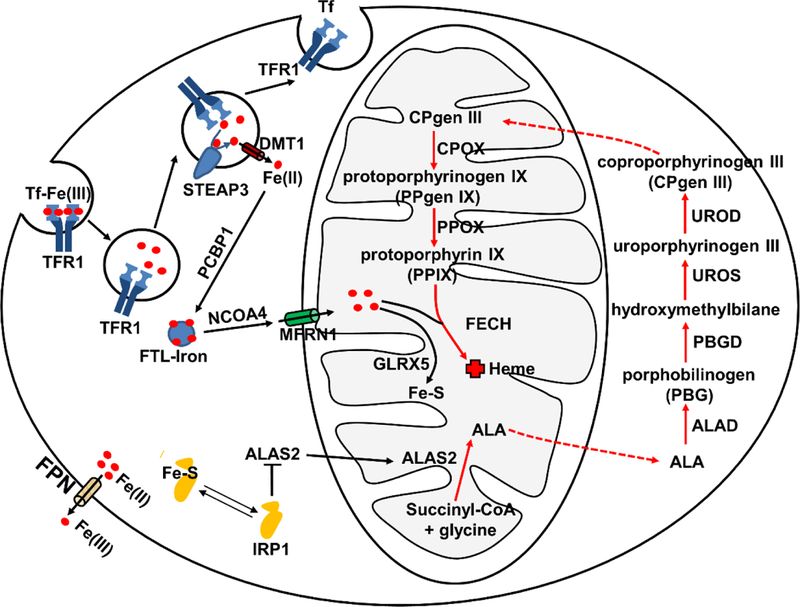
Iron acquisition in erythroid cells is dependent on endocytosis of Tf-bound Fe (Tf-Fe(III)) via the transferrin receptor (TFR1). Iron (Fe(III)) is imported into the cytoplasm by DMT1 after reduction by STEAP3. Iron can be stored in the ferritin (FTL) (FTL-Iron) mediated by PCBP1 and can be released from FTL promoted by NCOA4. MFRN1 is responsible for bringing iron to the mitochondria for heme synthesis. The red arrow represents overall heme synthetic pathway. Heme biosynthesis initiates in the mitochondrial matrix and is catalyzed by δ-aminolevulinic acid synthase (ALAS2) to synthesize δ-aminolevulinic acid (ALA) from glycine and succinyl-coenzyme A. ALA is subsequently transported out of the mitochondria to the cytosol for the following four enzymatic reaction steps: ALA to porphobilinogen (PBG) catalyzed by ALA dehydratase (ALAD); PBG to an unstable polymer hydroxymethylbilane by porphobilinogen deaminase (PBGD); hydroxymethylbilane to uroporphyrinogen III (UROgen III) by uroporphyrinogen synthase (UROS); and UROgen III to coproporphyrinogen III (CPgen III) by uroporphyrinogen decarboxylase (UROD). CPgen III is then transported into the mitochondria where coproporphyrinogen oxidase (CPOX), a mitochondrial intermembrane space enzyme, catalyzes the formation of protoporphyrinogen IX (PPgen IX). The inner mitochondrial membrane enzyme protoporphyrinogen oxidase (PPOX) catalyzes the formation of protoporphyrin IX (PPIX) from PPgen IX in the mitochondrial matrix. On the last step, ferrochelatase (FECH) catalyzes the insertion of ferrous iron (Fe2+) into PPIX to form heme. Iron is also used for Fe-S cluster synthesis with involvement of GLRX5. IRP1 binding can inhibit translation of ALAS2 to prevent the accumulation of toxic heme intermediates. Cellular iron efflux is mediated by FPN and requires iron oxidation on the extracellular side.
Since free iron is cytotoxic due to fenton chemistry, iron is either stored or utilized upon transport into the cytosol [24]. Iron is stored in the cytosol by binding to ferritin with the aid of Poly r(C)-binding protein (PCBP) [25]. PCBP1 and PCBP2 exhibit high affinity for ferritin in vitro and PCBP1 and its paralog PCBP2 co-immuoprecipitates with ferritin in HEK293 cells. Co-expression of PCBP1 and PCBP2 together with human ferritins in yeast causes iron deficiency response by increasing iron deposition into ferritin. While PCBP1 mediates delivery and integration of iron into ferritin, nuclear receptor coactivator 4 (NCOA4) promotes release of iron from ferritin by directing the ferritin complex to autophagosomes for degradation [26]. Binding of PCBP1 to ferritin precedes NCOA4-ferritin interaction and coincides with globin synthesis during erythroid maturation. NCOA4-deficient cells exhibit massive accumulation of iron in ferritin with impaired hemoglobinization and enhanced erythroid cell death by ferritinophagy, since ferritin is an essential source of iron for heme production during terminal erythroid differentiation.
Upon release from ferritin, iron is transported into the mitochondria mediated by SLC25A37 (mitoferrin1, MFRN1), a protein belonging to the family of mitochondrial solute carrier proteins. MFRN1 is expressed in the inner mitochondrial membrane and transports iron across mitochondrial membranes [27]. Mouse erythroblasts derived from Mfrn1-deficient embryonic stem cells show a complete block of iron incorporation into heme. Defect in Mfrn1 results in profound hypochromic anemia and erythroid maturation arrest owing to insufficient mitochondrial iron uptake in zebrafish. Deletion of two yeast Mfrn homologs, Mrs3 and Mrs4 impairs incorporation of iron into PPIX and formation of Fe-S cluster assembly, collectively resulting in poor growth under low iron conditions. Deletion of Mfrn1 in mice is embryonic lethal and mice with targeted deletion of Mfrn1 in adult hematopoietic tissues show severe anemia owing to deficits in erythroblast formation [28].
Large amount of heme is synthesized during differentiation and maturat ion of RBCs and therefore must be coupled with iron acquisition. The efficiency of converting iron to heme in maturing erythroid cells is extremely high, resulting in heme-iron concentrations to be over 40,000-fold greater than non-heme iron in mature RBCs [29, 30]. One mode of tackling this large requirement for heme would be upregulating heme synthesis in the mitochondria and then mobilizing heme out of the mitochondria for insertion into cytoplasmic globin to couple heme synthesis with increasing globin production. In contrast, it is also essential to downregulate heme synthesis and decrease intracellular iron content when globin production is low in the early stage of erythropoietic development, since both free heme and iron are toxic to erythroid cells by inducing oxidative stresses. Failure to regulate heme synthesis during erythropoiesis will cause either iron-deficient or iron-overload anemia [31].
Although there are eight enzymatic steps for heme biosynthesis, the rate-limiting step is the synthesis of ALA from glycine and succinyl-coenzyme A, catalyzed by ALAS [32] (Fig.1). Two forms of ALAS exist, ALAS1 and ALAS2. ALAS1 is ubiquitously expressed in all tissues, while ALAS2 (or ALAS-E), is exclusively expressed in developing erythroid cells [33, 34]. ALAS2 is activated by the transcription factor GATA1, a master regulator for erythropoiesis. Chip-Seq analysis using G1E-ER4 erythroid progenitor cell line derived from Gata1 mutant mice identified two GATA-1 cis elements in the first and eighth introns of Alas2 [35–37]. Disruption of these two GATA1-binding elements abrogates expression of ALAS2 and subsequent inhibition of heme synthesis, which can be rescued by supplementing cells with high concentrations of ALA, the product of ALAS [36, 37]. Another regulation of ALAS2 occurs at the post-transcriptional level, modulated by iron responsive elements (IREs) in the 5’UTR. IREs interact with iron regulatory proteins (IRPs), linking the regulation of heme biosynthesis in erythroid cells to iron availability [38]. Under iron-deficient conditions, IRPs bind to IREs and inhibit Alas2 mRNA translation. Conversely, when intracellular iron levels increase, IRPs are either degraded or converted to an aconitase by an iron-sulfur [4Fe-4S] and Alas2 mRNA translation resumes to promote heme synthesis. The regulation of ALAS2 expression is coordinated with the cellular iron levels to tightly control cellular heme content. Another rate-limiting enzyme in the heme biosynthetic pathway is FECH. Transcription of Fech spikes during terminal erythroid differentiation and is controlled by the transcription factors SP1, NFE2 and GATA1[39]. Furthermore, the enzymatic activity of FECH is dependent on the presence of [4Fe–4S] cluster, again linking iron levels to heme synthesis [40]. Distinct erythroid-specific elements have been identified in the promoter region of Fech, together with erythroid-specific alternative splicing in the 3′ noncoding region of Fech mRNA in the mouse genome [41, 42], underscoring the unique regulatory mechanisms for heme synthesis in RBC maturation. Beside regulation of heme synthesis in erythroid cells, globin synthesis is controlled by the heme/BACH1 axis, in which heme binds to BACH1, a transcription suppressor, to relieve the depression for globin gene expression [43]. These regulations collectively coordinate cellular iron levels, heme synthesis, and globin protein expression, in order to maintain heme and iron homeostasis in erythroid cells.
4. Heme Transport and erythropoiesis
The hydrophobicity and cytotoxicity of free heme suggests the existence of heme trafficking pathways [44] (Fig.2). The final step of heme biosynthetic pathway occurs in the inner mitochondria. Thus heme must be exported out of the mitochondria for incorporation into hemoproteins located in various subcellular compartments [45]. Studies have shown that ABCB10 may facilitate transport of heme out of the mitochondria in erythroid cells [46]. ABCB10 is a mitochondrial ABC transporter located in the inner mitochondrial membrane. ABCB10 interacts with MFRN1 and FECH and stabilizes the complex [47]. Expression of ABCB10 is highly induced during erythroid differentiation and ABCB10 overexpression strengthens hemoglobin synthesis in erythroid cells. It has been shown that ABCB10-null mice display defective erythropoiesis and lack of hemoglobinized RBCs, indicating that ABCB10 is essential for erythropoiesis in vivo [46]. However, conclusive evidence for direct heme transport by ABCB10 is still lacking.
Fig.2. The ins and outs of heme transport.
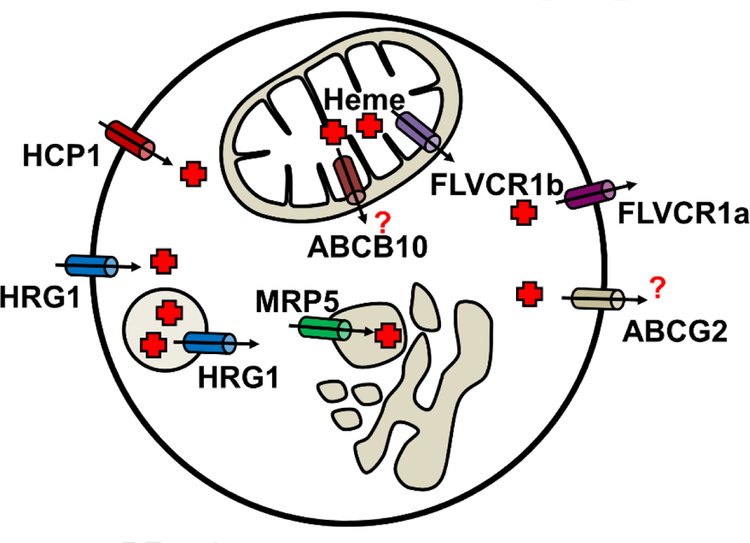
Mitochondrial isoform FLVCR1b transports heme into the cytosol. ABCB10 is reported to forms complex with FECH and MFRN1. It is not clear whether ABCB10 transport heme. The cell surface FLVCR1a and the ABC transporter ABCG2 have been implicated in heme e xport. MPR5 is a heme exporter which can transport heme from cytosol to the secretory pathway. HCP1 is a folate importer as well as a low-affinity heme importer. HRG-1 is a heme importer that localizes to endosomal/lysosomal compartments, but also traffics to the plasma membrane.
The Feline leukemia virus subgroup C receptor-related protein 1 (FLVCR1) was identified as a heme exporter [48]. FLVCR1 belongs to the family of MFS (major facilitator superfamily) proteins which transport small solutes across membranes facilitated by a counter ion gradient. Overexpression of FLVCR1 significant reduces cellular heme content, suggesting that FLVCR1 is involved in heme export [48]. FLVCR1-null mice are embryonic lethal with deficiencies in definitive erythropoiesis and suffer from craniofacial and limb deformities resembling those of patients with Diamond-Blackfan anemia (DBA). Flvcr1−/− mice develop a severe macrocytic anemia with proerythroblast maturation arrest, suggesting that erythroid precursors may be exporting excess heme to avoid heme toxicity [49]. Two isoforms of FLVCR has been identified, FLVCR1a and FLVCR1b. While FLVCR1a encodes a plasma membrane-localized heme transporter, FLVCR1b was identified to be a mitochondrial isoform e ncoded from an alternate transcription start-site, resulting in a shortened N-terminus containing a mitochondrial-targeting signal [50]. Thus, FLVCR1a and FLVCR1b contributes to intercellular and intracellular heme transport respectively. FLVCR1a is expressed in different hematopoietic cells and shows weak expression in the fetal liver, pancreas and kidney [51]. FLVCR1a may export heme during erythrophagocytosis (EP), a process in which macrophage phagocytose senescent RBCs, as FLVCR1a has been showed to interact with the extracellular heme-binding protein hemopexin and mediate heme export that is at least 100-fold more efficient than in the absence of hemopexin [52]. FLVCR1a expression is increased during erythropoiesis and is at its greatest during intermediate stages of RBC maturation when HMOX1 expression is low, implying that FLVCR1a helps to maintain stoichiometric amounts of heme and globin by exporting excess heme and preventing heme toxicity to RBCs [53]. Depletion of FLVCR1b in HeLa cells results in accumulation of mitochondrial heme, indicating that FLVCR1b may play a role in heme export from the mitochondria [50]. It is not known whether FLVCR1b resides on the inner or outer mitochondrial membrane. Moreover, if mitochondrial heme export by FLVCR1b is indispensable and indeed attenuated in Flvcr1−/− mice, it cannot explain why embryos from Flvcr1−/− null mice can survive until E14.5. Yeast does not appear to have an obvious FLVCR homolog, yet is able to export heme from the mitochondria indicating that alternate mechanisms must exist for mitochondrial heme export.
ABCG2, also known as breast cancer resistance protein (BCRP) has been identified as a heme exporter in mammals [54]. ABCG2 is expressed in hematopoietic stem cells (HSCs) and erythroid progenitors. Compared to high FLVCR1 expression during erythropoietic differentiation, the expression levels of ABCG2 are particularly high at the early stages of hematopoiesis [55]. ABCG2 binds to heme directly through an extracellular loop 3 with a porphyrin-binding domain [56]. Ectopically expressed ABCG2 exports ZnMP, a heme analog, in K562 cells. However, direct evidence that ABCG2 exports heme is still lacking [56]. Whether FLVCR1 and ABCG2 can function synergistically to export heme during erythropoiesis is not clear. ABCG2-null human patients are defined as Jr(a–) blood group with a unique side population of HSCs, but no apparent deficiencies in erythropoiesis [57, 58].
MRP-5 was identified as a heme exporter in Caenorhabditis elegans. This roundworm is a unique model for uncovering heme trafficking pathways because it cannot synthesize heme but acquires environmental heme for sustenance. Thus, worms need to acquire dietary heme via the intestine and distribute heme from the intestine to other tissues [59, 60]. C. elegans MRP-5 (CeMRP-5) localizes to the basolateral membrane of intestinal cells and loss of MRP-5 results in accumulation of ZnMP in intestinal cells and growth retardation. Overexpression of CeMRP −5 and human MRP5 in yeast retards growth owing to heme depletion, which can be rescued by co-expressing C. elegans heme importers. Interestingly, overexpression of CeMRP-5 and human MPR5 causes heme levels to increase in the secretory pathway in both yeast and mammalian cells, as measured by a secretory pathway hemoprotein reporter. However, the physiological role of MRP5 in vertebrates and its involvement in erythropoiesis is unclear.
Heme carrier protein 1 (HCP1, SLC46A1) is a membrane protein expressed in enterocytes and was proposed to be an intestinal heme transporter [61]. However, subsequent studies revealed that HCP1 is a proton-coupled folate transporter (PCFT) rather than a heme transporter [62]. Erythroblasts from HCP1 knockout mice showed deficiency in differentiation and high apoptosis rate resulting in severe macrocytic normochromic anemia, which was ascribed to folate but not heme deficiency [63]. Moreover, knockdown of HCP1 by shRNA in Caco-2 cells attenuated both heme and folate uptake but increased heme oxygenase expression, suggesting HCP1 could potentially function as a low affinity heme importer [62, 64].
The Heme Responsive Gene −1 (HRG1, SLC48A1) was identified as a heme importer in the intestine of C. elegans [65]. Four HRG1 homologs, CeHRG-1, CeHRG-4, CeHRG-5 and CeHRG-6 were found in the C. elegans genome. Significant heme-induced inward currents were observed in Xenopus oocytes injected with Cehrg-1, Cehrg-4, and human HRG1 mRNA, indicating heme-dependent transport across cell membranes [65]. Human HRG1 (SLC48A1) mRNA is abundant in the brain, kidney, heart, skeletal muscle, in addition to cell lines derived from duodenum and bone marrow [65]. HRG1 localizes to acidic endosomal and lysosomal organelles in HEK293 cells, and its binding affinity to heme increases as pH decreases. Human HRG1 interacts with the C subunit of the vacuolar proton ATPase (V-ATPase) pump and enhances endosomal acidification [66]. These studies collectively suggest that HRG1 transports heme from the exoplasmic space or the lumen of acidic endosome–lysosomal compartments into the cytoplasm. Global transcriptomic expression profiling shows that Hrg1 mRNA is expressed during erythroblast maturation [67]. Why a heme importer would be expressed during erythropoiesis since developing RBCs are capable of de novo heme synthesis is puzzling.
5. Zebrafish as a genetic model to study heme and iron metabolism
The complete sequencing of the zebrafish genome makes it a powerful tool for gene discovery research as the zebrafish genome largely resembles the human genome [68]. Large-scale forward genetic studies have been carried-out in some invertebrate model organisms, particularly in the nematode and fruit fly. However, it is extremely expensive to perform this approach in vertebrate animal model like mice. Zebrafish was the first vertebrate organism established for large-scale forward genetic screening. Chemical mutagenesis is achieved by exposing adult male fish to N-ethyl-N-nitrosourea (ENU) for several days which induces point mutations in the spermatogonia. These male fish are then mated with wildtype (WT) female fish to propagate the mutation to F1 progenies [4, 69]. Screening methods such as antibody staining, whole mount in situ hybridization (WISH) and behavioral analysis can be employed to identify morphological or genetic phenotypes. By contrast, gene-specific knockdown mediated by morpholino-injection serves as an efficient tool for reverse genetic study [70]. Mutagenesis and targeted gene disruptions have contributed to uncovering the genes involved in heme and iron metabolism in zebrafish. Recent development of gene-editing tools such as TALENs and CRISPR/cas9 for targeted gene disruption complements transient morpholino gene knockdowns that were typically used for reverse genetic studies [71, 72].
Several key genes which are involved in heme synthesis and metabolism have been elucidated in zebrafish by both forward and reverse genetic manipulation. In Table 1, we summarize the genes and corresponding mutants identified in zebrafish together with related disease in humans. We have also compiled a list of mammalian heme-iron metabolism genes and performed Bulk-BLAST homology alignments to identify potential orthologs in the zebrafish genome [73] (Supplemental Table 1). With the advent of current gene-editing tools for targeted gene disruptions, generating new zebrafish mutants and alleles will be a powerful approach to model human disorders of heme and iron metabolism in a facile vertebrate model.
Table 1.
Summary of zebrafish mutants related to heme -iron metabolism
| Genes | Mutant Name or Morpholino Knockdown | Potential Human Disease | References |
|---|---|---|---|
| ALAS2 | sauternes (sau) | Congenital Sideroblastic Anemia (CSA) | [74] |
| ALAD | Alad−/− | ALA dehydratase deficient porphyria (ADP) | [76] |
| CPOX | Cpox−/− | Hereditary coproporphyria (HCP) | [76] |
| UROD | yquem (yqe) | Porphyria cutanea tarda (PCT) / hepato-erythropoietic porphyria (HEP) | [77] |
| PPOX | montalcino (mno) | Human variegate porphyria | [78] |
| FECH | Freixenet (frx) / dracula (drc) | Erythropoietic protoporphyria | [4] [78] |
| GRX5 | shiraz (sir) | Sideroblastic anemia | [80] [81] |
| MFRN1 | frascati (frs) | Erythropoietic protoporphyria | [27] |
| DMT-1 | chardonnay (cdy) | Hypochromic microcytic anemia | [85] [86] |
| FPN1 | weissherbst (wei) | Iron-deficient anemia | [87] [88] |
| TF | gavi | congenital hypotransferrinemia | [90] |
| TFR1 | chianti (cia) | hypochromic anemia | [92] |
| HRG1 | hrg1a;hrg1b (CRISPR) | unknown | [73] |
| FLVCR | Morpholino Knockdown | Heme toxicity | [96] |
5. 1. ALAS2
Alas2 is the erythroid-specific enzyme for heme synthesis, while Alas1 is found in all other tissues. Zebrafish mutant sauternes (sau) was identified from ENU mutagenesis and positional cloning revealed that sau contains mutation in alas2 [74]. The sau mutants have a microcytic, hypochromic anemia, delayed erythroid maturation, and abnormal globin gene expression, suggesting that defects in heme synthesis can affect globin protein production. Interestingly, sau mutants have normal RBC numbers and show anemia around 33 hpf, possibly because maternal heme may permit early RBC development or genetic compensation by Alas1. As mutations in alas2 cause congenital sideroblastic anemia (CSA) in humans, sau represents the first animal model of this disease in zebrafish [74]. Most interestingly, sau zebrafish mutants are viable even though it has only one-tenth of overall heme content compared to WT zebrafish. Sau mutants can be fully rescued by supplementing developing embryos with ALA, which further enhances its value as a model to study the pathology and possible evaluate the cure methods for CSA in humans [75].
5. 2. ALAD and CPOX
Unlike ALAS, only one form of ALAD and CPOX was found in both human and zebrafish. By using targeted TALEN and CRISPR/Cas9, zebrafish alad−/− and cpox−/− were recently generated to model the ALA-dehydratase-deficient porphyria (ADP) and hereditary coproporphyria (HCP) [76]. alad−/− and cpox−/− mutants suffer from hypochromic anemia, owing to deficiency in heme synthesis, with accumulation of ALA and coproporphyrinogen-III. The abnormal morphology of early RBCs was possibly due by accumulated globin without heme, representing sickle-cell anemia in humans with globin protein aggregations.
5. 3. UROD
Zebrafish mutant yqe contains a nonsense mutation in the gene encoding UROD, which converts uroporphyrinogen to coproporphyrinogen. Homozygous mutation in urod leads to two forms of porphyrias, porphyria cutanea tarda (PCT) and hepatoerythropoietic porphyria (HEP) in human, similar to the phenotypes in zebrafish with photosensitive porphyria syndrome [77]. Excessive amounts of uroporphyrinogens and 7-carboxylate porphyrin accumulate in yqe embryos, representing human HEP patients characterized by photosensitive skin and excessive excretion of heme biosynthesis byproducts, uroporphyrin and 7-decarboxylate porphyrin in their urine. The zebrafish yqe mutant phenocopies human patients facilitating studying of HEP pathogenesis and development of new therapeutics.
5. 4. PPOX
Zebrafish porphyria mutant, montalcino (mno), contains mutation in ppox, which catalyzes the oxidation of protoporphyrinogen, representing human variegate porphyria [78]. Initially, at the onset of circulation, mno displays normal numbers of RBCs but are o-dianisidine negative. A visible decrease in circulating erythrocytes can be observed after 36 hpf and the RBCs in the mutant embryos are fluorescent. The mno mutant zebrafish could survive to approximately 25 dpf. Accordingly, human PPOX could partially rescue the hypochromia in homozygous mutants, revealing functional conservation. The zebrafish mno mutant will be very useful for further elucidating the pathophysiology of variegate porphyria and identifying chemical modifiers of this disease.
5. 5. FECH
Two different mutant alleles of fech have been identified in zebrafish, freixenet (frx) and dracula (drc) [4, 79]. Protoporphyrin IX accumulates in fech mutant embryos owing to a deficiency in the activity of ferrochelatase, the terminal enzyme in the pathway for heme biosynthesis. The mutants show light-dependent hemolysis and liver diseases, similar to that seen in humans with heredity erythropoietic protoporphyria (HEP) resulting from a disorder of ferrochelatase.
5. 6. GRX5
Phenotypic analysis of shiraz (sir) mutant zebrafish revealed an intimate connection between heme biosynthesis and [4Fe-4S] formation, connecting two main uses for iron in the mitochondria which are previously usually thought to be independent processes [80]. In sir mutants, hypochromic anemia, in the context of normal mitochondrial iron and oxidative stress levels, was shown to be caused by a deletion in the glutaredoxin 5 (grx5) gene. GRX5 is required for the synthesis of Fe-S clusters in the mitochondria [81]. The zebrafish protein also localizes to the mitochondria and is capable of rescuing grx5-deficient yeast strain. Fe-S clusters are known to negatively regulate binding of IRP1 to IREs. Decreased Fe-S cluster assembly in sir mutant leads to increased IRP1 activity, which inhibits the expression of IRE-regulated target genes involved in heme biosynthesis, such as alas2. Indeed, alas2 expression is absent in sir mutants. Deletion of IREs in the alas2 mRNA rescued the anemic phenotype while overexpression of full-length alas2 mRNA did not, suggesting that the function of GRX5 in regulating ALAS2 expression is through Fe-S clusters and IRP/IRE activity. An evolutionary conserved role for GRX5 in regulating heme synthesis was confirmed in human patients with GRX5 mutation [82]. Thus, shiraz mutant will be a good tool for searching effective therapeutics for diseases related to Fe-S deficiencies.
5. 7. MFRN1
Two mitoferrin homologs are found in the zebrafish genome, Mfrn1 and Mfrn2. Positional cloning of frascati (frs) zebrafish mutants identified a missense mutation in the mfrn1 (slc25A37) gene, an erythroid-specific form [27]. Frs mutants develop hypochromic anemia and show defects in erythroid maturation. Mouse Mfrn1 rescues the phenotypes in zebrafish frs mutants. The same anemic phenotype was observed in a mouse model with Mfrn1-knockout [28, 83]. The MFRN2 (Slc25A28) paralog functions in mitochondrial iron import in non-erythroid tissues.
5. 8. DMT1
DMT1 is upregulated by dietary iron deficiency and the Belgrade rat carries a mutation in dmt1 suffer from iron deficient anemia [23, 84]. Zebrafish mutants chardonnay (cdy) carry a nonsense mutation in dmt1 with reduced hemoglobin levels and delayed erythrocyte maturation [85]. The Dmt1 protein is expressed in erythroid cells and the duodenum suggesting its role in erythropoiesis and intestinal iron absorption. Cells with overexpression of WT zebrafish dmt1 uptake nearly ten-times the amount of iron as non-transfected control cells, whereas the cdy mutant protein is not functional. However, cdy mutants can survive to adulthood despite severe anemia, again suggesting an alternate path for iron absorption in zebrafish. In humans, mutations in DMT1 cause a phenotype of hypochromic microcytic anemia combined with iron overload, further supporting the possible existence of alternative mechanisms for duodenal iron absorption that bypasses DMT1 [86].
5. 9. FPN1
The first identified iron exporter Fpn1 (Slc40A1) was found by position-cloning of zebrafish weissherbst (weh) mutant [87]. weh mutant embryos show hypochromic anemia with decreased hemoglobin levels, blocked erythroid maturation, and reduced numbers of erythrocytes. Erythroid cells of mutant embryos have significantly lower iron concentrations compared to WT embryos, suggesting iron deficiency. Microinjection of iron-dextran rescues the anemic phenotype of weh mutants and continued injection of iron-dextran rescues the embryonic lethality. These rescued fish, however, are only normal until 6 months of age and eventually develop hypochromic anemia by 12 months, suggesting that Fpn1 is also involved in adult red cell function, specifically iron-recycling in adult zebrafish. Compared with iron injected WT fish, the rescued mutants had increased iron staining in kidney macrophages, as well as increased staining in intestinal villi, suggesting that fpn1 mutation impairs iron export in these tissues. The iron-rescued weh mutants also have hepatic iron overload, with particularly high iron levels in the liver [88]. FPN1 also localizes to the yolk-syncytial layer (YSL) during embryonic development, suggesting that FPN1 may transport maternal iron from the yolk for embryogenesis. Both mice and humans have homologs of FPN1with high similarities to zebrafish Fpn1. Mammalian FPN1 is robustly expressed in the placenta, duodenum, and liver. At the protein level, human FPN1 is concentrated on the basal surface of syncytiotrophoblasts in the placenta, an organ that is functionally like zebrafish YSL, indicating that human FPN1 plays a role in maternal-fetal iron export. In mice, FPN1 is expressed on the basolateral surface of enterocytes, suggesting a role in intestinal iron transport [89].
5. 10. TF
The zebrafish mutant gavi (gav) was shown to have mutations in transferrin-a (Tf-a), which encodes the principal serum iron carrier [90]. Gav mutant embryos exhibit reduced tf-a expression and impaired hemoglobin production with hypochromic anemia and embryonic lethality by 14 dpf. In humans, phenotype of congenital hypotransferrinemia caused by TF mutation is highly similar to those of gav mutants, including hypochromic anemia and embryonic death [91]. The gav mutant is thus an ideal whole vertebrate anemia model for studying symptoms corresponding to human pathologies related to Tf.
5. 11. TFR1
Transferrin-bound iron is taken up into cells by binding to the transferrin receptor 1 (TFR1). Four different zebrafish chianti (cia) mutants with varying degrees of hypochromic anemia and defective erythroid differentiation were ascribed to mutations in tfr1a gene [92]. During early development, tfr1a transcripts are expressed specifically in erythrocytes. Importantly, cytoplasmic delivery of iron by microinjection at one-cell stage - but not intravenous iron injections rescue the hypochromia in cia mutants, indicating that tfr1a mutation prevents erythrocytes from taking-up and utilizing circulating iron. Intriguingly, a second tfr1 gene, tfr1b was identified together with tfr1a, a typical feature in zebrafish genome which has undergone whole genome duplication [93, 94]. Whereas tfr1a is expressed in erythrocytes during early development and cia mutants are anemic, tfr1b is ubiquitously expressed throughout embryogenesis and knockdown of tfr1b by morpholinos do not affect hemoglobinization. Tfr1b morphants have retarded growth and develop brain necrosis, a phenotype that is similar to the neurologic defects observed in the mouse model [95], indicating that tfr1b may be involved in iron uptake in non-erythroid tissues. Tfr1a (cia) and tfr1b deficient zebrafish embryos recapitulate the phenotype of TFR1−/− mice [92].
5. 12. FLVCR and HRG1
Currently no zebrafish mutants have been reported with perturbation of Flvcr. However, the function of zebrafish Flvcr is related to erythroid differentiation and maturation as determined by morpholino gene knockdowns [96]. Two splicing isoforms have been identified: flvcr1a and flvcr1b. Flvcr1a is required for the expansion of committed erythroid progenitors but cannot drive their terminal differentiation, while Flvcr1b contributes to the expansion phase and is required for differentiation [96]. The coordinated expression of flvcr1a and flvcr1b contributes to controlling the cytosolic heme pool required to sustain regulation of erythroid progenitors and hemoglobin synthesis for hemoglobinization during terminal maturation. Interestingly, treatment with succinylacetone (SA), an inhibitor of heme synthesis, rescues the phenotype of flvcr1a morphants while heme supplementation restores hemoglobinzation of flvcr1b morphants, suggesting that the intracellular heme pool during erythropoiesis is tightly regulated. As Flvcr−/−mice are embryonically lethal, zebrafish flvcr mutants will be useful to dissect the embryonic functions of FLVCR.
Recently, zebrafish mutants for hrg1 (Slc48a1), a heme transporter with homologs in mammals, was generated by CRISPR/cas9 [73, 97]. Zebrafish genome contains two hrg1 paralogs - hrg1a (slc48a1b) and hrg1b (slc48a1a). Both are ubiquitously expressed throughout developing embryos and in the one-cell embryo. More importantly, hrg1a;hrg1b double mutants show accumulation of heme in the kidney macrophages due to defects in heme-iron recycling from damaged RBCs. RNAseq results show significant perturbation in heme-iron metabolism and immune-related genes in the absence of hrg1. Altogether, the fish studies affirm that Hrg1 is essential for recycling RBCs and that this function is performed by the kidney marrow, which is the adult hematopoietic organ in zebrafish [73].
6. Conclusions
Zebrafish has proven to be an indispensable vertebrate model to study heme and iron metabolism with unique genetic advantages not afforded by mammalian models. The advantages of ex utero genetic manipulation and embryonic transparency allow convenient functional validation of both zebrafish and human genes. Moreover, chemical-genetics drug screens in zebrafish provide a more comprehensive whole animal in-a-dish analyses for drug tests than typical cell-culture based assays [98]. Given the high degree of conservation in the heme and iron acquisition pathways in zebrafish and humans, it would be prudent to develop diseased models of humanized zebrafish for targeted high throughput screens to identify more efficacious therapeutic drugs against rare diseases.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Lieschke GJ, Currie PD, Animal models of human disease: zebrafish swim into view Nat Rev Genet 8 (2007) 353–367. [DOI] [PubMed] [Google Scholar]
- [2].Davidson AJ, Zon LI, The ‘definitive’ (and ‘primitive’) guide to zebrafish hematopoiesis Oncogene 23 (2004) 7233. [DOI] [PubMed] [Google Scholar]
- [3].Galloway JL, Zon LI, 3 Ontogeny of hematopoiesis: Examining the emergence of hematopoietic cells in the vertebrate embryo, in: B.T.-C.T.i.D. Biology (Ed.), Academic Press, 2003, pp. 139–158. [DOI] [PubMed] [Google Scholar]
- [4].Ransom DG, Haffter P, Odenthal J, Brownlie a., Vogelsang E, Kelsh RN, Brand M, van Eeden FJ, Furutani-Seiki M, Granato M, Hammerschmidt M, Heisenberg CP, Jiang YJ, Kane D.a., Mullins MC, Nüsslein-Volhard C, Characterization of zebrafish mutants with defects in embryonic hematopoiesis. Development (Cambridge, England) 123 (1996) 311–319. [DOI] [PubMed] [Google Scholar]
- [5].Zon J.L.O.d.J.a.L.I., Use of the Zebrafish System to Study Primitive and Definitive Hematopoiesis Annual Review of Genetics 39 (2005) 481–501. [DOI] [PubMed] [Google Scholar]
- [6].Lyons SE, Lawson ND, Lei L, Bennett PE, Weinstein BM, Liu PP, A nonsense mutation in zebrafish gata1 causes the bloodless phenotype in vlad tepes Proceedings of the National Academy of Sciences 99 (2002) 5454–5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Long Q, Meng a., Wang H, Jessen JR, Farrell MJ, Lin S, GATA-1 expression pattern can be recapitulated in living transgenic zebrafish using GFP reporter gene. Development (Cambridge, England) 124 (1997) 4105–4111. [DOI] [PubMed] [Google Scholar]
- [8].Cumano A.a.I.G., Ontogeny of the Hematopoietic System Annual Review of Immunology 25 (2007) 745–785. [DOI] [PubMed] [Google Scholar]
- [9].Burns CE, DeBlasio T, Zhou Y, Zhang J, Zon L, Nimer SD, Isolation and characterization of runxa and runxb, zebrafish members of the runt family of transcriptional regulators Experimental Hematology 30 (2002) 1381–1389. [DOI] [PubMed] [Google Scholar]
- [10].Willett CE, Cortes A, Zuasti A, Zapata AG, Early hematopoiesis and developing lymphoid organs in the zebrafish Developmental Dynamics 214 (1999) 323–336. [DOI] [PubMed] [Google Scholar]
- [11].Murayama E, Kissa K, Zapata A, Mordelet E, Briolat V, Lin H-F, Handin RI, Herbomel P, Tracing Hematopoietic Precursor Migration to Successive Hematopoietic Organs during Zebrafish Development Immunity 25 (2006) 963–975. [DOI] [PubMed] [Google Scholar]
- [12].Jin H, Xu J, Wen Z, Migratory path of definitive hematopoietic stem/progenitor cells during zebrafish development Blood 109 (2007) 5208 LP–5214. [DOI] [PubMed] [Google Scholar]
- [13].Liao EC, Trede NS, Ransom D, Zapata A, Kieran M, Zon LI, Non-cell autonomous requirement for the <em>bloodless</em> gene in primitive hematopoiesis of zebrafish Development 129 (2002) 649 LP–659. [DOI] [PubMed] [Google Scholar]
- [14].Chan F-Y, Robinson J, Brownlie A, Shivdasani RA, Donovan A, Brugnara C, Kim J, Lau B-C, Witkowska HE, Zon LI, Characterization of Adult α- and β-Globin Genes in the Zebrafish Blood 89 (1997) 688 LP–700. [PubMed] [Google Scholar]
- [15].Brownlie A, Hersey C, Oates AC, Paw BH, Falick AM, Witkowska HE, Flint J, Higgs D, Jessen J, Bahary N, Zhu H, Lin S, Zon L, Characterization of embryonic globin genes of the zebrafish Developmental Biology 255 (2003) 48–61. [DOI] [PubMed] [Google Scholar]
- [16].Traver D, Paw BH, Poss KD, Penberthy WT, Lin S, Zon LI, Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants Nat Immunol 4 (2003) 1238–1246. [DOI] [PubMed] [Google Scholar]
- [17].Traver D, Winzeler A, Stern HM, Mayhall EA, Langenau DM, Kutok JL, Look AT, Zon LI, Effects of lethal irradiation in zebrafish and rescue by hematopoietic cell transplantation Blood 104 (2004) 1298 LP–1305. [DOI] [PubMed] [Google Scholar]
- [18].Carradice D, Lieschke GJ, Zebrafish in hematology: sushi or science? Blood 111 (2008) 3331–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lieschke GJ, Oates AC, Crowhurst MO, Ward AC, Layton JE, Morphologic and functional characterization of granulocytes and macrophages in embryonic and adult zebrafish Blood 98 (2001) 3087 LP–3096. [DOI] [PubMed] [Google Scholar]
- [20].Menke AL, Spitsbergen JM, Wolterbeek APM, Woutersen R.a., Normal anatomy and histology of the adult zebrafish. Toxicologic pathology 39 (2011) 759–775. [DOI] [PubMed] [Google Scholar]
- [21].Aisen P, Leibman A, Zweier J, Stoichiometric and site characteristics of the binding of iron to human transferrin. Journal of Biological Chemistry 253 (1978) 1930–1937. [PubMed] [Google Scholar]
- [22].Ohgami RS, Campagna DR, Greer EL, Antiochos B, McDonald A, Chen J, Sharp JJ, Fujiwara Y, Barker JE, Fleming MD, Identification of a ferrireductase required for efficient transferrin-dependent iron uptake in erythroid cells Nat Genet 37 (2005) 1264–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gunshin H, Mackenzie B, Berger UV, Gunshin Y, Romero MF, Boron WF, Nussberger S, Gollan JL, Hediger MA, Cloning and characterization of a mammalian proton-coupled metal-ion transporter Nature 388 (1997) 482–488. [DOI] [PubMed] [Google Scholar]
- [24].Winterbourn CC, Toxicity of iron and hydrogen peroxide: the Fenton reaction Toxicology Letters 82–83 (1995) 969–974. [DOI] [PubMed] [Google Scholar]
- [25].Leidgens S, Bullough KZ, Shi H, Li F, Shakoury-Elizeh M, Yabe T, Subramanian P, Hsu E, Natarajan N, Nandal A, Stemmler TL, Philpott CC, Each Member of the Poly-r(C)-binding Protein 1 (PCBP) Family Exhibits Iron Chaperone Activity toward Ferritin Journal of Biological Chemistry 288 (2013) 17791–17802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ryu M.-s., Zhang D, Protchenko O, Shakoury-Elizeh M, Philpott CC, PCBP1 and NCOA4 regulate erythroid iron storage and heme biosynthesis Journal of Clinical Investigation (2017) 1–12. [DOI] [PMC free article] [PubMed]
- [27].Shaw GC, Cope JJ, Li L, Corson K, Hersey C, Ackermann GE, Gwynn B, Lambert AJ, Wingert R.a., Traver D, Trede NS, Barut B.a., Zhou Y, Minet E, Donovan A, Brownlie A, Balzan R, Weiss MJ, Peters LL, Kaplan J, Zon LI, Paw BH, Mitoferrin is essential for erythroid iron assimilation. Nature 440 (2006) 96–100. [DOI] [PubMed] [Google Scholar]
- [28].Troadec M-BB, Warner D, Wallace J, Thomas K, Spangrude GJ, Phillips J, Khalimonchuk O, Paw BH, Ward DM, Kaplan J, Targeted deletion of the mouse Mitoferrin1 gene: from anemia to protoporphyria. Blood 117 (2011) 5494–5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Richardson DR, Ponka P, Vyoral D, Distribution of iron in reticulocytes after inhibition of heme synthesis with succinylacetone: examination of the intermediates involved in iron metabolism. Blood 87 (1996) 3477–3488. [PubMed] [Google Scholar]
- [30].Zhang A-S, Sheftel AD, Ponka P, Intracellular kinetics of iron in reticulocytes: evidence for endosome involvement in iron targeting to mitochondria Blood 105 (2005) 368–375. [DOI] [PubMed] [Google Scholar]
- [31].Muñoz M, García-Erce JA, Remacha ÁF, Disorders of iron metabolism. Part II: iron deficiency and iron overload. Journal of clinical pathology 64 (2011) 287–296. [DOI] [PubMed] [Google Scholar]
- [32].Hunter GA, Ferreira GC, Molecular enzymology of 5-Aminolevulinate synthase, the gatekeeper of heme biosynthesis, Biochimica et Biophysica Acta - Proteins and Proteomics, 2011, pp. 1467–1473. [DOI] [PMC free article] [PubMed]
- [33].May BK, Dogra SC, Sadlon TJ, Bhasker CR, Cox TC, Bottomley SS, Molecular Regulation of Heme Biosynthesis in Higher Vertebrates Progress in Nucleic Acid Research and Molecular Biology 51 (1995) 1–51. [DOI] [PubMed] [Google Scholar]
- [34].Sadlon TJ, Dell’Oso T, Surinya KH, May BK, Dell’Oso T, Surinya KH, May BK, Regulation of erythroid 5-aminolevulinate synthase expression during erythropoiesis International Journal of Biochemistry and Cell Biology 31 (1999) 1153–1167. [DOI] [PubMed] [Google Scholar]
- [35].Weiss MJ, Yu C, Orkin SH, Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Molecular and cellular biology 17 (1997) 1642–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Tanimura N, Miller E, Igarashi K, Yang D, Burstyn JN, Dewey CN, Bresnick EH, Mechanism governing heme synthesis reveals a GATA factor/heme circuit that controls differentiation. EMBO reports 17 (2015) 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhang Y, Zhang J, An W, Wan Y, Ma S, Yin J, Li X, Gao J, Yuan W, Guo Y, Engel JD, Shi L, Cheng T, Zhu X, Intron 1 GATA site enhances ALAS2 expression indispensably during erythroid differentiation Nucleic Acids Research (2016). [DOI] [PMC free article] [PubMed]
- [38].Wilkinson N, Pantopoulos K, The IRP/IRE system in vivo: insights from mouse models Frontiers in Pharmacology 2014, pp. 176. [DOI] [PMC free article] [PubMed]
- [39].Tugores A, Magness ST, Brenner DA, A single promoter directs both housekeeping and erythroid preferential expression of the human ferrochelatase gene. Journal of Biological Chemistry 269 (1994) 30789–30797. [PubMed] [Google Scholar]
- [40].Crooks DR, Ghosh MC, Haller RG, Tong W-H, Rouault TA, Posttranslational stability of the heme biosynthetic enzyme ferrochelatase is dependent on iron availability and intact iron-sulfur cluster assembly machinery Blood 115 (2010) 860 LP–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Magness ST, Tugores A, Brenner DA, Analysis of ferrochelatase expression during hematopoietic development of embryonic stem cells Blood 95 (2000) 3568 LP–3577. [PubMed] [Google Scholar]
- [42].Chan RYY, Schulman HM, Ponka P, Expression of ferrochelatase mRNA in erythroid and non-erythroid cells Biochemical Journal 292 (1993) 343 LP–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Tahara T, Sun J, Nakanishi K, Yamamoto M, Mori H, Saito T, Fujita H, Igarashi K, Taketani S, Heme Positively Regulates the Expression of β-Globin at the Locus Control Region via the Transcriptional Factor Bach1 in Erythroid Cells Journal of Biological Chemistry 279 (2004) 5480–5487. [DOI] [PubMed] [Google Scholar]
- [44].Severance S, Hamza I, Trafficking of Heme and Porphyrins in Metazoa (2009) 4596–4616. [DOI] [PMC free article] [PubMed]
- [45].Schultz IJ, Chen C, Paw BH, Hamza I, Iron and Porphyrin Trafficking in Heme Biogenesis The Journal of Biological Chemistry 285 (2010) 26753–26759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Liesa M, Qiu W, Shirihai OS, Mitochondrial ABC transporters function: The role of ABCB10 (ABC-me) as a novel player in cellular handling of reactive oxygen species Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1823 (2012) 1945–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Chen W, Dailey HA, Paw BH, Ferrochelatase forms an oligomeric complex with mitoferrin-1 and Abcb10 for erythroid heme biosynthesis Blood 116 (2010) 628 LP–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Quigley JG, Yang Z, Worthington MT, Phillips JD, Sabo KM, Sabath DE, Berg CL, Sassa S, Wood BL, Abkowitz JL, Identification of a human heme exporter that is essential for erythropoiesis. Cell 118 (2004) 757–766. [DOI] [PubMed] [Google Scholar]
- [49].Keel SB, Doty RT, Yang Z, Quigley JG, Chen J, Knoblaugh S, Kingsley PD, De Domenico I, Vaughn MB, Kaplan J, Palis J, Abkowitz JL, A heme export protein is required for red blood cell differentiation and iron homeostasis Science (New York, NY) 319 (2008) 825–828. [DOI] [PubMed] [Google Scholar]
- [50].Chiabrando D, Marro S, Mercurio S, Giorgi C, Petrillo S, Vinchi F, Fiorito V, Fagoonee S, Camporeale A, Turco E, Merlo GR, Silengo L, Altruda F, Pinton P, Tolosano E, The mitochondrial heme exporter FLVCR1b mediates erythroid differentiation 122 (2012) 4569–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Quigley JG, Burns CC, Anderson MM, Lynch ED, Sabo KM, Overbaugh J, Abkowitz JL, Cloning of the cellular receptor for feline leukemia virus subgroup C (FeLV-C), a retrovirus that induces red cell aplasia Blood 95 (2000) 1093 LP–1099. [PubMed] [Google Scholar]
- [52].Yang Z, Philips JD, Doty RT, Giraudi P, Ostrow JD, Tiribelli C, Smith A, Abkowitz JL, Kinetics and Specificity of Feline Leukemia Virus Subgroup C Receptor (FLVCR) Export Function and Its Dependence on Hemopexin The Journal of Biological Chemistry 285 (2010) 28874–28882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Alves LR, Costa ES, Sorgine MHF, Nascimento-Silva MCL, Teodosio C, B??rcena P, Castro-Faria-Neto HC, Bozza P.T.P.c.T., Orfao A, Oliveira PL, Maya-Monteiro CM, Bárcena P, Castro-Faria-Neto HC, Bozza P.T.P.c.T., Orfao A, Oliveira PL, Maya-Monteiro CM, Heme-oxygenases during erythropoiesis in K562 and human bone marrow cells PLoS ONE 6 (2011) e21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Krishnamurthy P, Schuetz JD, The ABC Transporter Abcg2/Bcrp: Role in Hypoxia Mediated Survival Biometals 18 (2005) 349–358. [DOI] [PubMed] [Google Scholar]
- [55].Yamamoto K, Suzu S, Yoshidomi Y, Hiyoshi M, Harada H, Okada S, Erythroblasts highly express the ABC transporter Bcrp1/ABCG2 but do not show the side population (SP) phenotype. Immunology letters 114 (2007) 52–58. [DOI] [PubMed] [Google Scholar]
- [56].Desuzinges-Mandon E, Arnaud O, Martinez L, Huché F, Di Pietro A, Falson P, ABCG2 Transports and Transfers Heme to Albumin through Its Large Extracellular Loop The Journal of Biological Chemistry 285 (2010) 33123–33133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Saison C, Helias V, Ballif BA, Peyrard T, Puy H, Miyazaki T, Perrot S, Vayssier-Taussat M, Waldner M, Le Pennec P-Y, Cartron J-P, Arnaud L, Null alleles of ABCG2 encoding the breast cancer resistance protein define the new blood group system Junior Nat Genet 44 (2012) 174–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Zelinski T, Coghlan G, Liu X-Q, Reid ME, ABCG2 null alleles define the Jr(a-) blood group phenotype Nat Genet 44 (2012) 131–132. [DOI] [PubMed] [Google Scholar]
- [59].Rao AU, Carta LK, Lesuisse E, Hamza I, Lack of heme synthesis in a free-living eukaryote Proceedings of the National Academy of Sciences of the United States of America 102 (2005) 4270–4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Korolnek T, Zhang J, Beardsley S, Scheffer George L.L., Hamza I, Control of metazoan heme homeostasis by a conserved multidrug resistance protein Cell Metabolism 19 (2014) 1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Shayeghi M, Latunde-Dada GO, Oakhill JS, Laftah AH, Takeuchi K, Halliday N, Khan Y, Warley A, McCann FE, Hider RC, Frazer DM, Anderson GJ, Vulpe CD, Simpson RJ, McKie AT, Identification of an Intestinal Heme Transporter Cell 122 (2005) 789–801. [DOI] [PubMed] [Google Scholar]
- [62].Le Blanc S, Garrick MD, Arredondo M, Heme carrier protein 1 transports heme and is involved in heme-Fe metabolism AJP: Cell Physiology 302 (2012) C1780–C1785. [DOI] [PubMed] [Google Scholar]
- [63].Salojin KV, Cabrera RM, Sun W, Chang WC, Lin C, Duncan L, Platt KA, Read R, Vogel P, Liu Q, Finnell RH, Oravecz T, A mouse model of hereditary folate malabsorption: deletion of the <em>PCFT</em> gene leads to systemic folate deficiency Blood (2011). [DOI] [PubMed]
- [64].Laftah AH, Latunde-Dada GO, Fakih S, Hider RC, Simpson RJ, McKie AT, Haem and folate transport by proton-coupled folate transporter/haem carrier protein 1 (SLC46A1) British Journal of Nutrition 101 (2008) 1150–1156. [DOI] [PubMed] [Google Scholar]
- [65].Rajagopal A, Rao AU, Amigo J, Tian M, Upadhyay SK, Hall C, Uhm S, Mathew MK, Fleming MD, Paw BH, Krause M, Hamza I, Haem homeostasis is regulated by the conserved and concerted functions of HRG-1 proteins. Nature 453 (2008) 1127–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].O’Callaghan KM, Ayllon V, O’Keeffe J, Wang Y, Cox OT, Loughran G, Forgac M, O’Connor R, Heme-binding Protein HRG-1 Is Induced by Insulin-like Growth Factor I and Associates with the Vacuolar H(+)-ATPase to Control Endosomal pH and Receptor Trafficking The Journal of Biological Chemistry 285 (2010) 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].An X, Schulz VP, Li J, Wu K, Liu J, Xue F, Hu J, Mohandas N, Gallagher PG, Global transcriptome analyses of human and murine terminal erythroid differentiation Blood 123 (2014) 3466 LP–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, McLaren S, Sealy I, Caccamo M, Churcher C, Scott C, Barrett JC, Koch R, Rauch G-J, White S, Chow W, Kilian B, Quintais LT, Guerra-Assunção J.a., Zhou Y, Gu Y, Yen J, Vogel J-H, Eyre T, Redmond S, Banerjee R, Chi J, Fu B, Langley E, Maguire SF, Laird GK, Lloyd D, Kenyon E, Donaldson S, Sehra H, Almeida-King J, Loveland J, Trevanion S, Jones M, Quail M, Willey D, Hunt A, Burton J, Sims S, McLay K, Plumb B, Davis J, Clee C, Oliver K, Clark R, Riddle C, Elliot D, Eliott D, Threadgold G, Harden G, Ware D, Mortimore B, Mortimer B, Kerry G, Heath P, Phillimore B, Tracey A, Corby N, Dunn M, Johnson C, Wood J, Clark S, Pelan S, Griffiths G, Smith M, Glithero R, Howden P, Barker N, Stevens C, Harley J, Holt K, Panagiotidis G, Lovell J, Beasley H, Henderson C, Gordon D, Auger K, Wright D, Collins J, Raisen C, Dyer L, Leung K, Robertson L, Ambridge K, Leongamornlert D, McGuire S, Gilderthorp R, Griffiths C, Manthravadi D, Nichol S, Barker G, Whitehead S, Kay M, Brown J, Murnane C, Gray E, Humphries M, Sycamore N, Barker D, Saunders D, Wallis J, Babbage A, Hammond S, Mashreghi-Mohammadi M, Barr L, Martin S, Wray P, Ellington A, Matthews N, Ellwood M, Woodmansey R, Clark G, Cooper JD, Cooper J, Tromans A, Grafham D, Skuce C, Pandian R, Andrews R, Harrison E, Kimberley A, Garnett J, Fosker N, Hall R, Garner P, Kelly D, Bird C, Palmer S, Gehring I, Berger A, Dooley CM, Ersan-Ürün Z, Eser C, Geiger H, Geisler M, Karotki L, Kirn A, Konantz J, Konantz M, Oberländer M, Rudolph-Geiger S, Teucke M, Osoegawa K, Zhu B, Rapp A, Widaa S, Langford C, Yang F, Carter NP, Harrow J, Ning Z, Herrero J, Searle SMJ, Enright A, Geisler R, Plasterk R.H.a., Lee C, Westerfield M, de Jong PJ, Zon LI, Postlethwait JH, Nüsslein-Volhard C, Hubbard TJP, Roest Crollius H, Rogers J, Stemple DL, Begum S, Lloyd C, Lanz C, Raddatz G, Schuster SC, The zebrafish reference genome sequence and its relationship to the human genome. Nature 496 (2013) 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, Stainier DY, Zwartkruis F, Abdelilah S, Rangini Z, Belak J, Boggs C, A genetic screen for mutations affecting embryogenesis in zebrafish Development 123 (1996) 37 LP–46. [DOI] [PubMed] [Google Scholar]
- [70].Sumanas S, Larson JD, Morpholino phosphorodiamidate oligonucleotides in zebrafish: a recipe for functional genomics? Briefings in functional genomics & proteomics 1 (2002) 239–256. [DOI] [PubMed] [Google Scholar]
- [71].Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B, Heritable gene targeting in zebrafish using customized TALENs. Nature biotechnology 29 (2011) 699–700. [DOI] [PubMed] [Google Scholar]
- [72].Jao L-E, Wente SR, Chen W, Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proceedings of the National Academy of Sciences of the United States of America 110 (2013) 13904–13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Zhang J, Chambers I, Yun S, Phillips J, Krause M, Hamza I, Hrg1 promotes heme-iron recycling during hemolysis in the zebrafish kidney PLOS Genetics 14 (2018) e1007665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Brownlie a., Donovan a., Pratt SJ, Paw BH, Oates AC, Brugnara C, Witkowska HE, Sassa S, Zon LI, Positional cloning of the zebrafish sauternes gene: a model for congenital sideroblastic anaemia. Nature genetics 20 (1998) 244–250. [DOI] [PubMed] [Google Scholar]
- [75].Kardon JR, Yien YY, Huston NC, Branco DS, Hildick-Smith GJ, Rhee KY, Paw BH, Baker TA, Mitochondrial ClpX activates a key enzyme for heme biosynthesis and erythropoiesis Cell 161 (2015) 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Sakamoto D, Kudo H, Inohaya K, Yokoi H, Narita T, Naruse K, Mitani H, Araki K, Shima A, Ishikawa Y, Imai Y, Kudo A, A mutation in the gene for delta-aminolevulinic acid dehydratase (ALAD) causes hypochromic anemia in the medaka, Oryzias latipes. Mechanisms of development 121 (2004) 747–752. [DOI] [PubMed] [Google Scholar]
- [77].Wang H, Long Q, Marty SD, Sassa S, Lin S, A zebrafish model for hepatoerythropoietic porphyria Nature Genetics 20 (1998) 239–243. [DOI] [PubMed] [Google Scholar]
- [78].Dooley K.a., Fraenkel PG, Langer NB, Schmid B, Davidson AJ, Weber G, Chiang K, Foott H, Dwyer C, Wingert R.a., Zhou Y, Paw BH, Zon LI, montalcino, A zebrafish model for variegate porphyria. Experimental hematology 36 (2008) 1132–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Childs S, Weinstein BM, Mohideen M.a., Donohue S, Bonkovsky H, Fishman MC, Zebrafish dracula encodes ferrochelatase and its mutation provides a model for erythropoietic protoporphyria. Current biology : CB 10 (2000) 1001–1004. [DOI] [PubMed] [Google Scholar]
- [80].Wingert R.a., Galloway JL, Barut B, Foott H, Fraenkel P, Axe JL, Weber GJ, Dooley K, Davidson AJ, Schmid B, Paw BH, Shaw GC, Kingsley P, Palis J, Schubert H, Chen O, Kaplan J, Zon LI, Deficiency of glutaredoxin 5 reveals Fe-S clusters are required for vertebrate haem synthesis. Nature 436 (2005) 1035–1039. [DOI] [PubMed] [Google Scholar]
- [81].Rodríguez-Manzaneque MT, Tamarit J, Bellí G, Ros J, Herrero E, Grx5 Is a Mitochondrial Glutaredoxin Required for the Activity of Iron/Sulfur Enzymes Molecular Biology of the Cell 13 (2002) 1109–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Camaschella C, Campanella A, De Falco L, Boschetto L, Merlini R, Silvestri L, Levi S, Iolascon A, The human counterpart of zebrafish shiraz shows sideroblastic-like microcytic anemia and iron overload Blood 110 (2007) 1353–1358. [DOI] [PubMed] [Google Scholar]
- [83].Chung J, Anderson SA, Gwynn B, Deck KM, Chen MJ, Langer NB, Shaw GC, Huston NC, Boyer LF, Datta S, Paradkar PN, Li L, Wei Z, Lambert AJ, Sahr K, Wittig JG, Chen W, Lu W, Galy B, Schlaeger TM, Hentze MW, Ward DM, Kaplan J, Eisenstein RS, Peters LL, Paw BH, Iron Regulatory Protein-1 Protects against Mitoferrin-1-deficient Porphyria Journal of Biological Chemistry 289 (2014) 7835–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Fleming MD, Trenor CC III, Su MA, Foernzler D, Beier DR, Dietrich WE, Andrews NC, Microcytic anaemia mice have a mutation in Nramp2, a candidate iron transporter gene Nature Genetics 16 (1997) 383–386. [DOI] [PubMed] [Google Scholar]
- [85].Donovan A, Brownlie A, Dorschner MO, Zhou Y, Pratt SJ, Paw BH, Phillips RB, Thisse C, Thisse B, Zon LI, The zebrafish mutant gene chardonnay (cdy) encodes divalent metal transporter 1 (DMT1) Blood 100 (2002) 4655–4659. [DOI] [PubMed] [Google Scholar]
- [86].Mims MP, Guan Y, Pospisilova D, Priwitzerova M, Indrak K, Ponka P, Divoky V, Prchal JT, Identification of a human mutation of DMT1 in a patient with microcytic anemia and iron overload Blood 105 (2005) 1337–1342. [DOI] [PubMed] [Google Scholar]
- [87].Donovan a., Brownlie a., Zhou Y, Shepard J, Pratt SJ, Moynihan J, Paw BH, Drejer a., Barut B, Zapata a., Law TC, Brugnara C, Lux SE, Pinkus GS, Pinkus JL, Kingsley PD, Palis J, Fleming MD, Andrews NC, Zon LI, Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature 403 (2000) 776–781. [DOI] [PubMed] [Google Scholar]
- [88].Fraenkel PG, Traver D, Donovan A, Zahrieh D, Zon LI, Ferroportin1 is required for normal iron cycling in zebrafish Journal of Clinical Investigation 115 (2005) 1532–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Donovan A, Lima CA, Pinkus JL, Pinkus GS, Zon LI, Robine S, Andrews NC, The iron exporter ferroportin/Slc40a1 is essential for iron homeostasis Cell Metabolism 1 (2005) 191–200. [DOI] [PubMed] [Google Scholar]
- [90].Fraenkel PG, Gibert Y, Holzheimer JL, Lattanzi VJ, Burnett SF, Dooley KA, Wingert RA, Zon LI, Transferrin-a modulates hepcidin expression in zebrafish embryos Blood 113 (2009) 2843–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Goldwurm S, Casati C, Venturi N, Strada S, Santambrogio P, Indraccolo S, Arosio P, Cazzola M, Piperno A, Masera G, Blondi A, Biochemical and genetic defects underlying human congenital hypotransferrinemia Hematology Journal 1 (2000) 390–398. [DOI] [PubMed] [Google Scholar]
- [92].Wingert R.a., Brownlie A, Galloway JL, Dooley K, Fraenkel P, Axe JL, Davidson AJ, Barut B, Noriega L, Sheng X, Zhou Y, Zon LI, The chianti zebrafish mutant provides a model for erythroid-specific disruption of transferrin receptor 1. Development (Cambridge, England) 131 (2004) 6225–6235. [DOI] [PubMed] [Google Scholar]
- [93].Postlethwait JH, Yan Y-LL, Gates M.a., Horne S, Amores A, Brownlie A, Donovan A, Egan ES, Force A, Gong Z, Goutel C, Fritz A, Kelsh R, Knapik E, Liao E, Paw B, Ransom D, Singer A, Thomson M, Abduljabbar TS, Yelick P, Beier D, Joly J-SS, Larhammar D, Rosa F, Westerfield M, Zon LI, Johnson SL, Talbot WS, Vertebrate genome evolution and the zebrafish gene map Nat Genet 18 (1998) 345–349. [DOI] [PubMed] [Google Scholar]
- [94].Amores A, Force A, Yan Y-L, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang Y-L, Westerfield M, Ekker M, Postlethwait JH, Zebrafish hox clusters and vertebrate genome evolution Science 282 (1998) 1711 LP–1714. [DOI] [PubMed] [Google Scholar]
- [95].Levy JE, Jin O, Fujiwara Y, Kuo F, Andrews NC, Transferrin receptor is necessary for development of erythrocytes and the nervous system Nature Genetics 21 (1999) 396–399. [DOI] [PubMed] [Google Scholar]
- [96].Mercurio S, Petrillo S, Chiabrando D, Bassi ZI, Gays D, Camporeale A, Vacaru A, Miniscalco B, Valperga G, Silengo L, Altruda F, Baron MH, Santoro MM, Tolosano E, The heme exporter Flvcr1 regulates expansion and differentiation of committed erythroid progenitors by controlling intracellular heme accumulation Haematologica 100 (2015) 720–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].White C, Yuan X, Schmidt PJ, Bresciani E, Samuel TK, Campagna D, Hall C, Bishop K, Calicchio ML, Lapierre A, Ward DM, Liu P, Fleming MD, Hamza I, HRG1 is essential for heme transport from the phagolysosome of macrophages during erythrophagocytosis Cell metabolism 17 (2013) 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Kaufman CK, White RM, Zon L, Chemical Genetic Screening in the Zebrafish Embryo Nature protocols 4 (2009) 1422–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


