Introductory paragraph
The protozoan parasite Toxoplasma gondii has co-evolved with its homeothermic hosts, human included, strategies that drive its quasi asymptomatic persistence in hosts, hence optimizing the chance of transmission to new hosts. Persistence which starts with a small subset of parasites that escape from the host immune killing and colonize the so-called immune privileged tissues where they differentiate into a low replicating stage, is driven by the IL-12 and IFN-γ axis. The recent characterization of a Toxoplasma effector family delivered into the host cell where it rewires the host cell gene expression has allowed identifying regulators of the IL-12-IFN-γ axis including repressors. We now report on the dense granule-resident effector, called TEEGR (Toxoplasma E2F4-associated EZH2-inducing Gene Regulator) that counteracts the NF-κB signaling pathway. Once exported in the host cell TEEGR ends up in the nucleus where it not only complexes with E2F3 and E2F4 host transcription factors to induce gene expression but also promotes shaping of a nonpermissive chromatin through its capacity to switch on EZH2. Remarkably, EZH2 fosters the epigenetic silencing of a subset of NF-κB-regulated cytokines thereby strongly contributing to the host immune equilibrium that influencing the host immune response and in mice promotes parasite persistence.
Keywords: Toxoplasma gondii, E2F/DP transcription factor, EZH2, parasite-host interaction, NF-κB signaling
Introduction
Caused by the protozoan Apicomplexa Toxoplasma gondii, toxoplasmosis is one of the most widespread foodborne parasitic zoonosis on earth, and poses a significant risk to public health mostly in case of immune dysfunction. Triggered by the T. gondii initial colonization of the intestinal mucosa, the IL-12/IFN-γ axis plays a prominent role in anti-parasite immunity as it halts the acute expansion of the T. gondii “tachyzoite” population within host cells and throughout the host, yet it also promotes establishment of the long-term persistent “bradyzoite” stages in cells of deep tissues1. At the heart of this immune context lies the remarkable ability of tachyzoites to actively reshape gene expression of the hosting cell owing to a large number of effector molecules pre-stored in secretory organelles2. First, effectors contained in the apical rhoptry organelles (ROP proteins) are injected directly in the host cell cytoplasm at the very onset of cell invasion prior the tachyzoite gets enclosed within a Parasitophorous Vacuole (PV). Post PV formation, effectors from Dense Granule (DG), namely the GRA proteins, are exocytosed by tachyzoite in the lumen of the PV; they further reside either at the host-parasite interface or are exported in the host cell3. Therefore, some GRAs are featured to cross the PV membrane, and a subset of these can even traffic to the host cell nucleus where they gather into hyper-stable complexes of proteins that usually do not assemble in uninfected cells. Founder members include GRA164, GRA245 and TgIST6,7 which all contribute to the building of functional networks in infected cells by interfacing with the host signaling pathways or co-opting host transcription factors8.
Here, we identified TEEGR as a dense granule-resident effector and demonstrated its unique regulatory function on E2F/DP transcription factors activity and consequently on expression of the epigenetic silencer EZH2 in the host cell. These functional features provide TEEGR with a pivotal role in mediating a regulatory control loop that antagonizes the NF-κB-driven pro-inflammatory responses to T. gondii infection.
Results
TEEGR is a dense granule-resident protein exported to the host cell nucleus
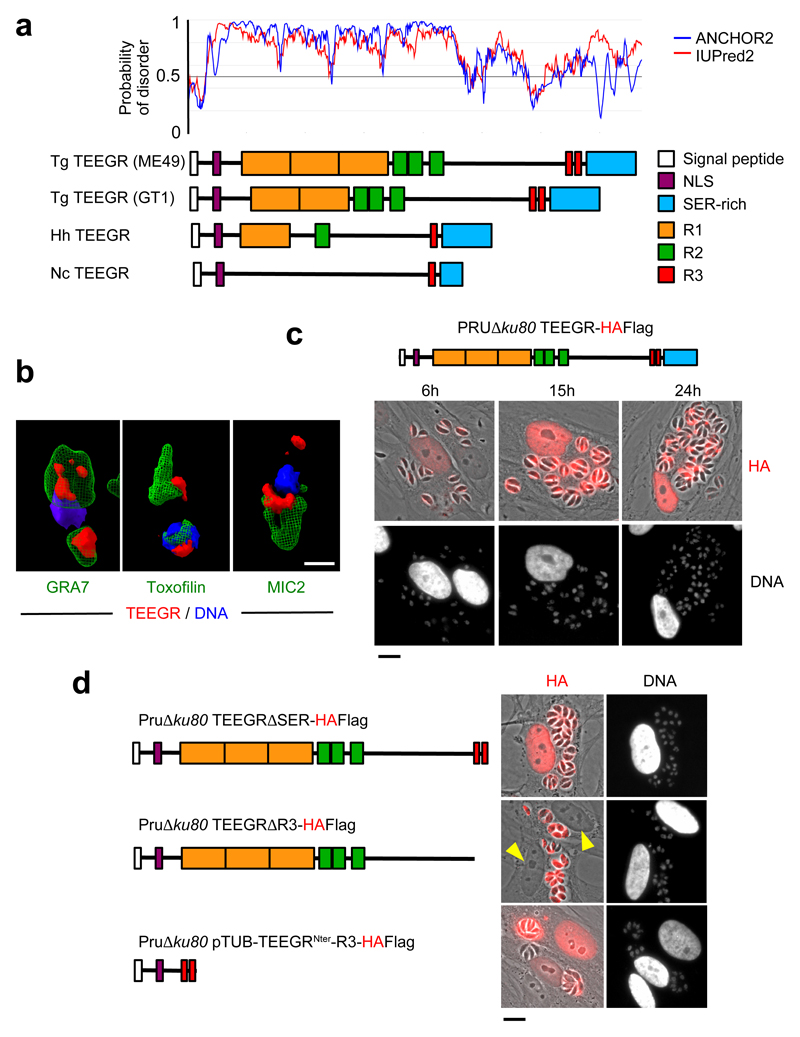
TEEGR was originally identified in a repertoire of intrinsically disordered Toxoplasma proteins (Fig. 1a) that are singularly exported into the nucleus of the infected cell4. The protein exhibited a characteristic punctate distribution pattern in the parasite cytoplasm and occasionally overlapped with the dense granule GRA7 but remained excluded from the apical rhoptry and microneme organelles as verified with the canonical toxofilin and MIC2 markers, respectively (Fig. 1b and Supplementary Fig. 1). Once parasites were enclosed in the PV, TEEGR was detected in the PV space and beyond the PVM in the host cell nuclei while PVs continue to enlarge along with parasite multiplication. Its localization in the host cell nucleus was seen as early as 6 hours post invasion and thereafter (Fig. 1c). In agreement with a shared export mechanism between GRAs8, the translocation of TEEGR across the PVM was dependent on the translocon protein MYR1 and, even on the Aspartyl Protease ASP5 in spite of any detectable TEXEL motif (Supplementary Fig. 2). Peculiarly, TEEGR export was prevented by the deletion of the Repeat 3 (R3)-containing C-terminal domain, which presumably impacted the step of secretion into the PV space (Fig. 1d). In the same line, the addition of R3 repeats to the extreme N-terminus region of TEEGR was sufficient to drive the chimeric protein to the host cell nucleus, its final destination (Fig. 1d). Regardless of its atypical mechanism of delivery, TEEGR is a genuine member of the subfamily of dense granule-resident proteins that are singularly exported into the host cell nuclei8.
Fig. 1. The export of TEEGR in the host cell nucleus.
The gene TGME49_239010 encoding TEEGR was originally identified in silico together with GRA164, GRA245 and TgIST6,7 as strong parasite candidate genes with attributes to both target the parasite secretory pathway and to reach the host cell nuclei. a, TEEGR is a highly disordered 80 kDa protein (predicted disorder score > 0.5) that carries a signal peptide, a putative nuclear localization sequence (NLS) and multiple repeated domains (R1, R2 and R3) whose repeat number and distribution have evolved through coccidian subclasses and among T. gondii lineages. Tg: Toxoplasma gondii, Nc: Neospora caninum, Hh: Hammondia hammondia. b, HAFlag (HF)-tagged TEEGR (in red) in Pru∆ku80 extracellular parasites is contained in cytoplasmic organelles distinct from the apical micronemes (MIC2, in green) and rhoptries (Toxofilin, in green), and partially co-localizing with the dense granule protein GRA7 (in green). Cells were co-stained with Hoechst DNA-specific dye (in blue). Scale bars, 1 μm. c, Time course of TEEGR secretion and export to the host cell nucleus. Human Foreskin Fibroblasts (HFF) were infected with a Pru∆ku80 TEEGR-HF and stained with anti-HA antibodies (red). Scale bars, 10 μm. d, in situ subcellular localization of TEEGR chimeric proteins (in red) in HFF infected with parasites expressing endogenously HF-tagged TEEGR truncated proteins (TEEGR∆SER and TEEGR∆R3) or TEEGRNter-R3 expressed under the control of a TUB8 promoter. Scale bars, 10 μm. All data are representative of 3 independent biological experiments.
TEEGR activates expression of E2F3/4-regulated host genes
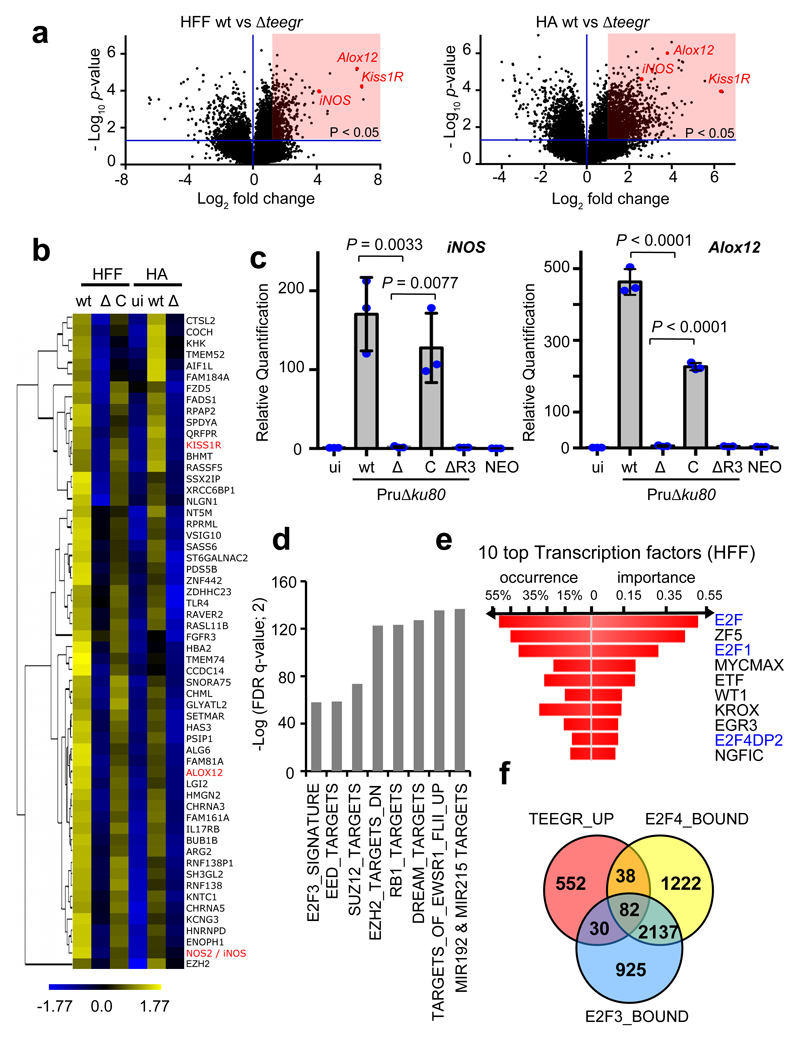
The detection of TEEGR in the host cell nucleus prompted us to investigate whether the protein could contribute to the re-orchestration of gene expression carried out by type II T. gondii genotypes in both non-hematopoietic and hematopoietic lineages. While comparing the ∆teegr mutant with the parental strain, we found indeed the expression of 784 and 1529 human genes to be up-regulated in a TEEGR-dependent fashion in human fibroblasts and astrocytes, respectively (Fig. 2a and Supplementary Table 1). 215 of these were common to both cell types and hierarchical clustering of the 58 top-ranked genes (more than fourfold, FDR <1%) delineated specific clusters of TEEGR-induced transcripts upon infection (Fig. 2b). Consistently, re-introduction of the TEEGR gene in the ∆teegr strain mutant restored the expression pattern to levels observed with WT parasites, thereby demonstrating the TEEGR-dependent induction of those genes (Fig. 2b). Furthermore, these results were unequivocally validated with the significant increase in mRNA levels of three specifically TEEGR-activated genes (iNOS, Alox12 and Kiss1R) and this, independently of the parasite type I/II strains and the host cell type analyzed. Indeed, reintroduction of a copy of the gene in teegr-deficient parasites was enough to restore expression of these genes (Fig. 2c and Supplementary Fig. 3). While the extent to which gene expression was induced varied with the starting T. gondii Multiplicity of Infection (MOI) and the time course of infection (Supplementary Fig. 3b), it is noteworthy that none of the TEEGR-induced gene products were detected in human cells infected by Neospora caninum, one of the closest T. gondii relative (Fig. 2c), which however harbors a poorly conserved TEEGR homologous gene (Fig. 1a). Finally, the strict requirement of TEEGR as transcriptional regulator of specific target genes was further evidenced by the absence of iNOS and Alox12 gene induction in cells infected by either Pru∆ku80∆R3, ∆myr1 or ∆asp5 strains in which TEEGR export was impaired (Fig. 2c and Supplementary Fig. 2c).
Fig. 2. TEEGR activates gene expression in human cells in a E2F3 and E2F4-dependent manner.
Genomewide expression profiling of human fibroblasts (HFF) and astrocytes (HA) infected with the ∆teegr mutant (∆), the parental type II strain Pru∆ku80 (wt) and the trans-complemented strain (C). a, Results of tests for differential expression are presented in a volcano plot which plots statistical significance against fold change for each gene. Genes (HFF, n=784; HA, n=1529, in red) with adjusted p-values (Bonferroni corrected P-value ≤ 0.05) and absolute fold changes of ≥1.5 are deemed of interest for in silico pathway analyses (Supplementary Table 1). b, Heat map of expression values for differentially expressed genes in human cells infected with WT, Δ and C parasites. For the 472 genes (more than fourfold, p < 0.05, unpaired t-test) that were defined as core TEEGR-regulated genes in HFF and HA, mean log2 gene expression values were median centered, genes were clustered by hierarchical clustering based on Pearson correlation, and a heat map with 58 genes is presented. The complete set of genes is listed in Supplementary Table 1. c, HFF cells were left uninfected (ui) or infected for 24 h with WT, Δ, C or TEEGR protein missing R3 (ΔR3). Levels of iNOS and Alox12 mRNAs were determined by RT-qPCR. Values were normalized to the amount of β2-microglobulin. Data are mean value ± SD of three biological replicates. The p-values were calculated using two-tailed unpaired Student’s t-test. d, Gene Set Enrichment Analysis (GSEA) of the 784 genes positively regulated by TEEGR in HFF cells (Supplementary Table 1) highlighted eight gene expression signatures of chemical and genetic perturbations (CGP) that were significantly and selectively enriched (p < 0.05, False Discovery Rate (FDR)-corrected, two-sided Welch t-test). e, Transcription Factor Binding Sites (TFBS) analysis of the 784 TEEGR-upregulated genes was performed by DIRE and the most significant transcription factors are listed. f, Venn diagram illustrating the overlap between the number of genes up-regulated by TEEGR (> twofold, p < 0.05, unpaired t-test) in HFF cells and the number of genes identified as E2F3- or E2F4-bound by Julian et al., (2016)39 and Marson et al., (2007)10 respectively.
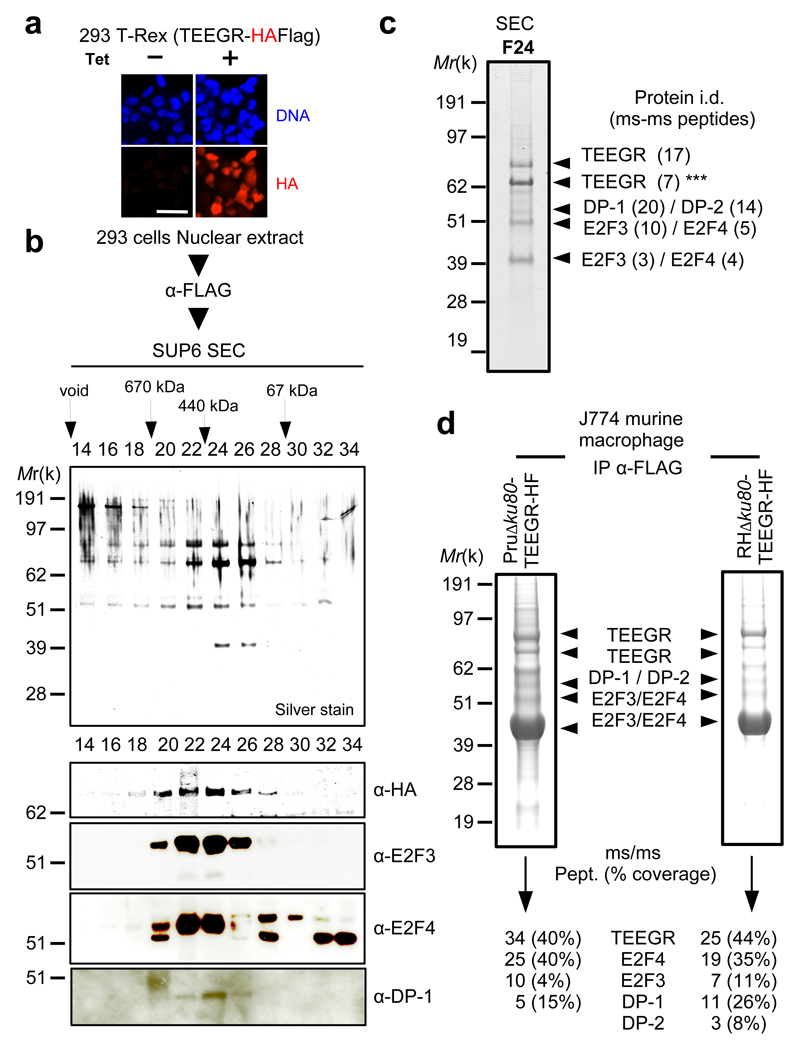
Fig. 3. TEEGR forms partnership with E2F/DP host transcription factors.
a, IFA of TEEGR-HAFlag ectopically and stably expressed in the 293-TRex cell line. Cells were either left untreated (-) or treated (+) with tetracycline for 20h and processed for IFA using anti-HA antibodies (red) and Hoechst DNA-specific dye (blue). Scale bar, 2.5 μm. b, TEEGR-associated polypeptides were purified from nuclear extracts of 293-TRex cells that had been tetracycline-induced to express TEEGR-HF. Size exclusion chromatography (SEC) of TEEGR-containing complexes after Flag affinity selection. Fractions were analyzed on silver stained SDS PAGE gels and blots processed to detect TEEGR-HF (anti-HA), E2F3, E2F4 and DP-1. c, Mass spectrometry-based proteomic analysis of SEC fraction 24 identified the transcription factors E2F3 and E2F4 associated with DP-1/DP-2 as the only TEEGR relevant partners. Identity of the proteins with their respective number of peptides is indicated on the right. Asterisks indicate degradation products. d, TEEGR-associated proteins were purified by Flag chromatography from protein extracts of murine J774 macrophage (MØ) line infected by Pru∆ku80 TEEGR-HAFlag or RH∆ku80 TEEGR-HAFlag. Fractions were analyzed on gels by silver staining and then by mass spectrometry-base proteomics to detect TEEGR and aforementioned partners. Identity of the proteins with their respective number of peptides and percentage of coverage are indicated. All data are representative of two independent biological experiments.
Applying Gene Set Enrichment Analysis (GSEA) allowed identifying eight gene expression signatures of chemical and genetic perturbations (CGP) amongst the 784 genes positively regulated by TEEGR in infected HFFs (Fig. 2d) while the Distant Regulatory Elements (DiRE) analysis pointed to enrichment in specific Transcription Factor Binding Sites (TFBS) especially for E2F3 and E2F4 Transcription Factors (TFs) (Fig. 2e). For instance, 87 genes were identified as bound by DREAM (DP, RB-like, E2F4 and MuvB), a complex gathering Retinoblastoma (RB)-like proteins and E2F4/DP transcription factor9. TEEGR showed similar activity in non-human cells with (i) 1129 genes up-regulated in a TEEGR-dependent manner (Supplementary Fig. 4a and Supplementary Table 2) and (ii) comparable CGP signatures and TFBS enrichments were picked up by GSEA and DiRE analysis, respectively in T. gondii infected murine bone marrow-derived macrophages (BMDMs) (Supplementary Fig. 4b-c). Given the putative prominent role of E2F3 and E2F4 in TEEGR-mediated gene activation in both human fibroblasts and murine macrophages, we next checked whether TEEGR and E2F3/4 regulate a shared set of genes by intersecting our transcriptomic data with ChIP-seq datasets that had previously allowed defining genomewide the position of E2F3 and E2F410,11. We found that about 20% and 37% of the TEEGR induced-genes in HFFs and BMDMs respectively (Fig. 2f and Supplementary Fig. 4d) had been previously identified as exclusive or overlapping E2F3- and E2F4-bound genes by ChIP-seq10,11, therefore emphasizing the implication of these two TFs in the TEEGR pathway.
TEEGR forms distinct complexes gathering E2F/DP transcription factors
Mapping of transcriptomic changes triggered by intracellular tachyzoites in conjunction with analysis of TFs occupancy brought strong evidence for cooperation between TEEGR and E2F/DP at the vicinity of shared genes, yet TEEGR has no detectable DNA binding domain. However, TEEGR in fusion with Gal4 DNA binding domain strongly activated the reporter gene His3 in yeast, which favored growth under selective conditions (up to 200 mM of 3-AT), therefore documenting the TEEGR intrinsic transcriptional transactivation properties. This is why we hypothesized that TEEGR-mediated gene induction may be achieved by co-opting a host transcription factor similarly to the T. gondii TgIST GRA effector6.
While TEEGR is predicted to be intrinsically unstructured (Fig. 1a), it does not carry any recognizable protein Short linear motifs (SLiM) that could suggest dedicated interacting partner(s) in the host cell nucleus where it accumulated post-infection. To search for putative TEEGR partners, we performed a systematical mapping of the TEEGR interactome when ectopically expressed in uninfected but T. gondii permissive cells or in the context of cell infection. In 293 epithelial cells, TEEGR was found in a high molecular-weight complex (~500-kD, fractions 20-26) that resisted to stringent salt conditions (Fig. 3a, b and Supplementary Fig. 5a). Mass spectrometry-based proteomic and western blotting allowed confirming the partnership between TEEGR and the E2F3 and E2F4 TFs in association with their Dimerization Partners (DP) DP-1 and DP-2 (Fig. 3c and Supplementary Fig. 5b)12. While ectopic expression of TEEGR in 293 cells proved informative, it did not allow capturing either the dynamic state of the TEEGR interactome over the tachyzoite intracellular growth period (i.e. involving co-exported effectors), or the complexity brought by specific combination of given strains and host cell types. Importantly, the TEEGR/E2F/DP-containing complexes were purified from murine J774 macrophages when infected with tachyzoites from either type I and II strains that expressed epitope-tagged TEEGR (Fig. 3d and Supplementary Fig. 5c). Taken together, these data uncovered a multivalent partnership of TEEGR with host E2F/DP transcription factors and validated the relevance of these interactions in the context of not only human non-hematopoietic but also murine hematopoietic lineages and regardless of the T. gondii infecting strains.
TEEGR induces the E2F3/4-dependent expression of the PRC2 subunit Ezh2
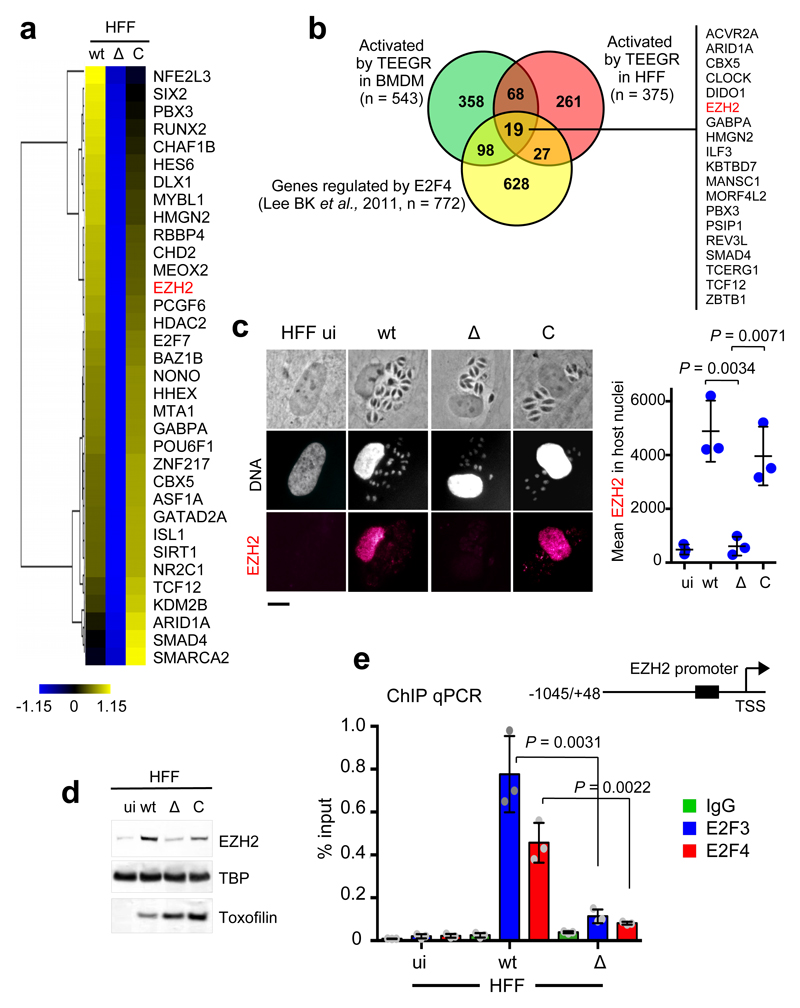
Apart from the TEEGR-induced genes whose expression has been associated with E2F3 and E2F4 TF activities, a significantly wider set of up-regulated genes is likely controlled by other TFs (Fig. 2f and Supplementary Fig. 4d). Indeed, transcriptomic analysis identified miscellaneous TFs and chromatin-modifying enzymes among which the Polycomb group (PcG) gene EZH2 whose expression was TEEGR-induced in both murine and human cells, regardless of the parasite strain (Fig. 4a-b). EZH2 is the catalytic subunit of the Polycomb repressive complex 2 (PRC2) that drives the addition of methyl groups to histone H3 at lysine 27 (H3K27me3), and ultimately mediates epigenetic transcriptional silencing during embryonic development, tumorigenesis and differentiation of immune cells13. Consistent with transcript profiling, EZH2 protein was never detected in primary and fully differentiated fibroblasts but clearly rose up and accumulated in nuclei of these cells once infected by T. gondii tachyzoites that expressed a functional version of TEEGR (Fig. 4c-d and Supplementary Fig. 5d).
Fig. 4. E2F3 and E2F4 DNA association to EZH2 promoter is enhanced by T. gondii infection in a TEEGR-dependent manner.
a, Heat map representation of host transcription factors and chromatin-modifying enzymes up-regulated in a TEEGR-dependent fashion in HFF cells infected for 24 h with WT, Δ and C strains. EZH2 is indicated in red. b, Venn diagram illustrating the overlap between the number of genes up-regulated by TEEGR in HFF cells as well as BMDM (> twofold, p < 0.05, unpaired t-test) and the number of genes identified as E2F4-bound by Lee BK et al., (2011). c, IFA of TEEGR-dependent induction of EZH2 (red) in uninfected (ui) or 24 h infected HFFs with WT, Δ and C strains. In situ quantification of nuclear EZH2 using IFA. Horizontal bars represent the mean nuclear EZH2 intensity ± s.d. of three independent experiments (n=100 nuclei per dot). Scale bars, 10 μm. d, Nuclear extracts from HFF cells left uninfected (ui) or infected (24 h) by the indicated strains were analyzed by Western blotting using indicated antibodies. TBP (host-specific) and toxofilin (parasite-specific) are shown as loading controls. Data are representative of three experiments. e, Schematic presentation of the genomic regions corresponding to the promoter of EZH2. The black box in the EZH2 promoter represents a region of four putative E2F sites. HFFs were left uninfected (ui) or infected for 24 h with WT or Δ strains. Samples were analyzed by ChIP assay with specific antibodies to E2F3 and E2F4. IgGs were used as negative control. Bound DNA corresponding to EZH2 promoter was quantified by qPCR-ChIP, and signals were normalized with the input DNA. Data are mean value ± SD of three biological replicates. The p-values were calculated using two-tailed unpaired Student’s t-test, unless otherwise stated in the figure.
Since the modus operandi by which TEEGR achieves host gene activation likely depends on the E2F3 or E2F4 DNA targeting, we then investigated whether EZH2 gene expression may be regulated in a TEEGR/E2F-dependent manner. In line with previous reports specifying that E2F3 and E2F4 can control EZH29,11,14, we found that both transcription factors were enriched at the EZH2 promoter in a TEEGR-dependent manner using ChIP-scanning analysis (Fig. 4e). Overall, TEEGR in partnership with E2F/DP positively regulates EZH2 expression, which in turn may repress host gene expression post-T. gondii infection.
The EZH2 switch controlled by TEEGR counteracts NF-κB-dependent gene induction
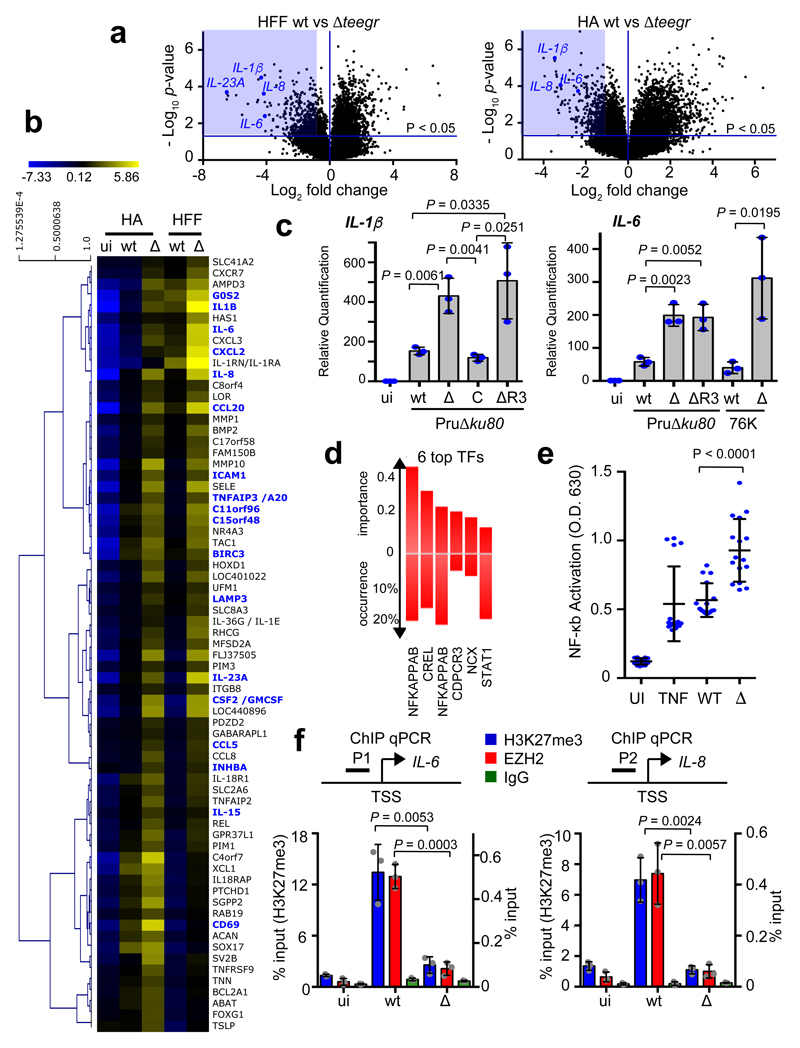
While transcriptomics unambiguously identified TEEGR as a transcriptional activator in infected cells (Fig. 2 and Supplementary Fig 4), it also pointed out that TEEGR triggered the repression of a sizeable fraction of host genes (n=494 and n=571 for human fibroblasts and astrocytes, respectively with a twofold change and p<0.05) (Supplementary Table 1 and Fig. 5a). Hierarchical clustering of the 69 top-ranked genes (FDR <1%) delineated clusters of TEEGR-down regulated transcripts that were moderately induced by WT parasites but culminated with teegr-deficient tachyzoites (Fig. 5b). Quantitative RT-PCR on mRNA from six of the repressed genes (i.e. IL-1β, IL-6, IL-8, IL-23A, Ptgs2 and CCL20) documented a robust and sustained pattern of TEEGR-mediated cytokine repression, which required TEEGR nuclear localization along with intracellular tachyzoite development (Fig. 5c and Supplementary Fig 6). Accordingly, the Pru∆ku80∆R3 mutant strain for which TEEGR was retained inside the parasite (Fig. 1d), failed to induce gene repression whereas reintroduction of the teegr gene in the mutant restored mRNA synthesis to basal levels, and consequently the repressive gene activity driven by wild-type TEEGR (Fig. 5c and Supplementary Fig. 6).
Fig. 5. TEEGR-dependent repression of NF-κB-regulated genes is mediated by EZH2.
a, Genomewide expression profiling of HFF and HA cells infected with wt and ∆. Results of tests for differential expression between wt and ∆ are presented in a volcano plot which plots statistical significance against fold change for each gene. Genes colored in blue (HFF, n=494; HA, n=571) have Bonferroni corrected p-values of ≤ 0.05 and absolute fold changes of ≤ 2 and are deemed of interest for in-silico pathway analyses (Supplementary Table 1). b, Heat map of expression values for differentially expressed genes in human cells infected with WT and Δ parasites. For the 69 (more than threefold, P < 0.05, unpaired t-test) that are defined as core TEEGR-repressed genes in HFF and HA, mean log2 gene expression values were median centered, genes were clustered by hierarchical clustering based on Pearson correlation, and a heat map is presented. c, HFF cells were left uninfected (ui) or infected for 24 h with WT, Δ, C or ΔR3 strains. Levels of IL-1β and IL-6 mRNAs were determined by RT-qPCR. Values were normalized to the amount of β2-microglobulin. Data are mean value ± SD of three biological replicates. d, TFBS analysis of the 69 TEEGR-repressed genes was performed by DIRE and the most significant transcription factors are listed. e, THP-1 murine cells were treated with TNFα or infected with Pru∆ku80 WT or Δteegr strains and then followed by determination of NF-kB activity using a reporter gene assay. Data are mean value ± SD of twenty biological replicates. f, HFFs were left uninfected (ui) or infected for 24 h with WT or Δ strains. Samples were analyzed by ChIP assay with specific antibodies to EZH2 and H3K27me3. IgG was used as negative control. Bound DNA corresponding to IL-6 and IL-8 promoters (respectively P1 and P2) was quantified by qPCR-ChIP, and signals were normalized with the input DNA. Data are mean value ± SD of three biological replicates. The p-values were calculated using two-tailed unpaired Student’s t-test, unless otherwise stated in the figure.
A majority of the significantly and selectively repressed pathways by TEEGR clustered in biological processes related to immunity in particular inflammation as emphasized with the pole position of the NF-κB dependent TNF-α signaling pathway (Supplementary Fig. 7a-c). The NF-κB transcription factor was in fact predicted with the highest probability to associate with the TEEGR-silenced genes (Fig. 5d), consistent with the NF-κB targeting of the vast majority of these genes already identified using ChIP-seq analysis15–17. NF-kB is a family of dimeric transcription factors consisting of homo- or heterodimers of different subunits18. While TEEGR neither altered the subunit
mRNA expression (Supplementary Fig. 7d), nor the nuclear translocation of either RelA (p65) (Supplementary Fig. 7e) or c-Rel upon T. gondii infection, it selectively repressed the dimer transcriptional activity (Fig. 5e). The TEEGR property to selectively inhibit TNF-α-induced transcription of several NF-κB-dependent genes led us testing whether TEEGR could counteract NF-κB activation in response to exogenous TNF-α. While tachyzoites carrying type II allele of GRA15 and TNF-α were shown separately to increase IL-6 and CCL20 mRNA levels, a cumulative effect was observed when tachyzoite-infected cells were stimulated with TNF-α (Supplementary Fig. 6d). Importantly, IL-6 and CCL20 levels were enhanced by teegr-deficient parasites highlighting the TEEGR ability to interfere with cell responsiveness to exogenous TNF-α (Supplementary Fig. 6d). Yet, TEEGR-mediated repression did not fully shutdown the activation by GRA15-II of NF-κB since WT parasites, even though they have TEEGR, still enhance TNF-α-induced transcription (Supplementary Fig. 6d).
We next interrogated whether the TEEGR-dependent EZH2-associated gene repression could act by thwarting the NF-κB response induced by T. gondii. We first identified 71 TEEGR-repressed genes that were previously described as silenced by EZH2 (underlined in blue in Fig. 5b and Supplementary Table 1)19–21. These included IL-6 and IL-8 which both re-gained activity in presence of GSK126, a highly selective inhibitor of EZH2 methyltransferase activity (Supplementary Fig. 7f)22. We applied ChIP-scanning to determine whether upon infection, EZH2 was recruited to cytokine genes such as IL-6 and IL-8, which in breast cancer cells are co-regulated by EZH2 and RelA/RelB20. In cells infected with wild-type TEEGR-expressing tachyzoites (type II), EZH2 was found markedly enriched on IL-6 and IL-8 promoters concomitantly to a high level of H3K27me3 (Fig. 5f). This was no longer true for cells infected by teegr-deficient type II tachyzoites (Fig. 5f), that instead produced increased amounts of IL-6 and IL-8 mRNAs (Fig. 5c and Supplementary Fig. 6). These data provide the evidence that by promoting EZH2 recruitment to NF-κB target genes, TEEGR acts upstream of NF-κB and drives gene down regulation, thereby counterbalancing the early typical induction of TNF-α signaling by T. gondii type II strain.
teegr deficiency associates with the control of parasite burden in mice
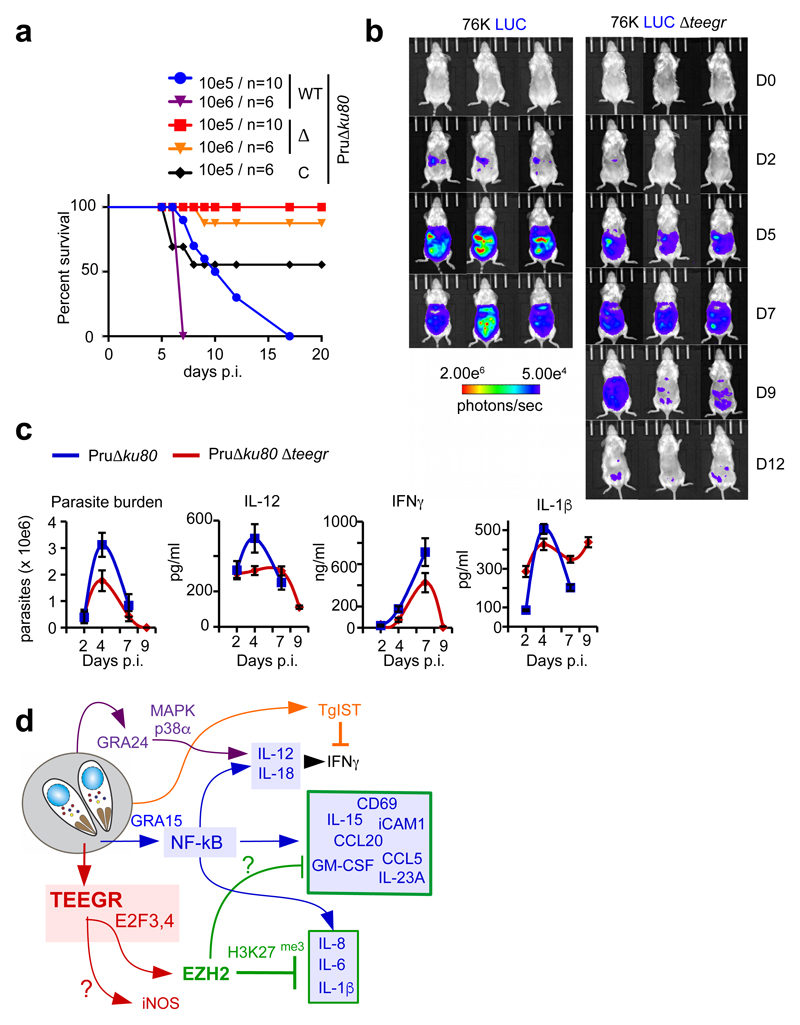
To explore in vivo how TEEGR could counterbalance the type II-specific activation of NF-κB, we monitored parasite multiplication and dissemination as well as BALB/c mice survival following intra-peritoneal (i.p.) delivery of WT or teegr-deficient tachyzoites. While all mice inoculated with 105 or 106 WT tachyzoites succumbed within 6–17 days post-inoculation (p.i.), those infected with the same doses of teegr-deficient strains survived (Fig. 6a and Supplementary Fig. 8a). Mice inoculated with tachyzoites from the Pru∆ku80∆teegr complemented with a TEEGR-HF-tagged version showed a significant but incomplete gain in virulence (Fig. 6a). The partial phenotype rescue could be caused by a decrease activity or/and a lower expression level of the chimeric version of TEEGR when compared to the endogenous one. Noninvasive bioluminescence imaging highlighted a significant gap in WT and ∆teegr parasite population sizes as early as day 5 p.i., that further increased with time (Fig. 6b and Supplementary Fig. 8b). The WT parasites continued proliferating until the mice died i.e. at day 8 p.i. whereas the teegr-deficient population expanded in the first week p.i., albeit less extensively than the WT one, and then retracted to few foci, compatible with both host survival (Fig. 6b and Supplementary Fig. 8b) and parasite persistence. When delivered per os with low dose of cyst-containing bradyzoites, mice chronically infected with teegr-deficient parasites had however a lower number of cysts in their brain when compared to the parental strain (Supplementary Fig. 8c), suggesting therefore a positive contribution of TEEGR to T. gondii persistence.
Fig. 6. In vivo control of teegr-deficient T. gondii tachyzoite population is likely mediated by NF-kB-regulated pro-inflammatory cytokines.
a, Mice were given intraperitoneally (i.p.) either 105 (n=10) or 106 (n=6) tachyzoites of WT, Δ and C strains and survival was monitored. At 5 days’ post inoculation all mice display clinical signs (weight loss, ruffled fur). Significance was tested using Log-rank (Mantel-Cox) test (p-value = 0.0161) and Gehan-Breslow-Wilcoxon test (p-value = 0.0154). A two-sided P value of <0.05 was considered statistically significant. b, Luminescent parasites were imaged with an IVIS imaging system from day 0 to 12 after the i.p. inoculation to BALB/c of 5.104 tachyzoites per condition. The data are representative of two experiments (three mice per group). c, BALB/c mice were given i.p. a dose of 105 Pru∆ku80 or Pru∆ku80 Δteegr tachyzoites. Peritoneal lavage fluids were collected on days 2, 5, 7, and 9 post inoculation. Number of tachyzoites was estimated within the collected samples by parasite DNA PCR and concentrations of IL-1β, IL-12, and IFN-γ were determined by ELISA. Data shown are means ± SD with n= 3 individual mice per parasite genotype at each time point. d, Schematic representation of the consequences of TEEGR delivery in the host cell by Toxoplasma tachyzoites.
Given the drastic retraction of the teegr-deficient T. gondii population, we then assessed whether ∆teegr deficiency could make intracellular tachyzoites more susceptible to immune killing. A major defense mechanism of innate immunity against T. gondii is mediated by the IFN-γ–inducible IRG pathway23. Interestingly in murine BMDMs pre-stimulated with low-dose of IFNγ prior to T. gondii infection, teegr-deficiency correlated with increased parasite clearance (Supplementary Fig. 8d) while it did not in unstimulated human (Supplementary Fig. 8e) or mouse BMDMs (Supplementary Fig. 8d). Although our data do not support a role for Irgb6 coating of the PVM in the clearance of ∆teegr tachyzoites in stimulated macrophages (Supplementary Fig. 8f), they suggest that parasite killing could be due to either IRG evasion24 or the recruitment of other IRG family members at the PVM. Overall, they certainly provide some explanation of the strong teegr-deficient T. gondii phenotype observed in vivo.
To better explain the in vivo loss of virulence shown by teegr-deficient T. gondii, we monitored the early recruitment of myeloid cells in the peritoneal cavity of mice inoculated with either TEEGR or teegr-deficient type II tachyzoites. The homing of neutrophils (CD11b+ Gr1high Ly6G+) and activated inflammatory monocytes (CD11b+ Gr1int Ly6G−), the latter playing a critical role downstream of IL-12–IFN-γ induction to control the first amplification wave of tachyzoites in mice25,26, was kept similar in mice inoculated with a high dose of teegr-deficient tachyzoites, yet those mice showed reduced myeloid cell infiltration (Supplementary Fig. 9a). This suggests that the restriction of the teegr-deficient population was not driven by an increase in Gr1+ monocyte recruitment.
Lethal infections with type II strains are usually associated with massive overstimulation of T-helper (Th) 1 (TH1) cytokines, which contribute to tissue destruction27. While, extremely high serum levels of inflammatory cytokines (e.g. IL-12, IFN-γ, and IL-18) were induced during lethal infections caused by a high dose of WT strain, ∆teegr infections were typified by a low level of the Th1-polarizing cytokine IL-12 and less circulating IL-18 and IFN-γ at day 7 p.i., likely as a consequence of parasite load control (Fig. 6c and Supplementary Fig. 9b). Intriguingly, IL-1β has a peculiar pattern of secretion in vivo throughout acute infection. When a high dose of Pru∆ku80 was inoculated, IL-1β levels declined at day 4 p.i. after an initial rise, whereas ∆teegr tachyzoites inoculation caused a sustained induction of the cytokine even when the parasite burden became barely detectable, indicating that TEEGR is also able to repress IL-1β expression in vivo at least in the early times of infection (Fig. 6c). In vitro, IL-1β levels were also slightly higher in the medium of BMDM infected by ∆teegr. In the case of oral infection with ∆teegr cysts, the tissue damages were reduced (Supplementary Fig. 10a-b), while the parasite burden stayed low (Supplementary Fig. 10c) and associated with a similar cytokine signature typified by a lower level of IFN-γ and an increased level of IL-1β in the ileum (Supplementary Fig. 10d). These data are consistent with those reporting a protective role for IL-1 against lethal T. gondii infection in vivo28,29, yet more data are needed to support a direct role of IL-1β in the control of ∆teegr parasite burden.
Discussion
The highly polarized TH1-type host response to a cystogenic T. gondii strain - typified by IL-12 secretion and IFN-γ induction - accounts for the restriction of the tachyzoite population through IFN-γ signaling. Meanwhile, the T. gondii counterdefenses offset this IFN-γ regulated innate immune defenses, allowing colonization of, and persistence in deep tissues. As proof of concept is the TgIST effector recently reported to co-opt host chromatin repressors and dampen STAT1-dependent IFN-γ signaling6,7. Our present findings reveal another path taken by T. gondii to block the TH1 response, which relies on the effector TEEGR. TEEGR teams up with the E2F/DP transcription factors to promote EZH2 activation which eventually antagonizes the positive regulation of NF-kB activity by tachyzoites.
The diversity of the NF-kB family members, which in mammals include five proteins, that form numerous homo- and heterodimers, provides high selectivity in the NF-kB-mediated transcriptional response. Remarkably, T. gondii can differentially modulate the NF-kB pathway depending on the parasite genotype and the host cell lineage. Thus, GRA15 from the type II background contributes to a sustained NF-kB p65 activation hence to the release of proinflammatory cytokines such as IL-1232 and IL-1β33. In addition, type II strains also elicit c-Rel nuclear translocation in a Myr1-dependent but GRA15-independent manner, thereby suggesting that another GRA effector could account for the activation of c-Rel. Since mice deficient in c-Rel are highly susceptible to the acute phase of T. gondii i.p. infection, this pathway certainly contributes to mount the resistance to the tachyzoite expansion34.
Importantly, because unchecked cytokine activation has dire consequences on tissue integrity, maintenance of immune homeostasis requires a negative feedback system. NF-kB negative regulatory mechanisms include amongst others the de novo synthesis IkB-α which shuts off the signal18. With TEEGR, we have discovered a negative regulator in the NF-kB signaling system that by restraining the inflammatory response sets the proper conditions for parasite persistence. Intriguingly, TEEGR selectively repressed the transcription of a subset of NF-kB-regulated cytokines (IL-1β, IL-6, IL-23A, IL-15) and chemokines (IL-8, CCL20) without altering the expression of other NF-kB-regulated cytokines (IL-12, IL-18) (see model in Fig. 6d). A possible explanation is that TEEGR counteracts the combined activities of both GRA1532 and still unidentified factors that direct NF-kB nuclear translocation without impacting the sustained p38a phosphorylation mediated by GRA24 that similarly shifts the macrophage polarization toward a M1 phenotype35. Alternatively, this would fit with the recurrent observation that chromatin state at different promoter classes dictates the kinetics of the NF-kB response. In this respect, lineage-defining transcription factors prime the chromatin and thereby govern the NF-kB-controlled transcriptional programs36. In line with this model, the TEEGR-induced EZH2 may contribute to generate a silenced chromatin landscape that would restrict NF-kB accessibility in a cell-specific manner. Interestingly, it was recently reported that whereas tachyzoites induce IL-1β in primary human monocytes, they inhibit IL-1β production in human neutrophils from the same blood donors by impairing the activation of the NF-kB signaling pathway37. Whether TEEGR plays a role in this human neutrophil-restricted phenotype deserves to be investigated.
In conclusion, TEEGR can be seen as an ‘epigenator signal’38 in the host cell that timely triggers the EZH2-mediated epigenetic phenotype but not subsequent events. Whether TEEGR is able to sustain a “transcriptional memory” in tachyzoite-loaded cells (macrophage, dendritic and T cells…) over the immunological tachyzoite clearance has yet to be determined. Clearly, TEEGR contributes to the diversity of GRA effectors that may reflect the large number of different hosts infected by T. gondii. In this respect, T. gondii has evolved a distinct set of effectors that target central hubs of immune signaling pathways and act synergically or antagonistically to govern the fate of T. gondii parasitism8.
Methods
Mouse and Experimental Infection
BALB/cJRj mice were obtained from Charles River Laboratories. Mouse care and experimental procedures were performed under pathogen-free conditions in accordance with established institutional guidance (European Directive 2010/63/EU) and approved protocols from the institutional Animal Care (PHTA) and Use Committee of University Grenoble Alpes (agreement # C3851610006 / APAFIS # 4536-2016031017075121). BALB/cJRj female mice between 6 and 8 weeks of age were used in all experiments. Mice were challenged by intraperitoneal (i.p.) injection of T. gondii tachyzoites or by per os with cysts. The percent cumulative mortality was defined as the number of animals that succumbed to infection/the total number of infected animals (i.e., deaths plus seropositive survivors). We used randomization to reduce bias in mice selection and outcome assessment. Blinding was used during data collection by assigning numbers in place of genotype and infection status to mice in the studies. We have carefully chosen the sample size based on empirical evidence of what is necessary for interpretation of the data and statistical significance. For statistical analysis of mice survival data, the log-rank (or Mantel-Haenszel) and the Peto & Peto modification of the Gehan-Wilcoxon test were used.
Parasites and host cells
Human primary fibroblasts (ATCC® CCL-171™), Human primary Astrocytes (Science Cell Laboratories, cat #1800), T-Rex-293 cell line (RRID:CVCL_D585), L929 cell line (Sigma-Aldrich Cat# 85011425), J774 cell line (J774A.1, Sigma-Aldrich Cat# 91051511), RAW264.7 cell line (ATCC Cat# TIB-71, RRID:CVCL_0493) and THP1-Blue™ NF-κB Cells cell line (InvivoGen Cat # thp-nfkb) were cultured in DMEM (Invitrogen) supplemented with 10% heat inactivated FBS (Invitrogen), 10 mM HEPES buffer pH 7.2, 2 mM L-glutamine, 50 μg/ml of penicillin and streptomycin (Invitrogen). Human Astrocytes (ScienCell Research Laboratories) were cultured in Astrocyte Medium (ScienCell Research Laboratories). Cells were incubated at 37°C in 5% CO2. The Toxoplasma strains used in this study are listed in Table S1. All T. gondii strains were maintained in vitro by serial passage on monolayers of HFFs. T. gondii strains were maintained in vitro by serial passage on monolayers of HFFs. The strains used in this study were Pru∆ku80, RH∆ku80 and 76K-GFP-LUC (gift of M. Grigg, National Institutes of Health, Bethesda, MD). The cultures were free of mycoplasma, as determined by qualitative PCR.
Generation of Bone Marrow Derived Macrophages (BMDM)
BMDMs were obtained from female C57BL/6 mice. Bone marrow was isolated by flushing hind tibias and femurs using a 25-gauge needle followed by passages through an 18-gauge needle to disperse cell clamps. Cells were suspended in DMEM supplemented with 10% heat-inactivated FBS and 20 ng/mL recombinant M-CSF (Invitrogen), and incubated in a tissue culture-treated flask for 8 to 12 hours. Then, non-adherent cells were harvested and transferred in 55 cm2 non tissue culture-treated plates (Corning) at 4 to 6×106 cells per plate and further incubated at 37°C, with 5% CO2 in humidified air. After 6 days, cells were washed with PBS to remove non-adherent cells, harvested by dislodging with a cell scraper in ice-cold PBS, and replated for the assay. This method yielded a highly pure population of F4/80+ macrophages by IFA.
Reagents
Primary antibodies mouse anti-HA (Roche, RRID:AB_2314622), MIC2 (provided by D. Sibley, Washington University School of Medicine, St Louis, MO), Toxofilin (provided by I. Tardieux, IAB, Grenoble, France), GRA7, E2F3 (Sigma-Aldrich, RRID:AB_1841394), E2F4 (Sigma-Aldrich, RRID:AB_1841395), E2F3 (Santa Cruz Biotechnology, RRID:AB_2096807), E2F4 (Santa Cruz Biotechnology, RRID:AB_2097106 and AB_2097104), TFDP-1 (Sigma-Aldrich, RRID:AB_259233), EZH2 (Cell Signaling Technology, RRID:AB_10694683 and AB_10694383), EZH2 (BD, RRID:AB_2102429), H3K27me3 (Diagenode, RRID:AB_2616049) and TBP1 (Abcam, RRID:AB_306337) were used in the immunofluorescence, immunoblotting and/or ChIP assays. Immunofluorescence secondary antibodies were coupled with Alexa Fluor 488 or Alexa Fluor 594 (Thermo Fisher Scientific). Secondary antibodies used in Western blotting were conjugated to alkaline phosphatase (Promega) or horseradish peroxidase.
Immunofluorescence microscopy
T. gondii infecting HFF cells grown on coverslips were fixed in 3% formaldehyde for 20 min at room temperature, permeabilized with 0.1% (v/v) Triton X-100 for 15 min and blocked in Phosphate buffered saline (PBS) containing 3% (w/v) BSA. The cells were then incubated for 1 hour with primary antibodies followed by the addition of secondary antibodies conjugated to Alexa Fluor 488 or 594 (Molecular Probes). Nuclei were stained for 10 min at room temperature with Hoechst 33258. Coverslips were mounted on a glass slide with Mowiol mounting medium, and images were acquired with a fluorescence ZEISS ApoTome.2 microscope and images were processed by ZEN software (Zeiss).
Western blot
Proteins were separated by SDS-PAGE, transferred to a polyvinylidene fluoride membrane (Immobilon-P; EMD Millipore) by liquid transfer, and Western blots were probed using appropriate primary antibodies followed by alkaline phosphatase or horseradish peroxidase-conjugated goat secondary antibodies. Signals were detected using NBT-BCIP (Amresco) or enhanced chemiluminescence system (Thermo Scientific).
Toxoplasma transfection
Vectors were transfected into RH∆ku80, Pru∆ku80, 76K-GFP-LUC tachyzoites by electroporation. Electroporation was done in a 2-mm cuvette in a BTX ECM 630 (Harvard Apparatus) at 1100 Volts, 25 Ohms, and 25 μF. Stable integrants were selected in media with 1 μM pyrimethamine and cloned by limiting dilution.
293-TRex transfection
24 h before transfection, the cells were plated in 6-well tissue culture dishes. 3 mg HAFlag-fusion protein-expressing plasmids and 0.3 mg puromycin selection plasmid were co-transfected into 293-TRex cells with Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. 72 h later, cells were diluted in the presence of 5 mg/ml puromycin (Sigma-Aldrich) for selection. Individual drug-resistant clones were expanded and tested for tetracycline-inducible gene expression.
Quantitative real-time PCR
Cells were left uninfected or infected with the Pru∆ku80, Pru∆ku80 Δteegr, and Pru∆ku80 Δteegr, TEEGR+ strains (MOI = 6) for 18 hours and subsequently subjected or not to TNFα stimulation (50 ng/mL; Sigma-Aldrich) for 6 hours. Total RNA was isolated using TRIzol reagent (Thermo Fisher Scientific). cDNA was synthetized with random hexamers by using the High Capacity RNA-to-cDNA kit (Applied Biosystem). Samples were analyzed by real time quantitative PCR for iNOS, Alox12, Kiss1R, IL-1β, IL-6, IL-8, IL-23A, CCL20 using TaqMan Gene Expression Master Mix (Applied Biosystem) according to the manufacturer’s instructions. C2-microglobulin was used as an internal control gene for normalization. The statistical significance of differences between data means was evaluated using an unpaired, two-tailed Student’s t test and by nonparametric ANOVA with multiple-comparisons.
Quantifying Numbers of Toxoplasma Cysts in the Brains of Mice
10 weeks’ post-infection, a half brain of each of the recipient mice was homogenized in 1 ml of PBS. Numbers of cysts in ten aliquots (20 μl each) of the brains suspensions were counted microscopically.
Bioluminescence imaging
Noninvasive bioluminescence imaging was performed 0, 2, 5, 7 and 10 days after Toxoplasma infection (5 x 104 tachyzoites). Five minutes before imaging, vigil mice received an intraperitoneal injection of 150 µg/g of D-luciferin (Promega, France) and were then anesthetized (isoflurane 4% for induction and 1,5% thereafter) and placed in the optical imaging system (IVIS Kinetic; PerkinElmer, USA). This allowed localization of luciferase-positive Toxoplasma and evaluation of the abdominal load. Bioluminescence signal was expressed as photons/seconds (p/s).
Flow Cytometry
6-week-old BALB/cJRj mice (Charles River) were i.p. infected with 5 x 105 parasites and sacrificed on days 2, 4, and 7 p.i. Immediately after being killed, mice were peritoneally lavaged with PBS, and recovered lavage fluid was centrifuged at 500 g for 8 min. Peritoneal cells were washed once with PBS. Harvested cells were suspended in stain buffer (PBS containing 2% FBS and 0.1 mM EDTA), seeded in 96-well plates (106 cells/100 μl), and pretreated with FcBlock (clone 2.4G2; BD Biosciences) for 30 min at 4°C. Cells were then incubated with fluorescently conjugated antibodies for cell surface markers from BD Biosciences: PE-conjugated anti-CD11b, PE-conjugated anti-Gr1 (clone RB6-8C5), PE-conjugated anti-F4/80, and APC-conjugated anti-Ly6G. Isotype controls consisted of PE-conjugated rat IgG2b, PE-conjugated rat IgG2a, and APC-conjugated rat IgG2a, provided by BD Biosciences. Analysis of stained cells was performed with a FACSCalibur flow cytometer (BD Biosciences). For all samples, 100,000 cells were analyzed for plot generation.
Harvest of peritoneal contents, in vivo cytokine ELISA, and determination of parasite load
6-week-old BALB/cJRj mice (Charles River) were i.p. infected with 105 parasites and sacrificed on days 2, 4, 7, and 9 p.i. Immediately after being killed, mice were peritoneally lavaged with 3 ml of physiological serum. 2 ml of recovered lavage fluid was centrifuged at 20, 000 g for 15 min at 4°C, and the clarified supernatant stored at -70°C until analyzed by ELISA. IFN-γ, IL-18, IL-1β, TNFα, IL-12, IL-23 and IL-22 protein concentrations were determined using commercially available ELISA kits (IL-18 ELISA Kit [MBL International], IL-1β Quantikine ELISA Kit [R&D Systems], IFN-γ ELISA Kit [BD Biosciences], TNFα, IL-12, IL-23 and IL-22 ELISA Kits [Thermo Fisher Scientific]) according to the manufacturer’s instructions. The parasite loads in peritoneal content were quantified after DNA extraction (QIAamp DNA mini kit; QIAGEN) using the quantitative PCR targeting of the T. gondii–specific 529-bp repeat element.
Histological analysis of ileum
Fragments of small intestine of mice infected with WT or teegr-deficient cysts were harvested on day 8 post-infection, fixed in 10% buffered formalin and paraffin processed. Tissue sections of 5µm thickness were mounted on slides and stained with hematoxylin and eosin. The histological score was analyzed with the following parameters: intensity of lamina propria (LP) inflammatory infiltration, thickening of LP, destruction of the villi and necrosis were evaluated by intensity, represented as arbitrary units ranging of 0 (less intense or absent) to 6 (highly intense) for each parameter. The histological final score is a sum of each parameter for each mouse.
Microarray hybridization and data analysis
Human (primary fibroblasts and astrocytes) and murine (BMDM) cells were left uninfected or infected with the Pru∆ku80, Pru∆ku80∆TEEGR and Pru∆ku80∆teegr, TEEGR strains (MOI = 6) for 18 hours. Total RNAs were extracted and purified using TRIzol (Invitrogen, Carlsbad, CA, USA). RNA quantity and quality were measured by Nano Drop 2000 (Thermo Scientific). Transcripts were obtained from three biological replicates each. RNA Labeling and Array Hybridization were done by Array Star Inc. Briefly, RNA quantity and quality were measured by Nano Drop ND-1000. RNA integrity was assessed by standard denaturing agarose gel electrophoresis. Transcripts were obtained from three biological replicates each. Total RNA from each sample was linearly amplified and labeled with Cy3-UTP. The Labeled cRNAs were purified by RNeasy Mini Kit (Qiagen). The concentration and specific activity of the labeled cRNAs (pmol Cy3/μg cRNA) were measured by Nano Drop ND-1000. 1 μg of each labeled cRNA was fragmented by adding 11 μl 10 × Blocking Agent and 2.2 μl of 25×Fragmentation Buffer, then heated at 60 °C for 30 min, and finally 55 μl 2 × GE Hybridization buffer was added to dilute the labeled cRNA. 100μl of hybridization solution was dispensed into the gasket slide and assembled to the gene expression microarray slide. The slides were incubated for 17 hours at 65°C in an Agilent Hybridization Oven. Agilent Feature Extraction software (version 11.0.1.1) was used to analyze the acquired array images. Quantile normalization and subsequent data processing were performed with using the GeneSpring GX v12.1 software (Agilent Technologies). After quantile normalization of the raw data, genes that at least 3 out of 27 samples have flags in Detected (“All Targets Value”) were chosen for further data analysis. Differentially expressed genes with statistical significance were identified through Volcano Plot filtering. Hierarchical Clustering was performed using the R software (version 2.15). GO analysis and Pathway analysis were performed in the standard enrichment computation method. The threshold are Fold Change ≥ 2.0, P-value ≤ 0.05. GO analysis and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis were performed using the DAVID Bioinformatics Resources. Microarray data has been uploaded to GEO Datasets under accession number (GSE113618 and GSE113626). Gene set enrichment analysis (GSEA, http://software.broadinstitute.org/gsea/index.jsp) was used to find candidate transcription factors and canonical pathways that are modulated differently between Toxoplasma infections. This program uses a priori defined sets of genes and determines whether the members of these sets of genes are randomly distributed throughout a ranked list or primarily found at the top or bottom. As GSEA is generally used to generate hypotheses, gene sets enriched with a false discovery rate (FDR) < 0.25 were considered significant. Both transcription factor and canonical pathway gene sets from the Molecular Signatures Database were evaluated for enrichment. Distant Regulatory Elements of co-regulated genes (DiRE, https://dire.dcode.org/) was also used. For every gene in a list, DiRE detects regulatory elements throughout the entire gene locus, and looks for enrichment of TFBSs.
Plasmid constructs
The plasmids and primers used in this work are listed in Table S1, respectively. To construct vectors pLIC-TEEGR-HF the coding sequence of TEEGR was amplified using primers LIC-239010-F and LIC-239010-R and genomic DNA of RH∆ku80 or Pru∆ku80 strains of Toxoplasma as DNA template. The resulting PCR product was cloned into the above pLIC vectors using the Ligation Independent Cloning (LIC) cloning method. The truncated versions of TEEGR were cloned as described above in the pLIC-HF-dhfr vector. Forward primer LIC-TEEGR-F was used with reverse primers LIC-TEEGR-QPV-REV or LIC-TEEGR-TNR-REV to construct the vectors pLIC-TEEGR-∆SER-HF, and pLIC-TEEGR-∆R3-HF, respectively. pcDNA-TEEGR-HF was constructed by amplifying TEEGR coding sequence from residue 29 to 744 with primers TEEGR-pcDNA4-Cter-FWD and TEEGR-pcDNA4-Cter-REV. The resulting PCR products were cloned into the pcDNA-LIC-HF plasmid using the LIC cloning method. The engineered LIC cassette of this vector fuses the C-terminal end of the protein of interest with an AGAGAGA linker followed by a HAFlag tag. The vectors pDEST14 KO TEEGRI and pDEST14 KO TEEGRII were generated to construct the deletion/insertion mutation of TEEGR in Toxoplasma strains RH∆ku80 (type I) and Pru∆ku80 (type II), respectively. The Multisite Gateway Pro 3-fragment Recombination system was used to clone the dhfr cassette flanked by the 5’ and 3’ surrounding regions of TEEGR coding sequence of type I and type II genomic DNA. The dhfr cassette, which confers resistance to pyrimethamine, was amplified by PCR using primers attB4r-5’dhfr F and attB3r-3’dhfr R and the plasmid pDHFR-TSc3 as DNA template. The resulting PCR product was cloned in pDONR221 P4r-P3r according to the manufacturer’s instructions, yielding the vector pDONR221/DHFR. The 5’ flanking region of TEEGR was amplified using primers attB1-TEEGR_F and attB4-TEEGR_R, and was cloned into the plasmid pDONR221 P1-P4. The 3’ flanking region of TEEGR was amplified using primers attB3-TEEGR_F and attB2-TEEGR_R, and was cloned into the plasmid pDONR221 P3-P2. The resulting vectors, pDONR221/5'TEEGR and pDONR221/3'TEEGR, respectively, were then recombined with the pDONR221/DHFR into the destination vector pDEST14, yielding the pDEST14 KO TEEGR. TEEGRNter-R3-HAFlag was DNA synthesis and cloned in pLIC-DHFR. To generate a complemented Pru∆ku80∆teegr strain, vector pLIC-PTEEGR-TEEGR-HF was co-electroporated with the pMiniHX at 1:10 ratio followed by selection with mycophenolic acid and xanthine, and cloned by limiting dilution. The resulting strain is named Pru∆ku80 ∆teegr, pLIC-PTEEGR-TEEGR-HF (Supplementary Table 3). The plasmid pTOXO_Cas9-CRISPR::sgTEEGR vector was generated as previously described6. Briefly, primers TEEGR-CRISP-FWD and TEEGR-CRISP-REV containing the sgRNA targeting TEEGR genomic sequence were phosphorylated, annealed and ligated in the pTOXO_Cas9-CRISPR plasmid linearized with BsaI, yielding pTOXO_Cas9-CRISPR-GFP_sgTEEGR.
Chromatographic purification of TEEGR-containing complex
Nuclear extracts from 293-TRex cells stably expressing HAFlag-tagged protein or J774 cells infected with Pru∆ku80 or RH∆ku80 expressing HAFlag-tagged TEEGR, were incubated with anti-FLAG M2 affinity gel (Sigma-Aldrich) for 1 hour at 4°C. Beads were washed with 10-column volumes of BC500 buffer (20 mM Tris-HCl, pH 8.0, 500 mM KCl, 20% glycerol, 1 mM EDTA, 1 mM DTT, 0.5% NP-40, and protease inhibitors). Bound polypeptides were eluted stepwise with 250 μg/ml FLAG peptide (Sigma Aldrich) diluted in BC100 buffer. For size-exclusion chromatography, protein eluates were loaded onto a Superose 6 HR 10/30 column equilibrated with BC500. Flow rate was fixed at 0.35 ml/min, and 0.5-ml fractions were collected.
Mass spectrometry-based proteomics
Protein bands were excised from colloidal blue–stained gels (Thermo Fisher Scientific), treated with DTT and iodoacetamide to alkylate the cysteines before in-gel digestion using modified trypsin (Promega, sequencing grade). Resulting peptides from individual bands were analyzed by online nanoLC-MS/MS (UltiMate 3000 coupled to LTQ-Orbitrap Velos, Thermo Scientific) using a 25-min gradient. Peptides and proteins were identified using Mascot (Matrix Science) and filtered using IRMa software: only rank 1 peptides exhibiting a query identity threshold above 0.02 and a score superior to 20 were selected before protein grouping.
Chromatin immunoprecipitation assay
HFF cells were left uninfected or infected for 24 hours with Pru∆ku80 TEEGR-HAFlag or Pru∆ku80 ∆teegr. Cells were then cross-linked with 1% formaldehyde for 10 min before quenching with 125 mM glycine for 5 min. The ChIP assay was performed by using Transcription Factor Chromatin Immunoprecipitation Kit (Diagenode) according to the manufacturer's protocol. In brief, fixed cells were sonicated to shear the cross-linked chromatin into an average DNA fragment size of 200–600 bp. We used 40 million sorted nuclei in 300 μl of immunoprecipitation buffer supplemented with fresh proteinase inhibitors. By using a Diagenode Bioruptor precooled to 4 °C, shearing was achieved in 1.5-ml low binding tubes in the appropriate tube adapter with 18 high-energy cycles of 30 s ON/30 s OFF. The aforementioned antibodies were used for immunoprecipitation. After overnight incubation, DNA-protein-antibody complex was eluted. The crosslinks were reversed by heating the samples at 65°C for 4 hours. DNA was purified by using IPure kit (Diagenode) according to the manufacturer's protocol. For validation, quantitative real-time ChIP-PCR was performed by SYBR Green (Applied Biosystems) using the StepOnePLus (Applied Biosystems). Real-time PCR was performed then quantitated using the delta–delta CT (ΔΔCT) method. Primer sequences are listed in Supplementary Table 3.
NF-kB reporter assay
THP1-Blue™ NF-kB cells (InvivoGen, San Diego, CA, USA) were grown in RPMI 1640 supplemented with 10% (v/v) FBS, 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 25 mM HEPES, 100 μg/ml Normocin™, 10 μg/ml Blasticidin. THP1-Blue™ NF-kB cells were treated with TNFα or infected with Pru∆ku80 WT or Δteegr strains and then followed by determination of NF-kB activity using a reporter gene assay. QUANTI-Blue™ (InvivoGen) was added to cells, plates were incubated for a further 2 h and the optical density at 655 nm was recorded. Graphs show results from averaged technical replicates and at least three independent experiments.
Supplementary Material
Acknowledgments
This work was supported by the Laboratoire d'Excellence (LabEx) ParaFrap [ANR-11-LABX-0024], the Agence Nationale pour la Recherche [Project HostQuest, ANR-18-CE15-0023] and the European Research Council [ERC Consolidator Grant N°614880 Hosting TOXO to M.A.H]. Proteomic experiments were partly supported by the Agence Nationale pour la Recherche (Investissement d’Avenir Infrastructures, ProFi project ANR-10-INBS-08-01).
Footnotes
Data availability
Correspondence and requests for materials should be addressed to M.A.H. The microarray data have been deposited to the GEO Datasets under accession number GSE113618 and GSE113626.
Mohamed-ali Hakimi: 0000-0002-2547-8233
Alexandre Bougdour: 0000-0002-5895-0020
Contributions
M.A.H., L.B. and A.B. conceived the project. L.B., M.P.B-P., P.M.H., D.C., S.K-J., J.V., V.J., B.T., Y.C. I.T., and A.B. designed, performed and interpreted the experimental work. M.A.H. supervised the research. M.A.H. wrote the paper with editorial support from I.T., L.B., and A.B.
Competing interests,
The authors declare no competing interests.
References
- 1.Jeffers V, Tampaki Z, Kim K, Sullivan WJ. A latent ability to persist: differentiation in Toxoplasma gondii. Cell Mol Life Sci. 2018 doi: 10.1007/s00018-018-2808-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Melo MB, Jensen KDC, Saeij JPJ. Toxoplasma gondii effectors are master regulators of the inflammatory response. Trends Parasitol. 2011;27:487–495. doi: 10.1016/j.pt.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunter CA, Sibley LD. Modulation of innate immunity by Toxoplasma gondii virulence effectors. Nat Rev Microbiol. 2012;10:766–778. doi: 10.1038/nrmicro2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bougdour A, et al. Host cell subversion by Toxoplasma GRA16, an exported dense granule protein that targets the host cell nucleus and alters gene expression. Cell Host Microbe. 2013;13:489–500. doi: 10.1016/j.chom.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Braun L, et al. A Toxoplasma dense granule protein, GRA24, modulates the early immune response to infection by promoting a direct and sustained host p38 MAPK activation. J Exp Med. 2013;210:2071–2086. doi: 10.1084/jem.20130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gay G, et al. Toxoplasma gondii TgIST co-opts host chromatin repressors dampening STAT1-dependent gene regulation and IFN-γ-mediated host defenses. J Exp Med. 2016;213:1779–1798. doi: 10.1084/jem.20160340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olias P, Etheridge RD, Zhang Y, Holtzman MJ, Sibley LD. Toxoplasma Effector Recruits the Mi-2/NuRD Complex to Repress STAT1 Transcription and Block IFN-γ-Dependent Gene Expression. Cell Host Microbe. 2016;20:72–82. doi: 10.1016/j.chom.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hakimi M-A, Olias P, Sibley LD. Toxoplasma Effectors Targeting Host Signaling and Transcription. Clin Microbiol Rev. 2017;30:615–645. doi: 10.1128/CMR.00005-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer M, Grossmann P, Padi M, DeCaprio JA. Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Res. 2016;44:6070–6086. doi: 10.1093/nar/gkw523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marson A, et al. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature. 2007;445:931–935. doi: 10.1038/nature05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Julian LM, et al. Tissue-specific targeting of cell fate regulatory genes by E2f factors. Cell Death Differ. 2016;23:565–575. doi: 10.1038/cdd.2015.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng N, Fraenkel E, Pabo CO, Pavletich NP. Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F–DP. 10 doi: 10.1101/gad.13.6.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bracken AP, et al. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. 13 doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garber M, et al. A High-Throughput Chromatin Immunoprecipitation Approach Reveals Principles of Dynamic Gene Regulation in Mammals. Mol Cell. 2012;47:810–822. doi: 10.1016/j.molcel.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xing Y, Zhou F, Wang J. Subset of genes targeted by transcription factor NF-κB in TNFα-stimulated human HeLa cells. Funct Integr Genomics. 2013;13:143–154. doi: 10.1007/s10142-012-0305-0. [DOI] [PubMed] [Google Scholar]
- 17.Luo X, Chae M, Krishnakumar R, Danko CG, Kraus W. Dynamic reorganization of the AC16 cardiomyocyte transcriptome in response to TNFα signaling revealed by integrated genomic analyses. BMC Genomics. 2014;15:155. doi: 10.1186/1471-2164-15-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Afonina IS, Zhong Z, Karin M, Beyaert R. Limiting inflammation—the negative regulation of NF-κB and the NLRP3 inflammasome. Nat Immunol. 2017;18:861–869. doi: 10.1038/ni.3772. [DOI] [PubMed] [Google Scholar]
- 19.Sun F, et al. Combinatorial pharmacologic approaches target EZH2-mediated gene repression in breast cancer cells. Mol Cancer Ther. 2009;8:3191–3202. doi: 10.1158/1535-7163.MCT-09-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee ST, et al. Context-Specific Regulation of NF-κB Target Gene Expression by EZH2 in Breast Cancers. Mol Cell. 2011;43:798–810. doi: 10.1016/j.molcel.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, et al. Epithelial EZH2 serves as an epigenetic determinant in experimental colitis by inhibiting TNFα-mediated inflammation and apoptosis. Proc Natl Acad Sci. 2017;114:E3796–E3805. doi: 10.1073/pnas.1700909114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCabe MT, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 23.Howard JC, Hunn JP, Steinfeldt T. The IRG protein-based resistance mechanism in mice and its relation to virulence in Toxoplasma gondii. Curr Opin Microbiol. 2011;14:414–421. doi: 10.1016/j.mib.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 24.Virreira Winter S, et al. Determinants of GBP Recruitment to Toxoplasma gondii Vacuoles and the Parasitic Factors That Control It. PLoS ONE. 2011;6:e24434. doi: 10.1371/journal.pone.0024434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robben PM, LaRegina M, Kuziel WA, Sibley LD. Recruitment of Gr-1 + monocytes is essential for control of acute toxoplasmosis. J Exp Med. 2005;201:1761–1769. doi: 10.1084/jem.20050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunay IR, et al. Gr1(+) inflammatory monocytes are required for mucosal resistance to the pathogen Toxoplasma gondii. Immunity. 2008;29:306–317. doi: 10.1016/j.immuni.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mordue DG, Monroy F, La Regina M, Dinarello CA, Sibley LD. Acute toxoplasmosis leads to lethal overproduction of Th1 cytokines. J Immunol Baltim Md 1950. 2001;167:4574–4584. doi: 10.4049/jimmunol.167.8.4574. [DOI] [PubMed] [Google Scholar]
- 28.Chang HR, Grau GE, Department, J. C. P Role of TNF and IL-1 in infections with Toxoplasma gondii. 5 [PMC free article] [PubMed] [Google Scholar]
- 29.Hunter CA, Chizzonite R, Remington JS. IL-1 beta is required for IL-12 to induce production of IFN-gamma by NK cells. A role for IL-1 beta in the T cell-independent mechanism of resistance against intracellular pathogens. 9 [PubMed] [Google Scholar]
- 30.Lieberman LA, et al. IL-23 Provides a Limited Mechanism of Resistance to Acute Toxoplasmosis in the Absence of IL-12. J Immunol. 2004;173:1887–1893. doi: 10.4049/jimmunol.173.3.1887. [DOI] [PubMed] [Google Scholar]
- 31.Muñoz M, et al. Interleukin (IL)-23 mediates Toxoplasma gondii–induced immunopathology in the gut via matrixmetalloproteinase-2 and IL-22 but independent of IL-17. J Exp Med. 2009;206:3047–3059. doi: 10.1084/jem.20090900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosowski EE, et al. Strain-specific activation of the NF-κB pathway by GRA15, a novel Toxoplasma gondii dense granule protein. J Exp Med. 2011;208:195–212. doi: 10.1084/jem.20100717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorfu G, et al. Dual Role for Inflammasome Sensors NLRP1 and NLRP3 in Murine Resistance to Toxoplasma gondii. mBio. 2014;5 doi: 10.1128/mBio.01117-13. e01117-13-e01117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mason NJ, Liou H-C, Hunter CA. T Cell-Intrinsic Expression of c-Rel Regulates Th1 Cell Responses Essential for Resistance to Toxoplasma gondii. J Immunol. 2004;172:3704–3711. doi: 10.4049/jimmunol.172.6.3704. [DOI] [PubMed] [Google Scholar]
- 35.Braun L, et al. A Toxoplasma dense granule protein, GRA24, modulates the early immune response to infection by promoting a direct and sustained host p38 MAPK activation. J Exp Med. 2013;210:2071–2086. doi: 10.1084/jem.20130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhatt D, Ghosh S. Regulation of the NF-κB-Mediated Transcription of Inflammatory Genes. Front Immunol. 2014;5 doi: 10.3389/fimmu.2014.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lima TS, Gov L, Lodoen MB. Evasion of Human Neutrophil-Mediated Host Defense during Toxoplasma gondii Infection. mBio. 2018;9 doi: 10.1128/mBio.02027-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Julian LM, et al. Tissue-specific targeting of cell fate regulatory genes by E2f factors. Cell Death Differ. 2016;23:565–575. doi: 10.1038/cdd.2015.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.