Abstract
Until recently, our understanding of the cellular tropism of human norovirus (HuNoV), a major cause of viral gastroenteritis, has been limited. Immune cells and intestinal epithelial cells (IECs) have been proposed as targets of HuNoV replication in vivo, although the contribution of each to pathogenesis and transmission is unknown. Murine norovirus (MNV) is widely used as a surrogate model for HuNoV, as it replicates in cultured immune cells. The importance of the complete MNV immune cell tropism in vivo has not been determined. Recent work has linked replication in IECs to viral persistence in vivo. MNV provides a model to assess the relative contribution of each cell tropism to viral replication in immunocompetent native hosts. Here we exploited cell-specific microRNAs to control MNV replication, through insertion of microRNA target sequences into the MNV genome. We demonstrated the utility of this approach for MNV in vitro by selectively reducing replication in microglial cells, using microglial-specific miR-467c. We then showed that inserting a target sequence for the haematopoietic-specific miR-142-3p abrogated replication in a macrophage cell line. The presence of a target sequence for either miR-142-3p or IEC miR-215 significantly reduced viral secretion during the early stages of a persistent infection in immunocompetent mice, confirming that both cell types support viral replication in vivo. This study provides additional evidence that MNV shares the IEC tropism of HuNoVs in vivo, and now provides a model to dissect the contribution of replication in each cell type to viral pathogenesis and transmission in a native host.
Keywords: Norovirus, tropism, microRNA, epithelial cells, haematopoetic cells
Introduction
Human noroviruses (HuNoVs) are a major cause of viral gastroenteritis worldwide, across all age groups (1), and cause between 2-6 million infections each year in the UK alone (2). HuNoVs represent a particular problem in healthcare settings, with 4000 hospital outbreaks reported in a 2 year period in the UK, resulting in over 9000 ward closures and a significant economic cost to the National Health Service (3). Whilst most infections are acute, chronic infections in immunocompromised individuals are a significant cause of morbidity and mortality (4). In low- and middle-income countries (LMICs), HuNoVs account for over 200,000 deaths in children under 5 each year (5), although a full picture of the impact and prevalence in LMICs has only recently begun to emerge (6, 7).
Despite the global burden, there are still no licenced antivirals or a vaccine. Although HuNoV was identified in 1972 (8), robust in vitro propagation systems have only been developed in the past three years (9, 10). A major hurdle in the development of a permissive cell culture system was that, until recently, the cellular tropism of HuNoVs in vivo remained unclear. Both immune cells and epithelial cells have been implicated as the primary cellular targets for HuNoVs (9, 10) (11). Evidence of HuNoV antigens in immune cells has been provided by numerous animal infection models, including chimpanzees, piglets and immunocompromised mice (12–14). These studies led to the first demonstration of HuNoV replication in vitro, in a cultured B cell line (BJAB) (10). Replication in BJAB cells is modest and requires the presence of enteric bacteria expressing histo-blood group antigens (HBGAs) or soluble HBGAs, which serve as attachment factors for HuNoVs to facilitate entry. Intestinal epithelial cells (IECs) are targeted by a variety of enteric pathogens that cause gastroenteritis, and as such have also been considered candidate target cells for HuNoVs. However, many studies tried and failed to find epithelial cell lines permissive for infection (15–17). Evidence of HuNoV replication in IECs in vivo came from studies of symptomatic HuNoV infection in gnotobiotic pigs, where viral non-structural proteins, markers of active replication, were observed in enterocytes, the most abundant type of IEC (18). A recent seminal study using intestinal biopsies from chronically infected patients revealed the presence of non-structural proteins and genomic replication intermediates in IECs, suggesting they are a target cell type for HuNoV in vivo (11), at least in immunocompromised individuals. This study paved the way for development of a robust in vitro replication system using stem-cell derived enteroids, multicellular monolayers that recapitulate the natural intestinal epithelium (9). This system supports the replication of diverse HuNoV strains in enterocytes, present in the enteroids, and allows dissection of virus-host interactions (9). The relative contribution of HuNoV replication in enterocytes versus B cells in vivo and how this links to pathogenesis and transmission in immunocompetent hosts has not yet been established. Likewise, the possibility that specialised tissue-resident immune cell subsets may also serve as target cells in vivo cannot be fully excluded (11).
Prior to the development and use of the B cell and enteroid propagation systems for HuNoV, understanding of norovirus replication has mainly come from studies using murine norovirus (MNV) as a surrogate model, due to the availability of efficient cell culture and reverse genetics systems (19–22). Like HuNoV, MNV is enteric, spread predominantly via the faecal-oral route and can cause both acute and persistent infections in the natural host, depending on the strain (23). MNV replicates in macrophage, DC, B cells and T cells in vitro (10, 19, 22). Genomic replication intermediates were recently identifed predominantly in these cell types in the gut-associated lymphoid tissue of immunocompetent mice infected with an acute strain of MNV (22). Replication in DC and B cells is important for acute and persistent MNV infections in vivo respectively (22, 24), however the importance of the immune cell tropism as a whole has not been determined.
Specialised microfold (M-cell) IECs have also been shown to be important for MNV replication in vivo by transcytosis of viral particles across the intestinal epithelium without active replication (25, 26). Likewise, cultured IEC cell lines do not support MNV replication in vitro (10). However recent work, published during this study, has established a correlation between the ability to replicate in IEC, sensitivity to interferon lambda and viral persistence in vivo (27). Given its use a surrogate model, particularly for dissecting host-pathogen interactions, it is important to determine whether MNV shares the cellular tropism of HuNoV. MNV also provides the opportunity to evaluate the contribution of different cell types to norovirus replication in vivo in a native immunocompetent host.
To further probe the role of both immune cells and IECs in viral persistence we sought to explore use of the cellular microRNA (miRNA) machinery to control MNV tropism. This approach exploits the cell-specific nature and regulatory power of host miRNAs and has recently been used to investigate the tropism of a number of RNA viruses. miRNAs are short non-coding RNAs (18-22nt) that post-transcriptionally regulate gene expression by binding to complementary sequences known as miRNA response elements (MREs) present in the open reading frame (ORF) or 3’ UTR of messenger RNAs (mRNAs). Binding of a miRNA targets an RNA-induced silencing complex (RISC) onto the mRNA, which results in either translational silencing or mRNA cleavage depending on whether the miRNA has partial or perfect homology respectively. Engineering cell-specific MREs into a viral genome has been demonstrated to regulate replication in a cell-specific manner, as viral replication is targeted by the RISC complex in cells expressing the cognate miRNA (28–33). This approach has been successfully used for poliovirus, Japanese encephalitis virus, measles virus, influenza virus and dengue virus to rationally generate live attenuated vaccines candidates, enhance existing vaccine safety and to improve the safety of oncolytic viruses by preventing replication in non-tumour cell lineages and tissues where it would lead to pathogenesis (28–37). Here, we first explored whether this approach could be applied to MNV in vitro. We then exploited haematopoetic- and IEC-specific miRNAs to probe the importance of each cell tropism to MNV replication during persistent infection of and immunocompetent host. The presence of a MRE for either haematopoetic miR-142-3p or IEC miR-215, but not scrambled control sequences, significantly reduced replication in vivo, suggesting that both cell types represent cellular targets for MNV during the early stages of persistent infection in vivo.
Results
Insertion of miR-467c target sequences into the MNV genome restricts replication in microglial cells and BMDM cells in vitro
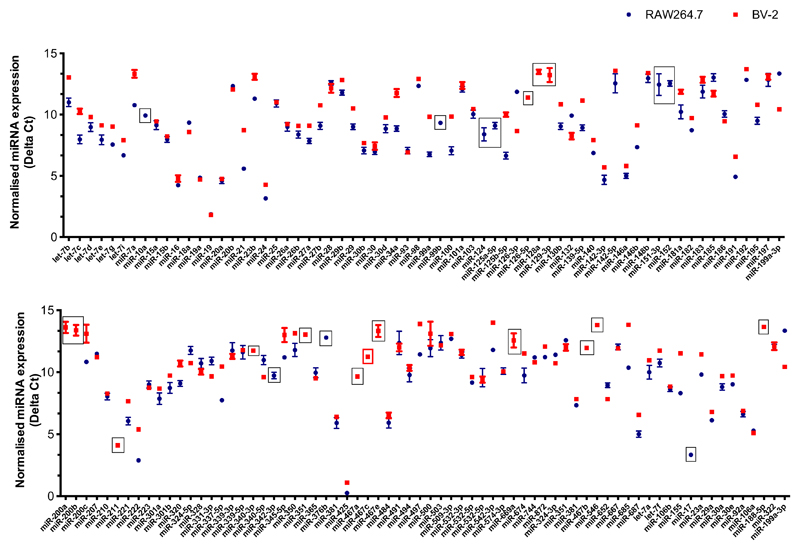
To determine the potential of targeting the miRNA machinery against MNV in vitro, as a proof-of-concept that this approach can be applied to norovirus, we set out to specifically restrict replication in one of the two permissive cell lines. We began by comparing miRNA expression in MNV-1-infected murine macrophage RAW264.7 cells and murine microglial BV-2s cells, with the aim of identifying a miRNA that was expressed in only one of these cell lines. As many microglial and macrophage miRNAs play a role in regulating the innate immune response (38–41), we analysed miRNA expression at 20 h following MNV-1 infection at low MOI, to coincide with the peak of interferon stimulated gene induction in RAW264.7 cells, as we have previous identified (42). Using Taqman-based miRNA arrays, we compared the expression of 136 detectable miRNAs and identified 15 miRNAs that were expressed only in BV-2 cells and 6 that were detected solely in RAW264.7 cells (Figure 1). We then validated three of these from each cell line: miR-10a, miR-124 and miR-125a-5p for RAW264.7 cells and miR-126-5p, miR-467a and miR-467c for BV-2 cells, using RT-qPCR. Cell-specific expression was validated for all of these miRNAs except miR-124, which was detected in both cell types (Table 1), suggesting amplification of this miRNA failed on the arrays.
Figure 1. Comparison of miRNA expression profiles in MNV1-infected murine macrophage RAW264.7 cells and microglial BV-2 cells.
RAW264.7 and BV-2 cells were infected with MNV-1 at an MOI of 0.1 TCID50/cell. The small RNA fraction was harvested at 20 hpi. Reverse transcription was performed using a set of primers for 380 miRNAs and control small RNAs, followed by qPCR analysis using TLDA miRNA cards. Results for the 136 detectable miRNAs are shown. The Ct for each specific miRNA is normalised against the Ct of the U6 control RNA to give the Delta Ct value shown on the Y-axis. The higher the Delta Ct the lower the expression. Boxed points indicate miRNAs that are expressed in only one of the cell lines during MNV-1 infection, the red box indicated miR-467c.
Table 1. Validation of the miRNA array data for a panel of differentially expressed miRNAs.
| Array cell line | miRNA | RT-qPCR validation |
|
|---|---|---|---|
| RAW264.7 | BV-2 | ||
| RAW264.7 | miR-10a | + | - |
| miR-124 | + | + | |
| miR-125-5p | + | - | |
| BV-2 | miR-126-5p | - | + |
| miR-467a | - | + | |
| miR-467c | - | + | |
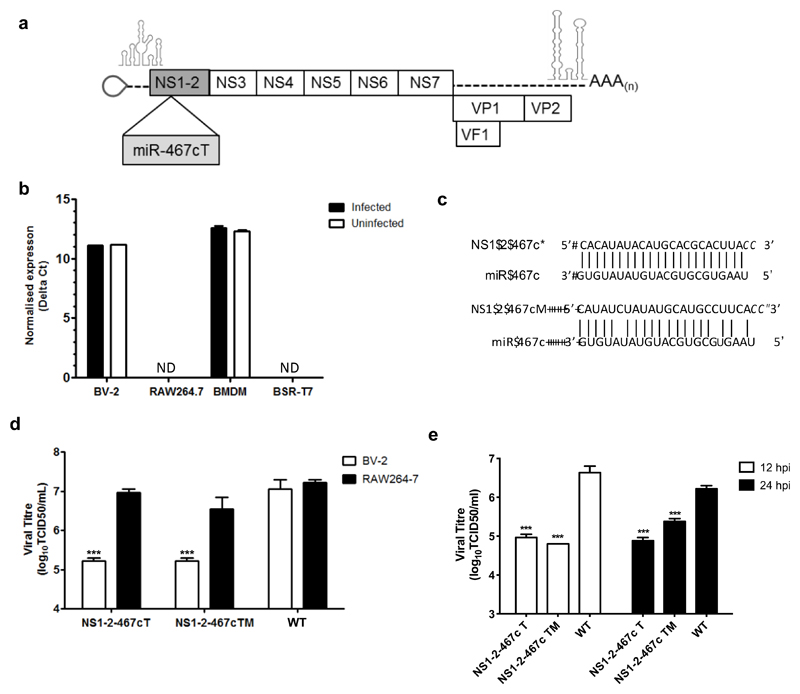
Of the validated miRNAs, we selected miR-467c to target MNV replication, on the basis that it was one of the most highly expressed BV-2-specific miRNAs. As the cellular expression profile of miR-467c is not known we also confirmed that it is expressed in bone-marrow derived macrophage (BMDMs), but importantly not in BSR-T7 cells, which are used for viral recovery using our DNA-based reverse genetics system, Figure 2b.
Figure 2. Insertion of miR-467c target sequences into the MNV genome restricts replication in microglial cells and BMDMs.
(a) miR-467c target sequences were inserted into in the coding region for NS1-2. (b) miR_467c is expressed in BV-2 and BMDM cells but not RAW264.7 and BSTR-T7 cells. ND: not detected, indicates samples that fell below the limit of detection. (c) The miR-467c target sequence was inserted to be complementary to miR-467c in the positive sense gRNA (NS1-2-467cT) and with modified (M) sequence complementarity (NS1-2-467cM), but with the same coding sequence. (d) miR-467c T or M sequences specifically reduce replication in BV-2 cells expressing miR-467c. RAW267.4 and BV-2 cells were infected with each virus at low MOI (0.01 TCID50/cell), titrated on RAW264.7 cells, and virus was harvested at 24 hpi. Significance was tested using Two Way ANOVA with a Dunnett post-test to compare replication of each virus between the two cell lines. ***: p< 0.001, n=3. (e) miR-467c target and modified target sequences reduce replication in BMDM cells. BMDMs were infected with each virus at an MOI of 10 TCID50/cell, titrated on RAW264.7 cells, and virus was harvested at 12 and 24 hpi. Significance was tested using Two-Way ANOVA, with Sidak’s multiple comparison test to compare replication of each virus between the two cell lines. ***: p<0.001, n=3.
To target miR-467c towards the MNV genomic RNA, it was necessary to insert a cognate target sequence into the genome without compromising replication. We have previously used transposon-mediated insertional mutagenesis to identify positions across the norovirus genome that tolerate the insertion of short nucleotide sequences without compromising virus replication in cell culture (43). Based on this study, we inserted MREs for miR-467c into the MNV-1 genome after nt 383 in the NS1-2 coding region of ORF1 (Figure 2a and c), which we have previously shown tolerates insertion of a FLAG epitope tag coding sequence, the same size as the MRE insertion (24 nt) (43, 44). We introduced a single MRE with either perfect or partial homology, to direct different effector functions of RISC against the genome. The partial homology was limited to 3 mismatches in order to to maintain the same amino acid sequence as the perfect MRE sequence. The viruses were designated as NS1-2-467cT or NS1-2-467cTM, where T indicates target and M indicates a modified partial complementary target sequence (Figure 2).
As expected, replication of NS1-2-467cT carrying a perfect MRE was reduced by nearly 100-fold in BV-2 cells compared to RAW264.7 cells, whereas WT-MNV replicated to the same level in both cell lines (Figure 2c). Replication of the virus carrying the partially complementary target sequence (NS1-2-467cM) was also reduced in BV-2 cells compared to RAW264.7 cells, indicating that the restriction applies for both perfect and imperfect MREs. Replication of NS1-2-467cT and NS1-2-467cTM was also significantly reduced by up to 100-fold in BMDM cells (Figure 2e), with the NS1-2-467cT virus displaying the largest defect at the later time point.
Insertion of a MRE for haematopoetic-specific miR-142-3p into the MNV genome inhibits replication in macrophage cells in vitro
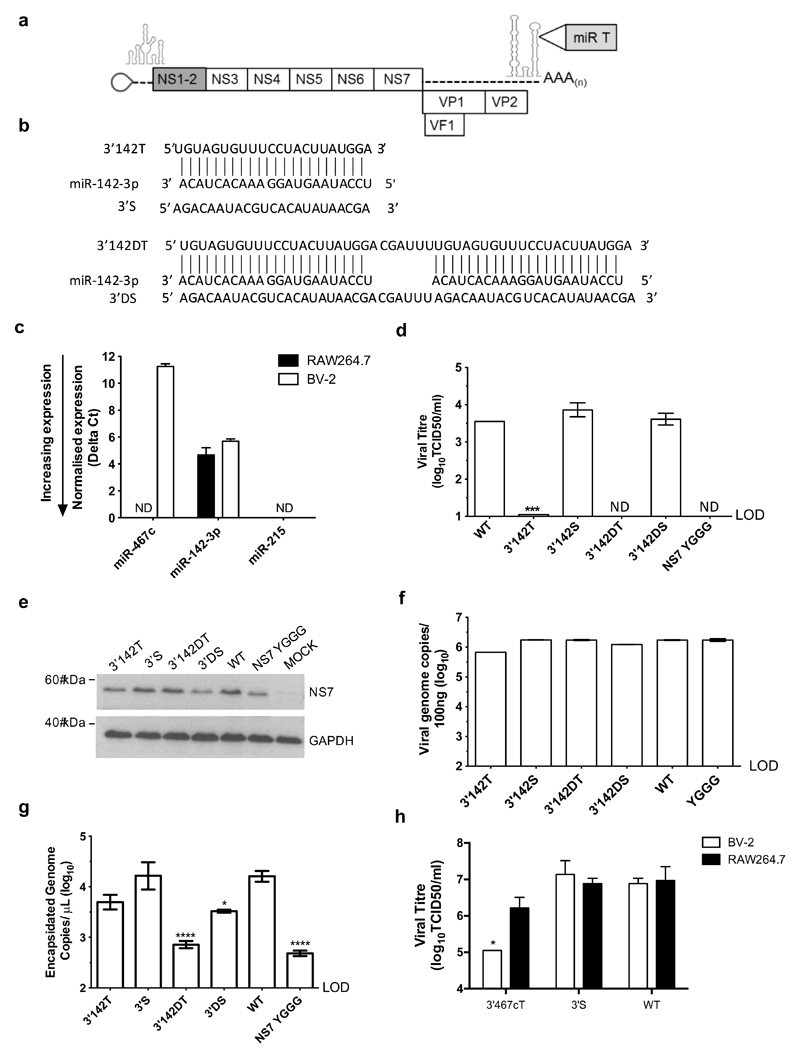
To use this approach to probe the importance of the immune cell tropism in vivo we selected a miRNA with a well-defined hematopoietic-specific expression profile. As targeting the genome with miR-467c did not achieve complete suppression in vitro (Figure 2d and e) and its cellular expression is not well characterized, we employed one of the most highly abundant haematopoetic-specific miRNAs, miR-142-3p (45). This miRNA has previously been used as a tool to study dengue virus replication in hematopoietic cells in a similar approach (33).
Examination of the array data (Figure 1) and subsequent RT-qPCR (Figure 3c) confirmed that miR-142-3p is expressed in both RAW264.7 and BV-2 cells, and at higher levels than miR-467c in BV-2 cells. To assess the importance of tropism during persistent infections in immunocompetent mice, we inserted the MREs into the genome of the persistent MNV-3 strain (21). To avoid any impact of having the MRE in a coding sequence, this time we inserted miR-142-3p MREs into a site in the 3’UTR, which we have previously identified as being able to tolerate nucleotide insertions (43). We inserted one (3’142T) or two (3’142DT) perfectly complementary MREs into this site, alongside scrambled control sequences of the same length and base composition (3’S and 3’DS), Figure 3b.
Figure 3. Insertion of haematopoetic-specific miR-142-3p target sequences into the MNV genome inhibits replication in macrophage cells.
(a) miR-142-3p target sequences were inserted into a site in the MNV-3 3’UTR, previously found to tolerate nt insertions. (b) Single and double perfectly complementary target sequences were inserted into the MNV-3 genomes, alongside single or double scrambled control sequences. (c) miR-142-3p is expressed in RAW264.7 and BV-2 cells at higher levels that miR-467c. ND: not detected, indicates samples that fell below the limit of detection. (d) Insertion of single or double miR-142-3p target sequences reduces replication in RAW264.7 cells. Recovery supernatants for each virus were titred on RAW264.7 cells to determine TCID50/ml. The polymerase active site mutant (NS7 YGGG) was included as a replication-defective control. ND: not detected, indicates samples that fell below the limit of detection.. (e) Production of the MNV-3 polymerase (NS7) or intracellular genomes (f) were not limiting during virus recovery of 3’142T or 3’142DT, compared to WT and scrambled controls. (g) Encapsidated genome copies, indicating infectious virion production, were detected at WT-like levels for for 3’142T and 3’S, but were significantly reduced for 3’142DT and 3’D. (h) Insertion of a single microglial miR-467c target sequence into the 3’UTR site selectively reduces replication in BV-2 cells. RAW264.7 nd BV-2 cells were infected with each virus at 0.01 TCID50/ml (titred on RAW264.7 cells) and virus was harvested at 24 hpi. (d) and (g) Significance was tested using OneWay ANOVA with Dunnett’s post-test to compare replication of each virus to the WT. ****: p<0.0001, *: p<0.05, n=3. LOD: limit of detection. (h) Significance was tested using Two-Way ANOVA, using Sidak’s multiple comparison test to compare replication of each virus between the two cell lines. *: p<0.05, n=3.
The presence of a single miR-142-3p MRE (3’142T) reduced replication to the limit of detection and a double MRE (3’142DT) completely prevented detectable viral replication in murine macrophage RAW264.7 cells (Figure 3d). In comparison, viruses carrying the single and double scrambled control sequences (3’S and 3’DS) replicated to the level of WT MNV-3, (Figure 3d). To determine that the loss of observed replication was due to restriction in the RAW264.7 target cells and was not a result of lower production or stability of the RNA during virus recovery, we measured the level of the viral polymerase protein (NS7) and intracellular genomes produced during virus rescue in BSR-T7 cells. NS7 levels and intracellular genomes levels were not limiting for 3’142T or 3’142DT (Figure 3e and f), indicative of comparable transfection efficiency and no major impact on RNA stability. To confirm that infectious virions were produced for 3’142T and 3’142DT, we determined the amount of nuclease-resistant encapsidated viral genomes produced during virus recovery. Comparable encapsidated genome copies were detected for the 3’142T virus as for WT MNV-3 and 3’S, however the double MRE insertion resulted in a significant reduction for 3’142DT, and to a lesser extent for the double scrambled control (Figure 3g). As we could therefore not be sure that the loss of 3’142DT replication in RAW264.7 cells was solely due to miR-142-3p targeting and not an unknown effect on genome encapsidation and viral assembly due to the size of the insertion, we did not take the 3’142DT virus forward for further characterization. We made multiple attempts to deplete miR-142-3p, using miRNA antagonirs and CRIPSR-Cas9 for miR-142-3p and Dicer, a key component of the miRNA processing machinery, in order to further confirm that miR-142-3p was responsible for the restriction and to generate a cell line to be able to titre the targeted viruses in the absence of miR-142-3p. However due to macrophage cell lines being refractory to transfection and the necessity of the miRNA machinery for regular cellular function, it was not possible to obtain a reduction in miR-142-3p levels using these approaches (data not shown).
We also compared the effect of inserting a single perfect MRE for miR-467c into the same site in the 3’UTR, alongside a single scrambled control sequence of the same length and base composition. The presence of the miR-467c MRE reduced replication in BV-2 cells by nearly 10-fold in comparison to the scrambled control virus and WT-MNV-3, but did not affect replication in RAW264.7 cells (Figure 3h). The differences in restriction resulting from each MRE at the same site correlates with the expression level of the cognate miRNA, where miR-142-3p is most highly expressed and results in the greatest restriction.
MNV carrying a MRE for IEC-specific miR-215 replicates in macrophage cells
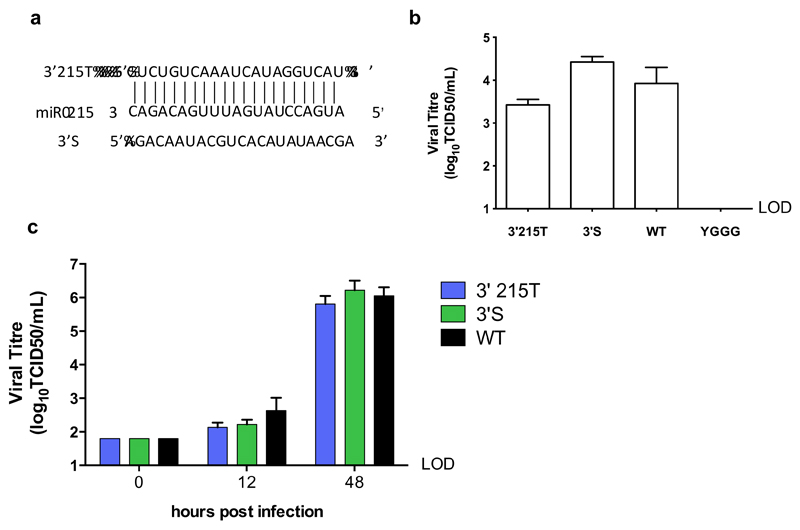
As IECs have recently been shown to be a target cell for HuNoVs in vivo (11), to be able to confirm whether they support MNV-3 replication in vivo, we next inserted a perfect MRE for the most abundant miRNA in murine IECs, miR-215 (46) (Figure 4a). miR-215 is not expressed in either RAW264.7 or BV-2 cells (Figure 2c) and there are no reports of its expression in immune cells. Accordingly, the 3’215T virus recovered to comparable levels as the WT and 3’S control during reverse genetics recovery from BSR-T7 cells (Figure 4b) and replication was not restricted in RAW 264.7 cells (Figure 4c). As MNV does not replicate in cultured IEC lines (10), and as we could not achieve sufficient transfection of either macrophage or microglial cell lines to exogenously express miR-215 (data not shown), we were not able to further characterise the restriction by miR-215 in vitro.
Figure 4. MNV carrying a target sequences for intestinal epithelial cell-specific miR-215 replicates in macrophage cells.
(a) A perfect target sequence for highly abundant IEC miR-215 was inserted into the 3’UTR site in the MNV-3 genome alongside the scrambled control sequence. (b) A virus carrying the miR-215 MRE (3’215T) recovered to similar levels as the WT and scrambled control virus. (c) 3’215T replicates with WT-like kinetics in RAW264.7 cells, which do not express miR-214. RAW264.7 cells were infected with each virus at low MOI (0.01 TCID50/cell) and virus was harvested at 12 and 48 hpi. LOD: limit of detection.
Carrying MREs for haematopoietic miR-142-3p and IEC miR-215 reduces MNV-3 replication in the initial phase of a persistent infection in vivo
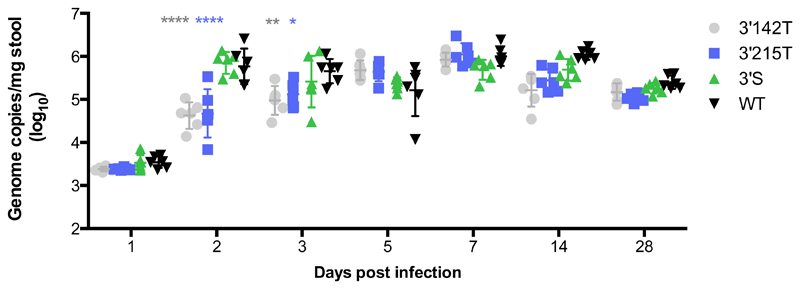
To determine the effect of carrying a MRE for either miR-142-3p or miR-215 on replication in vivo, we infected immunocompetent mice with equal doses of MNV-3 WT, 3’142T, 3’215T and 3’S and monitored viral secretion. Equivalent levels of viral genomes were secreted 1 day post infection (dpi), which reflects equal inoculation doses and shedding of the excess viral inoculum prior to the peak of replication at 2 to 3 dpi, as we have previously confirmed in this infection model (21). At 2 dpi both the 3’142T and 3’215T viruses were secreted in the faeces at significantly lower levels than the WT and 3’S viruses, with over a 10-fold reduction in the average genome copies/mg stool (Figure 5). There was no significant difference in the level of the genomes secreted between the 3’142T and 3’215T viruses, suggesting that the replication of both were equally restricted (Figure 5). A significant defect in replication of these of these viruses was also observed at 3 dpi albeit to a lesser extent. The defect was fully recovered by 5 dpi, both viruses established persistent infections and were then secreted at comparable levels to the WT and 3’S for the duration of the experiment (Figure 5). To determine if there were any mutations in the two MREs, we sequenced the 3’UTR of the secreted virus population from each group of infected mice at 5 dpi. We observed no mutations in the WT or 3’S 3’UTR and could not detect any changes in the 3’142 or 3’215 MREs that would compromise binding of the cognate miRNAs (data not shown).
Figure 5. Carrying a target sequence for haematopoietic miR-142-3p and intestinal epithelial miR-215 reduces MNV replication in the initial phase of a persistent infection in vivo.
Mice were infected with 100ul of each virus recovery supernatant containing equivalent genome copies and secreted viral genome copies were measured on the indicated days by RT-qPCR. Significance was tested using Two-Way Anova with Dunnett’s multiple comparison test to compare each group to the WT control group. ****: p< 0.001. n=6.
Discussion
Here we have exploited the cell-specific regulatory nature of host miRNA expression to further define the cellular tropism of MNV during persistent infection of immunocompetent hosts. Having demonstrated that this approach could be applied to MNV in vitro, selectively targeting replication in microglial cell lines, we then employed hematopoietic-specific miR-142-3p and IEC miR-215 to probe the importance of replication in these cell types in vivo. The presence of a single target sequence for either miRNA reduced viral secretion during the early phase of infection in immunocompetent mice, indicative of reduced replication. This provides additional evidence that MNV actively replicates in IECs in vivo, and that in addition to having an immune cell tropism, it also shares the HuNoV tropism for IECs (11), at least during chronic infections. MNV therefore now provides a surrogate model to dissect and understand the contribution of each cell tropism to pathogenesis and transmission, which remains unknown for HuNoV infections.
During preparation of this manuscript, evidence emerged that supports our finding of MNV replication in IECs in vivo. Lee et al. have recently demonstrated that IECs serve as a reservoir for MNV during persistent infections in vivo (27). Using a highly sensitive flow cytometry assay to detect two non-structural proteins, indicative of active viral replication, they found that MNV maintains low level infection of IECs during persistent infection, at a rate of approximately 20 infected cells per million IECs. These low infected cell numbers, combined with the high background signals they observed using single staining for viral proteins, suggests why infection of IECs has not been detected before in similar studies (27). It also shows the utility of exploiting the cellular miRNA machinery and the virus itself as a tool to probe tropism as a complementary approach that does not reply on staining protocols for viral detection.
Lee et al. demonstrated that the ability of MNV to infect IECs correlated with an ability to persist in vivo, and was determined by a combination of the action of MNV non-structural (NS) protein NS1-2 and host cytokine interferon lambda (IFNλ) (27). Studies from the same group have previously demonstrated that both of these factors can determine the persistence of MNV in vivo (47–49), which now neatly pin-points IECs as the cell type in which interplay between MNV and components of host innate immune response influences the outcome of persistent infections. Clearance of MNV replication in IECs by IFNλ treatment correlated with a loss of viral secretion (27), suggesting that replication in IECs results in release of virus into the intestinal lumen, and may therefore contribute to transmission, although this has yet to be demonstrated. It is striking that the very low number of infected IECs may be solely responsible for production of the high numbers of viruses secreted during persistent MNV infection. This study did not quantify the number of infected macrophage, DC and B cells in the intestinal lamina propria, which would provide an important comparison. This is especially relevant as another recently published study identified immune cells of the gut-associated lymphoid tissue as the predominant cell target during acute MNV infection in immunocompetent hosts (22). This study used RNAScope in situ hybridization to detect both positive and negative sense MNV genomic RNA, which were also observed at very low frequency in the follicule-associated epithelium and the contribution of these cell types in acute infection is therefore not known (22). These apparently contradictory reports may be explained by the use of different strains, suggesting that acute and persistent strains of MNV may have predominantly different tropisms (27). It will be interesting to determine if this is also the case for HuNoV. Our findings suggest that restricting replication in both cells types results in lower viral secretion of a persistent MNV strain. Therefore, further studies are required to determine whether newly synthesised virus in cells of the lamina propria could be secreted directly back into the intestinal lumen through the epithelium, or whether replication in immune cells indirectly affects secretion by providing more virus to infect IECs.
Our approach could be used to further investigate whether norovirus tropism is the same during acute infections and address the reported discrepancies. Using this approach in a model where the host develops a symptomatic infection would also allow assessment of the contribution of replication in each cell type to pathogenesis and disease, although symptomatic infection models involve the use of innate or adaptive immunocompromised mice which may remove natural barriers to replication (50, 51). It would be interesting to determine how replication in each cell type affects viral dissemination through the intestines and into extra-intestinal tissues in these models, such as the spleen and MLN. In our MNV-3 model of persistent infection, replication in these tissues peaks between day 3 and 5 pi (21), by which time viral replication had overcome restriction by each miRNA.
Although this approach of exploiting miRNAs has been successfully applied to selectively control viral tropism and attenuate a number of other RNA viruses (28, 33, 35), we encountered several limitations in using it as a tool to to study MNV tropism. The first was that given MNV only replicates in immune cells in vitro, which all express miR-142-3p and are resistant to transfection and genetic manipulation, there was no alternative permissive cell type to titre the miR-142-3p-restricted viruses and demonstrate replication in the absence of miR-142-3p. However, we confirmed that equal levels of encapsidated viral genomes, and therefore infectious virions, were produced for the 3’142T virus, and an equivalent genome copy numbers were shed as the inoculation dose on day 1 post infection in vivo. Conversely, as cultured IEC lines do not support MNV replication in vitro (10, 27), we did not have a permissive cell line with endogenous miR-215 expression to enable in vitro characterisation of the miR-215 restriction.
A second major challenge was the difficulty in making large insertions in the compact MNV genome, which has short UTRs, without disrupting essential genomic elements or protein domains that would compromise viral replication. We were guided here by our previous studies identifying positions across the genome that tolerated 15nt insertions (43, 44). However whether the same site tolerates larger insertions is size- and sequence-specific, and we have found that many sites that tolerate small epitope tags or random 15nt insertions do not tolerate an insertions of larger sizes, such as genes for fluorescent proteins of various sizes (unpublished data). We encountered the same challenge here with insertion of MREs into a previously identified site in the 3’UTR (43). Insertions of various single MREs (23 nt) were tolerated at this position, without replication being compromised by the insertion itself, evidenced by WT-like replication of the scrambled control viruses, and differential replication of the miRNA-targeted viruses correlating with differences in expression of the cognate miRNA. However insertion a double miR-142-3p MRE with spacer region (51 nt) into the same site reduced the production of infectious virions, measured as encapsidated viral genomes. The norovirus 3’UTR is known to contain structures important for viral replication (52). This finding suggests that the 3’UTR may also impact genome encapsidation and viral production, however further studies are required to confirm this. In the majority of other studies using this approach, restriction has been achieved in vivo by inserting at least a double MRE, and in some cases by insertion of a cassette containing up to 6 MREs (28, 31–33, 53). A greater number of MREs likely facilitates complete restriction early in infection and in the face of high levels of RNA virus genome replication during infection. It has been suggested that a minimum threshold of miRNA copy number must be present for gene silencing to occur (54). Therefore if the restriction is not complete then it is likely that the number of newly synthesised genomes could soon outnumber the copy number of the miRNA in each cell and thereby overcome restriction. As such, replication of MNV-3 carrying single MREs overcame each miRNA restriction in the early stages of infection in vivo. The lack of resistance mutations in the MRE sequences at the point at which restriction was overcome (5 dpi), suggests that a single MRE did not exert sufficient selection pressure on each virus, which is also in line with high levels of secreted virus production from relatively few infected cells in vivo, at least for IECs (27). To exert more selective pressure, some studies have inserted MREs at multiple sites in the genome (32), which could be a possibility for taking this approach further for MNV, however given its short UTRs, additional insertions would need to be in coding regions, which can be complicated by effects at the protein level.
As both the miR-142-3p and miR-215-targeted viruses overcame restriction during the acute phase of infection (21), it meant that we could not examine the importance of each cellular tropism to maintaining persistent infection and shedding. This may also reflect the fact that several cell types are permissive in vivo so restriction in one cell type may only have a minor impact. This approach has so far only been applied to the study of tropism in acute viral infections and it also suggests that maintaining restriction in the face of high rates of mutagenic RNA virus replication may not applicable to persistent viruses.
Finally, the finding that MNV replicates in IECs in vivo raises a number of interesting questions and possibilities. Firstly, a proteinaceous receptor for MNV has recently been identified as CD300lf (55), an Ig domain-containing molecule expressed on myeloid cells with low expression on epithelial cells (22). This suggests that MNV may use a different proteinaceous receptor to enter IECs. As in vitro replication in cell culture of MNV and HuNoVs is predominantly at the level of entry (55, 56), identification of this receptor could by comparison shed light on the long-sought-after receptor for HuNoV and facilitate development of further in vitro propagation systems for both viruses.
Methods
Cells
The murine macrophage cell line RAW264.7 was maintained in Dulbecco’s modified Eagles Medium (DMEM, Gibco) containing 10% fetal calf serum (FCS), penicillin (P) (100 SI units/mL) and streptomycin (S) (100 μg/mL) and 10 mM HEPES (pH7.6) at 37°C with 10% CO2. The murine microglial BV-2 cell line (45) was provided by Jennifer Pocock (University College London). BV-2 cells were maintained in DMEM supplemented with 10% FCS (Biosera), 2 mM L-glutamine, 0.075% sodium bicarbonate (Gibco) and antibiotics penicillin and streptomycin as above. BHK cells engineered to express T7 RNA polymerase (BSR-T7 cells, obtained from Karl-Klaus Conzelmann, Ludwid Maximillians University, Munich, Germany) were maintained in DMEM containing 10% FCS, penicillin (100 SI units/mL) and streptomycin (100 μg/mL) and 0.5 mg/mL G418. For preparation of bone marrow derived macrophage (BMDM), bone marrow cells were harvested from female C57BL/6 mice and were cultured in DMEM containing 10% fetal calf serum (FCS), penicillin (100 SI units/mL) and streptomycin (100μg/mL). For differentiation the supernatant from CMG14 cells, which contains macrophage colony stimulating factor (M-CSF), was added to the media for 5-7 days.
Viral reverse genetics
The cDNA clone pT7:MNV 3’ RZ, containing the WT MNV-1 genome under control of a truncated T7 polymerase promoter were previously constructed (20). The cDNA clone pT7:MNV3 3’RZ (MNV3-WT) contains the MNV-3 sequence under control of a the T7 promoter as is described in (21). Insertion of MREs into each genome was achieved by overlap mutagenic PCR (primer details available upon request). Virus was rescued from cDNA clones using the reverse genetics system based on recombinant Fowlpox expressing T7 RNA polymerase, as previously described (20). Briefly, 1 μg of each cDNA clone was transfected, using Lipofectamine 2000 transfection reagent (Invitrogen), into BSR-T7 cells previously infected with recombinant Fowlpox virus expressing T7 RNA polymerase at an MOI of approximately 0.5 pfu per cell. After 48 h post transfection, cells were freeze-thawed at -80°C to release virus particles.
Viral TCID50 and growth analysis
Fifty percent tissue culture infectious dose (TCID50) titrations were performed on BV-2 or RAW264.7 cells using 10-fold serial dilutions typically over a range of neat to 10-7. The viral TCID50/mL was determined by scoring for signs of cytopathic effect (CPE) at 5 days post infection by visual inspection. Multistep growth curve analysis of miRNA-targeted viruses was performed by infecting RAW264.7 cells with sequence-verified virus at an MOI of 0.01 TCID50/cell. BMDM cells were infected with an MOI of 10 TCID50/cell, as titred on RAW 264.7 cells. Virus was harvested at various time points post infection by hpi a single freeze-thaw cycle and virus titre was determined by TCID50 on RAW264.7 cells.
miRNA extraction and expression analysis
To analyse miRNA expression RAW264.7 and BV-2 cells were infected with MNV-1 WT at an MOI of 0.1 TCID50/cell or were mock infected, and BMDM cells were infected at an MOI of 10 TCID50/cell, and the small cellular RNA fraction was harvested at 20 hpi. A DNA-based recovery of MNV-1 was performed in BSR-T7 cells and small RNA species extracted at 24 h post transfection. In each case the small cellular RNA fraction (<200bp) was harvested using the miRVana RNA isolation kit, as per manufacturer’s instructions. This retains RNA molecules as small as 10 nt. For cDNA synthesis the Taqman miRNA reverse transcription kit was used (Life Technologies), according to manufacturer’s instructions. 1000 ng of RNA extracted from infected and uninfected RAW264.7 cells and BV-2 cells were used with Megaplex RT primers, Rodent Pool A (Life Technologies). The pool contains primers specific to 335 and 238 mature unique mouse and rat miRNAs respectively, alongside primers for 4 specific endogenous controls. The cDNA was then used for qPCR using Taqman Rodent miRNA Array A cards (TDLA, Life Technologies). The Taqman Rodent miRNA Array A is a pre-configured 384 well card that can amplify and detect 381 mature rodent miRNAs and 3 control small RNAs (that are common to those in the Megaplex primer pools). As recommended in the manufacturer’s instructions, 6 μL of cDNA was mixed with Taqman Universal PCR 2X Master Mix (no UNG) (Life Technologies) into a final volume of 900 μL. 100 μL of each mix was loaded into a port on the TLDA card which covered two rows of the 384 well card. The TLDA cards were run on the 7900HT T Fast Real-Time PCR System and the data was visualised and analysed for the Ct value of each miRNA by RQ manager software (Life Technologies). Further analysis was performed using Microsoft Excel. The Ct value for each miRNA was normalised against the Ct value for the endogenous control small non-coding nuclear RNA U6, which did not appear to be altered with infection. RT-qPCR validation was performed using RT and PCR primer-probes specific to a selection of miRNAs. The RT-qPCR was performed using the same enzymes and master mixes but with individual primers for miR-467a, miR-467c, miR-126, miR-125-5p, miR-124, miR-10a and the control RNA U6.
Quantification of viral RNA by RT-qPCR
Quantification of viral genome copies from infected cells or faecal samples was performed by two-step RT-qPCR. Nuclease-resistant encapsidated genomes were produced as previously described (20), prior to RNA extraction using 100uL of treated recovery supernatants. From infected cell samples, 20 ng of RNA was used for reverse transcription. The amount of RNA used from faecal samples corresponded to that extracted from 0.5 mg of stool. Viral RNA of known copy number was generated by in vitro transcription of the MNV-1 or MNV-3 cDNA clones and was serially diluted to generate a standard curve. Reverse transcription was performed with M-MLV using random hexamers, as per manufacturer’s instructions. qPCR was performed on the cDNA using a Taqman Low Rox qPCR mastermix (Primer design), with an initial denaturation step of 8 min at 95 °C, followed by 50 cycles of 95 °C for 10 s and 60 °C for 1 min. The genome copy number was interpolated from the standard curve and was calculated per ng of RNA or per mg of stool depending on the sample, using Microsoft Excel. All graphs were produced using Graph Pad Prism V.5 Software.
In vivo infections
Three-to-four week old female C57BL/6 mice were inoculated by oral gavage with 100ul of recovery supernatant of MNV-3 3’142T, 3’215T, 3’S and WT viruses, which contained equivalent encapsidated viral genome copy numbers. Each group contained 6 mice and a control group was mock infected. Mice were weighed and faecal samples were collected on days 0-7, 14, 21, and 28 post infection and mice were euthanized on day 28. Faecal pellets were homogenised in PBS (100mg/mL), followed by centrifugation at 4000 rpm for 5 min, 4 °C. RNA was extracted from 100 μL of supernatant using the GenElute mammalian total RNA kit and viral genome copy numbers were determined as described.
Acknowledgments
Funding statement
This work was supported by a Sir Henry Wellcome Postdoctoral Fellowship to LT [106079/Z/14/Z] and Wellcome Trust Senior Fellowships to IG [097997/Z/11/Z and 207498/Z/17/Z].
Abbrieviations used
- 3’UTR
- BMDM
bone marrow derived macrophage
- CPE
cytopathic effect
- DC
dendritic cell
- Dpi
days post-infection
- HBGA
histo-blood group antigens
- Hpi
hours post-infection
- HuNoV
human norovirus
- IEC
intestinal epithelial cells
- miR/miRNA
microRNA
- MNV
murine norovirus
- MOI
multiplicity of infection
- MRE
microRNA response element
- NS
non-structural
- ORF
open reading frame
- Pfu
plaque forming units
- TCID50
tissue culture infectious dose 50
Footnotes
Conflict of interest statement
The authors declare no conflicts of interest or competing financial interests.
References
- 1.Ahmed SM, Hall AJ, Robinson AE, Verhoef L, Premkumar P, Parashar UD, et al. Global prevalence of norovirus in cases of gastroenteritis: a systematic review and meta-analysis. The Lancet Infectious diseases. 2014;14(8):725–30. doi: 10.1016/S1473-3099(14)70767-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phillips G, Tam CC, Conti S, Rodrigues LC, Brown D, Iturriza-Gomara M, et al. Community Incidence of Norovirus-associated Infectious Intestinal Disease in England: Improved Estimates Using Viral Load for Norovirus Diagnosis. Am J Epidemiol. 2010;171(9):1014–22. doi: 10.1093/aje/kwq021. [DOI] [PubMed] [Google Scholar]
- 3.Harris JP, Adams NL, Lopman BA, Allen DJ, Adak GK. The development of Web-based surveillance provides new insights into the burden of norovirus outbreaks in hospitals in England. Epidemiol Infect. 2014;142(8):1590–8. doi: 10.1017/S0950268813002896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee LY, Ladner DP, Ison MG. Norovirus infection in solid organ transplant recipients: a single-center retrospective study. Transplant infectious disease : an official journal of the Transplantation Society. 2016 doi: 10.1111/tid.12622. [DOI] [PubMed] [Google Scholar]
- 5.Patel MM, Widdowson M-A, Glass RI, Akazawa K, Vinjé J, Parashar UD. Systematic Literature Review of Role of Noroviruses in Sporadic Gastroenteritis. Emerging Infectious Diseases. 2008;14(8):1224–31. doi: 10.3201/eid1408.071114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rouhani S, Penataro Yori P, Paredes Olortegui M, Siguas Salas M, Rengifo Trigoso D, Mondal D, et al. Norovirus Infection and Acquired Immunity in 8 Countries: Results From the MAL-ED Study. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2016;62(10):1210–7. doi: 10.1093/cid/ciw072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopman BA, Grassly NC. Editorial Commentary: Pediatric Norovirus in Developing Countries: A Picture Slowly Comes Into Focus. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2016;62(10):1218–20. doi: 10.1093/cid/ciw078. [DOI] [PubMed] [Google Scholar]
- 8.Kapikian AZ, Wyatt RG, Dolin R, Thornhill TS, Kalica AR, Chanock RM. Visualization by immune electron microscopy of a 27-nm particle associated with acute infectious nonbacterial gastroenteritis. Journal of virology. 1972;10(5):1075–81. doi: 10.1128/jvi.10.5.1075-1081.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, et al. Replication of human noroviruses in stem cell–derived human enteroids. Science (New York, NY) 2016 doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, Grau KR, et al. Enteric bacteria promote human and mouse norovirus infection of B cells. Science (New York, NY) 2014;346(6210):755–9. doi: 10.1126/science.1257147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karandikar UC, Crawford SE, Ajami NJ, Murakami K, Kou B, Ettayebi K, et al. Detection of human norovirus in intestinal biopsies from immunocompromised transplant patients. The Journal of general virology. 2016;97(9):2291–300. doi: 10.1099/jgv.0.000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taube S, Kolawole AO, Hohne M, Wilkinson JE, Handley SA, Perry JW, et al. A mouse model for human norovirus. mBio. 2013;4(4):00450–13. doi: 10.1128/mBio.00450-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bok K, Parra GI, Mitra T, Abente E, Shaver CK, Boon D, et al. Chimpanzees as an animal model for human norovirus infection and vaccine development. Proc Natl Acad Sci U S A. 2011;108(1):325–30. doi: 10.1073/pnas.1014577107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seo DJ, Jung D, Jung S, Ha SK, Ha SD, Choi IS, et al. Experimental miniature piglet model for the infection of human norovirus GII. Journal of medical virology. 2018;90(4):655–62. doi: 10.1002/jmv.24991. [DOI] [PubMed] [Google Scholar]
- 15.Duizer E, Schwab KJ, Neill FH, Atmar RL, Koopmans MPG, Estes MK. Laboratory efforts to cultivate noroviruses. The Journal of general virology. 2004;85(1):79–87. doi: 10.1099/vir.0.19478-0. [DOI] [PubMed] [Google Scholar]
- 16.Leung WK, Chan PK, Lee NL, Sung JJ. Development of an in vitro cell culture model for human noroviruses and its clinical application. Hong Kong Med J. 2010;16(5 Suppl 4):18–21. [PubMed] [Google Scholar]
- 17.Herbst-Kralovetz MM, Radtke AL, Lay MK, Hjelm BE, Bolick AN, Sarker SS, et al. Lack of norovirus replication and histo-blood group antigen expression in 3-dimensional intestinal epithelial cells. Emerg Infect Dis. 2013;19(3):431–8. doi: 10.3201/eid1903.121029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheetham S, Souza M, Meulia T, Grimes S, Han MG, Saif LJ. Pathogenesis of a genogroup II human norovirus in gnotobiotic pigs. Journal of virology. 2006;80(21):10372–81. doi: 10.1128/JVI.00809-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wobus C, Karst S, Thackray L, Chang K, Sosnovtsev S, Belliot G, et al. Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2004;2:e432. doi: 10.1371/journal.pbio.0020432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaudhry Y, Skinner MA, Goodfellow IG. Recovery of genetically defined murine norovirus in tissue culture by using a fowlpox virus expressing T7 RNA polymerase. The Journal of general virology. 2007;88(8):2091–100. doi: 10.1099/vir.0.82940-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arias A, Bailey D, Chaudhry Y, Goodfellow I. Development of a reverse-genetics system for murine norovirus 3: long-term persistence occurs in the caecum and colon. The Journal of general virology. 2012;93(Pt 7):1432–41. doi: 10.1099/vir.0.042176-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grau KR, Roth AN, Zhu S, Hernandez A, Colliou N, DiVita BB, et al. The major targets of acute norovirus infection are immune cells in the gut-associated lymphoid tissue. Nature Microbiology. 2017 doi: 10.1038/s41564-017-0057-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thackray LB, Wobus CE, Chachu KA, Liu B, Alegre ER, Henderson KS, et al. Murine Noroviruses Comprising a Single Genogroup Exhibit Biological Diversity despite Limited Sequence Divergence. J Virol. 2007;81(19):10460–73. doi: 10.1128/JVI.00783-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elftman MD, Gonzalez-Hernandez MB, Kamada N, Perkins C, Henderson KS, Nunez G, et al. Multiple effects of dendritic cell depletion on murine norovirus infection. The Journal of general virology. 2013;94(Pt 8):1761–8. doi: 10.1099/vir.0.052134-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez-Hernandez MB, Liu T, Blanco LP, Auble H, Payne HC, Wobus CE. Murine norovirus transcytosis across an in vitro polarized murine intestinal epithelial monolayer is mediated by M-like cells. Journal of virology. 2013;87(23):12685–93. doi: 10.1128/JVI.02378-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzalez-Hernandez MB, Liu T, Payne HC, Stencel-Baerenwald JE, Ikizler M, Yagita H, et al. Efficient norovirus and reovirus replication in the mouse intestine requires microfold (M) cells. Journal of virology. 2014;88(12):6934–43. doi: 10.1128/JVI.00204-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S, Wilen CB, Orvedahl A, McCune BT, Kim KW, Orchard RC, et al. Norovirus Cell Tropism Is Determined by Combinatorial Action of a Viral Non-structural Protein and Host Cytokine. Cell Host Microbe. 2017;22(4):449–59.e4. doi: 10.1016/j.chom.2017.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barnes D, Kunitomi M, Vignuzzi M, Saksela K, Andino R. Harnessing Endogenous miRNAs to Control Virus Tissue Tropism as a Strategy for Developing Attenuated Virus Vaccines. Cell Host & Microbe. 2008;4(3):239. doi: 10.1016/j.chom.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heiss BL, Maximova OA, Pletnev AG. Insertion of microRNA targets into the flavivirus genome alters its highly neurovirulent phenotype. Journal of virology. 2011;85(4):1464–72. doi: 10.1128/JVI.02091-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yen LC, Lin YL, Sung HH, Liao JT, Tsao CH, Su CM, et al. Neurovirulent flavivirus can be attenuated in mice by incorporation of neuron-specific microRNA recognition elements into viral genome. Vaccine. 2011;18:18. doi: 10.1016/j.vaccine.2011.09.102. [DOI] [PubMed] [Google Scholar]
- 31.Ylosmaki E, Martikainen M, Hinkkanen A, Saksela K. Attenuation of Semliki Forest virus neurovirulence by microRNA-mediated detargeting. Journal of virology. 2013;87(1):335–44. doi: 10.1128/JVI.01940-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez JT, Pham AM, Lorini MH, Chua MA, Steel J, tenOever BR. MicroRNA-mediated species-specific attenuation of influenza A virus. Nature biotechnology. 2009;27(6):572–6. doi: 10.1038/nbt.1542. [DOI] [PubMed] [Google Scholar]
- 33.Pham AM, Langlois RA, tenOever BR. Replication in Cells of Hematopoietic Origin Is Necessary for Dengue Virus Dissemination. PLOS Pathogens. 2012;8(1):e1002465. doi: 10.1371/journal.ppat.1002465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelly EJ, Hadac EM, Greiner S, Russell SJ. Engineering microRNA responsiveness to decrease virus pathogenicity. Nat Med. 2008;14(11):1278–83. doi: 10.1038/nm.1776. [DOI] [PubMed] [Google Scholar]
- 35.Kelly EJ, Nace R, Barber GN, Russell SJ. Attenuation of vesicular stomatitis virus encephalitis through microRNA targeting. Journal of virology. 2010;84(3):1550–62. doi: 10.1128/JVI.01788-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee CYF, Rennie PS, Jia WWG. MicroRNA Regulation of Oncolytic Herpes Simplex Virus-1 for Selective Killing of Prostate Cancer Cells. Clinical Cancer Research. 2009;15(16):5126–35. doi: 10.1158/1078-0432.CCR-09-0051. [DOI] [PubMed] [Google Scholar]
- 37.Ylosmaki E, Lavilla-Alonso S, Jaamaa S, Vaha-Koskela M, af Hallstrom T, Hemminki A, et al. MicroRNA-mediated suppression of oncolytic adenovirus replication in human liver. PloS one. 2013;8(1):22. doi: 10.1371/journal.pone.0054506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proceedings of the National Academy of Sciences. 2007;104(5):1604–9. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang P, Hou J, Lin L, Wang C, Liu X, Li D. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J Immunol. 2010;185(10):6226–33. doi: 10.4049/jimmunol.1000491. [DOI] [PubMed] [Google Scholar]
- 40.Rom S, Rom I, Passiatore G, Pacifici M, Radhakrishnan S, Del Valle L, et al. CCL8/MCP-2 is a target for mir-146a in HIV-1-infected human microglial cells. The FASEB Journal. 2010;24(7):2292–300. doi: 10.1096/fj.09-143503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Witwer KW, Sisk JM, Gama L, Clements JE. MicroRNA regulation of IFN-beta protein expression: rapid and sensitive modulation of the innate immune response. J Immunol. 2010;184(5):2369–76. doi: 10.4049/jimmunol.0902712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McFadden N, Bailey D, Carrara G, Benson A, Chaudhry Y, Shortland A, et al. Norovirus Regulation of the Innate Immune Response and Apoptosis Occurs via the Product of the Alternative Open Reading Frame 4. PLoS Pathog. 2011;7(12):e1002413. doi: 10.1371/journal.ppat.1002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thorne L, Bailey D, Goodfellow I. High-resolution functional profiling of the norovirus genome. Journal of virology. 2012;86(21):11441–56. doi: 10.1128/JVI.00439-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCune BT, Tang W, Lu J, Eaglesham JB, Thorne L, Mayer AE, et al. Noroviruses Co-opt the Function of Host Proteins VAPA and VAPB for Replication via a Phenylalanine-Phenylalanine-AcidicTract-Motif Mimic in Nonstructural Viral Protein NS1/2. mBio. 2017;8(4) doi: 10.1128/mBio.00668-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKenna LB, Schug J, Friedman JR, McKenna JB, Kaestner KH. MicroRNAs Control Intestinal Epithelial Differentiation, Architecture, and Barrier Function. Gastroenterology. 2010;139(5):1654–64.e1. doi: 10.1053/j.gastro.2010.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nice TJ, Baldridge MT, McCune BT, Norman JM, Lazear HM, Artyomov M, et al. Interferon lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science (New York, NY) 2014;27 doi: 10.1126/science.1258100. 1258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baldridge MT, Nice TJ, McCune BT, Yokoyama CC, Kambal A, Wheadon M, et al. Commensal microbes and interferon-lambda determine persistence of enteric murine norovirus infection. Science (New York, NY) 2015;347(6219):266–9. doi: 10.1126/science.1258025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nice TJ, Strong DW, McCune BT, Pohl CS, Virgin HW. A single-amino-acid change in murine norovirus NS1/2 is sufficient for colonic tropism and persistence. Journal of virology. 2013;87(1):327–34. doi: 10.1128/JVI.01864-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karst S, Wobus C, Lay M, Davidson J, Virgin H. STAT1-dependent innate immunity to a Norwalk-like virus. Science (New York, NY) 2003;299:1575–8. doi: 10.1126/science.1077905. [DOI] [PubMed] [Google Scholar]
- 51.Mumphrey SM, Changotra H, Moore TN, Heimann-Nichols ER, Wobus CE, Reilly MJ, et al. Murine norovirus 1 infection is associated with histopathological changes in immunocompetent hosts, but clinical disease is prevented by STAT1-dependent interferon responses. Journal of virology. 2007;81(7):3251–63. doi: 10.1128/JVI.02096-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bailey D, Karakasiliotis I, Vashist S, Chung LMW, Reese J, McFadden N, et al. Functional Analysis of RNA Structures Present at the 3' Extremity of the Murine Norovirus Genome: the Variable Polypyrimidine Tract Plays a Role in Viral Virulence. J Virol. 2010;84(6):2859–70. doi: 10.1128/JVI.02053-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langlois RA, Albrecht RA, Kimble B, Sutton T, Shapiro JS, Finch C, et al. MicroRNA-based strategy to mitigate the risk of gain-of-function influenza studies. Nature biotechnology. 2013;31(9):844–7. doi: 10.1038/nbt.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brown BD, Gentner B, Cantore A, Colleoni S, Amendola M, Zingale A, et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nature biotechnology. 2007;25(12):1457–67. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 55.Orchard RC, Wilen CB, Doench JG, Baldridge MT, McCune BT, Lee YC, et al. Discovery of a proteinaceous cellular receptor for a norovirus. Science (New York, NY) 2016;353(6302):933–6. doi: 10.1126/science.aaf1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guix S, Asanaka M, Katayama K, Crawford SE, Neill FH, Atmar RL, et al. Norwalk virus RNA is infectious in mammalian cells. Journal of virology. 2007;81(22):12238–48. doi: 10.1128/JVI.01489-07. [DOI] [PMC free article] [PubMed] [Google Scholar]