Abstract
Kwashiorkor and marasmus are considered to be two different clinical diseases resulting from severe malnutrition, but this distinction has been questioned. In a previous study comparing children with kwashiorkor and healthy children from Niger and Senegal, we found a dramatic gut microbiota alteration with a predominant depletion of anaerobes and enrichment in Proteobacteria and Fusobacteria in kwashiorkor. However, it remained unknown whether this association was related to malnutrition or was a specific feature of kwashiorkor. In this continuation study, we added 7 new marasmus subjects and 71,162 new colonies from the same countries. Our results showed that, compared to marasmus, the kwashiorkor gut microbiota was characterized by an increased proportion of Proteobacteria (culturomics, Marasmus 5.0%, Kwashiorkor 16.7%, p < 0.0001; metagenomics, Marasmus 14.7%, Kwashiorkor 22.0%, p = 0.001), but there was a decreased proportion of Bacteroidetes in marasmus (culturomics, Marasmus 0.8%, Kwashiorkor 6.5%, p = 0.001; metagenomics, Marasmus 5.4%, Kwashiorkor 7.0%, p = 0.03). Fusobacterium was more frequently cultured from kwashiorkor. All detected potential pathogenic species were enriched in the kwashiorkor gut microbiota. These results provide a biological basis to support the usage of an antibiotic therapy more effective in suppressing the overgrowth of bacterial communities resistant to penicillin, combined with antioxidants and probiotics for nutritional recovery therapies, particularly for kwashiorkor.
Subject terms: Microbiome, Nutrition
Introduction
Child malnutrition is a global health challenge and despite recent progress, is responsible for 45% of under-five mortality1. In addition to adequate water, protein and energy intake, the discovery of the critical importance of micronutrients and the generalization and application of World Health Organization (WHO) standardized protocols, including ready-to-use therapeutic foods and antibiotics, have largely contributed to recent achievements. However, new therapeutic options are warranted because 10–30% of children with complicated severe acute malnutrition (SAM) do not respond to optimal WHO-compliant management in randomized controlled studies2,3.
Among the various diseases associated with malnutrition, there is controversy over the definition of marasmus and kwashiorkor as two distinct diseases. Indeed, the WHO criteria no longer distinguish between the treatment offered for the two diseases and considers that although the different forms of malnutrition can be distinguished by the presence of oedema, the treatment does not address the pathophysiological differences. According to the WHO, SAM is defined in children under five years of age by a weight-for-height z-score (WHZ) below −3 standard deviations (SD) and/or bilateral oedema and/or, only in children over six months of age, a mid-upper arm circumference (MUAC) < 115 mm4. While an epidemiological approach is effective for identifying public health issues to guide international policy decisions and humanitarian financial investments, it may have limited ability to resolve different disease entities. A clinical approach may more efficiently detect different syndromes associated with different mechanisms, prognosis and most importantly require different treatment for optimum case management.
Kwashiorkor is such a clinical disease characterised by oedema and first named by Cicely D. Williams in 19335. Studies have consistently reported distinctive clinical features supporting the notion that kwashiorkor is different from marasmus (Supplementary Table S1A), but children can have both conditions (marasmic-kwashiorkor). Kwashiorkor appears with an acute onset four to twelve months after the onset of defective feeding, while the patient appears to develop normally. Irritability then begins, accompanied by attacks of diarrhoea, swelling of the hands and feet, ‘moon face’, depigmented hair and skin changes (depigmentation, thickened black and crumpled patches). Mucous membranes are frequently inflamed and ulcerated, and infection is probably ubiquitous. There is usually extreme fatty infiltration of the liver. Meanwhile marasmus can occur at any age and is characterized by a loss of muscle mass and subcutaneous fat5.
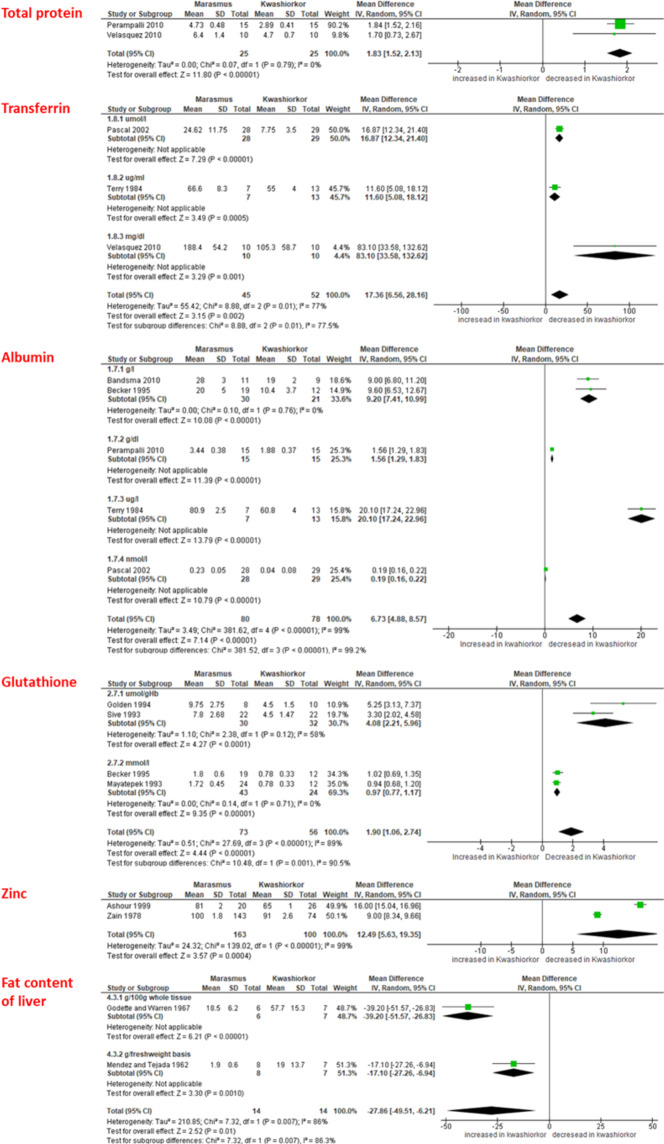
In a quantitative meta-analysis of published comparative studies of marasmus and kwashiorkor6–18 (Fig. 1, Supplementary Table S1B, Supplementary Fig. S1), we found that kwashiorkor was consistently associated with a decrease in zinc, glutathione, total protein, albumin and transferrin, but ferritin consistently tended to increase (Supplementary Fig. 1B). While a descending gradient from controls to patients with marasmus and kwashiorkor could be observed for minerals, antioxidants and protein levels; the massive increase in liver fat was only observed with kwashiorkor, confirmed by the meta-analysis. This suggests a specific metabolic process is at play in kwashiorkor, but not in marasmus, in the context of a severely restricted and inadequate diet.
Figure 1.
Meta-analysis results of studies analysing the differences in intermediary metabolism patterns between KW and MRS (only significant results are shown).
Based on recent findings19, gut dysbiosis may be a discriminant feature between kwashiorkor and marasmus. Gut microbiota alteration was found to be instrumental in weight loss and malnutrition20. An impoverished microbiota and a predominant loss of strict anaerobes associated with an increased gut redox potential was observed in children with kwashiorkor21. Strikingly, non-alcoholic steatohepatitis (NASH) in adults is another disease associated with both an unhealthy diet and fatty liver. In this disease, fatty liver has been linked to gut dysbiosis; specifically, a Proteobacteria, Enterobacteriaceae, and particularly Escherichia enrichment associated with endogenous ethanol22. Similar mechanisms may be at play in kwashiorkor pathogenesis and could impact the choice of antibiotics used in kwashiorkor management.
In this study, we investigated whether alterations to the gut microbiota in healthy children, and in children diagnosed with marasmus or kwashiorkor exhibited a gradient that paralleled the decreasing mineral, antioxidant, and protein levels observed in our meta-analysis. We also expected that gut dysbiosis in kwashiorkor could be a potential key factor in fatty liver pathogenesis, as previously reported for NASH.
As no children with marasmus were included in our previous study23, it was not clear whether this association was related to malnutrition or if this was a specific feature of kwashiorkor. In this culturomics study, we added 7 new marasmus subjects corresponding to 71,162 new colonies to clarify the gut microbiota variations between marasmus and kwashiorkor. Metagenomics was used as a confirmatory technique.
Results
Gut microbial diversity revealed a decreasing gradient from controls to children with marasmus and kwashiorkor
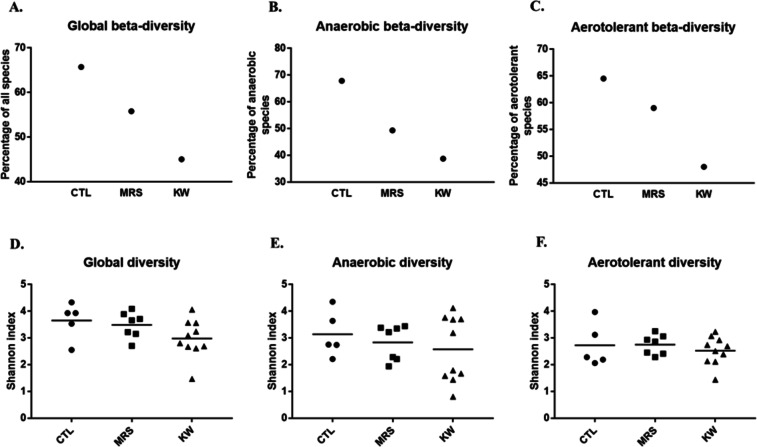
A total of 22 samples from five healthy controls, seven marasmus and ten kwashiorkor cases were analysed by using 18 culture conditions. Culturomics yielded 276,070 colonies, allowing us to identify 281 bacterial species in healthy controls (CTL), 224 species in marasmus (MRS) and 335 species in kwashiorkor (KW). The β-diversity dramatically decreased from controls (ratio of the number of unique species on total species in each group (U/T ratio) – see methods) at 65.7%, to marasmus at 55.8% and kwashiorkor at 45.0%. The differences between the three groups were significant (uncorrected chi-square test: CTL-MRS, p = 0.02; CTL-KW, p = 2.5*10−7; MRS-KW, p = 0.013; Table 1). In addition, we identified 44 species that were previously unknown to be present in the human gut, including 20 putative new species (Supplementary Table S3), 12 species previously unknown to be present in humans and only two species that were previously known to be present in humans but were not gut-associated (Table 1). A decreasing gradient was observed from CTL to MRS and KW, significant for β-diversity (U/T ratio - CTL > MRS > KW; Cochran-Armitage trend test, p = 0.0002; Fig. 2A), but not significant for the hitherto unknown bacteria’s diversity (Supplementary Fig. S2).
Table 1.
Descriptive results of culturomics in healthy controls and patients with marasmus and kwashiorkor.
| Mean ± SD/Proportions | Comparison (p-value) | ||||||
|---|---|---|---|---|---|---|---|
| CTL (n = 5) | MRS (n = 7) | KW (n = 10) | CTL-MRS | CTL-KW | MRS-KW | CTL-MRS-KW | |
| β diversity (proportions) | |||||||
| All species (Ratio U/T) | 185/281 | 125/224 | 151/335 | 0.02d | 2.5*10−7d | 0.013d | 0.0002c |
| 65.71% | 55.80% | 45% | |||||
| Anaerobic species | 76/112 | 36/73 | 43/111 | 0.012d | 0.00001d | 0.15d | 0.0002c |
| 67.85% | 49.31% | 38.73% | |||||
| Aerotolerant species | 109/169 | 89/151 | 108/224 | 0.3d | 0.001d | 0.041d | 0.001c |
| 64.50% | 59% | 48% | |||||
| Global diversity (mean ± SD) | |||||||
| Number of phyla | 3.8 ± 0.4 | 3.4 ± 0.8 | 4.2 ± 0.6 | 0.14a | 0.2a | 0.03b | No gradient |
| Number of genera | 34 ± 12 | 22 ± 5 | 36 ± 7 | 0.06b | 0.67a | 0.003b | No gradient |
| Total number of species | 92 ± 20 | 67 ± 14 | 89 ± 22 | 0.07a | 0.82a | 0.03b | No gradient |
| Number of putative new species | 5 ± 3 | 3 ± 2 | 2 ± 1 | 0.22a | 0.009b | 0.28a | 0.06e |
| Number of non-human species | 2.8 ± 0.4 | 1.6 ± 1.7 | 1.5 ± 1.2 | 0.23a | 0.02a | 0.92a | 0.28e |
| Phyla distribution (proportions of each phylum in a group) | |||||||
| Actinobacteria | 42/281 | 12/224 | 47/335 | 0.001d | 0.74d | 0.001d | No gradient |
| 15% | 5.40% | 13.70% | |||||
| Bacteroidetes | 21/281 | 2/224 | 21/335 | 0.0004d | 0.55d | 0.001d | No gradient |
| 7.50% | 0.80% | 6.50% | |||||
| Proteobacteria | 25/281 | 11/224 | 56/335 | 0.082d | 0.004d | 2.5*10−5d | No gradient |
| 8.90% | 5% | 16.70% | |||||
| Firmicutes | 192/281 | 198/224 | 208/335 | <10−7d | 0.1d | <10−7d | No gradient |
| 68.30% | 88.40% | 61.70% | |||||
| Fusobacteria | 0/281 | 1/224 | 3/335 | 0.88f | 0.32f | 0.53d | No gradient |
| 0% | 0.40% | 0.80% | |||||
aTwo-tailed Student’s t-test; bMann-Whitney test; cCochran-Armitage trend test; duncorrected chi-square test; elinear regression test; fFisher’s exact test. U/T: unique/total species ratio. New species: unknown species identified for the first time. Non-human species: previously known species identified in human for the first time.
Figure 2.
Analysis of gut microbial diversity revealed a decreasing gradient from controls to children with marasmus and kwashiorkor. Scatter plots visually display gradients across the 3 groups in descending order, from controls (CTL) to children with marasmus (MRS) to kwashiorkor (KW). (A–C) Culturomics: A/global β-diversity (U/T ratio, see methods), Cochran-Armitage trend test, p = 0.0002; B/anaerobic β-diversity, p = 0.0002; C/aerotolerant β-diversity, p = 0.001. (D–F) Metagenomics. The decreasing gradient was significant for culturomics results (A–C) but not for metagenomics results (D–F).
With metagenomics, a total of 1,842,831 reads, 1,960,894 reads and 2,933,416 reads were generated in control, marasmus and kwashiorkor samples, respectively. There was no significant difference in the number of raw reads between the 3 groups (Mean ± SD, CTL 368,566 ± 148,491; MRS 280,128 ± 28,855; KW 293,342 ± 145,673, one-way ANOVA test, p = 0.44; Supplementary Table S2). The number of reads not assigned to a prokaryotic species was 101,848 ± 82,799 for controls, 24,134 ± 20,848 for marasmus and 14,954 ± 12,661 for kwashiorkor (Supplementary Table S2). The difference was significant (one-way ANOVA test, p = 0.003) with a linear trend (CTL > MRS > KW, post-test for linear trend, p = 0.001). In addition, a clear gradient (CTL > MRS > KW) was observed for the mean Shannon index (Mean ± SD): CTL 3.65 ± 0.67, MRS 3.48 ± 0.48 and KW 2.97 ± 0.7 (Fig. 2D); however, no significant differences were found between the three groups and the test for trend was not significant (Table 2). This may be due to the limited sample size compared with the culturomics analysis.
Table 2.
Descriptive results of metagenomics in healthy controls and patients with marasmus and kwashiorkor.
| Mean ± SD/Proportions | Comparison (p-value) | ||||||
|---|---|---|---|---|---|---|---|
| CTL (n = 5) | MRS (n = 7) | KW (n = 10) | CTL-MRS | CTL-KW | MRS-KW | CTL-MRS-KW | |
| Global diversity (mean Shannon index by group) | |||||||
| Global diversity | 3.65 ± 0.67 | 3.48 ± 0.48 | 2.97 ± 0.7 | 0.63a | 0.1a | 0.12a | 0.12e |
| Anaerobic diversity | 3.1 ± 0.8 | 2.8 ± 0.6 | 2.57 ± 1.2 | 0.49a | 0.37a | 0.61a | 0.58e |
| Aerotolerant diversity | 2.7 ± 0.8 | 2.7 ± 0.36 | 2.5 ± 0.5 | 0.94a | 0.57a | 0.34a | 0.67e |
| Phylum distribution (proportions of each phylum in a group) | |||||||
| Actinobacteria | 61/486 | 75/482 | 86/589 | 0.68d | 0.37d | 0.55d | No gradient |
| 12% | 16% | 14% | |||||
| Bacteroidetes | 46/486 | 26/482 | 44/589 | 0.0011d | 0.24d | 0.03d | No gradient |
| 9% | 5.40% | 7% | |||||
| Proteobacteria | 75/486 | 86/482 | 131/589 | 0.76d | 0.004d | 0.001d | No gradient |
| 15% | 14.77% | 22% | |||||
| Firmicutes | 296/486 | 285/482 | 319/589 | 9.6*10−5d | 0.026d | 0.075d | No gradient |
| 61% | 49% | 54% | |||||
| Fusobacteria | 1/486 | 2/482 | 7/589 | 0.67d | 0.062d | 0.097d | No gradient |
| 0.20% | 0.34% | 1.20% | |||||
| Cyanobacteria | 0/486 | 1/482 | 0/589 | 0.54f | NA | 0.49f | No gradient |
| 0% | 0.17% | 0% | |||||
| Verrucomicrobia | 1/486 | 1/482 | 1/589 | 0.89d | 0.89d | 0.99d | No gradient |
| 0.20% | 0.17% | 0.16% | |||||
| Chloroflexi | 0/486 | 0/482 | 1/589 | NA | 0.36f | 0.50f | No gradient |
| 0% | 0% | 0.16% | |||||
| Euryarchaeota | 4/486 | 0/482 | 0/589 | 0.085f | 0.082f | NA | No gradient |
| 0.80% | 0% | 0% | |||||
| Lentisphaerae | 1/486 | 0/482 | 0/589 | 0.91f | 0.90f | NA | No gradient |
| 0.20% | 0% | 0% | |||||
| Tenericutes | 1/486 | 0/482 | 0/589 | 0.91f | 0.9f | NA | No gradient |
| 0.20% | 0% | 0% | |||||
aTwo-tailed Student’s t-test; bMann-Whitney test; cCochran-Armitage trend test; duncorrected chi-square test; elinear regression test; fFisher’s exact test; NA: non-available.
The gradient was more pronounced for anaerobic than for aerotolerant diversity
By culturomics, anaerobic β-diversity assessed by the U/T ratio was 67.85% for CTL, 49.31% for MRS and 38.73% for KW, with a significant difference between groups (uncorrected chi-square test: CTL-MRS, p = 0.012; CTL-KW, p = 0.00001). In parallel, for aerotolerant species, the U/T ratio was 64.5% for CTL, 59% for MRS and 48% for KW, with a significant difference between groups (uncorrected chi-square test: CTL-KW, p = 0.001; MRS-KW, p = 0.041) (Table 1). Interestingly, we also found a significant decreasing trend in anaerobic diversity (Cochran-Armitage trend test, p = 0.0002), which was greater and more significant than the decrease in aerotolerant diversity (Cochran-Armitage trend test, p = 0.001) (Fig. 2B,C).
The data obtained from our metagenomic results showed the same decreasing gradient (Fig. 2E,F) in anaerobic diversity (mean Shannon index: CTL 3.1 ± 0.8, MRS 2.8 ± 0.6 and KW 2.57 ± 1.2). For aerotolerant diversity, no difference was noted (CTL 2.7 ± 0.8, MRS 2.7 ± 0.36 and KW 2.5 ± 0.5). None of the differences were statistically significant (Table 2).
Globally, the decrease in diversity was more pronounced and significant for anaerobic than for aerotolerant species in both the culturomics and metagenomics results (Supplementary Fig. S3A,B).
Principal coordinate analysis
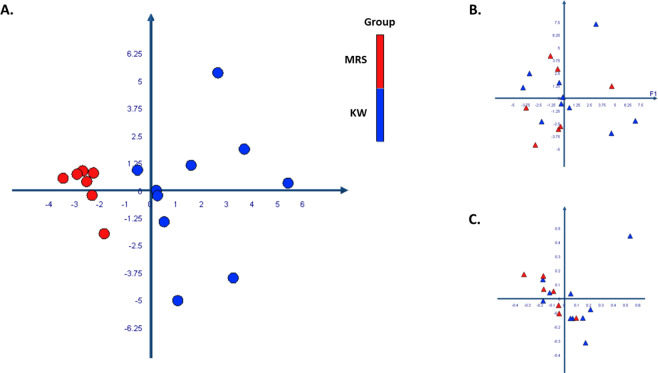
We have performed a PCoA analysis to test whether the microbiota profile analysed by culturomics or metagenomics discriminated individuals based on their nutritional status, age, gender and geographical origin. The results showed that only culturomics clearly distinguished between children with marasmus and kwashiorkor (Fig. 3A), regardless of their age, sex or geographical origin (Supplementary Fig. S4). On the other hand, the analysis of microbiota by metagenomics was not able to discriminate between the different groups regarding the detection or the relative abundance of each species (Fig. 3B,C). For other clinical variables, metagenomics yielded age and geographical origin clustering but not the nutritional status. This was found only by the detection of each species but not by relative abundance (Supplementary Fig. S4). By adding the controls in the analysis, we observed that in culturomics, the CTL formed a distinct group from MRS but was mixed with KW (Supplementary Fig. S5).
Figure 3.
PCoA analysis of gut microbiota diversity among kwashiorkor and marasmus subjects. The 3 charts are allowing us to visualize the proximities between kwashiorkor and marasmus subjects. For culturomics (A), there was a clear distance between 2 groups (KW and MRS) In metagenomics, analysis based on the relative abundance (B) and on the detection (C) of each species in each group showed that the clustering was not observed. The colour coded as red for MRS group and blue for KW group.
Paucity of Bacteroidetes in marasmus samples and bloom of Proteobacteria and Fusobacteria in kwashiorkor samples
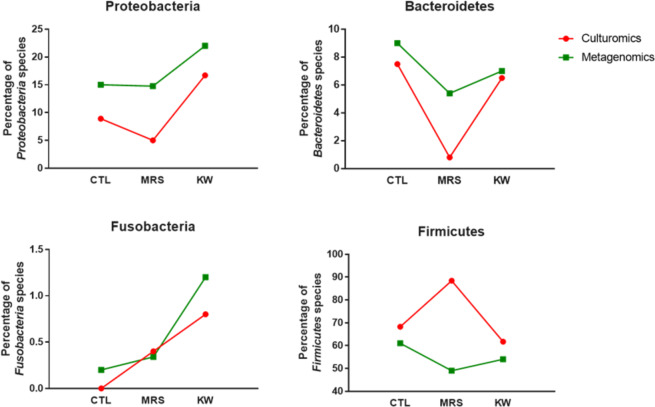
The culturomics results for marasmus samples identified a total of 5 bacterial phyla containing 224 species, including 198 Firmicutes (88.40%), 12 Actinobacteria (5.40%), 11 Proteobacteria (5%), 2 Bacteroidetes (0.80%) and only 1 Fusobacteria (0.40%). Focusing on Proteobacteria (CTL 8.90%, MRS 5% and KW 16.70%) and Bacteroidetes (CTL 7.50%, MRS 0.80% and KW 6.50%), we found a significant increase in Proteobacteria in kwashiorkor (uncorrected chi-square test: CTL-KW, p = 0.004; MRS-KW, p = 2.5*10−5) and a significant decrease in Bacteroidetes in marasmus (uncorrected chi-square test: CTL-MRS, p = 0.0004; MRS-KW, p = 0.001; Table 1). Strikingly, no Fusobacteria species were isolated from CTL, although one and three Fusobacteria species were isolated from MRS and KW, respectively.
The metagenomics results confirmed the changes in Proteobacteria and Bacteroidetes found by culturomics (Fig. 4). There was a significant increase in Proteobacteria in kwashiorkor samples (CTL 15%, MRS 14.77% and KW 22%; uncorrected chi-square test: CTL-KW, p = 0.004; MRS-KW, p = 0.001) and a significant decrease in Bacteroidetes in marasmus samples (CTL 9%, MRS 5.40% and KW 7%; uncorrected chi-square test: CTL-MRS, p = 0.0011; MRS-KW, p = 0.03; Table 2). Metagenomics also confirmed the increase in Fusobacteria species in kwashiorkor samples (CTL 1/486 versus KW 7/589, unilateral mid-P exact test: p = 0.034) but not in marasmus samples (Table 2).
Figure 4.
Phylum distribution in culturomics and metagenomics. Culturomics and metagenomics results confirmed a significant increase in Proteobacteria in kwashiorkor (uncorrected chi-square test, CTL-KW, MRS-KW, p < 0.001) as well as a significant decrease in Bacteroidetes in marasmus (uncorrected chi-square test, CTL-MRS, MRS-KW, p < 0.0005). Strikingly, the increased Fusobacteria diversity in kwashiorkor observed by metagenomics was also confirmed in kwashiorkor, but not in marasmus, by culturomics (uncorrected chi-square test, CTL-KW, p = 0.004).
Kwashiorkor microbial diversity demonstrated greater numbers of pathogenic species than marasmus microbial diversity
To more accurately characterize the species that were enriched or depleted in patients with kwashiorkor versus marasmus, we choose 30 bacterial species exhibiting the largest differences in relative frequency (culturomics) and relative abundance (metagenomics) between these two groups.
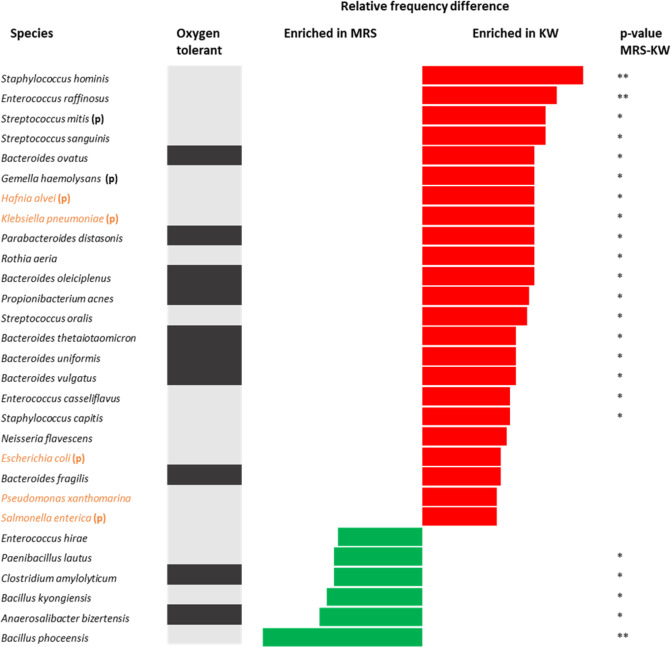
With culturomics, Fig. 5 show that there were 5 Proteobacteria species four of which were potential pathogenic and two aerobic pathogens enriched in kwashiorkor. All the potential pathogenic species were aerobic. There were no potential pathogenic species among the bacteria enriched in the marasmic children (One-Sample test for binomial proportion: p = 0.025 for the Proteobacteria proportion, p = 0.014 for the potential pathogenic species proportion).
Figure 5.
Increased frequency of species in kwashiorkor and marasmus samples identified by the culturomics approach. Each bar represents the relative difference for each species, with red bars representing an increased frequency in patients with kwashiorkor and green bars representing an increased frequency in patients with marasmus. The grey bars represent oxygen tolerant species and the black bars represent obligate anaerobes. Proteobacteria (in orange) and potential pathogenic bacteria (p) were enriched in kwashiorkor but not in marasmus. **p-value ranging from 0.001–0.01; *p-value ranging from 0.01–0.05 (two-tailed chi-square test, Fisher’s exact test).
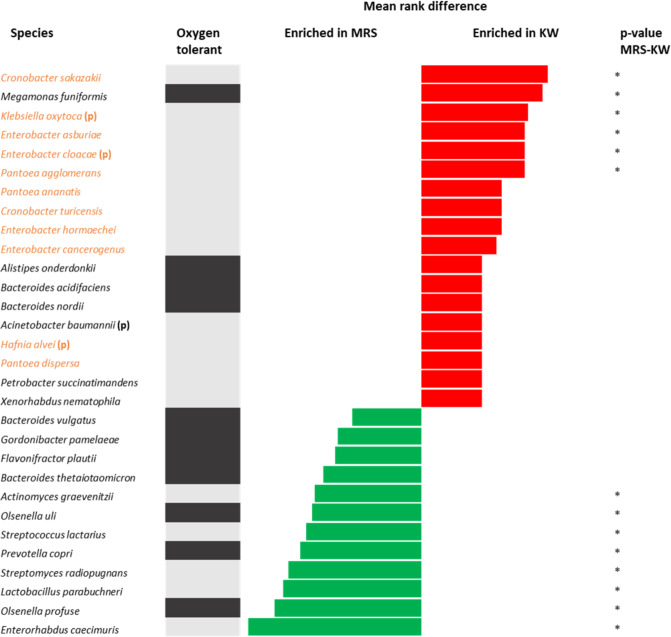
According to the metagenomics results, 18 species were enriched in kwashiorkor samples and 12 species in marasmus samples (Fig. 6). There were 11 Proteobacteria species three of which were potential pathogenic and one further aerobic pathogen enriched in kwashiorkor. There were no Proteobacteria species and no potential pathogenic bacteria enriched in marasmus (One-Sample test for binomial proportion: p = 0.00091 for Proteobacteria, p = 0.045 for pathogenic species). Accordingly, culturomics (6 species: Escherichia coli, Gemella haemolysans, Hafnia alvei, Klebsiella pneumoniae, Salmonella enterica, Streptococcus mitis) was better than metagenomics (4 species: Acinetobacter baumanii, Enterobacter cloacae, Hafnia alvei, Klebsiella oxytoca) for the detection of pathogenic bacterial enrichment in kwashiorkor. In marked contrast no potential pathogens were enriched by either technique from marasmus. Thus, both culturomics and metagenomics results confirmed higher numbers of Proteobacteria and potential pathogenic species, mainly belonging to the Gammaproteobacteria and Bacilli families, in the guts of children with kwashiorkor than in children with marasmus.
Figure 6.
Increased frequency of species in kwashiorkor and marasmus samples identified by the metagenomics approach. Mean rank values of species representing differences in relative abundance were compared between kwashiorkor and marasmus samples. The red bars represent an increased abundance in patients with kwashiorkor, while the green bars represent an increased abundance in Marasmus. The grey bars represent oxygen tolerant species and the black bars represent obligate anaerobes. Proteobacteria (in orange) and potential pathogenic bacteria (p) were enriched in kwashiorkor but not in marasmus. *p-value ranging from 0.01–0.05 (Mann-Whitney test).
In addition, a similar comparison at species level between marasmus and control groups (Supplementary Figs S6, S7) confirmed the proliferation of Proteobacteria and potential pathogenic species was only observed in children with kwashiorkor.
Discussion
Whether kwashiorkor and marasmus are two different diseases, or a continuum of a single entity has, in the past been subject to controversy. However, two separate diseases, like many other conditions, can occur simultaneously in the same child. A mixed clinical presentation is then referred to as marasmic-kwashiorkor. In general, such children have a worse prognosis (23% mortality without antibiotics) than that of children with isolated marasmus (8% mortality) or isolated kwashiorkor (5% mortality)2. Despite the differences in clinical features (Supplementary Table S1A), there is a marked overlap between the biological features of kwashiorkor and marasmic patients (Supplementary Table S1B), making it hard to pin-point specific features that are unique to kwashiorkor or marasmus. In addition, inconsistent results have led to confusion in differentiating kwashiorkor and marasmus. Our quantitative meta-analysis of published comparative studies of marasmus and kwashiorkor (Fig. 1, Supplementary Table S1B, Supplementary Fig. S1) showed that, compared to marasmus, kwashiorkor was consistently associated with the depletion of several plasmatic components, including proteins (total protein, albumin, transferrin), antioxidant molecules (glutathione), and minerals (zinc). For the first time, to our knowledge, these results reconcile two opposing visions of protein deficiency and oxidative stress. Moreover, albumin and transferrin are major antioxidants in plasma24,25. The depletion of other antioxidant molecules such as vitamin A and C in kwashiorkor was demonstrated as early as 195826. Although oedema is the main clinical feature of kwashiorkor, fatty liver is not included in the clinical definition, nevertheless it is a ubiquitous feature26. Based on recent findings in NASH, a candidate mechanism for alcoholic fatty liver is the gut microbiota alteration22. Overall, gut dysbiosis appears to be an instrumental factor in the hitherto unknown pathophysiological process resulting in both fatty liver and oedema in kwashiorkor. This prompted us to investigate the gut microbiota by both culturomics and metagenomics, which are the two major approaches for characterization of the human microbiota27 in both marasmus and kwashiorkor.
In this study, we observed a depletion in global diversity due to anaerobic and aerotolerant diversity depletion in the gut microbiota of children with both kwashiorkor and marasmus. The depletion was more profound in kwashiorkor than marasmus and affected anaerobic species to a great extent than aerobic species. It is noteworthy that Methanobrevibacter smithii, the most oxygen-sensitive prokaryote of the human gut, was not detected in children with SAM23. Our results not only confirmed alteration of the gut microbiota, especially anaerobic diversity, in SAM children, as reported in previous studies21,23, but also showed a decreasing gradient of diversity (CTL > MRS > KW). The unknown microbial abundance (unassigned, not yet cultivated) was 5 times higher in controls than in children with marasmus. This confirms the loss of diversity. This had already been found in our previous study comparing controls and children with kwashiorkor23. We have shown here a gradient that confirms a loss of diversity in malnutrition and that it is even more important in the case of kwashiorkor.
Our findings led us to propose a theory linking oxidative stress seen in children with kwashiorkor, and to a lesser extent in marasmus, to the depletion of anaerobic diversity in children with marasmus and kwashiorkor. These results were consistent with the lower levels of glutathione, minerals and protein observed in our meta-analysis. On the one hand the gut microbiome is a potential source of hepato-toxins, other noxious products and pro-oxidants. On the other hand, a lack of dietary anti-oxidants would make intestinal anaerobic species vulnerable to oxidative damage.
In our study, children were included and assigned to the marasmus and kwashiorkor groups mainly based on the clinical assessment made during the hospital admission (Supplementary Table S4). Indeed, we deliberately choose to assess the pathophysiological mechanisms of these clinically different diseases and not malnutrition severity (moderate versus severe). However, we observed a decreasing gradient from controls to marasmus and kwashiorkor in our meta-analysis for minerals, antioxidants and protein levels (Fig. 1, Supplementary Table S1B, Supplementary Fig. S1). All these findings suggested that nutritional status in children with marasmus may be intermediate to that of healthy children and those with kwashiorkor.
Furthermore, the clustering analysis of culturomics results (Fig. 3A) showed that there was a microbiota specific to each form of malnutrition, and confirmed the difference between kwashiorkor and marasmus. Particularly, the group of 7 children with marasmus gathered in culturomics (Supplementary Fig. S4) included children of different ages, genders and origins. However, the clustering was not observed in metagenomics (Figs 3B,C, S4). These results suggest that culturomics can be a discriminant tool to distinguish the microbiota between kwashiorkor and marasmus, without being biased, neither by age, sex nor by origin.
To better describe the difference between culturomics and metagenomics approaches, we have performed a series of Venn diagram comparing the taxa identified in each group (CTL, MRS, and KW) by culturomics and metagenomics (Supplementary Fig. S8). The result showed there were only 16.5%, 11.4% and 18.5% of species (for all species identified in each CTL, MRS and KW groups, respectively) that were shared between culturomics and metagenomics approaches.
On the other hand, both our culturomics and metagenomics results showed that the potentially pathogenic taxa Proteobacteria and Fusobacteria were specifically increased in kwashiorkor, while the suppression of beneficial taxa in Bacteroidetes occurred only in marasmus. Thus, we confirmed the complementarity of culturomics and metagenomics approaches (Fig. 4). We observed the proliferation of many Enterobacteriaceae species in kwashiorkor, such as Escherichia coli, Klebsiella oxytoca, Klebsiella pneumoniae and Hafniaceae, including Hafnia alvei (Figs 5, 6). These species are able to produce β-lactamase to resist penicillin28,29, suggesting that penicillin may be suboptimal for the management of kwashiorkor30. Indeed, Trehan et al. demonstrated that children receiving cefdinir (a third-generation cephalosporin) had a higher nutritional recovery rate and lower mortality rate than those who received amoxicillin2. Further studies are necessary to confirm the results of Trehan; if confirmed, cephalosporin should replace amoxicillin in the standard care regimen for severe acute malnutrition, particularly in children with kwashiorkor. Strikingly, a meta-analysis of studies analysing the treatment with antibiotics in malnourished children2,3 showed that antibiotics had a different effect on survival in children with kwashiorkor or without kwashiorkor (Supplementary Fig. S9). More specifically, antibiotics highly reduced mortality in children with kwashiorkor and marasmic kwashiorkor but had no effect on the mortality in children with marasmus. The results suggested a difference in ecosystems between children with kwashiorkor and those without kwashiorkor.
In addition, we observed the abnormal depletion of Bacteroidetes in marasmus. This phylum includes the enterotype leaders Bacteroides and Prevotella, which are positively correlated with the fiber and polysaccharides intake in the diet of rural populations31–38.
Our results not only helped us pinpoint a potential specific feature to distinguish kwashiorkor and marasmus but also may contribute to the development of nutritional therapies for children with severe malnutrition. Undoubtedly, diet components such as fibre, fat, proteins and micronutrients have a strong influence in shaping the gut microbiota39,40. On the other hand, the gut microbiota notably contributes to the absorption of minerals and nutrient uptake40. In addition, Kismul41 showed that children with kwashiorkor consume fewer sweet potatoes and papaya, which are characterized by high β-carotene content that provides powerful antioxidant capacities for the host42,43.
One of the limits of this study is the small sample size with respect to the time necessary to analyse each sample by microbial culturomics (see Methods). While a larger number of samples can be easily and quickly analysed by amplicon sequencing (v3v4 rRNA 16S gene amplicon sequencing in this study), culturomics has the unbeatable advantage of isolating bacterial strains and identifying them much more reliably, thanks to MALDI-TOF44. Taxonomic assignment on less than 500 bp (v3v4 16S gene hypervariable region between the FwOvAd_341F and the RevOvAd_785R primers include 444 bp) is not accurate for all species. Different species may have the same v3v4 sequence. Here, we voluntarily carried out a culturomics study. Molecular methods were used only to test the reproducibility of our culturomics results. New attempts to shorten the time of culturomics analysis from 6 to 3 weeks per sample are ongoing using new protocols, which are currently being evaluated. This will increase the power and make clinical case-control culturomics studies easier, allowing to increase the sample size. Moreover, we demonstrated here by principal coordinate analysis that only culturomics was able to discriminate between marasmus and kwashiorkor.
The incomplete information about clinical parameters and comorbidities is another limitation of our study. As we mainly focused on the gut microbial repertoire and its variation in different forms of malnutrition, we neglected the assessment of many covariables of interest that could be potential confounders. Indeed, we put huge efforts on culturomics (18 standardized conditions, >10,000 colonies for each sample so that about 70,000 colonies were analysed only for this new study) and we mainly tried to compare the different repertoire of the digestive microbiota in marasmus, kwashiorkor and controls. In addition, we propose in the Supplementary Data a new comprehensive questionnaire for future studies on microbiota and malnutrition to discover the specific links between clinical, biological and microbiological features in severe acute malnutrition. Such a questionnaire should include all information on medical history (history of malnutrition, previous hospitalization, malaria, meningitis); co-morbidities (malaria, HIV, tuberculosis, diarrhoea, parasites (amoebiasis, giardiasis)); breastfeeding (duration, mixed or exclusive); number of siblings; complete nutritional survey (specifically assessing maize, carotene, zinc, fibre consumption), nutritional status when faecal samples were collected (anthropometric and clinical parameters, onset of malnutrition, first entry in the hospital or during therapy), but also biological parameters (plasmatic concentration of protein, transferrin, albumin, glutathione, zinc) and liver characteristic (fat content of the liver assessable by non-invasive means such as ultrasound). This new questionnaire is provided in the Supplementary Data (Supplementary File 1). Future studies with comprehensive questionnaire identifying the environmental parameters linked to dysbiosis and nutritional features identified in this study (total protein, transferrin, albumin, zinc, glutathione and fat content of the liver) are urgently needed to guide future preventive measures against marasmus and kwashiorkor. Such biological parameters should also be assessed in NASH since common mechanisms are suspected.
For the available data: age, gender, geographical origin and period of sampling, there was no significant difference between the 3 groups (Supplementary Table S5). This suggests that our results are not biased by any of these usual potential confounders. The inclusion of individuals from different countries has made it possible to analyse a greater microbial diversity. In addition, each country is represented in each group (CTL, MRS, KW) by at least 2 children, and the proportion of children from each country is not different among groups (Supplementary Table S5), which limits the risk of geographical bias.
In conclusion, our meta-analysis results identified four consistent plasmatic markers that were depleted in kwashiorkor compared to marasmus: albumin, transferrin, glutathione and zinc. Future studies are needed to confirm the association between biological parameters (albumin, transferrin, glutathione and zinc) with gut microbiota in children with kwashiorkor. Fatty liver was confirmed to be a specific feature of kwashiorkor in our meta-analysis. Alteration of the gut microbiota, including a bloom of Proteobacteria and Fusobacteria and pathogenic invasion, was identified as a specific, instrumental factor in kwashiorkor, stressing the need for non-dietary treatments including broad-spectrum antibiotics such as cephalosporins (amoxicillin is inadequate for Enterobacteriaceae due to β-lactamase) and/or probiotic mixtures exhibiting antagonistic and regulatory activities to restore host-microbial mutualism.
Methods
Population and samples
This study continues the previously published study by Tidjani Alou et al.23, which recruited ten children with kwashiorkor and five healthy children without any anthropometric deficit (no wasting, no stunting and not underweight)23. A further seven children diagnosed with marasmus were recruited in Senegal and Niger for our present study. Diagnosis of marasmus was made by medical staff on admission based on clinical presentation and WHO criteria4. All characteristics of study participants are detailed in Supplementary Tables S4, S5. The number of samples was limited because of the time requirement for culturomics analysis of each sample (time to pre-incubated faecal sample after inoculation, 30 days to follow up with total 18 different culture conditions were all performed corresponding to 6 weeks for each sample). All analysed stool samples were stored at −80 °C. All the parents of the children involved in this study gave an informed consent for the participation of their children. The study was approved by the ethics committee of the Institute Fédératif de Recherche 48 (IFR48), Marseille, France under agreement number 09-022. All experiments were performed in accordance with relevant guidelines and regulations of the institution.
Microbial culturomics
A total of 18 standard culture conditions (Supplementary Table S6) were used to test each sample, as previously described by Lagier et al.45. Among the 212 different culture conditions analysed, Lagier et al. selected the 18 culture conditions allowing to capture the maximal diversity with the minimal number of conditions23,27,45. These 18 culture conditions are a combination of 8 preincubation methods (blood culture bottle, blood culture bottle supplemented with rumen, blood culture bottle supplemented with sheep blood, marine broth, brain heart infusion, trypticase soy broth, Columbia-like broth supplemented with sheep blood) and 4 culture conditions (5 µ active filtration, thermic shock, temperature and atmosphere). For each condition, 1 gram of faeces was diluted in 9 mL of DPBS (Thermo Fisher Scientific, Illkirch, France); the suspension was then inoculated in the various liquid medium and incubated at a chosen atmospheric temperature. On days 1, 3, 7, 10, 15, 21 and 30, culture dilution series ranging from 1/10 to 1/1010 were inoculated on 5% sheep’s blood agar (Becton Dickinson, Le Pont de Claix, France).
MALDI-TOF identification
The identification of isolated colonies was performed using a MICROFLEX LT spectrometer (Bruker Daltonics) and an MSP 96 MALDI-TOF target plate (Bruker Daltonics, Brenen, Germany), as previously described44. The obtained spectra were imported into MALDI Biotyper 3.0 software (Bruker) and compared with databases from Bruker and the base-specific laboratory at the La Timone hospital in Marseille, France. The resulting score permitted the identification (or not) of tested species as follows: species were labelled as correctly identified at the species level with a score ≥2.0 and at the genus level with a score ≥1.7 and <2.0, while no identification was made with a score <1.7.
16S rRNA sequencing
All strains that were not identified by MALDI-TOF were identified by 16S rRNA amplification and sequencing. The DNA was previously extracted by the EZ1 DNA Tissue Kit using a BioRobot EZ1 Advanced XL (Qiagen, Courtaboeuf, France). The 16S rRNA gene was amplified via PCR by using the universal primer pair consisting of fD1 and rP2 (Eurogentec, Angers, France)23. The obtained sequences were assembled and corrected by using Codon Code Aligner software (http://www.codoncode.com) and were compared with the sequences available in the GenBank database by using BLASTn (http://blast.ncbi.nlm.nih.gov.gate1.inist.fr/Blast.cgi). A similarity threshold <98.7% was used to detect a putative new species, whereas a threshold <95% was used to detect a putative new genus23. The confirmation and description of these putative new species is in progress according to taxonogenomics46,47.
High-throughput sequencing: metagenomics
Two protocols were used to extract DNA from stool samples48. The first protocol was based on a physical lysis process with glass powder, followed by an overnight chemical lysis with proteinase K. The resulting solution was then washed and eluted according to the instructions for the Macherey-Nagel DNA Tissue Extraction Kit. The second protocol was based on glycoprotein lysis with the addition of de-glycosylation steps. Sequencing of the DNA obtained via these two protocols was then performed targeting the V3V4 region of the 16S rRNA49. Microbial culturomics helped us decipher the various biases of metagenomics, including DNA extraction, primer bias, DNA amplification, and difference in bio-informatic methods and libraries27. In a previous study, we showed that glycoproteins affect DNA extraction, and that a protocol including a deglycosylation step yields the maximal diversity as observed with rarefaction curves48. This protocol was named P5 and was used with the standard QIAGEN protocol (P1), which is more rapid and yields different microbes. These protocols have been chosen once and all samples analysed in our centre use the same protocols, for reproducibility48. Raw sequencing data was available from Nucleotide Sequence Database EMBL-EBI (Reference: PRJEB31774). Further information on the accession number of each sample is described in Supplementary Table S2.
Diversity assessment
β-diversity is the number of taxa that are unique to each ecosystem and is used to compare the species diversity between ecosystems along environmental gradients50. In each group (CTL, MRS, KW) beta diversity was assessed by comparing the number of species found in a single individual with the number of species found in the entire group. Thus, the unique /total ratio (U/T ratio) was 0 if all individuals had an identical microbiota and 1 (100%) if each individual in the group had specific species without any species shared between the different individuals in the group23. For the metagenomics results, we first created a workflow, which is depicted in Supplementary Fig. S10. To reduce the number of false-positive hits and increase the accuracy of abundance estimates, we used a normalization method (rarefying of counts)51. An analysis of the rarefaction curves based on OTUs (Supplementary Fig. S11) showed a clear difference between the marasmus and kwashiorkor groups, and was almost constant from 5,000 reads up to 60,000 reads. This suggested that a rarefaction at 60,000 reads was relevant to compare the gut microbiota diversity of children with marasmus and kwashiorkor. Therefore, we picked 60,0000 reads as a threshold value after reaching the minimum total number of reads in each sample, which allowed us to maintain all 22 individual patient’s samples after normalizing. Consequently, all relative abundances <1/60,0000 were considered 0. All OTUs detected in each sample of each group were described in Supplementary Table S7. At the species level, the comparison between two groups was assessed by calculating and comparing the mean rank in each group52. Second, the Shannon index was also calculated to assess global, anaerobic and aerotolerant diversity by using the following formula: H’ = ∑pi*log2pi, where pi is the proportion of each species in the sample for which diversity is being estimated53.
Both culturomics and metagenomics results were compared to each other to confirm our results and highlight the complementarity between these two methods.
Statistical analysis
RevMan v5.3 (Copenhagen: The Nordic Cochrane Centre, The Cochrane Collaboration, 2014) was used to perform a meta-analysis, with random-effects model. Tests for heterogeneity is used to demonstrate the consistency/inconsistency between the several studies analysing the differences between kwashiorkor and marasmus.
To compare the means between two populations, two-tailed Student’s t-test and two-tailed Mann-Whitney U-test were used after testing for normality (Gaussian distribution). The bilateral exact Fisher test and uncorrected Chi-squared test were used to compare proportions. Correction for the false discovery rate was not necessary since this was an exploratory study54. As our prespecified hypothesis included a gradient, and we expected children with marasmus to have intermediate values between those of controls and children with kwashiorkor, we sought to test this gradient by statistical analysis. A linear regression model (ANOVA test) was used to test gradients for quantitative variables, and group value was the independent variable. The Cochran-Armitage trend test was used for proportions55,56.
Statistical analyses were performed with SPSS v22.0 (IBM, Paris, France) and XLSTAT Biomed v19.5 (Addinsoft, Paris, France).
Ethics committee approval
The study was approved by the ethics committee of the Institute Fédératif de Recherche 48 (IFR48), Marseille, France under agreement number 09-022.
Supplementary information
Acknowledgements
We would like to thank Dr. Sylvain BUFFET for all of his generous supports in the bioinformatic analysis. This work was supported by the French Government under the « Investissements d’avenir » (Investments for the Future) program managed by the Agence Nationale de la Recherche (ANR, fr: National Agency for Research), (reference: Méditerranée Infection 10-IAHU-03). Funding source had no role in the writing of the manuscript or the decision to submit it for publication.
Author Contributions
T.P.T.P., M.T.A. performed the culturomics analysis. D.B., A.L. performed the bioinformatics analysis. S.B., D.A., C.S., A.D. included children and collected samples and data. F.W. review the manuscript as an expert in childhood malnutrition. T.P.T.P., M.M. and D.R. supervised the study and analysed the data. D.R. designed the study.
Competing Interests
The authors declare no competing interests.
Footnotes
This article has been retracted. Please see the retraction notice for more detail:https://doi.org/10.1038/s41598-023-44593-7
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
10/30/2023
This article has been retracted. Please see the Retraction Notice for more detail: 10.1038/s41598-023-44593-7
Contributor Information
Matthieu Million, Email: matthieumillion@gmail.com.
Didier Raoult, Email: didier.raoult@gmail.com.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-45611-3.
References
- 1.WHO | UNICEF-WHO-The World Bank Group: Joint child malnutrition estimates - levels and trends in child malnutrition. WHO Available at, http://www.who.int/nutrition/publications/jointchildmalnutrition_2017_estimates/en/(Accessed: 20th February 2018).
- 2.Trehan I, et al. Antibiotics as part of the management of severe acute malnutrition. Malawi Med. J. 2016;28:123–130. [PMC free article] [PubMed] [Google Scholar]
- 3.Isanaka S, et al. Routine Amoxicillin for Uncomplicated Severe Acute Malnutrition in Children. N. Engl. J. Med. 2016;374:444–453. doi: 10.1056/NEJMoa1507024. [DOI] [PubMed] [Google Scholar]
- 4.WHO | WHO child growth standards and the identification of severe acute malnutrition in infants and children. Available at, http://www.who.int/nutrition/publications/severemalnutrition/9789241598163/en/(Accessed: 18th October 2018). [PubMed]
- 5.Williams CD. A nutritional disease of childhood associated with a maize diet. Arch. Dis. Child. 1933;8:423–433. doi: 10.1136/adc.8.48.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perampalli, T., Swami, S. C., Kumbar, K. M., Suryakar, A. N. & Shaikh, A. K. Possible role of oxidative stress in malnourished children. Curr. Pediatr. Res. 14 (2010).
- 7.Velasquez C, Navarro C, Muñoz C, Gonzalez Á. Inflammatory response in Colombian children with severe protein-energymalnutrition before and after nutritional intervention. Colomb. Médica. 2010;41:121–128. [Google Scholar]
- 8.Pascal N, et al. Serum concentrations of sex hormone binding globulin are elevated in kwashiorkor and anorexia nervosa but not in marasmus. Am. J. Clin. Nutr. 2002;76:239–244. doi: 10.1093/ajcn/76.1.239. [DOI] [PubMed] [Google Scholar]
- 9.Unterman TG, Vazquez RM, Slas AJ, Martyn PA, Phillips LS. Nutrition and somatomedin. XIII. Usefulness of somatomedin-C in nutritional assessment. Am. J. Med. 1985;78:228–234. doi: 10.1016/0002-9343(85)90431-0. [DOI] [PubMed] [Google Scholar]
- 10.Bandsma RHJ, et al. Mechanisms behind decreased endogenous glucose production in malnourished children. Pediatr. Res. 2010;68:423–428. doi: 10.1203/PDR.0b013e3181f2b959. [DOI] [PubMed] [Google Scholar]
- 11.Becker K, Leichsenring M, Gana L, Bremer HJ, Schirmer RH. Glutathione and association antioxidant systems in protein energy malnutrition: results of a study in Nigeria. Free Radic. Biol. Med. 1995;18:257–263. doi: 10.1016/0891-5849(94)E0131-2. [DOI] [PubMed] [Google Scholar]
- 12.Golden M. Free radicals and the aetiology of Kwashiorkor. The Biochemist. 1994;16:12–15. [Google Scholar]
- 13.Sive AA, Subotzky EF, Malan H, Dempster WS, Heese HD. Red blood cell antioxidant enzyme concentrations in kwashiorkor and marasmus. Ann. Trop. Paediatr. 1993;13:33–38. doi: 10.1080/02724936.1993.11747622. [DOI] [PubMed] [Google Scholar]
- 14.Mayatepek E, Becker K, Gana L, Hoffmann GF, Leichsenring M. Leukotrienes in the pathophysiology of kwashiorkor. Lancet Lond. Engl. 1993;342:958–960. doi: 10.1016/0140-6736(93)92003-C. [DOI] [PubMed] [Google Scholar]
- 15.Ashour MN, Salem SI, El-Gadban HM, Elwan NM, Basu TK. Antioxidant status in children with protein-energy malnutrition (PEM) living in Cairo, Egypt. Eur. J. Clin. Nutr. 1999;53:669–673. doi: 10.1038/sj.ejcn.1600830. [DOI] [PubMed] [Google Scholar]
- 16.Zain K, Haquani B, Iffat-un-Nisa AH. Serum copper and zinc levels in P-CM. J. Trop. Pediatr. Environ. Child Health. 1978;24:198–9. doi: 10.1093/tropej/24.4.198. [DOI] [PubMed] [Google Scholar]
- 17.Godette L, Warren PJ. The concentration of copper in the liver of African children with marasmus. Br. J. Nutr. 1967;21:419–423. doi: 10.1079/BJN19670043. [DOI] [PubMed] [Google Scholar]
- 18.Méndez J, Tejada C. Liver composition in kwashiorkor and marasmus. Exp. Mol. Pathol. 1962;1:344–352. doi: 10.1016/0014-4800(62)90029-1. [DOI] [PubMed] [Google Scholar]
- 19.Million M, Diallo A, Raoult D. Gut microbiota and malnutrition. Microb. Pathog. 2017;106:127–138. doi: 10.1016/j.micpath.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 20.Smith MI, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Million M, et al. Increased Gut Redox and Depletion of Anaerobic and Methanogenic Prokaryotes in Severe Acute Malnutrition. Sci. Rep. 2016;6:26051. doi: 10.1038/srep26051. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Zhu L, et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatol. Baltim. Md. 2013;57:601–609. doi: 10.1002/hep.26093. [DOI] [PubMed] [Google Scholar]
- 23.Tidjani Alou, M. et al. Gut Bacteria Missing in Severe Acute Malnutrition, Can We Identify Potential Probiotics by Culturomics? Front. Microbiol. 8 (2017). [DOI] [PMC free article] [PubMed]
- 24.Loban, A., Kime, R. & Powers, H. Iron-binding antioxidant potential of plasma albumin. Clin. Sci. Lond. Engl. 1979 93, 445–451 (1997). [DOI] [PubMed]
- 25.Brieland JK, Clarke SJ, Karmiol S, Phan SH, Fantone JC. Transferrin: a potential source of iron for oxygen free radical-mediated endothelial cell injury. Arch. Biochem. Biophys. 1992;294:265–270. doi: 10.1016/0003-9861(92)90167-U. [DOI] [PubMed] [Google Scholar]
- 26.Raoult A. Aspects of malnutrition in older children in French West Africa. Bull. Société Pathol. Exot. 1958;51:762–792. [Google Scholar]
- 27.Lagier J-C, et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PubMed] [Google Scholar]
- 28.Rawat D, Nair D. Extended-spectrum β-lactamases in Gram Negative Bacteria. J. Glob. Infect. Dis. 2010;2:263–274. doi: 10.4103/0974-777X.68531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stock I, Rahman M, Sherwood KJ, Wiedemann B. Natural antimicrobial susceptibility patterns and biochemical identification of Escherichia albertii and Hafnia alvei strains. Diagn. Microbiol. Infect. Dis. 2005;51:151–163. doi: 10.1016/j.diagmicrobio.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 30.Million M, Lagier J-C, Raoult D. Meta-analysis on efficacy of amoxicillin in uncomplicated severe acute malnutrition. Microb. Pathog. 2017;106:76–77. doi: 10.1016/j.micpath.2016.06.025. [DOI] [PubMed] [Google Scholar]
- 31.De Filippo C, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Filippo, C. et al. Diet, Environments, and Gut Microbiota. A Preliminary Investigation in Children Living in Rural and Urban Burkina Faso and Italy. Front. Microbiol. 8 (2017). [DOI] [PMC free article] [PubMed]
- 33.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schnorr SL, et al. Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 2014;5:3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Obregon-Tito AJ, et al. Subsistence strategies in traditional societies distinguish gut microbiomes. Nat. Commun. 2015;6:6505. doi: 10.1038/ncomms7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martínez I, et al. The Gut Microbiota of Rural Papua New Guineans: Composition, Diversity Patterns, and Ecological Processes. Cell Rep. 2015;11:527–538. doi: 10.1016/j.celrep.2015.03.049. [DOI] [PubMed] [Google Scholar]
- 37.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gomez A, et al. Gut Microbiome of Coexisting BaAka Pygmies and Bantu Reflects Gradients of Traditional Subsistence Patterns. Cell Rep. 2016;14:2142–2153. doi: 10.1016/j.celrep.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 39.Requena T, Martínez-Cuesta MC, Peláez C. Diet and microbiota linked in health and disease. Food Funct. 2018;9:688–704. doi: 10.1039/C7FO01820G. [DOI] [PubMed] [Google Scholar]
- 40.Kane AV, Dinh DM, Ward HD. Childhood Malnutrition and the Intestinal Microbiome Malnutrition and the microbiome. Pediatr. Res. 2015;77:256–262. doi: 10.1038/pr.2014.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kismul, H., Van den Broeck, J. & Lunde, T. M. Diet and kwashiorkor: a prospective study from rural DR Congo. PeerJ2 (2014). [DOI] [PMC free article] [PubMed]
- 42.Fiedor J, Burda K. Potential role of carotenoids as antioxidants in human health and disease. Nutrients. 2014;6:466–488. doi: 10.3390/nu6020466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chew BP, Park JS. Carotenoid action on the immune response. J. Nutr. 2004;134:257S–261S. doi: 10.1093/jn/134.1.257S. [DOI] [PubMed] [Google Scholar]
- 44.Seng P, et al. Ongoing Revolution in Bacteriology: Routine Identification of Bacteria by Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry. Clin. Infect. Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 45.Lagier J-C, et al. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin. Microbiol. Infect. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 46.Pham T-P-T, et al. Noncontiguous finished genome sequences and descriptions of ‘Paenibacillus bouchesdurhonensis,’ ‘Paenibacillus rubinfantis,’ ‘Paenibacillus senegalimassiliensis’ and ‘Paenibacillus tuaregi’ identified by culturomics. New Microbes New Infect. 2017;20:1–13. doi: 10.1016/j.nmni.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tindall BJ, Rosselló-Móra R, Busse H-J, Ludwig W, Kämpfer P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int. J. Syst. Evol. Microbiol. 2010;60:249–266. doi: 10.1099/ijs.0.016949-0. [DOI] [PubMed] [Google Scholar]
- 48.Angelakis E, et al. Glycans affect DNA extraction and induce substantial differences in gut metagenomic studies. Sci. Rep. 2016;6:26276. doi: 10.1038/srep26276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coulthard MG. Oedema in kwashiorkor is caused by hypoalbuminaemia. Paediatr. Int. Child Health. 2015;35:83–89. doi: 10.1179/2046905514Y.0000000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anderson MJ, et al. Navigating the multiple meanings of β diversity: a roadmap for the practicing ecologist. Ecol. Lett. 2011;14:19–28. doi: 10.1111/j.1461-0248.2010.01552.x. [DOI] [PubMed] [Google Scholar]
- 51.McMurdie, P. J. & Holmes, S. Waste Not, Want Not: Why Rarefying Microbiome Data Is Inadmissible. PLoS Comput. Biol. 10 (2014). [DOI] [PMC free article] [PubMed]
- 52.Cassir N, et al. Clostridium butyricum Strains and Dysbiosis Linked to Necrotizing Enterocolitis in Preterm Neonates. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2015;61:1107–1115. doi: 10.1093/cid/civ468. [DOI] [PubMed] [Google Scholar]
- 53.Spellerberg IF, Fedor PJ. A tribute to Claude Shannon (1916–2001) and a plea for more rigorous use of species richness, species diversity and the ‘Shannon–Wiener’. Index. Glob. Ecol. Biogeogr. 2003;12:177–179. doi: 10.1046/j.1466-822X.2003.00015.x. [DOI] [Google Scholar]
- 54.Rothman KJ. No adjustments are needed for multiple comparisons. Epidemiol. Camb. Mass. 1990;1:43–46. doi: 10.1097/00001648-199001000-00010. [DOI] [PubMed] [Google Scholar]
- 55.Cochran WG. Some Methods for Strengthening the Common χ2 Tests. Biometrics. 1954;10:417–451. doi: 10.2307/3001616. [DOI] [Google Scholar]
- 56.Armitage P. Tests for Linear Trends in Proportions and Frequencies. Biometrics. 1955;11:375–386. doi: 10.2307/3001775. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.