Abstract
Long non-coding RNAs (lncRNAs) represent a class of epigenetic regulators implicated in a number of physiological and pathological conditions. Herein, we characterized the lncRNA expression portrait from 837 patients with invasive breast cancer and 105 normals from the cancer genome atlas (TCGA), which revealed eighteen upregulated and forty-six downregulated lncRNAs. Clustering analysis revealed distinct lncRNA profile for the triple negative breast cancer (TNBC) and normal breast tissue, while less separation was observed among the HER2+HR+, HER2+HR−, HER2−HR+ molecular subtypes. LINC01614, and LINC01235 correlated with worse disease-free survival (DFS), while the expression of lnc-LRR1–1, lnc-ODF3B-2, AC015712.5, lnc-LAMB3–1, lnc-SPP2–3, and lnc-MAP9–2 correlated with better DFS. The expression of LINC01235 correlated with worse overall survival (OS), while the expression of MIR205HG, lnc-MAP2K6–5, FGF14-AS2, lnc-SPP2–3 correlated with better OS. Highest expression of LINC01614 was observed in progesterone receptor (PR)+, Estrogen receptor (PR)+, and HER2+ tumors, while lowest expression was in TNBC. Concordantly, LINC01614 was highly expressed in the luminalB/HER2+ subtype from the SRP062132 dataset. Elevated expression of LINC01614 was subsequently validated in primary breast cancer tissue and breast cancer cell lines. Bioinformatics and pathway analyses on LINC01614high vs. LINC01614low BC tissue revealed TGFβ1 and ECM as the most activated networks in LINC01614high tumors. Concordantly, strong correlation between the expression of LINC01614 and COL10A1 (R2 = 0.6929), SPOCK1 (R2 = 0.5156), ZEB1 (R2 = 0.3372), TGFBI (R2 = 0.2978), TGFB1 (R2 = 0.1985), ACTA2 (R2 = 0.1833), and TAGLN (R2 = 0.1909) was observed. Mechanistically, exogenous TGFB1 induced LINC01614 expression in the BT474 triple positive BC model, while small-molecule inhibition of transforming growth factor β (TGFβ, SB-431542) or focal adhesion kinase (FAK, PF-573228) abrogated LINC01614 expression. Our data revealed the lncRNA transcription landscape in breast cancer and its molecular subtypes. Our data provide novel insight implicating LINC01614 as unfavorable prognostic marker in BC, its association with the HR+/HER2+ BC molecular subtype and its regulation by TGFβ and FAK signaling.
Introduction
Breast cancer (BC) is the most common cancer type in females worldwide1. The molecular mechanisms involved in BC pathogenesis have been thoroughly studied, leading to BC classification into three major subtypes: Luminal which is positive for estrogen (ER+) and progesterone receptors (PR+), human epidermal growth factor receptor 2 (HER2+), and basal-like tumors, which lacks hormone receptor and HER2 expression, called triple-negative breast cancer (TNBC)2–4. While this classification has led to better stratification of BC, this disease is still associated with high mortality rate underscoring a need to develop novel molecular signature for better stratification and for prediction of disease outcome.
Although a number of gene-expression signatures have been reported as diagnostic and prognostic biomarkers in BC, the role of non-coding RNAs in this domain is just beginning to unfold. Genome wide transcriptome studies have revealed the existence of large number of long non-coding RNAs (≥200 nucleotides)5,6. Current GENCODE database (version 30) revealed the existence of approximately 16,193 lncRNAs and 14,706 pseudogenes in the human genome.
LncRNAs are involved in regulating various biological processes, including tumor-suppressor and oncogenic pathways and may serve as prognostic markers in BC. A number of oncogenic (H19, SRA, LSINCT5, Zfas1, lncRNA-Smad7, LOC554202, HOTAIR, SOX2OT and FAL1) and tumor suppressor (GAS5 and XIST) lncRNAs have been identified in BC; however their regulation and the mechanisms of action for the majority of lncRNAs remains to be unraveled6,7.
In this study, we characterized the lncRNA expression portrait from 837 patients with invasive BC and 105 normal breast tissues from the cancer genome atlas (TCGA) breast cancer dataset.
Our comparative analysis identified eighteen upregulated and forty-six downregulated lncRNAs in breast cancer compared to normal tissue. Interestingly, we identified eleven lncRNAs: LINC01614, LINC01235, lnc-LRR1–1, lnc-ODF3B-2, AC015712.5, lnc-LAMB3–1, lnc-SPP2–3, lnc-MAP9–2, MIR205HG, lnc-MAP2K6–5 and FGF14-AS2 to be associated with BC patient outcome. In particular, our data provided novel insight implicating LINC01614 as unfavorable prognostic marker in BC, its association with the HR+/HER2+ BC molecular subtype and its regulation by TGFβ and FAK signaling.
Materials and methods
Data analyses and bioinformatics
Long noncoding RNA (lncRNA) expression from 837 invasive breast carcinoma and 105 normal subjects were retrieved from The Atlas of Noncoding RNAs in Cancer (TANRIC; http://bioinformatics.mdanderson.org/main/TANRIC:Overview) database. Expression data were subsequently imported into Altanalyze v.2.1.0 software as described before8. Hierarchical clustering was performed using cosine for columns and cosine for rows while principal component analysis was performed to assess the relatedness of samples. Gene expression for the same cohort was retrieved from the cBioPortal for Cancer Genomics (https://www.cbioportal.org/) database as we described before9.
RNA-Seq data analysis
Raw RNA sequencing data were retrieved from sequence read archive (SRA) database under accession no. SRP062132. Data were retrieved using the SRA toolkit version 2.9.2 as previously described10. Pair end reads were aligned to the hg19 human reference genome in CLC Genomics Workbench-12 (QIAGEN, Germany). The abundance of the expression of transcripts was measured as the score of TPM (Transcript Per Kilobase Million) mapped reads in CLC Genomics Workbench 12. Expression of LINC01614 in each molecular subtype was plotted using Graphpad Prism 6.0 software (Graphpad ® Software, San Diego, CA, USA)
Gene set enrichment and modeling of gene interactions networks
Upregulated genes in the LINC01614high BC group were imported into the Ingenuity Pathways Analysis (IPA) software (Ingenuity Systems; www.ingenuity.com/) and were subjected to functional annotations and regulatory network analysis using upstream regulator analysis (URA), downstream effects analysis (DEA), mechanistic networks (MN) and causal network analysis (CNA) prediction algorithms. IPA uses precise algorithm to predict functional regulatory networks from gene expression data and provides a significance score for each network according to the fit of the network to the set of focus genes in the database. The p value is the negative log of P and represents the possibility that focus genes in the network being found together by chance11.
Statistical and survival analysis
Kaplan-Meir survival analysis and plotting were conducted using IBM SPSS statistics version 24 software. For survival analysis, patients were grouped into high or low based on LINC01614 log2 gene expression. The log-rank test was used to compare the outcome between expression groups. Statistical analyses to compare specific gene expression and graphing were performed using Graphpad Prism 6.0 software. Unpaired two-tailed t-test and p value of <0.05 was considered significant as we described before12.
Cell culture, recombinant TGFβ treatment, and small molecule inhibition
Human breast cancer cell lines (BT474, T47D, MDAMB453, ZR751, MCF7, HCC70, HS578T, MDAMB468, BT549 and MDAMB231) were cultured in Dulbecco’s modified Eagle’s medium/RPMI 1640 supplemented with D-glucose 4500 mg/l, 4 mM L-glutamine and 110 mg/l sodium pyruvate, 10% fetal bovine serum and 1x penicillin–streptomycin (Pen-Strep) (all purchased from Gibco-Invitrogen, Waltham, MA, USA). The triple positive BC cell line (BT474) was treated with rhTGFβ (10 ng/ml, Peprotec, London, UK), TGFβ inhibitor (SB-431542; 10 μM, Selleckchem Inc., Houston, TX, USA), FAK inhibitor (PF-573228; 5 μM, Selleckchem Inc., Houston, TX, USA) and combination of rhTGFβ and TGFβ inhibitor. Pharmacological inhibition of TGF-β and FAK pathways were conducted as we previously described9,13. Briefly, 0.2 × 106 cells/well were cultured in 6 well plates (duplicate) and incubated for 48 hours and subsequently the expression of LINC01614 was measured using qRT-PCR. Assays were carried out with appropriate DMSO control.
LncRNA validation using qRT-PCR
Tumor tissue (TT) specimens from eight BC tissue and adjacent normal tissue (NT) were obtained from treatment-naive BC patients prior to surgery with a proper written informed consent. The study was approved by Qatar Biomedical Research Institute, Doha, Qatar (Protocol no. 2017–006). Total RNA was extracted from eight primary BC tissue, adjacent normal tissue, and from a panel of breast cancer cell lines using Norgon RNA/DNA/Protein Purification Plus Kit (Norgen Biotek Corp, Ontario, Canada) as per the manufacturer’s instructions. Expression level of LINC01614 was validated using SYBR Green-based quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR). The total RNA (500 ng) was reverse transcribed into complementary DNA (cDNA) using a High Capacity cDNA Reverse Transcript Kit (catalogue No. 4368814; ABI) according to the manufacturer’s protocol. Relative levels of lncRNA was determined using the cDNA as template in real-time PCR analysis using the Applied Biosystems QuantStudio 6/7 Flex Real-time PCR system. Primer sequences used in the current study were: LINC01614 F: 5′-AACCAAGAGCGAAGCCAAGA-3′; LINC01614 R: 5′-GCTTGGACACAGACCCTAGC-3′; GAPDH F: 5′-CTGGTAAAGTGGATATTGTTGCCAT-3′; and GAPDH R: 5′-TGGAATCATATTGGAACATGTAAACC-3′. The relative expression level was calculated using –ΔΔCT, GAPDH was used as an endogenous control.
Results
Expression profiling of lncRNAs from the TCGA BC dataset compared to normal breast tissue
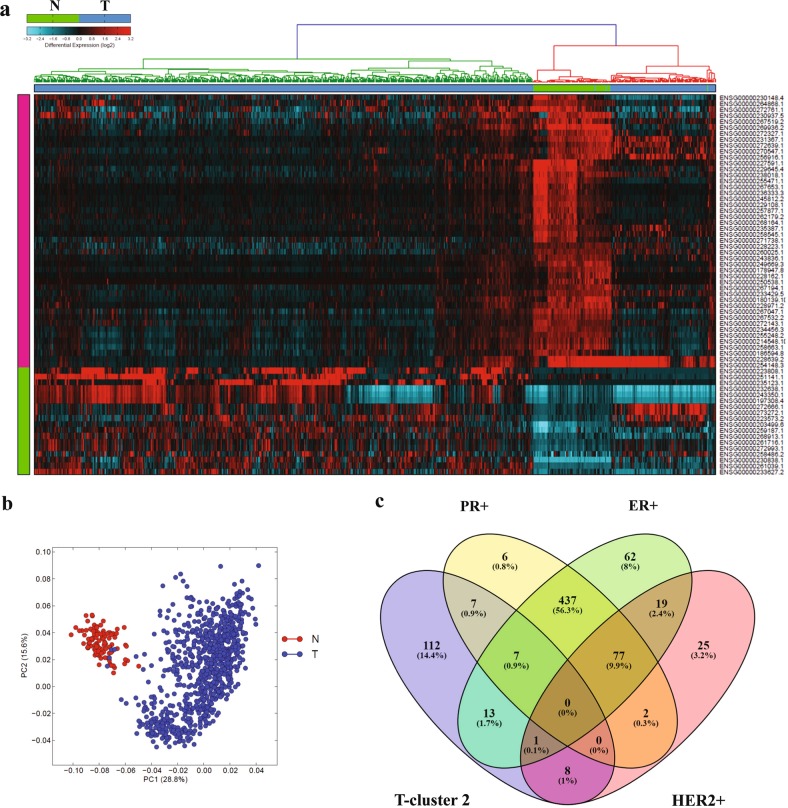
Expression data for 12727 lncRNAs from 837 patients with invasive BC and 105 normal breast tissue were retrieved from TANRIC database and were subjected to differential expression analysis, which identified 18 upregulated and 46 downregulated lncRNAs (≤2≥, FDR p ≤ 0.05; Table 1). Hierarchical clustering revealed three major clusters, where breast cancer samples clustered at both sides while normals clustered in the middle (Fig. 1a). Principle component analysis (PCA) also revealed clear separation of normal from breast cancer based on lncRNA expression (Fig. 1b). Interestingly, we observed majority of patients in the further right cluster (76%; Fig. 1a) to be of the TNBC molecular subtype (Fig. 1c).
Table 1.
Differentially expressed lncRNAs in breast cancer vs normal
| Ensembl Gene ID | LNCipedia gene ID | log fold Tum vs Nor | fold Tum vs Nor | Tum vs Nor p value (raw) | Tum vs Nor p value (adj) | Status |
|---|---|---|---|---|---|---|
| ENSG00000230838.1 | LINC01614 | 2.341821475 | 5.069422738 | 5.65E-81 | 3.76E-79 | Up |
| ENSG00000203499.6 | lnc-MAPK15–6 | 1.767788642 | 3.405315908 | 3.82E-82 | 2.61E-80 | Up |
| ENSG00000223808.1 | AC044784.1, lnc-GATA3–7 | 1.610083258 | 3.052694584 | 1.25E-16 | 1.13E-15 | Up |
| ENSG00000259187.1 | lnc-TRIM69–1 | 1.590879481 | 3.01232928 | 1.81E-44 | 4.73E-43 | Up |
| ENSG00000261039.1 | LINC02544 | 1.398432474 | 2.636150018 | 7.47E-43 | 1.85E-41 | Up |
| ENSG00000258486.2 | lnc-LRR1–1 | 1.341780081 | 2.534638644 | 9.58E-26 | 1.33E-24 | Up |
| ENSG00000235123.1 | DSCAM-AS1 | 1.326866563 | 2.508572377 | 6.20E-13 | 4.46E-12 | Up |
| ENSG00000232638.1 | lnc-TAF3–1 | 1.21241703 | 2.317255348 | 1.40E-12 | 9.85E-12 | Up |
| ENSG00000272993.1 | lnc-HIST2H2AA4–1 | 1.185802408 | 2.274898863 | 1.78E-37 | 3.74E-36 | Up |
| ENSG00000243350.1 | GATA3-AS1 | 1.148653771 | 2.217069154 | 3.27E-13 | 2.39E-12 | Up |
| ENSG00000233627.2 | C4A-AS1 | 1.105244735 | 2.1513537 | 7.23E-16 | 6.20E-15 | Up |
| ENSG00000272666.1 | lnc-ODF3B-2 | 1.077677457 | 2.110635509 | 1.89E-19 | 1.98E-18 | Up |
| ENSG00000273272.1 | lnc-KLHDC7B-2 | 1.048411607 | 2.06825147 | 4.74E-19 | 4.87E-18 | Up |
| ENSG00000197308.4 | GATA3-AS1 | 1.044585641 | 2.062773821 | 9.84E-14 | 7.48E-13 | Up |
| ENSG00000223573.2 | lnc-TINCR-1 | 1.044336743 | 2.062417976 | 1.83E-17 | 1.73E-16 | Up |
| ENSG00000261716.1 | lnc-HIST2H2AA3–1 | 1.026047258 | 2.036437106 | 3.92E-35 | 7.61E-34 | Up |
| ENSG00000251141.1 | MRPS30-DT | 1.02054162 | 2.028680429 | 1.58E-07 | 7.36E-07 | Up |
| ENSG00000268913.1 | lnc-KCNK6–1 | 1.001295226 | 2.001796371 | 8.85E-23 | 1.07E-21 | Up |
| ENSG00000228223.1 | HCG11 | −1.017371351 | −2.024227373 | 2.51E-53 | 8.53E-52 | Down |
| ENSG00000235387.1 | na | −1.020539284 | −2.028677144 | 1.96E-59 | 7.71E-58 | Down |
| ENSG00000228162.1 | lnc-SPP2–3 | −1.043000128 | −2.060508089 | 9.21E-115 | 1.41E-112 | Down |
| ENSG00000272639.1 | AC015712.5 | −1.05048961 | −2.071232647 | 1.95E-49 | 5.91E-48 | Down |
| ENSG00000256916.1 | lnc-BIRC2–4 | −1.069704648 | −2.09900361 | 2.70E-26 | 3.84E-25 | Down |
| ENSG00000264868.1 | lnc-STEAP4–1 | −1.080167322 | −2.114281279 | 8.30E-28 | 1.25E-26 | Down |
| ENSG00000250538.1 | lnc-MAP9–2 | −1.096920898 | −2.138976893 | 1.05E-104 | 1.21E-102 | Down |
| ENSG00000243836.1 | WDR86-AS1 | −1.151180613 | −2.220955695 | 1.05E-116 | 1.73E-114 | Down |
| ENSG00000178947.8 | SMIM10L2A | −1.161204888 | −2.236441294 | 2.98E-104 | 3.39E-102 | Down |
| ENSG00000267194.1 | lnc-MAP2K6–5 | −1.162956703 | −2.239158576 | 5.17E-82 | 3.52E-80 | Down |
| ENSG00000233429.5 | HOTAIRM1 | −1.179118648 | −2.264384021 | 1.06E-87 | 8.10E-86 | Down |
| ENSG00000271738.1 | lnc-TSPAN14–1 | −1.191983318 | −2.284666075 | 1.60E-57 | 6.01E-56 | Down |
| ENSG00000214548.10 | lnc-DLK1–35 | −1.199354187 | −2.296368526 | 4.55E-110 | 6.30E-108 | Down |
| ENSG00000268164.1 | na | −1.204434904 | −2.304469867 | 2.30E-93 | 2.01E-91 | Down |
| ENSG00000260025.1 | lnc-FEZ2–7 | −1.20586992 | −2.306763212 | 5.20E-63 | 2.24E-61 | Down |
| ENSG00000245812.2 | LINC02202 | −1.227351365 | −2.341367447 | 7.20E-190 | 9.16E-187 | Down |
| ENSG00000262179.2 | MYMX | −1.233789324 | −2.351839037 | 8.67E-99 | 8.49E-97 | Down |
| ENSG00000186594.8 | MIR22HG | −1.237175737 | −2.357365953 | 4.69E-86 | 3.43E-84 | Down |
| ENSG00000270547.1 | LINC01235 | −1.239824501 | −2.361698012 | 6.31E-42 | 1.50E-40 | Down |
| ENSG00000255248.2 | MIR100HG | −1.261015808 | −2.396644303 | 3.69E-100 | 3.82E-98 | Down |
| ENSG00000272143.1 | FGF14-AS2 | −1.285565793 | −2.437776382 | 8.61E-100 | 8.62E-98 | Down |
| ENSG00000236333.3 | TRHDE-AS1 | −1.286805825 | −2.439872611 | 8.25E-157 | 3.50E-154 | Down |
| ENSG00000257877.1 | lnc-MAPKAPK5–1 | −1.291455467 | −2.447748728 | 4.55E-139 | 1.16E-136 | Down |
| ENSG00000267532.2 | lnc-SLC16A11–7 | −1.332087847 | −2.517667645 | 7.47E-147 | 2.50E-144 | Down |
| ENSG00000234456.3 | MAGI2-AS3 | −1.339265343 | −2.530224405 | 1.04E-159 | 4.58E-157 | Down |
| ENSG00000272327.1 | lnc-NRG1–3 | −1.352076922 | −2.552793645 | 8.52E-68 | 4.13E-66 | Down |
| ENSG00000258545.1 | RHOXF1-AS1 | −1.356631857 | −2.560866164 | 6.26E-114 | 9.16E-112 | Down |
| ENSG00000267653.1 | lnc-ABCA5–6 | −1.372853753 | −2.589823452 | 1.55E-108 | 2.04E-106 | Down |
| ENSG00000229108.1 | LINC02587 | −1.395635686 | −2.631044566 | 6.69E-164 | 3.41E-161 | Down |
| ENSG00000230148.4 | HOXB-AS1 | −1.426672474 | −2.68825962 | 2.86E-42 | 6.89E-41 | Down |
| ENSG00000228971.2 | lnc-RWDD3–5 | −1.450759321 | −2.733518843 | 2.47E-108 | 3.21E-106 | Down |
| ENSG00000249669.3 | CARMN | −1.474915257 | −2.779673159 | 1.36E-234 | 1.73E-230 | Down |
| ENSG00000230937.5 | MIR205HG | −1.500142384 | −2.828706285 | 1.10E-27 | 1.65E-26 | Down |
| ENSG00000180139.10 | ACTA2-AS1 | −1.563395501 | −2.955486233 | 1.37E-118 | 2.33E-116 | Down |
| ENSG00000231367.1 | lnc-ATL2–1 | −1.570416598 | −2.969904619 | 4.73E-83 | 3.27E-81 | Down |
| ENSG00000258663.1 | lnc-RTL1–1 | −1.573129653 | −2.975494916 | 3.68E-96 | 3.40E-94 | Down |
| ENSG00000255471.1 | lnc-FZD4–1 | −1.665847283 | −3.172999476 | 1.39E-151 | 5.52E-149 | Down |
| ENSG00000267519.2 | lnc-C19orf57 | −1.684869673 | −3.215113511 | 2.96E-81 | 1.98E-79 | Down |
| ENSG00000267047.1 | lnc-SLC16A11–7 | −1.783333068 | −3.442205117 | 1.36E-109 | 1.85E-107 | Down |
| ENSG00000238018.1 | lnc-RTN4–3 | −1.826091508 | −3.545751691 | 7.48E-143 | 2.21E-140 | Down |
| ENSG00000272761.1 | lnc-CCDC80–5 | −1.868752854 | −3.652167299 | 2.80E-47 | 7.92E-46 | Down |
| ENSG00000229645.4 | lnc-SYNE3–1 | −2.025990744 | −4.072714669 | 5.48E-134 | 1.32E-131 | Down |
| ENSG00000228639.2 | lnc-SLC39A11–10 | −2.048436659 | −4.136574768 | 6.54E-44 | 1.68E-42 | Down |
| ENSG00000254148.3 | lnc-SLC39A11–10 | −2.181617437 | −4.536618794 | 6.46E-45 | 1.71E-43 | Down |
| ENSG00000227591.1 | lnc-LAMB3–1 | −2.192798277 | −4.571914035 | 1.67E-169 | 1.12E-166 | Down |
| ENSG00000269936.2 | CARMN | −2.778459057 | −6.861191136 | 5.33E-170 | 3.77E-167 | Down |
Fig. 1. lncRNA transcriptional portrait in breast cancer compared to normal tissue.
a Hierarchical clustering of breast cancer (n = 837) and normal (n = 105) breast tissue based on differentially expressed lncRNA transcripts. Each column represents one sample and each row represents a transcript. Expression level of each transcript (log2) in a single sample is depicted according to the color scale. b Principal component analysis (PCA) for the lncRNA transcriptome of breast cancer and normal breast tissue. c Venn diagram depicting the overlap between tumors samples from tumor cluster 2 (T-cluster 2) on the further right of the heatmap in panel (a) and the indicated breast cancer molecular subtype
Identification of lncRNAs associated with overall survival (OS) and disease-free survival (DFS) in the TCGA BC dataset
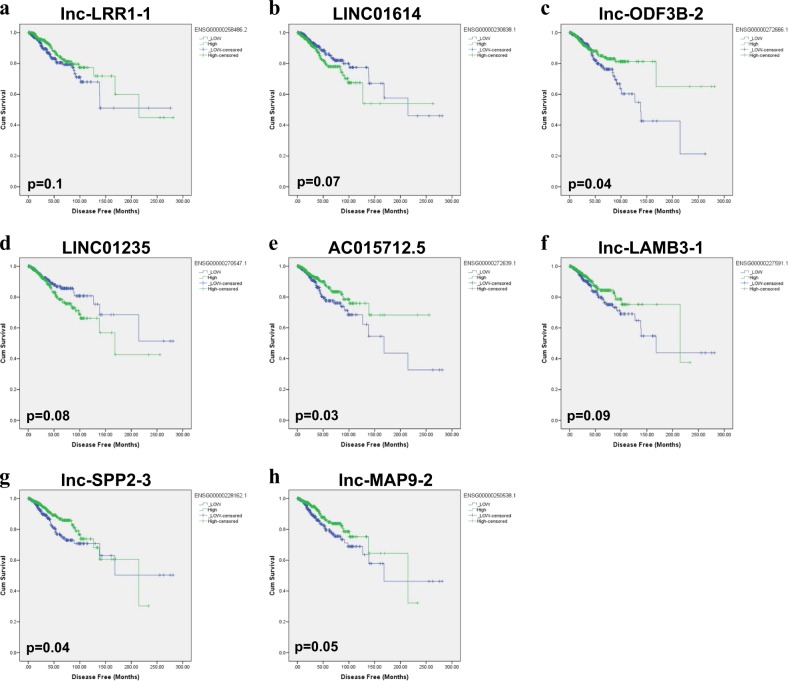
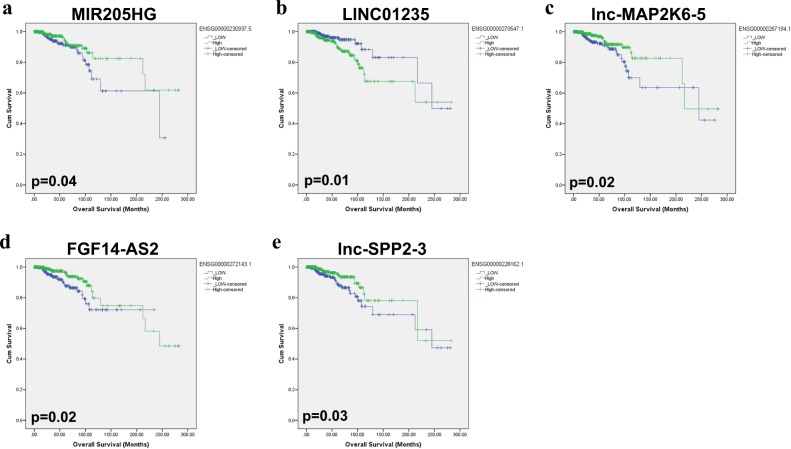
The differentially expressed lncRNAs from the profiling study were subsequently subjected to Kaplan-Meir survival analysis. Our data revealed LINC01614 and LINC01235 to predict worse DFS of BC patients (Fig. 2b and d; log-rank test = 0.07 and 0.08, respectively). On the other hand, lnc-LRR1–1, lnc-ODF3B-2, AC015712.5, lnc-LAMB3–1, lnc-SPP2–3, and lnc-MAP9–2 were associated with better DFS (Fig. 2a, c, e–h). Interestingly, the expression of LINC01235 correlated with worse DFS and worse OS (Figs. 2d and 3b), while the expression of MIR205HG, lnc-MAP2K6–5, FGF14-AS2, lnc-SPP2–3 correlated with better OS (Fig. 3a, c–e). Taken together, our data revealed LINC01614 as the only upregulated lncRNAs in BC and associated with worse DFS in the TCGA dataset.
Fig. 2. Disease-free survival (DFS) of breast cancer patients based on lncRNA expression.
Kaplan-Meir DFS analysis for lnc-LRR1–1 (a), LINC01614 (b), lnc-ODF3B-2 (c), LINC01235 (d), AC015712.5 (e), lnc-LAMB3–1 (f), lnc-SPP2–3 (g), and lnc-MAP9–2 (h) in the TCGA BC cohort. Significance was calculated using the log-rank test. p values are indicated on each plot
Fig. 3. Overall survival (OS) of breast cancer patients based on lncRNA expression.
Kaplan-Meir OS analysis for MIR205HG (a), LINC01235 (b), lnc-MAP2K6–5 (c), FGF14-AS2 (d), and lnc-SPP2–3 (e) in the TCGA BC cohort. Significance was calculated using the log-rank test. p values are indicated on each plot
LINC01614 expression correlates with HER2+HR+ invasive breast cancer molecular subtype
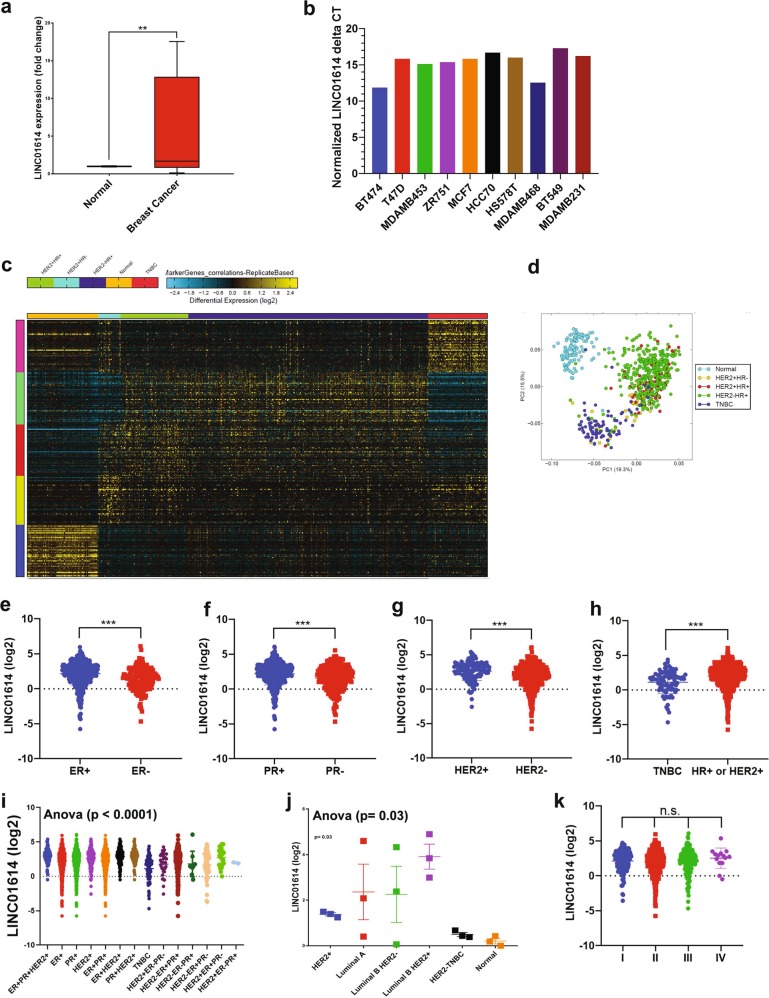
LINC01614 was the most highly expressed lncRNA (5.0 FC, p (adj) = 3.7 × 10–79) in breast cancer compared to normal tissue. We subsequently validated the expression of LINC01614 in a cohort of breast cancer patients, which revealed elevated expression of LINC01614 in BC compared to adjacent normal tissue (5.9 FC, p = 0.0007, Fig. 4a). Similarly, LINC01614 expression was detected in a panel of BC cell lines, where highest expression was observed in the BT474 triple positive BC cell line (Fig. 4b). We subsequently sought to determine if lncRNA expression can discriminate breast cancer with various molecular subtypes. To that end, the 837 BC samples were divided into HER2+HR+, HER2+HR−, HER2−HR+, and TNBC and were subjected to the marker finder algorithm in Altanalyze v.2.1.0 compared to 115 normal breast tissue. Clustering analysis revealed distinct molecular subtype for the TNBC and normal breast tissue, while less clear separation was observed among the three other molecular subtypes (HER2+HR+, HER2+HR−, HER2−HR+, Figs. 4c, d). The lncRNA profile distinctive for each molecular subtype is shown in Table 2. Interestingly, the expression of LINC01614 was highest in the HR+HER2+, while lowest expression was observed in TNBC molecular subtype (Fig. 4e–i). The expression of LINC01614 was subsequently validated in another cohort (SRP062132), which concordantly revealed highest expression in the luminal B/HER2+ molecular subtype (Fig. 4j). There was no difference in the expression of LINC01614 in relation to tumor stage (Fig. 4k).
Fig. 4. Correlation between the expression of LINC01614 and BC molecule subtype.
a Expression of LINC01614 in eight BC patients (red box) and adjacent normal tissue (blue box) using qRT-PCR. Data are presented as mean ± S.E., n = 8 for each group. b Expression of LINC01614 in a panel of BC cell lines. Data are presented as normalized delta CT expression value. (c) Heat map clustering of BC (n = 837) and normal (n = 105) breast tissue based on molecular subtypes of BC (HER2 + HR+, HER2+HR−, HER2-HR+, TNBC, and normal). Each row represents expression level of the indicated lncRNA (log2). Expression level of transcriptomes in a single sample is depicted according to the color scale (blue to yellow). (d) Principal component analysis (PCA) for the indicated BC molecular subtypes and normal breast tissue. Expression of LINC01614 in relation to estrogen receptor (ER, e), progesterone receptor (PR, f), and HER2 (g) status. h Expression of LINC01614 in TNBC vs non-TNBC BC patients. i Expression of LINC01614 in the indicated molecular subtype subgroups. j Expression of LINC01614 in the SRP062132 dataset in relation to the indicated molecular subtype. k Expression of LINC01614 in relation to the tumor stages
Table 2.
lncRNAs enriched according to breast cancer molecular subtype
| TNBC | HER2-HR+ | HER2+HR+ | HER2+HR− | Normal | |||||
|---|---|---|---|---|---|---|---|---|---|
| Ensembl gene ID | LNCipedia gene ID | Ensembl gene ID | LNCipedia gene ID | Ensembl Gene ID | LNCipedia gene ID | Ensembl gene ID | LNCipedia gene ID | Ensembl gene ID | LNCipedia gene ID |
| ENSG00000263680.2 | lnc-SOX9–4 | ENSG00000248008.2 | NRAV | ENSG00000266469.1 | lnc-CDK12–2 | ENSG00000232044.4 | LINC01105 | ENSG00000264016.2 | lnc-TMEM220–1 |
| ENSG00000227036.2 | lnc-SLC39A11–1 | ENSG00000249042.1 | lnc-ANKRD34B-2 | ENSG00000270145.1 | lnc-CDC6–1 | ENSG00000256940.1 | lnc-FKBP2–1 | ENSG00000234456.3 | MAGI2-AS3 |
| ENSG00000225177.1 | lnc-CCDC28A-1 | ENSG00000269619.1 | lnc-C5AR2–3 | ENSG00000266999.1 | lnc-RDM1–1 | ENSG00000261235.1 | lnc-MPHOSPH6–1 | ENSG00000241684.1 | ADAMTS9-AS2 |
| ENSG00000229124.2 | VIM-AS1 | ENSG00000267348.2 | GEMIN7-AS1 | ENSG00000272264.1 | lnc-TPD52–1 | ENSG00000251179.1 | TMEM92-AS1 | ENSG00000263400.2 | |
| ENSG00000258910.2 | LINC01956 | ENSG00000263214.1 | lnc-ZSCAN10–3 | ENSG00000263874.1 | ENSG00000249700.4 | SRD5A3-AS1 | ENSG00000267532.2 | lnc-SLC16A11–7 | |
| ENSG00000246334.2 | PRR7-AS1 | ENSG00000248360.3 | LINC00504 | ENSG00000230838.1 | LINC01614 | ENSG00000257086.1 | lnc-PPP1R14B-1 | ENSG00000262097.1 | LINC02185 |
| ENSG00000272524.1 | lnc-KLF11–3 | ENSG00000235106.4 | lnc-BRD3OS-1 | ENSG00000263466.1 | lnc-FBXO47–2 | ENSG00000273257.1 | lnc-SRD5A3–1 | ENSG00000267047.1 | lnc-SLC16A11–7 |
| ENSG00000258938.1 | lnc-RALGAPA1–1 | ENSG00000263011.1 | lnc-ZNF205–1 | ENSG00000228265.1 | RALY-AS1 | ENSG00000235491.1 | LINC01889 | ENSG00000235904.1 | RBMS3-AS3 |
| ENSG00000224167.1 | lnc-FAM19A3–2 | ENSG00000225361.3 | PPP1R26-AS1 | ENSG00000261005.1 | lnc-MLLT6–1 | ENSG00000204603.2 | LINC01257 | ENSG00000255090.1 | MIR100HG |
| ENSG00000227392.1 | HPN-AS1 | ENSG00000205913.2 | SRRM2-AS1 | ENSG00000228613.1 | lnc-PXDN-3 | ENSG00000234378.1 | lnc-LAPTM4A-2 | ENSG00000269936.2 | CARMN |
| ENSG00000215866.3 | LINC01356 | ENSG00000231439.3 | WASIR2 | ENSG00000250917.1 | lnc-AHCY-1 | ENSG00000257588.1 | lnc-RACGAP1–1 | ENSG00000267069.1 | lnc-CIDEA-1 |
| ENSG00000254035.1 | lnc-ZNF454–1 | ENSG00000261801.1 | LOXL1-AS1 | ENSG00000260228.1 | lnc-SLC38A8–2 | ENSG00000231298.2 | MANCR | ENSG00000254847.1 | lnc-DKK3–2 |
| ENSG00000237517.4 | lnc-DGCR6–6 | ENSG00000268913.1 | lnc-KCNK6–1 | ENSG00000203635.2 | lnc-PXDN-1 | ENSG00000236345.1 | lnc-PHF3–1 | ENSG00000215386.6 | MIR99AHG |
| ENSG00000254615.2 | lnc-OXR1–1 | ENSG00000259459.1 | LINC02568 | ENSG00000253917.3 | lnc-OTOP1–6 | ENSG00000248359.1 | lnc-CCDC125–6 | ENSG00000226005.3 | lnc-PFKP-16 |
| ENSG00000272192.1 | lnc-MTFR1–1 | ENSG00000261664.1 | TTC39A-AS1 | ENSG00000263975.1 | lnc-PCGF2–1 | ENSG00000266222.1 | lnc-HOXB1–1 | ENSG00000249669.3 | CARMN |
| ENSG00000229891.1 | LINC01315 | ENSG00000268204.1 | lnc-PCP2–1 | ENSG00000257702.3 | lnc-TTC31–4 | ENSG00000204832.5 | ST8SIA6-AS1 | ENSG00000244513.2 | lnc-ARL6IP5–1 |
| ENSG00000261175.1 | LINC02188 | ENSG00000260810.1 | lnc-DGLUCY-1 | ENSG00000226853.2 | lnc-SCRN3–1 | ENSG00000214546.3 | lnc-PPP1R1B-1 | ENSG00000228962.1 | HCG23 |
| ENSG00000234899.5 | lnc-SLC39A11–10 | ENSG00000230314.2 | ELOVL2-AS1 | ENSG00000213793.3 | lnc-ZNF320–1 | ENSG00000230033.1 | lnc-NOM1–1 | ENSG00000255248.2 | MIR100HG |
| ENSG00000251161.2 | lnc-VPS18–1 | ENSG00000267715.1 | lnc-REEP6–1 | ENSG00000234703.1 | lnc-CLIC6–3 | ENSG00000272663.1 | lnc-LHCGR-2 | ENSG00000226237.1 | GAS1RR |
| ENSG00000272430.1 | lnc-SYT15–3 | ENSG00000250101.1 | lnc-FAM153C-5 | ENSG00000232679.1 | LINC01705 | ENSG00000237883.1 | DGUOK-AS1 | ENSG00000231246.1 | lnc-ST7L-2 |
| ENSG00000257718.1 | CPNE8-AS1 | ENSG00000260219.1 | lnc-MYLPF-1 | ENSG00000259802.1 | lnc-CMBL-1 | ENSG00000236921.1 | lnc-CDKN2B-1 | ENSG00000248869.1 | LINC02511 |
| ENSG00000215190.4 | lnc-RAB23–53 | ENSG00000236703.1 | MYB-AS1 | ENSG00000266040.1 | lnc-EPOP-2 | ENSG00000255118.1 | lnc-SCGB1D4–1 | ENSG00000260947.1 | lnc-PRSS3–2 |
| ENSG00000272620.1 | lnc-SORCS2–1 | ENSG00000260136.1 | lnc-SCNN1B-1 | ENSG00000250081.1 | lnc-ADAMTS6–1 | ENSG00000262973.1 | lnc-ABI3–6 | ENSG00000237560.1 | LINC01497 |
| ENSG00000226419.2 | SLC16A1-AS1 | ENSG00000264589.1 | MAPT-AS1 | ENSG00000232940.1 | ENSG00000254966.1 | AC103974.1 | ENSG00000263586.1 | HID1-AS1 | |
| ENSG00000267287.1 | lnc-NFATC1–1 | ENSG00000249684.1 | lnc-FAM153C-5 | ENSG00000236591.1 | lnc-ZC3H12D-2 | ENSG00000259702.1 | lnc-PGPEP1L-60 | ENSG00000245812.2 | LINC02202 |
| ENSG00000253348.1 | lnc-KCNMB1–3 | ENSG00000254231.1 | lnc-SLC7A13–1 | ENSG00000263931.1 | lnc-MARCH10–1 | ENSG00000240497.2 | lnc-TMEM212–1 | ENSG00000238120.1 | LINC01589 |
| ENSG00000270170.1 | NCBP2-AS2 | ENSG00000268049.1 | AC012313.2 | ENSG00000228221.1 | LINC00578 | ENSG00000273328.1 | lnc-KRBOX1–1 | ENSG00000230587.1 | LINC02580 |
| ENSG00000233654.1 | lnc-MFSD6–1 | ENSG00000263105.1 | lnc-SRL-1 | ENSG00000244300.2 | GATA2-AS1 | ENSG00000273237.1 | lnc-HNRNPA2B1–14 | ENSG00000272316.1 | lnc-RAB23–53 |
| ENSG00000257271.1 | KIRREL3-AS1 | ENSG00000260954.1 | lnc-PTX4–2 | ENSG00000254854.1 | lnc-RNF26–1 | ENSG00000225243.1 | lnc-MYOC-1 | ENSG00000272143.1 | FGF14-AS2 |
| ENSG00000237976.1 | lnc-PSMB4–1 | ENSG00000228133.2 | lnc-DPH1–1 | ENSG00000231482.2 | lnc-PXDN-2 | ENSG00000226733.1 | lnc-C6orf141–2 | ENSG00000232079.2 | LINC01697 |
| ENSG00000253164.1 | lnc-ATP6V1B2–3 | ENSG00000228043.1 | lnc-NXPH2–3 | ENSG00000228950.1 | lnc-RHOB-1 | ENSG00000234072.1 | lnc-SNX17–1 | ENSG00000261625.1 | lnc-MRGPRF-1 |
| ENSG00000228109.1 | MELTF-AS1 | ENSG00000238122.1 | lnc-NBPF4–1 | ENSG00000261039.1 | LINC02544 | ENSG00000265282.1 | lnc-CYB561–1 | ENSG00000253864.1 | CARMN |
| ENSG00000267480.1 | lnc-MPPE1–10 | ENSG00000260009.1 | LINC02130 | ENSG00000227857.2 | lnc-TSPAN1–1 | ENSG00000261712.1 | lnc-KIAA1024–1 | ENSG00000243243.1 | lnc-TFEC-2 |
| ENSG00000263893.2 | lnc-SOX9–5 | ENSG00000253959.1 | LINC01863 | ENSG00000233818.1 | lnc-CHAF1B-2 | ENSG00000259933.2 | ENSG00000225731.1 | lnc-WDR4–5 | |
| ENSG00000254148.3 | lnc-SLC39A11–10 | ENSG00000251169.2 | LINC01843 | ENSG00000224189.2 | HAGLR | ENSG00000225833.1 | lnc-BMX-1 | ENSG00000229645.4 | lnc-SYNE3–1 |
| ENSG00000228639.2 | lnc-SLC39A11–10 | ENSG00000272933.1 | lnc-ARL3–1 | ENSG00000249917.2 | LINC00536 | ENSG00000258689.1 | LINC01269 | ENSG00000227467.3 | LINC01537 |
| ENSG00000245614.2 | DDX11-AS1 | ENSG00000271420.1 | lnc-TCEA3–1 | ENSG00000230479.1 | lnc-CHAF1B-3 | ENSG00000242540.2 | lnc-SOX11–1 | ENSG00000238018.1 | lnc-RTN4–3 |
| ENSG00000260420.1 | LINC02182 | ENSG00000248399.1 | lnc-CFAP99–1 | ENSG00000267493.2 | CIRBP-AS1 | ENSG00000261123.1 | lnc-NTHL1–1 | ENSG00000224318.1 | CHL1-AS2 |
| ENSG00000232803.1 | SLCO4A1-AS1 | ENSG00000268895.1 | A1BG-AS1 | ENSG00000223387.2 | LINC02068 | ENSG00000228288.2 | PCAT6 | ENSG00000272777.1 | lnc-ADH5–1 |
| ENSG00000229692.3 | SOS1-IT1 | ENSG00000262869.1 | lnc-SMG6–4 | ENSG00000264007.1 | lnc-SSH2–1 | ENSG00000245080.4 | MIR3150BHG | ENSG00000231943.3 | PGM5P4-AS1 |
| ENSG00000232445.1 | LNCPRESS1 | ENSG00000263244.1 | lnc-PMM2–6 | ENSG00000267013.1 | LINC01929 | ENSG00000249199.1 | lnc-MYO10–2 | ENSG00000178947.8 | SMIM10L2A |
| ENSG00000224930.2 | ENSG00000224698.1 | lnc-NBPF6–1 | ENSG00000260766.1 | lnc-PIGM-1 | ENSG00000254248.1 | lnc-TP53INP1–2 | ENSG00000226833.1 | lnc-TRMT61B-1 | |
| ENSG00000272825.1 | lnc-FAM207A-2 | ENSG00000267277.1 | lnc-EPOR-2 | ENSG00000267118.1 | lnc-CCL4–3 | ENSG00000253217.1 | lnc-SPAG1–1 | ENSG00000255850.1 | RXYLT1-AS1 |
| ENSG00000224717.1 | lnc-CDK18–1 | ENSG00000261742.1 | LINC00922 | ENSG00000264112.1 | lnc-VEZF1–1 | ENSG00000258616.1 | LINC02303 | ENSG00000180769.4 | WDFY3-AS2 |
| ENSG00000251136.4 | lnc-NBN-1 | ENSG00000234918.1 | lnc-ZNF33A-13 | ENSG00000260162.2 | lnc-BANP-14 | ENSG00000248161.1 | lnc-SLC39A8–1 | ENSG00000267107.2 | PCAT19 |
| ENSG00000254607.2 | lnc-ST3GAL4–4 | ENSG00000215256.3 | DHRS4-AS1 | ENSG00000263655.1 | lnc-FBXO15–1 | ENSG00000225077.2 | LINC00337 | ENSG00000259134.1 | LINC00924 |
| ENSG00000261189.1 | lnc-CAGE1–1 | ENSG00000272335.1 | lnc-MRPS30–8 | ENSG00000249942.1 | lnc-BTC-5 | ENSG00000226207.1 | lnc-MMS22L-1 | ENSG00000225039.1 | LINC01058 |
| ENSG00000248846.2 | LINC02065 | ENSG00000263280.1 | lnc-PRSS22–1 | ENSG00000238123.1 | MID1IP1-AS1 | ENSG00000230212.2 | CBR3-AS1 | ENSG00000228649.4 | SNHG26 |
| ENSG00000237857.2 | lnc-FANCC-1 | ENSG00000260793.2 | lnc-TMUB2–1 | ENSG00000196421.3 | ENSG00000229272.1 | lnc-EIF3A-2 | ENSG00000229694.2 | LINC00484 | |
| ENSG00000225096.1 | lnc-PRIM2–7 | ENSG00000224577.1 | LINC01117 | ENSG00000227543.3 | SPAG5-AS1 | ENSG00000260209.1 | lnc-PUS3–1 | ENSG00000225655.1 | PGM5P3-AS1 |
| ENSG00000185904.7 | LINC00839 | ENSG00000260526.1 | lnc-TIFA-1 | ENSG00000226876.2 | lnc-KIF26B-2 | ENSG00000253417.1 | LINC02159 | ENSG00000230747.1 | lnc-SNRNP200–1 |
| ENSG00000226101.1 | LINC02097 | ENSG00000233621.1 | LINC01137 | ENSG00000261327.3 | lnc-CA5A-4 | ENSG00000235939.1 | lnc-C10orf71–1 | ENSG00000253552.3 | HOXA-AS2 |
| ENSG00000203362.2 | POLH-AS1 | ENSG00000271916.1 | lnc-CHDH-1 | ENSG00000265908.1 | lnc-PIPOX-5 | ENSG00000272894.1 | lnc-COL28A1–1 | ENSG00000267082.1 | lnc-ANGPTL8–1 |
| ENSG00000236663.1 | FRGCA | ENSG00000231868.1 | lnc-TNFRSF25–1 | ENSG00000254239.1 | lnc-SLC25A48–1 | ENSG00000260231.1 | KDM7A-DT | ENSG00000226031.1 | FGF13-AS1 |
| ENSG00000241155.1 | ARHGAP31-AS1 | ENSG00000272913.1 | lnc-TRIM43B-1 | ENSG00000153363.8 | LINC00467 | ENSG00000233342.1 | lnc-MAP3K4–2 | ENSG00000227591.1 | lnc-LAMB3–1 |
| ENSG00000272048.1 | lnc-DNMT3A-1 | ENSG00000273472.1 | lnc-ELMOD2–4 | ENSG00000258457.1 | lnc-PSMB11–1 | ENSG00000258829.1 | lnc-DIO2–5 | ENSG00000231999.2 | LRRC8C-DT |
| ENSG00000239445.1 | ST3GAL6-AS1 | ENSG00000223813.2 | lnc-CPVL-4 | ENSG00000264083.1 | lnc-PSMD11–1 | ENSG00000261472.1 | lnc-WWOX-1 | ||
| ENSG00000261521.1 | lnc-DSC2–1 | ENSG00000255367.1 | lnc-ART5–1 | ENSG00000242136.1 | lnc-ASAP2–4 | ENSG00000186594.8 | MIR22HG | ||
| ENSG00000237594.2 | lnc-SOD1–3 | ENSG00000270177.1 | lnc-TCF7–4 | ENSG00000268686.1 | lnc-TEAD2–1 | ENSG00000255471.1 | lnc-FZD4–1 | ||
| ENSG00000234753.1 | FOXP4-AS1 | ENSG00000267009.2 | lnc-ARSG-3 | ENSG00000224468.3 | LAMC1-AS1 | ||||
LINC01614 elevated expression is associates with enhanced BC tumorigenenic molecular profile
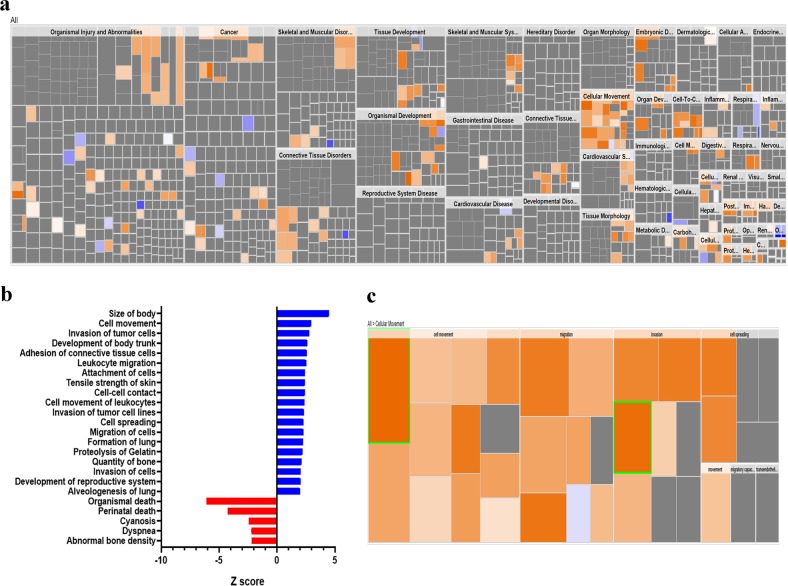
To gain more insight into plausible role for LINC01614 in BC pathology, the 837 BC patients were divided into LINC01614high and LINC01614low according to the median LINC01614 expression. The upregulated genes in the LINC01614high group were subsequently subjected to Ingenuity Pathway Analysis (IPA) and downstream effect analysis (DEA). Affected functional categories are illustrated as heat tree map, which clusters functionally associated categories together, therefore depicting a high-level outlook of enriched functional categories. Data presented in Figs. 5a, b revealed a number of enriched functional categories including those involved in cell movement and invasion, while functional categories associated with cell death were under presented. Illustration of the cellular movement functional category is shown in Fig. 5c.
Fig. 5. Enrichment in tumor cell migration and invasion functional categories in LINC01614high BC patients.
a Disease and function heat map depicting enrichment in the indicated functional and disease categories in the upregulated transcripts in LINC01614high vs LINC01614low BC patients based on IPA analysis. b Bar-graph depicting the most significantly affected factional categories in LINC01614high vs LINC01614low BC patients. c Heat map-illustrating enrichment in cellular movement functional category
Mechanistic network analysis predicts activation of TGFβ1 and ECM pathways in LINC01614high BC tissue
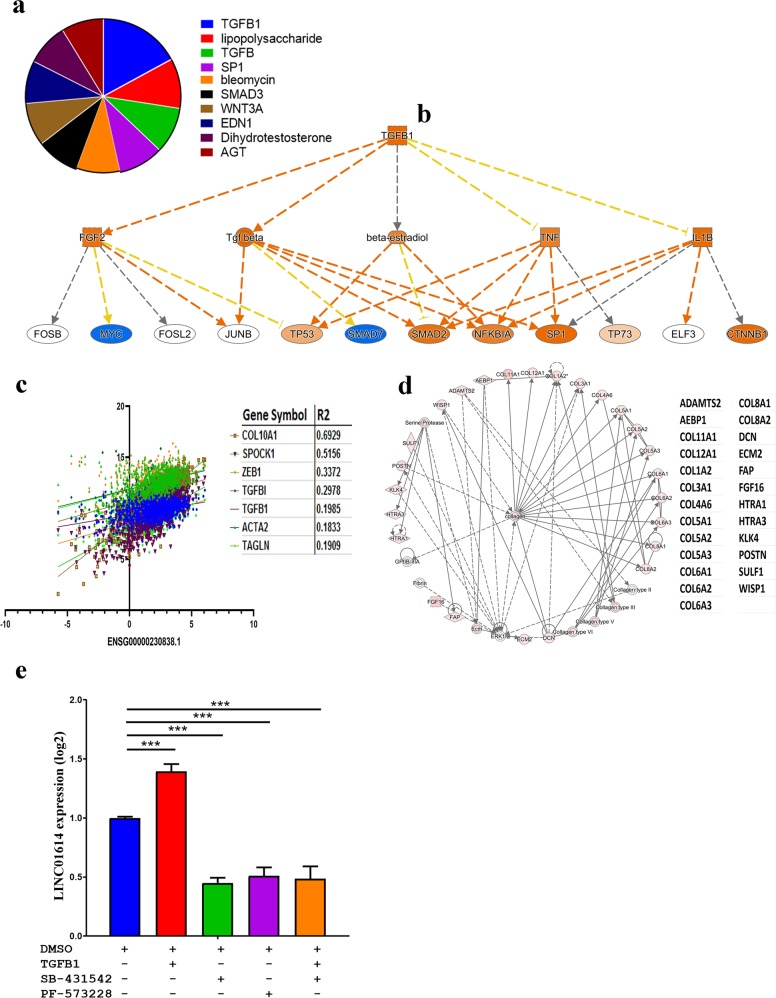
Upstream regulator analysis (URA) on the upregulated genes in LINC01614high revealed significant enrichment for a number of networks including TGFβ1, lipopolysaccharide, TGFβ, SP1, bleomycin, SMAD3, WNT3A, EDN1, dihydrotestosterone, and AGT (Fig. 6a). Highest enrichment was for the TGFβ1 network (Z score = 5.6; Fig. 6a). Mechanistic network analysis predicted TGFβ1 to directly activate the SMAD2, NFKB1A and SP1 through TGFβ (direct activation) and TNF (inconsistent sate), and to inhibit MYC through FGF2 (direct activation) and inhibit SMAD7 through TGFβ with higher confidence level (Fig. 6b). Concordantly, LINC01614 expression demonstrated significant positive correlation with various members of the TGFβ signaling pathways (COL10A1 (R2 = 0.7), SPOCK1 (R2 = 0.5156), ZEB1 (R2 = 0.3372), TGFBI (R2 = 0.2978), TGFB1 (R2 = 0.1985), ACTA2 (R2 = 0.1833), and TAGLN (R2 = 0.1909)) in the TCGA BC cohort (Fig. 6c). Moreover, we observed several collagen family members to be upregulated in LINC01614high BC, suggesting enhanced extracellular matrix (ECM) formation. Illustration of the ECM network in LINC01614high BC mapped by IPA is shown in Fig. 6d. The color shade intensity of the node correlates with the expression level of the indicated genes. Therefore, our molecular and network analyses revealed strong correlation between LINC01614 expression, TGFβ and ECM signaling. Mechanistically, recombinant TGFβ1 induced LINC01614 expression, while pharmacological inhibition of TGFβ signaling (using SB-431542) and FAK (using PF-573228) inhibited LINC01614 expression in BC cells (Fig. 6e), thus implication TGFβ and FAK signaling in regulating LINC01614 expression in BC cells.
Fig. 6. Mechanistic network analysis predicts predominant activation of the TGFB pathway in LINC01614high BC tumors.
a Pie chart illustrating the top activated mechanistic networks in LINC01614high BC tumors based on IPA analyses. Segment size corresponds to the activation Z score. b Illustration of the TGFβ1 signaling network. c Correlation between the expression of LINC01614 and the expression several members of the TGFβ family in BC tumors. d Extra cellular matrix functional enrichment in LINC01614high BC tumors. Color intensity indicates their activation state. Effect of recombinant TGFβ (10 ng/ml), SB-431542 (ββ inhibitor, 10 µM), and PF-573228 (FAK inhibitor, 5 µM) on LINC01614 expression measured by qRT-PCR. Data are presented as mean ± S.E. from two independent experiments, n = 6
Discussion
In recent years, lncRNAs have emerged as key players in regulating cellular functions, differentiation and disease progression, including cancer, through epigenetics, chromatin remodeling, transcriptional and post-transcriptional regulation5,14.
While the number of annotated lncRNAs in the human genome has increase dramatically, functional characterization of lncRNAs and their utilization as disease biomarkers is begging to unfold. In current study, we analyzed the lncRNA transcriptome from the TCGA breast cancer dataset and performed thorough survival and bioinformatics analyses which revealed eighteen upregulated and forty-six downregulated lncRNAs in BC compared to normal breast tissue. Additionally, our data identified different lncRNA signatures associated with various BC molecular subtypes (HER2+HR+, HER2+HR−, HER2−HR+, and TNBC) as well as those specific to normal breast tissue. Interestingly, our data revealed a distinct lncRNA cluster for the TNBC tumors, while such segregation was less evident among the other molecular subtypes (HER2+HR+, HER2+HR−, HER2−HR+). This could be attributed to the tissue of origin for various BC molecular subtypes, where TNBC originates from ductal cells, while the HR+ and HER2+ originate from luminal cells15.
Our analyses identified eleven lncRNAs (LINC01614, LINC01235, lnc-LRR1–1, lnc-ODF3B-2, AC015712.5, lnc-LAMB3–1, lnc-SPP2–3, lnc-MAP9–2, MIR205HG, lnc-MAP2K6–5 and FGF14-AS2) whose expression correlated with patient outcome. Among the identified lncRNAs, LINC01614 and LINC01235 correlated with worse DFS, while LINC01235 correlated with worse OS. Interestingly, LINC01235 was downregulated in in BC compared to normal tissue, while at the same time it predicted worse DFS and OS. It is plausible that due to the large heterogeneity of BC cases included the TCGA BC cohort, the expression pattern for LINC01235 did not correlate with survival data. Additionally, a previous study reported LINC01235 (also called FLJ41200; ENSG00000270547.1) as cancer-related genes that mapped telomeric and centromeric to CD274 (PDL-1) at 9p23 in small-cell lung carcinoma16, suggesting possible link between LINC01235 and immune regulation in cancer.
Interestingly, our data revealed over-expression of LINC01614 in BC compared to normal tissue and its elevated expression correlated with worse DFS. More in-depth analysis revealed LINC01614 to be highly expressed in ER+ (log2 exp = 2.1), in PR+ (log2 exp = 2.2) and HER2+ (log2 exp = 2.623), while TNBC exhibited lowest expression (log2 exp = 1.1). Those data were further validated in a second cohort where highest expression was observed in the luminal B/HER2+ molecular subtype while lowest expression was observed in the HER2-TNBC molecular subtype. The expression of LINC01614 did not correlate with BC disease stage, suggesting alteration in LINC01614 expression as an early feature during BC development and progression. Concordant with our data, LINC01614 expression has been linked to lung adenocarcinoma17 and the LINC01614-contaitng signature predicted OS and DFS in patients with esophageal squamous cell carcinoma18. Recently, LINC01614 has also been reported as one of the lncRNA associated survival of ER + BC patients19.
To gain more insight into plausible molecular mechanisms of LINC01614 expression and function, we dicatomized the TCGA BC cohort into LINC01614high and LINC01614low and subsequently retrieved and identified mRNA transcripts upregulated in the LINC01614high group, which revealed 187 upregulated transcripts. Interestingly, IPA analysis on the upregulated gene list suggested strong correlation between LINC01614 expression and enriched functional categories associated with tumor cell movement and invasion. Nonetheless, LINC01614high expression was most significantly associated with TGFβ signaling, suggesting possible induction of LINC01614 by TGFβ signaling. Additionally, LINC01614high tumors exhibited high expression of several collagens, suggesting possible association between LINC01614 expression and enhanced ECM formation. It is noteworthy that ECM itself could be regulated by TGFB signaling20. Mechanistic investigation validated induction of LINC01614 by TGFβ, while it’s expression was inhibited by small molecule inhibitor of TGFβ and FAK, suggesting its regulation by TGFβ and FAK signaling.
Our data also revealed elevated expression of LINC01614 in HER2+ BC tumors. Interestingly, we observed significant correlation between LINC01614 expression and HER2 mutation status in BC (supplementary figure 1). HER2+ (erbB2) represent 25 to 30 % of breast cancer patients and is elevated expression has been associated with more aggressive BC phenotype and shorter DFS and OS21,22. Additionally, activation of HER2 has been linked to Epithelial-to-mesenchymal transition (EMT), hence endowing cancer cells with a more aggressive and invasive phenotype23,24. Interestingly, HER2 and TGFβ signaling cooperated in the induction of cellular processes associated with tumorigenic development in immortalized mammary epithelial cell line25. Additionally, overexpression of HER2 activated the TGFβ/SMAD signaling pathway and induced SNAIL, SLUG and ZEB-1 expression and subsequent acquisition of mesenchymal phenotype24. These published reports are consistent with our current data linking LINC01614 to TGFβ signaling and HER2 + molecular subtype.
Conclusions
Our data revealed the lncRNA transcriptional landscape in breast cancer and identified the lcnRNA signatures associated with each molecular subtype. Specifically, our data provide novel insight implicating LINC01614 as unfavorable prognostic marker in BC, and its association with the HR+/HER2+ BC molecular subtype and its regulation by TGFβ and FAK signaling.
Supplementary information
Acknowledgements
This work was supported by a start-up grant [QB13] for Dr Nehad Alajez from Qatar Biomedical Research Institute, Qatar Foundation. The publication of this article was funded by the Qatar National Library.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by I. D'Agnano
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41420-019-0190-6) contains supplementary material, which is available to authorized users.
References
- 1.Polyak K. Heterogeneity in breast cancer. J. Clin. Investig. 2011;121:3786–3788. doi: 10.1172/JCI60534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl Acad. Sci. USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 4.Onitilo AA, Engel JM, Greenlee RT, Mukesh BN. Breast cancer subtypes based on ER/PR and Her2 expression: comparison of clinicopathologic features and survival. Clin. Med. Res. 2009;7:4–13. doi: 10.3121/cmr.2008.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beckedorff Felipe C., Amaral Murilo Sena, Deocesano‑Pereira Carlos, Verjovski‑Almeida Sergio. Long non-coding RNAs and their implications in cancer epigenetics. Bioscience Reports. 2013;33(4):667–675. doi: 10.1042/BSR20130054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu H, et al. Long non-coding RNAs as prognostic markers in human breast cancer. Oncotarget. 2016;7:20584–20596. doi: 10.18632/oncotarget.7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y, Sharma S, Watabe K. Roles of lncRNA in breast cancer. Front. Biosci. 2015;7:94–108. doi: 10.2741/s427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olsson A, et al. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature. 2016;537:698–702. doi: 10.1038/nature19348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vishnubalaji R, et al. Molecular profiling of ALDH1( + ) colorectal cancer stem cells reveals preferential activation of MAPK, FAK, and oxidative stress pro-survival signalling pathways. Oncotarget. 2018;9:13551–13564. doi: 10.18632/oncotarget.24420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leinonen R, Sugawara H, Shumway M. International Nucleotide Sequence Database C. The sequence read archive. Nucleic acids Res. 2011;39(Database issue):D19–D21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bredel M, et al. A network model of a cooperative genetic landscape in brain tumors. JAMA. 2009;302:261–275. doi: 10.1001/jama.2009.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alajez NM. Large-scale analysis of gene expression data reveals a novel gene expression signature associated with colorectal cancer distant recurrence. PloS ONE. 2016;11:e0167455. doi: 10.1371/journal.pone.0167455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-toub M, et al. Pleiotropic effects of cancer cells’ secreted factors on human stromal (mesenchymal) stem cells. Stem Cell Res. Ther. 2013;4:114. doi: 10.1186/scrt325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.George J, et al. Genomic amplification of CD274 (PD-L1) in small-cell lung cancer. Clin. Cancer Res. 2017;23:1220–1226. doi: 10.1158/1078-0432.CCR-16-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu M, et al. A novel lncRNA, LUADT1, promotes lung adenocarcinoma proliferation via the epigenetic suppression of p27. Cell Death Dis. 2015;6:e1858. doi: 10.1038/cddis.2015.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang GW, et al. A three-lncRNA signature predicts overall survival and disease-free survival in patients with esophageal squamous cell carcinoma. BMC Cancer. 2018;18:147. doi: 10.1186/s12885-018-4058-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong L, et al. A six-long non-coding RNAs signature as a potential prognostic marker for survival prediction of ER-positive breast cancer patients. Oncotarget. 2017;8:67861–67870. doi: 10.18632/oncotarget.18919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verrecchia F, Mauviel A. Transforming growth factor-beta signaling through the Smad pathway: role in extracellular matrix gene expression and regulation. J. Invest. Dermatol. 2002;118:211–215. doi: 10.1046/j.1523-1747.2002.01641.x. [DOI] [PubMed] [Google Scholar]
- 21.Hynes NE, Stern DF. The biology of erbB-2/neu/HER-2 and its role in cancer. Biochim. et. Biophys. Acta. 1994;1198:165–184. doi: 10.1016/0304-419x(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 22.Slamon DJ, et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 23.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 24.Gupta P, Srivastava SK. HER2 mediated de novo production of TGFbeta leads to SNAIL driven epithelial-to-mesenchymal transition and metastasis of breast cancer. Mol. Oncol. 2014;8:1532–1547. doi: 10.1016/j.molonc.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ueda Y, et al. Overexpression of HER2 (erbB2) in human breast epithelial cells unmasks transforming growth factor beta-induced cell motility. J. Biol. Chem. 2004;279:24505–24513. doi: 10.1074/jbc.M400081200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.