Abstract
Testicular germ cell tumour (TGCT) represents the most common malignancy in young men in large parts of the world, but the aetiology is yet unclear. Multiple TGCT susceptibility loci have been identified, and we have shown that one of these, SPRY4, may act as a TGCT oncogene. Furthermore, many of the loci are in non-coding regions of the genome. miRNAs, a class of non-coding RNAs may play a crucial role in cell proliferation, differentiation, and apoptosis, and alteration in their expression may lead to oncogenesis. Differential expression of miRNAs in TGCT and normal testis has been reported in previous studies. In this study, we used qPCR to analyse, in normal and malignant testis tissue, the expression of the ten miRNAs that we had previously identified by sequencing to be the most upregulated in TGCT. We found high expression of these miRNAs also by qPCR analysis. The levels of miR-302a-3p, miR-302b-3p, and miR-302c-3p were downregulated after treatment of the TGCT cell lines NT2-D1 and 833 K with the chemotherapy drug cisplatin. By using miRNA inhibitor-mediated transient transfection, we inhibited the expression of the three members of miR-302 family (miR-302s). Inhibition of miR-302s resulted in a decreased cell proliferation in NT2-D1 cells, but not in 833 K cells. In both cell lines, inhibition of miR-302s resulted in decreased expression of SPRY4, which we have previously shown to regulate MAPK/ERK and PI3K/Akt signalling pathways in these cells. Inhibition of miR-302b-3p and miR-302c-3p decreased phosphorylation of ERK1/2, whereas inhibition of miR-302a-3p and miR-302b-3p led to decreased expression of the apoptosis inhibitor, survivin. Our findings suggest that miR-302s act as TGCT oncogenes by inducing the expression of SPRY4 and activating MAPK/ERK pathway while inhibiting apoptosis via increased survivin expression.
Subject terms: Reverse transcription polymerase chain reaction, Testicular cancer, Cell growth, Cell invasion, miRNAs
Introduction
Testicular germ cell tumour (TGCT) represents the most common malignancy in young men in large part of the world, and the incidence is rising1,2. The mortality of TGCT has, however, declined over the last decades, mainly due to the introduction of cisplatin-based chemotherapy, and the survival rate is approximately 95% in the Western world3. The aetiology of TGCT is yet unclear, and both genetic and environmental factors are believed to contribute to the disease risk4,5. Genome-wide association studies (GWAS) have identified more than 50 TGCT susceptibility loci6,7, and SPRY4 is one of the risk genes with strong and consistent association7–10. Our results in a previous study, indicate that SPRY4 acts as a TGCT oncogene11. Many of the susceptibility loci identified by GWAS are in the non-coding regions of the genome suggesting that non-coding RNAs also influence the development of TGCT. Non-coding RNAs (ncRNAs) may also play a role in TGCT progression12.
MicroRNAs (miRNAs), a class of small non-coding RNAs (sncRNAs), play crucial roles in many physiological processes including proliferation, differentiation, and apoptosis, and alterations in expression of miRNAs have been associated with tumourigenesis13–16. In a recent study, we showed that miRNAs were one of the most common groups of sncRNAs in TGCT17. We also found a different miRNA expression pattern in malignant and normal testis tissue. The largest difference was among members of two clusters, miR-302/367 and miR-371-373, consisting of nine and four miRNAs, respectively. Other studies have also shown high expression of the miR-302/367 and miR-371-373 clusters in TGCT tissue as well as in serum samples from TGCT patients18–21. High expression of these miRNA clusters in TGCTs indicates that they may act as oncogenes. In a genetic screen-based study, miR-372 and miR-373 were shown to act as TGCT oncogenes through inhibition of a tumour suppressor gene, LATS222. The role of the miR-302/367 cluster in TGCTs is yet unknown, however, this cluster has been reported to act as tumour suppressor genes in several other cancers23–26.
The primary aim of the current study was to investigate the functional role of selected miRNAs in TGCT development by use of two metastatic TGCT (embryonal carcinoma) cell lines 833 K27 and NT2-D128. In our previous study, we analysed the expression pattern of miRNAs mainly by sequencing17. In the present study, by using a different approach, i.e. quantitative PCR (qPCR) analysis, we measured the levels of the ten most differentially expressed miRNAs identified in the previous study. We also investigated the effect of the cytotoxic drug cisplatin on the expression of these miRNAs. Subsequently, we inhibited the expression of the most cisplatin-sensitive miRNAs and studied the effect on cell growth, cell death, and cell signalling. We found that miR302s, like SPRY4, were highly expressed in TGCTs and also acted as oncogenes in the TGCT cell lines11. We further investigated if there was an association between miR302s and SPRY4 by studying the effect of inhibition of the most cisplatin-sensitive miR302s on SPRY4 expression.
Methods
Human tissue samples
For miRNA expression analysis, the TGCT subtypes embryonal carcinoma, seminoma, and mixed germ cell tumour, were bought from Origene (MD, USA), whereas normal adult testis samples were collected from adult organ transplant donors. According to the manufacturer, the mixed germ cell tumour was composed of a mixture of yolk sac tumour, immature teratoma, and mature teratoma. No definite embryonal carcinoma was seen.
The study has been approved by the Regional Committee for Medical and Health Research Ethics – South East Norway (2016/2006, REC South East), and all experiments were performed in accordance with approved guidelines and regulations. For the normal testis samples in connection with organ transplantation, informed consent was obtained according to the Norwegian legislation relating to transplantation, hospital autopsies and the donation of bodies.
Cell culture
Two TGCT cell lines NT2-D1 and 833 K representing the embryonal carcinoma (EC) were kindly provided by Dr Birgitte Lindeman (Norwegian Institute of Public Health, Oslo). NT2-D1 and 833 K were cultured in DMEM (ATCC, VA, USA) and RPMI-1640 medium (Thermo Fisher Scientific, Massachusetts, USA), respectively, supplemented with 10% foetal bovine serum (Thermo Fisher Scientific, Massachusetts, USA) and 1% Pen/Strep (Thermo Fisher Scientific, Massachusetts, USA) at 37 °C in a humidified 5% CO2 incubator. The morphology of both cell lines was regularly investigated, and for use in experiments, stocks of cell lines were passaged no more than ten times.
miRNA inhibition
miRNA inhibition was performed by following the manufacturer’s instruction (Ambion, CA, USA). Cells were seeded out in a six-well plate and grown overnight. Lipofectamine RNAiMAX (Invitrogen, CA, USA) transfection mix was prepared, and specific miRNA inhibitors (Supplementary Table S1) were used. After 48 hours of transfection, cells were harvested and stored at −70 °C until further use. Inhibition was verified using qPCR analysis.
Quantitative PCR
Total RNA from cell lines and tissue samples were extracted using RNeasy (Qiagen, CA, USA), and 200 ng of RNA was converted to cDNA using Qiagen miScript II RT Kit (Qiagen, Hilden, Germany). qPCR was performed using 1 ng of cDNA and the Qiagen miScript SYBR Green PCR Kit (Qiagen, Hilden, Germany) under recommended conditions on an AriaMx instrument (Agilent Technologies, Santa Clara, USA). All samples were run in triplicates, and the relative expression was calculated using the equation RQ = 2−ΔΔCT. CT values > 35 were regarded as negative. miR-25-3p has been shown to be stably expressed in TGCT cells29 and was used as a reference gene in our study. The primers used are listed in Supplementary Table S2.
Western blot
Proteins were isolated after 48-hour transfection using RIPA buffer (Sigma Aldrich, Missouri, USA) containing 150 mM NaCl, 1.0% IGEPAL® CA-630, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0, phosphatase inhibitors, and protease inhibitors. The total protein concentration was measured using a BCA protein assay kit (Thermofisher Scientific, CA, USA). 30 μg protein was loaded onto 10% Mini-PROTEAN® TGX™ Precast Gels (Bio-Rad Laboratories, CA, USA). After SDS-PAGE, the proteins were blotted onto a PVDF membrane, and the membrane was blocked in TBST with 5% skim milk before incubating with primary antibody overnight at 4 °C. An HRP conjugated secondary antibody was used, and the proteins were detected using the ImageLab machine (Bio-Rad Laboratories, CA, USA).
Cell survival
Cells (300,000) were seeded out in six-well plate and grown overnight. After 24 hours of incubation with various concentrations of cisplatin (Sigma Aldrich, Missouri, USA), cells were counted using a haemocytometer. The cells were stained with trypan blue before counting to exclude dead cells.
Cell proliferation/viability
The proliferative capacity of the cells was examined by XTT assay, containing a labelling reagent (XTT) and an electron-coupling reagent (PMS) (Roche, Basel, Switzerland)30. The absorbance was measured at 450 nm with a microplate reader after 24 hours of XTT treatment. Cells were incubated with various concentrations of cisplatin for 24 hours and then treated with 50 μl of a mixture of XTT and PMS. Cell proliferation assay was also performed after miRNA inhibition. After 48 hours of transfection with miRNA inhibitors, the cells were seeded out in a 96-well plate and cultured at a density of 3,000 cells/well. At the time points 1 hour, 12 hours, 24 hours, 48 hours, 60 hours, and 72 hours, the cells were treated with 50 μl of XTT solution.
Cell migration and invasion
For cell migration and invasion assays, the cells were seeded out and grown overnight, followed by transfection with miRNA inhibitors. The transfected cells were grown for 48 hours, followed by serum-deprivation for 24 hours. 50,000 cells were then assayed in each well of a 96-well Boyden Chamber (R&D Systems, Minnesota, USA) for migration and invasion according to the manufacturer’s protocol. 20% foetal bovine serum was used as the chemoattractant. Basement Membrane Extract (R&D Systems, Minnesota, USA) was used as the matrix barrier for the invasion assay.
Statistical analysis
The results were analysed by t-test using the PRISM software. Significant differences were defined by values of p < 0.05.
Results
Expression of selected miRNAs in TGCTs
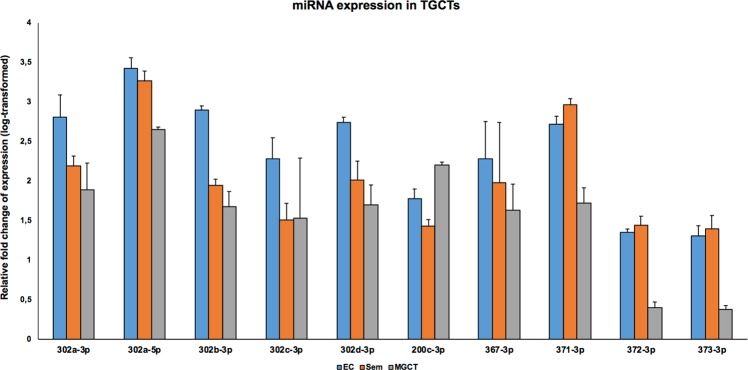
We first examined the expression levels of the ten selected miRNAs in normal and malignant testis tissue. These miRNAs were upregulated in all the three TGCT subtypes relative to normal tissue samples (Fig. 1). Notably, the expression of most of the miR-302/367 cluster members was highest in EC subtype. The expression of these miRNAs was also examined in 833 K and NT2-D1 cells representing EC subtype, in which the members of miR-302/367 cluster and miR-200c were found to be abundantly expressed, whereas the expression of miR-371-373 cluster members was not detected (data not shown).
Figure 1.
Relative expression levels of ten selected miRNAs in TGCTs. Expression levels of miRNAs in TGCTs were analysed by qPCR. The relative expression levels of the miRNAs were higher in all TGCTs after normalisation to normal testis. Particularly, the expression level of the miR-302/367 cluster was highest in embryonal carcinoma, the expression of miR-200c-3p was highest in mixed germ cell tumours, and the expression of miR-371-3p was highest in seminoma. Relative fold change of expression was determined using the equation RQ = 2−ΔΔCT; Relative fold change of expression was presented as log-transformed values and the y-axis indicates the exponents. EC: Embryonal carcinoma; Sem: Seminoma; MGT: Mixed germ cell tumours. The designation on the x-axis indicates the miRNA.
Effect of cisplatin on miRNAs in TGCT cells
IC50 dose (a concentration causing approximately 50% inhibition of the desired activity) of cisplatin was first determined to be 6 μM in 833 K and NT2-D1 cells by assessing cell survival and cell proliferation (Supplementary Fig. S1). Subsequently, the expression of the selected ten miRNAs was analysed by qPCR in both cell lines after 24-hour incubation with cisplatin (Fig. 2). Cisplatin treatment in 833 K and NT2-D1 cells resulted in a significant downregulation of miR-302a-3p, miR-302b-3p, and miR-302c-3p. The expression of miR-302d-3p was only significantly downregulated in NT2-D1 cells, whereas miR-302a-5p, miR-200c, and miR-367-3p showed no significant changes in either of the cell lines.
Figure 2.

TGCT specific miRNA expression in TGCT cells after cisplatin treatment. The expression of ten miRNAs was analysed by qPCR in both 833 K and NT2-D1 cells after 24-hour incubation with cisplatin (6 μM). Both in 833 K (a) and NT2-D1 cells (b), the expressions of miR-302a-3p, miR-302b-3p and miR-302c-3p were significantly downregulated while the downregulation of miR-302d-3p was only significant in NT2-D1 cells. The miR-302a-5p, miR-200c-3p and miR-367-3p showed no significant changes, and the expression of the miR-371-373 cluster was not detected in either cell line. Relative fold change of miRNA expression was determined using the equation RQ = 2−ΔΔCT. t-test: Control vs Cisplatin (6 μM), mean ± SD (calculated from three independent experiments), statistical significance p < 0.05. The designation on the x-axis indicates the miRNA.
Inhibition of miR302s in TGCT cells
To further study the functional role of the miRNAs shown to be differentially expressed upon cisplatin treatment, we selected miR302a-3p, miR-302b-3p and miR-302c-3p (miR-302s), and performed miRNA inhibitor-mediated transient transfection in 833 K and NT2-D1 cells. The transfection resulted in more than 80% inhibition of the expression for all three miRNAs (Fig. 3).
Figure 3.
Inhibition of miR302a-3p, miR-302b-3p, and miR-302c-3p in TGCT cells. miRNA inhibitor-mediated transient transfection reduced the expression levels of all three miRNAs by more than 80% in 833 K (a) and NT2-D1 cells (b), as analysed by qPCR. A non-targeting, miRNA inhibitor negative control with the same chemical modifications was used as a vehicle control. Relative fold change of expression was determined using the equation RQ = 2−ΔΔCT. The designation on the x-axis indicates the miRNA inhibitor.
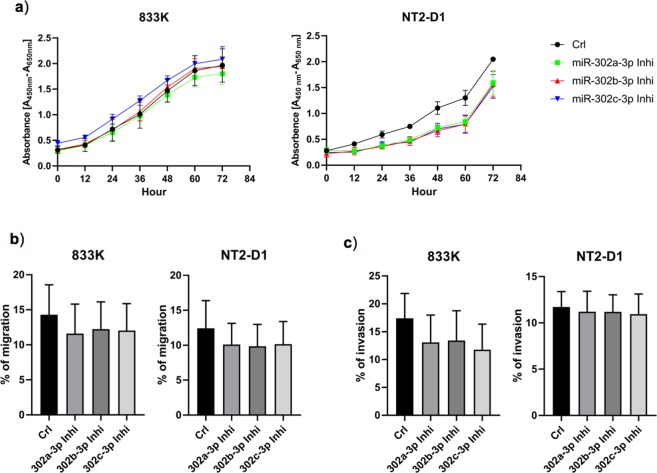
Effect of inhibition of the miR-302s on cell proliferation, migration, and invasion
Inhibition of miR-302s resulted in a significant reduction in cell proliferation in NT2-D1 cells in a time-dependent manner, whereas no significant change was observed in 833 K cells after miRNA inhibition (Fig. 4a). Inhibition of miR-302s in 833 K and NT2-D1 cells did not result in a significant reduction in cell migration and invasion, however, a tendency towards reduction in migration and invasion was observed (Fig. 4b,c).
Figure 4.
Cell proliferation, migration, and invasion in TGCT cells after inhibition of miR-302s. Inhibition of miR-302s resulted in a significant decrease of cell proliferation in NT2-D1 cells in a time-dependent manner, whereas no significant change was observed in 833 K cells (a). Inhibition of miR-302s did not result in a significant decrease in cell migration and invasion in either cell line (b,c). t-test: Control vs miRNA inhibitor, mean ± SD (calculated from three independent experiments), statistical significance p < 0.05. In figure b and c, the designation on the x-axis indicates the miRNA inhibitor.
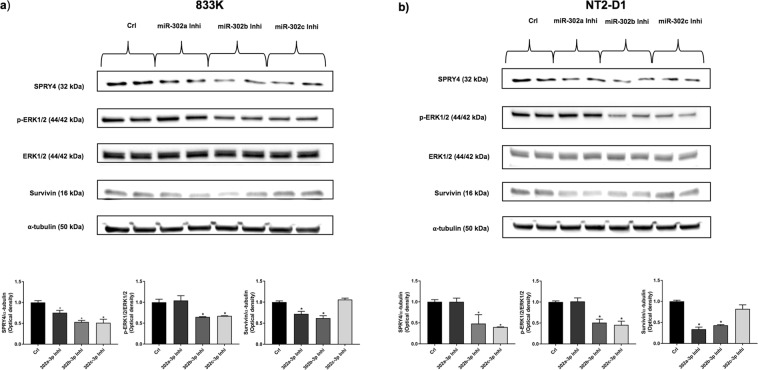
Effect of inhibition of the miR-302s on SPRY4, MAPK/ERK signalling, and apoptosis
Suppressing the expression of miR-302s significantly reduced the expression of SPRY4 (Fig. 5a,b). Inhibition of miR-302b-3p and miR-302c-3p significantly decreased the phosphorylation of ERK1/2 in NT2-D1 and 833 K cells, whereas inhibition of miR-302a-3p and miR-302b-3p resulted in a significant decrease in survivin protein expression, an inhibitor of apoptosis (Fig. 5a,b). We also examined the expression level of survivin in the tissue. We found survivin in all the TGCTs except yolk sac tumour, whereas no detection of survivin was observed in normal testis (Supplementary Fig. S4).
Figure 5.
Effect of inhibition of miR-302s on SPRY4, MAPK/ERK signalling and apoptotic inhibitor. Western blot was performed to investigate the expression of SPRY4, phosphorylation of ERK1/2 and the apoptotic inhibitor survivin after inhibition of miR-302s. Inhibition of miR-302s significantly reduced the expression of SPRY4 in both cell lines (a,b) while inhibition of miR-302b-3p and miR-302c-3p significantly decreased phosphorylation of ERK1/2 in 833 K (a) and NT2-D1 cells (b). After inhibition of miR-302a-3p and miR-302b-3p, the expression of survivin, an inhibitor of apoptosis, decreased significantly in 833 K (a) and NT2-D1 cells (b). ERK1/2 was used as endogenous control for phospho-ERK1/2 (p-ERK1/2), and α-tubulin was used as a loading control. The bar graphs show the corresponding densitometric analyses of the western blots where the ratio of p-ERK1/2/ERK1/2 was calculated after normalising to α-tubulin. SPRY4 and survivin were also normalised to α-tubulin. Samples were loaded as duplicates from two independent experiments. The cropped blots are used in the figure, and full-length blots are presented in Supplementary Figs S2 and S3. The experiments were repeated at least three times, and a representative experiment is shown. t-test: Control vs miRNA inhibitor, mean ± SD, statistical significance p < 0.05. The designation on the x-axis indicates the miRNA inhibitor.
Discussion
Several miRNAs have been reported to be associated with cancer development and are believed to function as either tumour suppressor genes or oncogenes31. Moreover, about 50% of annotated human miRNAs were found to be mapped in genomic areas, distinguished as cancer-related chromosomal fragile sites32. Though differential expression of miRNAs in TGCTs is well documented, only a few miRNAs have functionally been implicated in TGCT development17,18,22.
In the present study, we confirmed with qPCR analysis, that the ten most differentially expressed miRNAs found in our sequencing study were highly expressed in TGCTs compared to the normal testis. Particularly, expression of the miR-302/367 cluster members was highest in the embryonal carcinoma subtype. High expression of miR-302s has not been reported in cancers other than TGCTs. A low or barely detectable expression of miR-302s was reported in hepatocellular carcinoma23, gastric cancer33, colon cancer34, and cervical carcinoma24. Human embryonic stem cells (hESCs) and induced pluripotent stem cells, however, showed high expression of miR-302s, which declined rapidly after differentiation35. High expression of miR-302s in TGCTs may suggest that they act as oncogenes in TGCTs. After cisplatin treatment in 833 K and NT2-D1 cells, we found that the expressions of miR-302a-3p, miR-302b-3p, and miR-302c-3p were downregulated. It is evident that treatment with cisplatin in cancer cells alters the expression of genes which are functionally involved in the disease mechanism36. In our study, downregulation of miR-302s in TGCT cells upon cisplatin treatment as well as the high expression in TGCTs, could indicate that miR-302s play a role in TGCT pathogenesis. However, it is also possible that this result reflects merely a role in cisplatin response.
The effect of miR-302s on cell proliferation may vary depending on cell type. In our study, inhibition of miR-302s resulted in a decreased proliferation of NT2-D1 cells, but not of 833 K cells. A decreased cell proliferation was also observed with hESCs after suppressing the expression of miR-302s37. hESCs and human embryonal carcinoma cells (hECCs) have been reported to share overlapping metabolic signatures38, similar gene, protein, and miRNA expression profiles39,40. Li et al. showed that, like hECCs, hESCs expressing high levels of miR-302s could form teratomas in vivo, and suppressing the expression of miR-302s in hESCs resulted in reduced teratoma formation37. In contrast to the oncogenic properties of miR-302s as indicated in TGCT cells, overexpression of these miRNAs has been shown to suppress cell proliferation in other cancer cells23–26. Furthermore, overexpression of miR-302s inhibited cell invasion and migration in melanoma34, colon34, osteosarcoma41, and colorectal cancer cells42. Although inhibition of miR-302s in our study did not result in a significant decrease in cell migration and invasion, a tendency towards decline was observed. These differences may reflect a differing mode of action of miRNAs in different cell types, probably through regulating different target genes43. The same miRNA may have different roles in various cellular contexts44. For example, Chen et al. illustrated that ectopic expression of miR-181 had different effects on the differentiation of B cells and cytotoxic T cells45. miR-20a and miR-290 showed a pro-senescence role in mouse embryonic fibroblasts46,47, whereas in tumours and mouse ESCs, a proliferative role was observed48,49.
The miR-302-367 cluster has been demonstrated to target regulatory proteins associated with cell signalling, cell cycle, and cell death50,51. We found that inhibition of miR-302s reduced the expression of SPRY4, which is a regulator of MAPK/ERK and PI3K/Akt signalling pathways52. In our previous study, we showed that SPRY4 was highly expressed in TGCTs, but no expression was detected in normal testis, and suppression of SPRY4 in TGCT cells attenuated cell growth, migration, invasion, and phosphorylation of ERK1/2 and Akt11. Inhibition of miR-302b-3p and miR-302c-3p also resulted in a decreased phosphorylation of ERK1/2 in both cell lines. To our knowledge, our study is the first demonstrating that suppression of miR-302s inhibits the activation of MAPK/ERK signalling pathway in any cancer type. An opposite finding was reported by Wei et al., in which overexpression of miR-302a inhibited ERK1/2 phosphorylation in colorectal cancer cells42. They also showed that overexpression of miR-302a decreased Akt phosphorylation. However, we found no significant changes of Akt phosphorylation in 833 K and NT2-D1 cells after inhibition of miR-302s (data not shown). Inhibition of miR-302s in TGCT cells resulting in decreased expression of SPRY4 and reduced phosphorylation of ERK1/2 suggests that miR-302s may act as oncogene in TGCTs by targeting a tumour suppressor gene upstream SPRY4.
In our study, we also showed that inhibition of miR-302a-3p and miR-302b-3 suppressed the expression of survivin in NT2-D1 and 833 K cells. Survivin (encoded by BIRC5), an inhibitor of apoptosis, has been widely studied in malignancies, and high expression of survivin is a hallmark of virtually all human tumours including TGCTs53,54. We confirmed high expression of survivin in various TGCT samples. Decreased expression of survivin in TGCT cells upon suppression of miR-302s indicates that these miRNAs may inhibit apoptosis in TGCTs, possibly acting through the mechanism of increasing expression of survivin. Divergences in the findings in the two EC cell lines NT2-D1 and 833 K may reflect the heterogeneity of the cell lines. Both cell lines show the characteristics of EC cells, and NT2-D1 also comprises teratoma28, whereas 833 K comprises seminoma and choriocarcinoma cells in addition to EC and teratoma27. This histologic difference indicates that the 833 K cells may harbour more complex microenvironment than the NT2-D1 cells.
In conclusion, miR-302s may act as TGCT oncogenes via inducing the expression of SPRY4 and activating MAPK/ERK pathway while inhibiting apoptosis via increasing the expression of survivin. More mechanistic studies are needed to understand the role of miR-302s in TGCT pathogenesis.
Supplementary information
Acknowledgements
We are grateful to Dr Birgitte Lindeman (Norwegian Institute of Public Health, Oslo) for providing two TGCT cell lines. This study was funded by OsloMet-Oslo Metropolitan University.
Author Contributions
M.K.D., K.F. and T.B.H. are responsible for the conception of the study and the experimental design. M.K.D. and H.S.F.E. performed the experiments and all the authors interpreted data. M.K.D. was responsible for drafting the manuscript, and all authors critically revised the manuscript and approved of the final version to be published.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kari Furu and Trine B. Haugen jointly supervised.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-45573-6.
References
- 1.Znaor A, Lortet-Tieulent J, Jemal A, Bray F. International variations and trends in testicular cancer incidence and mortality. Eur Urol. 2014;65:1095–1106. doi: 10.1016/j.eururo.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 2.Trabert B, Chen J, Devesa SS, Bray F, McGlynn KA. International patterns and trends in testicular cancer incidence, overall and by histologic subtype, 1973-2007. Andrology. 2015;3:4–12. doi: 10.1111/andr.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shanmugalingam T, Soultati A, Chowdhury S, Rudman S, Van Hemelrijck M. Global incidence and outcome of testicular cancer. Clin Epidemiol. 2013;5:417–427. doi: 10.2147/CLEP.S34430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richiardi Lorenzo, Pettersson Andreas, Akre Olof. Genetic and environmental risk factors for testicular cancer. International Journal of Andrology. 2007;30(4):230–241. doi: 10.1111/j.1365-2605.2007.00760.x. [DOI] [PubMed] [Google Scholar]
- 5.Ferlin A, Foresta C. Testis cancer: genes, environment, hormones. Front Endocrinol (Lausanne) 2014;5:172. doi: 10.3389/fendo.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Litchfield Kevin, Levy Max, Orlando Giulia, Loveday Chey, Law Philip J, Migliorini Gabriele, Holroyd Amy, Broderick Peter, Karlsson Robert, Haugen Trine B, Kristiansen Wenche, Nsengimana Jérémie, Fenwick Kerry, Assiotis Ioannis, Kote-Jarai ZSofia, Dunning Alison M, Muir Kenneth, Peto Julian, Eeles Rosalind, Easton Douglas F, Dudakia Darshna, Orr Nick, Pashayan Nora, Bishop D Timothy, Reid Alison, Huddart Robert A, Shipley Janet, Grotmol Tom, Wiklund Fredrik, Houlston Richard S, Turnbull Clare. Identification of 19 new risk loci and potential regulatory mechanisms influencing susceptibility to testicular germ cell tumor. Nature Genetics. 2017;49(7):1133–1140. doi: 10.1038/ng.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Zhaoming, McGlynn Katherine A, Rajpert-De Meyts Ewa, Bishop D Timothy, Chung Charles C, Dalgaard Marlene D, Greene Mark H, Gupta Ramneek, Grotmol Tom, Haugen Trine B, Karlsson Robert, Litchfield Kevin, Mitra Nandita, Nielsen Kasper, Pyle Louise C, Schwartz Stephen M, Thorsson Vésteinn, Vardhanabhuti Saran, Wiklund Fredrik, Turnbull Clare, Chanock Stephen J, Kanetsky Peter A, Nathanson Katherine L. Meta-analysis of five genome-wide association studies identifies multiple new loci associated with testicular germ cell tumor. Nature Genetics. 2017;49(7):1141–1147. doi: 10.1038/ng.3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karlsson R, et al. Investigation of six testicular germ cell tumor susceptibility genes suggests a parent-of-origin effect in SPRY4. Hum Mol Genet. 2013;22:3373–3380. doi: 10.1093/hmg/ddt188. [DOI] [PubMed] [Google Scholar]
- 9.Kanetsky PA, et al. Common variation in KITLG and at 5q31.3 predisposes to testicular germ cell cancer. Nat Genet. 2009;41:811–815. doi: 10.1038/ng.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rapley EA, et al. A genome-wide association study of testicular germ cell tumor. Nat Genet. 2009;41:807–810. doi: 10.1038/ng.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das MK, Furu K, Evensen HF, Haugen OP, Haugen TB. Knockdown of SPRY4 and SPRY4-IT1 inhibits cell growth and phosphorylation of Akt in human testicular germ cell tumours. Sci Rep. 2018;8:2462. doi: 10.1038/s41598-018-20846-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pyle LC, Nathanson KL. Genetic changes associated with testicular cancer susceptibility. Semin Oncol. 2016;43:575–581. doi: 10.1053/j.seminoncol.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, Xu C. Role of microRNAs in mammalian spermatogenesis and testicular germ cell tumors. Reproduction. 2015;149:R127–137. doi: 10.1530/REP-14-0239. [DOI] [PubMed] [Google Scholar]
- 14.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 15.Calin GA, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. The New England journal of medicine. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 16.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rounge TB, et al. Profiling of the small RNA populations in human testicular germ cell tumors shows global loss of piRNAs. Mol Cancer. 2015;14:153. doi: 10.1186/s12943-015-0411-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmer RD, et al. Malignant germ cell tumors display common microRNA profiles resulting in global changes in expression of messenger RNA targets. Cancer Res. 2010;70:2911–2923. doi: 10.1158/0008-5472.CAN-09-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murray MJ, et al. Identification of microRNAs From the miR-371~373 and miR-302 clusters as potential serum biomarkers of malignant germ cell tumors. Am J Clin Pathol. 2011;135:119–125. doi: 10.1309/AJCPOE11KEYZCJHT. [DOI] [PubMed] [Google Scholar]
- 20.Gillis AJ, et al. High-throughput microRNAome analysis in human germ cell tumours. J Pathol. 2007;213:319–328. doi: 10.1002/path.2230. [DOI] [PubMed] [Google Scholar]
- 21.Murray MJ, et al. The two most common histological subtypes of malignant germ cell tumour are distinguished by global microRNA profiles, associated with differential transcription factor expression. Mol Cancer. 2010;9:290. doi: 10.1186/1476-4598-9-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voorhoeve PM, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 23.Wang L, et al. MicroRNA-302b suppresses cell proliferation by targeting EGFR in human hepatocellular carcinoma SMMC-7721 cells. BMC Cancer. 2013;13:448. doi: 10.1186/1471-2407-13-448. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Cai N, Wang YD, Zheng PS. The microRNA-302-367 cluster suppresses the proliferation of cervical carcinoma cells through the novel target AKT1. RNA. 2013;19:85–95. doi: 10.1261/rna.035295.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fareh M, et al. The miR 302-367 cluster drastically affects self-renewal and infiltration properties of glioma-initiating cells through CXCR4 repression and consequent disruption of the SHH-GLI-NANOG network. Cell Death Differ. 2012;19:232–244. doi: 10.1038/cdd.2011.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogawa H, et al. MicroRNAs Induce Epigenetic Reprogramming and Suppress Malignant Phenotypes of Human Colon Cancer Cells. PLoS One. 2015;10:e0127119. doi: 10.1371/journal.pone.0127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bronson DL, et al. Cell line derived from a metastasis of a human testicular germ cell tumor. Cancer Res. 1980;40:2500–2506. [PubMed] [Google Scholar]
- 28.Andrews PW, et al. Pluripotent embryonal carcinoma clones derived from the human teratocarcinoma cell line Tera-2. Differentiation in vivo and in vitro. Lab Invest. 1984;50:147–162. [PubMed] [Google Scholar]
- 29.Das MK, Andreassen R, Haugen TB, Furu K. Identification of Endogenous Controls for Use in miRNA Quantification in Human Cancer Cell Lines. Cancer Genomics Proteomics. 2016;13:63–68. [PubMed] [Google Scholar]
- 30.Bourgeois-Droin C, et al. Thymoma associated with myasthenia, erythroblastopenia, myositis and giant cell myocarditis. One case (author’s transl) Nouv Presse Med. 1981;10(2097–2098):2103–2094. [PubMed] [Google Scholar]
- 31.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 32.Calin GA, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu FY, et al. miR-302b regulates cell cycles by targeting CDK2 via ERK signaling pathway in gastric cancer. Cancer Med. 2016;5:2302–2313. doi: 10.1002/cam4.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maadi H, et al. Multimodal tumor suppression by miR-302 cluster in melanoma and colon cancer. Int J Biochem Cell Biol. 2016;81:121–132. doi: 10.1016/j.biocel.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 35.Lin SL, et al. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA. 2008;14:2115–2124. doi: 10.1261/rna.1162708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, Wood WH, III, Becker KG, Weeraratna AT, Morin PJ. Gene expression response to cisplatin treatment in drug-sensitive and drug-resistant ovarian cancer cells. Oncogene. 2007;26:2860–2872. doi: 10.1038/sj.onc.1210086. [DOI] [PubMed] [Google Scholar]
- 37.Li HL, et al. miR-302 regulates pluripotency, teratoma formation and differentiation in stem cells via an AKT1/OCT4-dependent manner. Cell Death Dis. 2016;7:e2078. doi: 10.1038/cddis.2015.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abu Dawud R, Schreiber K, Schomburg D, Adjaye J. Human embryonic stem cells and embryonal carcinoma cells have overlapping and distinct metabolic signatures. PLoS One. 2012;7:e39896. doi: 10.1371/journal.pone.0039896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Josephson R, et al. Qualification of embryonal carcinoma 2102Ep as a reference for human embryonic stem cell research. Stem Cells. 2007;25:437–446. doi: 10.1634/stemcells.2006-0236. [DOI] [PubMed] [Google Scholar]
- 40.Sperger JM, et al. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc Natl Acad Sci USA. 2003;100:13350–13355. doi: 10.1073/pnas.2235735100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie Y, et al. MiR-302b Suppresses Osteosarcoma Cell Migration and Invasion by Targeting Runx2. Sci Rep. 2017;7:13388. doi: 10.1038/s41598-017-13353-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei ZJ, et al. Up-regulation of microRNA-302a inhibited the proliferation and invasion of colorectal cancer cells by regulation of the MAPK and PI3K/Akt signaling pathways. Int J Clin Exp Pathol. 2015;8:4481–4491. [PMC free article] [PubMed] [Google Scholar]
- 43.Kim YJ, et al. MicroRNA 21 regulates the proliferation of human adipose tissue-derived mesenchymal stem cells and high-fat diet-induced obesity alters microRNA 21 expression in white adipose tissues. J Cell Physiol. 2012;227:183–193. doi: 10.1002/jcp.22716. [DOI] [PubMed] [Google Scholar]
- 44.Rizzo M, Mariani L, Pitto L, Rainaldi G, Simili M. miR-20a and miR-290, multi-faceted players with a role in tumourigenesis and senescence. J Cell Mol Med. 2010;14:2633–2640. doi: 10.1111/j.1582-4934.2010.01173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 46.Poliseno L, et al. The proto-oncogene LRF is under post-transcriptional control of MiR-20a: implications for senescence. PLoS One. 2008;3:e2542. doi: 10.1371/journal.pone.0002542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pitto L, et al. miR-290 acts as a physiological effector of senescence in mouse embryo fibroblasts. Physiol Genomics. 2009;39:210–218. doi: 10.1152/physiolgenomics.00085.2009. [DOI] [PubMed] [Google Scholar]
- 48.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marson A, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao Z, Zhu X, Dou Y. The miR-302/367 cluster: a comprehensive update on its evolution and functions. Open Biol. 2015;5:150138. doi: 10.1098/rsob.150138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim JY, et al. MicroRNA-302 induces proliferation and inhibits oxidant-induced cell death in human adipose tissue-derived mesenchymal stem cells. Cell Death Dis. 2014;5:e1385. doi: 10.1038/cddis.2014.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Masoumi-Moghaddam S, Amini A, Morris DL. The developing story of Sprouty and cancer. Cancer Metastasis Rev. 2014;33:695–720. doi: 10.1007/s10555-014-9497-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Altieri DC. Targeting survivin in cancer. Cancer Lett. 2013;332:225–228. doi: 10.1016/j.canlet.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weikert S, et al. The inhibitor of apoptosis (IAP) survivin is expressed in human testicular germ cell tumors and normal testes. Cancer Lett. 2005;223:331–337. doi: 10.1016/j.canlet.2004.10.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.