Abstract
Introduction
Nasal colonization with coagulase-negative staphylococci (CoNS) may be a preliminary risk factor for systemic infection. The aim of this study was to assess the frequency of ica A and D genes and biofilm formation among hospital-acquired nasal colonizing CoNS strains isolated from neonates in the neonatal intensive care units (NICUs). Antibiotic sensitivity patterns and some relevant risk factors were estimated.
Methods
This study assessed nasal colonization with CoNS among neonates at days one and three of admission to NICUs of Beni-Suef University Hospital and Beni-Suef General Hospital from November 2015 to May 2016. The isolates were screened and identified; susceptibility testing was performed. Biofilm formation was examined using the Congo red agar method. Isolates identified as CoNS were tested by polymerase chain reaction (PCR) for the presence of mecA and icaA and icaD genes.
Results
A total of 340 nasal swabs were collected from 170 neonates. The incidence of nasal colonization with CoNS was 50%. The species most frequently isolated were S. haemolyticus and S. epidermidis. Multidrug resistance (MDR) was detected in 86% of isolates. It was found that there was a strong association between the presence of mecA gene and phenotypic resistance to methicillin and also the presence of the icaA gene and biofilm formation.
Conclusions
Neonates admitted to NICUs can become reservoirs for CoNS strains, leading to potential dissemination of MDR strains into the community.
Keywords: Coagulase-negative Staphylococcus, icaA, icaD, mecA, neonatal intensive care units
Introduction
Coagulase-negative staphylococci (CoNS) have been associated with late onset sepsis in the neonatal intensive care unit (NICU), particularly in very low birth weight neonates.1 In a clinical setting, the most common sources of CoNS are the patient’s own bacterial microbiota and the hands of healthcare workers.2
Neonates are prone to developing infections, due to factors such as: an immature immune system, using invasive procedures, and aggressive antibiotic treatment. Consequently, CoNS strains disperse in NICUs environments. Invasive procedures favor the migration of CoNS from the skin to the catheter insertion point, leading to infection.3
Moreover, the ability of CoNS strains to form biofilm on polymeric surfaces make them more pathogenic.4 Biofilm consists of multilayered clusters of cells embedded in an extracellular polysaccharide (slime) matrix that facilitates adhesion and protects against the host immune system and antimicrobials.5 The intercellular adhesion (ica) operon is responsible for the production of slime, encoded by the genes of icaADBC; the gene icaA encodes N-acetylglucosaminyl transferase, the enzyme involved in the synthesis of polysaccharide intercellular adhesion (PIA). icaD plays an important role in maximum expression of N-acetyl glucosaminyl transferase, resulting in the capsular polysaccharide's full phenotype expression. It has been reported that icaA and icaD play a significant role among ica genes in the formation of biofilms.6,7
In addition, the presence of mecA gene on the mobile genetic element staphylococcal cassette chromosome (SCCmec), decreased its affinity to methicillin.8 The resistance of mecA-positive isolates to wide-spectrum β-lactams is spread globally in staphylococci from hospitals, farms as well as community environments, making it ineffective to treat staphylococcal infections.9
The aims of the present study were to evaluate the rate of hospital acquired nasal colonization by CoNS during hospitalization following the neonate's admission to NICUs, the potential risk factors for colonization, antibiotic susceptibility testing of the isolates to determine multidrug-resistant (MDR) organisms and the genetic relatedness with their phenotypic resistance to antibiotics. Moreover, the ability of the isolates to form biofilms was evaluated, as well as their relationships with certain genes (icaA and icaD).
Methods
The current study is a prospective cross-sectional one that was carried out in Beni-Suef University hospital and Beni-Suef General Hospital from November 2015 to May 2016. One hundred and seventy neonates were enrolled in the study. All neonates were eligible for nasal swab (Sterile swab ST, CK) collection at admittance and three days after. Informed written consents from guardian were obtained at enrollment. Blood cultures were done for suspected cases with sepsis.
Full history was taken, including different risk factors for CoNS colonization including: age, sex, weight, route of delivery, sepsis occurrence, prematurity, use of antimicrobial drugs during the period of hospitalization, period of hospitalization (in days), presence of external devices such as cannula, nasal cannula and use of continuous positive airway pressure (CPAP), and existence of comorbidities such as chronic diseases, genetic diseases, infectious diseases, or congenital malformations.
All samples were transported immediately in Stuart’s medium to the Microbiology Laboratory of Beni-Suef University hospital for processing.
Identification of bacterial isolates
All nasal swabs and blood cultures were inoculated onto blood agar, MacConkey’s agar and mannitol salt agar (Oxoid, Basingstoke, UK). After incubation, the suggestive colony was identified by the conventional microbiological standard tests. Further identification of CoNS isolates to the species level was performed by API Staph. (BioMérieux, Marcy-l'Étoile, France).10
Antibiogram, biological characteristics and biochemical tests were conducted for all isolates collected; we excluded the cases found on the first day and which continued to appear on the 3rd day, as they were not considered as hospital-acquired.
Susceptibility tests
CoNS isolates were submitted to a disc diffusion susceptibility test using the following antibiotics: penicillin (10 μg), oxacillin (1 μg), cefoxitin (30 μg), erythromycin (15 μg), clindamycin (2 μg), rifampicin (15 μg), gentamicin (15 μg), ciprofloxacin (5 μg), tetracycline (30 μg), vancomycin (30 μg), linezolid (30 μg), ceftazidime (30 μg), sulfamethoxazole-trimethoprim (23.75/1.25 μg) and novobiocin (Oxoid). Inhibition zones were interpreted according to Clinical Laboratory and Standards Institute (CLSI) guidelines.11 The D zone test for inducible resistance to clindamycin was performed. Resistance to at least three or more classes of antibiotics was considered as MDR.12
Detection of biofilm production
A phenotypic method was used, according to Freeman et al.,13 by the Congo red agar (CRA) medium. A positive result was indicated by black colonies, whereas nonproducing strains produced red colonies. Darkening of the colonies in the absence of a dry crystalline morphology indicated a moderate biofilm formation.14
Detection of mecA gene phenotypically
This was done by detecting the susceptibility of the isolates to cefoxitin by disc diffusion method as a preliminary screening test for the existence of mecA gene. Resistant strains were tested for the existence of mecA gene by PCR.
Detection of mecA, icaA and icaD genes by PCR
The bacterial DNA was extracted using QIAGEN DNA extraction kit (Qiagen, Manchester, UK) and the procedure was followed according to the manufacturer’s instructions.
PCR amplification of the mecA gene was performed using a primer set (salt-free 0.2 µmol Germany 49 bp). Two pairs of primers SA-1 (5' CGG TAA CAT TGA TCG CAA CGT TCA 3') were used as the forward primer and SA-2 (5' CTT TGG AAC GAT GCC TAA TCT CAT 3') was used as the reverse primer to amplify a 214-bp fragment of the mecA gene. The set included an internal control template. For amplification of mecA gene the protocol described in Rigatti et al.15 was used.
The amplified products were separated by electrophoresis on 2% agarose gel prepared in 0.5X TBE buffer and stained with SYBR Safe. The gels were photographed under UV transillumination.
PCR for icaA and icaD genes was done using the method described by Arciola et al.6 For detection of icaA gene 5-TCTCTTGCAGGAGCAATCAA-3 was used as the forward primer (corresponding to nucleotides 4796-4815) and 5-TCAGGCACTAACATCCAGCA-3 was used as the reverse primer (corresponding to nucleotides 4964-4983). For icaD, 5-ATGGTCAAGCCCAGACAGAG-3 was used as the forward primer (corresponding to nucleotides 5422–5441), and 5-CGTGTTTTCAACATTTAATGCAA-3 was used as the reverse primer (corresponding to nucleotides 5616–5597). The amplified product sizes were estimated by comparison with 100 bp DNA ladder.16
Two S. epidermidis reference strains were used, the slime-producing strain ATCC 35984 (RP62A), as positive control for icaA and icaD, and the non-slime-producing strain ATCC 12228 as a negative control for icaA and icaD.6 S. aureus strain ATCC 43300 was used as a positive control for the mecA gene.15
Statistical analysis
The collected data were coded then entered and analyzed using Statistical Package for Social Science SPSS v22.0 (IBM Corp, Armonk, NY, USA). Descriptive statistics were done for categorical variables by frequency and percentage, and for numerical variation in the form of mean and standard deviation (mean±SD). Suitable statistical tests of significance were used: Chi-square for categorical data and independent sample t-test for numerical data. P-values equal to or less than 0.05 were considered statistically significant
Results
During the period of the study, 340 nasal swabs were collected from 170 neonates (at days one and three of admission) of NICUs in Beni-Suef University Hospital and Beni-Suef General Hospital.
Using antibiograms, biological characteristics and biochemical tests, comparing the isolates found in the 1st and 3rd days, 20 of these isolates were discarded, as they were not considered as hospital-acquired colonization. Isolates that appeared only on the third day were included in the study as nosocomial colonization; they were 100 isolates, while 50 neonates revealed no growth finding.
The incidence of nosocomial colonization in the present study was (100/170) 58.8%. It was found that the most common isolates were Gram-positive organisms, reported in 86/100 isolates. CoNS were the most prevalent (50/100), followed by S. aureus (36/100) while Gram negative bacilli were reported in 14/100 cases. Klebsiella pneumoniae was the most common (8/100) followed by E. coli (4/100) and Pseudomonas aeruginosa (2/100).
In the current study, males predominated (64.2%). The age range was 1-38 days with a mean of 7.2±8.9 days. Furthermore, the most common cause of admission was respiratory distress (47.6%).
In the present study, an incidence of 50% of CoNS colonizers among isolates was recorded. Further identification of CoNS isolates was done using API Staph (BioMérieux) kit; it revealed that S. haemolyticus (24/50, 48%) and S. epidermidis (18/50, 36%) strains were the most prevalent yields, followed by S. xylosus (4/50, 8%), S. hominis (2/50, 4%) and S. chromogenes (2/50, 4%).
Based on the data obtained from the descriptive analysis of the important risk factors included, comparing CoNS colonized cases with no growth cases, there was a significant association with the use of intravenous cannula, patients with signs of sepsis and administration of empirical antimicrobials in cases colonized with CoNS (p<0.001) (Table 1). Furthermore, the age of the neonates showed statistical significance as a risk factor (Table 2).
Table 1. The potential risk factors for acquisition of nosocomial colonization by CoNS in neonatal intensive care units.
| Risk factors | CoNS positive | No growth | P value | Chi-square value | df |
|---|---|---|---|---|---|
| Sex | |||||
| Male N=63 (63%) | 32 (64%) | 31 (62%) | 0.500 | 0.043 | 1 |
| Female N=37 (37%) | 18 (36%) | 19 (38%) | |||
| Mode of delivery | |||||
| Caesarean section N=66 (66%) | 31 (62%) | 35 (70%) | 0.263 | 0.713 | 1 |
| Vaginal N=34 (34%) | 19 (38%) | 15 (30%) | |||
| Prematurity | |||||
| Preterm N=68 (68%) | 36 (72%) | 32 (64%) | 0.260 | 0.735 | 1 |
| Full term N=32 (32%) | 14 (28%) | 18 (36%) | |||
| Presence of signs of sepsis | |||||
| N=44 (44%) | 44 (88%) | 0 (0%) | <0.001* | 92.3 | 1 |
| Presence of intravenous cannula | |||||
| N=55 (55%) | 50 (100%) | 5 (10%) | <0.001* | 81.8 | 1 |
| Presence of other devices | |||||
| N=78 (78%) | 42 (84%) | 36 (72%) | 0.114 | 2.98 | 1 |
| Congenital anomalies | |||||
| N=17 (17%) | 11 (22%) | 6 (12%) | 0.356 | 6.6 | 6 |
| Empirical antimicrobial administration | |||||
| N=56 (56%) | 50 (100%) | 6 (12%) | <0.001* | 78.6 | 1 |
| Respiratory distress on admission | |||||
| N=77 (77%) | 39 (78%) | 38 (76%) | 0.500 | 0.056 | 1 |
Data is presented as number (percentage). CoNS – coagulase-negative staphylococci; df – degrees of freedom
p value <0.05 is considered significant by Chi-Square test.
Table 2. Age, weight and length of hospitalization as a risk factors for acquisition of nosocomial colonization in neonatal intensive care units.
| Number | Mean | SD | 95% confidence interval for mean | P value | |||
|---|---|---|---|---|---|---|---|
| Lower bound | Upper bound | ||||||
| Age (days) | CoNS positive | 50 | 10.5 | 10.2 | 1.7 | 5.8 | <0.001* |
| No growth | 50 | 3.8 | 7.1 | 7.6 | 13.4 | ||
| Weight (Kg) | CoNS | 50 | 2.4 | 782.3 | 2086.5 | 2535.1 | 0.716 |
| No growth | 50 | 2.3 | 789. 4 | 2145.9 | 2590.5 | ||
p value <0.05 is considered significant by independent sample t-test.
CoNS – coagulase-negative staphylococci.
In addition, a strong relationship was established between colonization with CoNS in neonates with cases suspected of having sepsis that revealed blood cultures positive for CoNS (p<0.001) (Table 3).
Table 3. Relation between blood culture positive with CoNS in neonates suspected with sepsis, neonates with nasal colonization by CoNS, and others showing no growth.
| No sepsis | Clinically suspected sepsis with negative blood culture | Clinically suspected sepsis and positive blood culture | P-value | Chi-square value | df | ||
|---|---|---|---|---|---|---|---|
| No growth | Number (50) | 49 | 0 | 1 | <0.001* | 85.1 | 1 |
| CoNS | Number (50) | 2 | 4 | 44 |
p value <0.05 is considered significant by Chi-Square test. df – degrees of freedom.
Detection of biofilm formation by Congo red agar (CRA)
Out of 50 CoNS isolates, 26 (52%) isolates were Congo red positive. Congo red positive strains appeared bright black or dry black colonies indicating biofilm formation, while Congo red negative were 24 (48%), their color ranged from pink to red indicating non-biofilm formers (Figure 1).
Figure 1. CoNS strains tested for biofilm formation by Congo red test (A) Congo red positive isolates (growth appears black in color). (B) Congo red negative isolates (growth appears red in color).

Table 3. Relation between blood culture positive with CoNS in neonates suspected with sepsis, neonates with nasal colonization by CoNS, and others showing no growth
Among CoNS isolates, the frequency of multidrug resistance detected was as high as (86%); the resistance was mostly demonstrated against the following antibiotics: penicillin (98%), followed by ceftriaxone (96%), oxacillin and cefoxitin (82%) (Table 4). Inducible clindamycin resistance between clindamycin and erythromycin was detected in only five cases (Figure 2).
Table 4. Antimicrobial susceptibility of CoNS to different antibiotics.
| Antimicrobial | Sensitive | Intermediate | Resistant | Total | |||
|---|---|---|---|---|---|---|---|
| Number | Percentage | Number | Percentage | Number | Percentage | Number | |
| Penicillin | 1 | 2 | 0 | 0 | 49 | 98 | 50 |
| Oxacillin | 9 | 18 | 0 | 0 | 41 | 82 | 50 |
| Vancomycin | 33 | 66 | 1 | 2 | 16 | 32 | 50 |
| Erythromycin | 18 | 36 | 0 | 0 | 32 | 64 | 50 |
| Clindamycin | 31 | 62 | 1 | 2 | 18 | 36 | 50 |
| Tetracycline | 16 | 32 | 0 | 0 | 34 | 68 | 50 |
| Sulfamethoxazole-trimethoprim | 23 | 46 | 1 | 2 | 26 | 52 | 50 |
| Rifampicin | 47 | 94 | 0 | 0 | 3 | 6 | 50 |
| Cefoxitin | 9 | 18 | 0 | 0 | 41 | 82 | 50 |
| Ceftazidime | 2 | 4 | 0 | 0 | 48 | 96 | 50 |
| Linezolid | 49 | 98 | 0 | 0 | 1 | 2 | 50 |
| Ciprofloxacin | 24 | 48 | 0 | 0 | 26 | 52 | 50 |
| Gentamicin | 32 | 64 | 0 | 0 | 18 | 36 | 50 |
| Novobiocin | 50 | 100 | 0 | 0 | 0 | 0 | 50 |
Figure 2. Inducible inhibitor D shape resistance.
All CoNS strains were tested for methicillin resistance using cefoxitin and for the presence of the mecA gene by PCR. Forty-one isolates out of 50 (82%) proved to be mecA gene positive and showed resistance to cefoxitin and other β-lactam antibiotics as well, with a total of 42/50 resistance rate for cefoxitin (84%). The relationship between the presence of mecA and phenotypic methicillin resistance in CoNS was statistically significant (p<0.001) (Table 5). There was no evident connection between biofilm formation and methicillin resistance among the tested isolates (p=0.704) (Table 6).
Table 5. Relation between phenotypic expression of methicillin resistance and genotypic mecA gene positivity (n=50).
| Presence of mecA gene | Methicillin phenotype, n (%) | P-value | Chi-square value | df | |
|---|---|---|---|---|---|
| Resistant | Sensitive | ||||
| Negative (n=9) | 1 (11.1%) | 8 (88.9%) | <0.001* | 43.4 | 1 |
| Positive (n=41) | 41 (100%) | 0 (0%) | |||
p value <0.05 is considered significant by Chi-Square test. df – degrees of freedom.
Table 6. The relation between biofilm formation and methicillin phenotype.
| Methicillin phenotype, n (%) | P value | Chi-square value | df | ||
|---|---|---|---|---|---|
| Sensitive N=8 | Resistant N=42 | ||||
| Biofilm positive 26 (52%) | 5 (62.5%) | 21 (50%) | 0.399 | 0.420 | 1 |
| Biofilm negative 24 (48%) | 3 (37.5%) | 21 (50%) | |||
df – degrees of freedom.
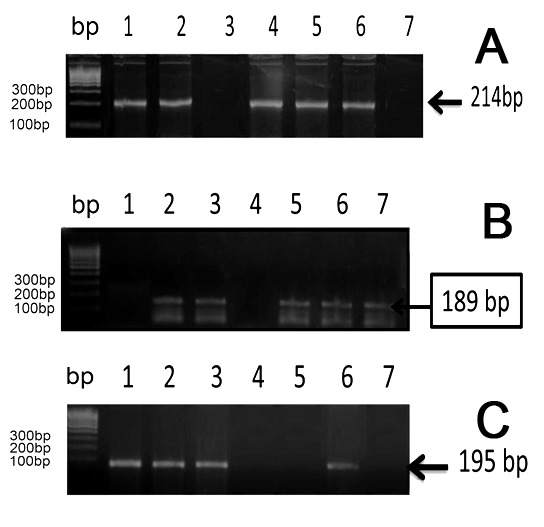
The presence of the icaA (189 bp) and icaD (198 bp) genes among CoNS isolates was demonstrated by the amplification of the corresponding fragments. There were 29 (58%) isolates positive for icaA, while 30 (60%) strains were positive for the icaD gene. A clear relationship was observed between biofilm formation and detection of icaA gene among the tested strains (p=0.025). On the other hand, the icaD gene was detected only in 43.3% of CoNS biofilm formers, statistically non-significant (p=0.133) as shown in Table 7 and Figure 3.
Table 7. Comparison between Congo red agar test and PCR test results for icaA and icaD gene.
| Congo red agar, n (%) | Total | P-value | Chi-square value | df | ||
|---|---|---|---|---|---|---|
| Positive N=26 | Negative N=24 | |||||
| icaA | ||||||
| Positive | 19 (73.1%) | 10 (41.7%) | 29 (58.0%) | 0.025* | 5.1 | 1 |
| Negative | 7 (26.9%) | 14 (58.3%) | 21 (42.0%) | |||
| icaD | ||||||
| Positive | 13 (50.0%) | 17 (70.8%) | 20 (40.0%) | 0.133 | 2.3 | 1 |
| Negative | 13 (50.0%) | 7 (29.2%) | 30 (60.0%) | |||
Figure 3. PCR detection of icaA, icaD and mecA. Lane 1, 100 bp DNA molecular size marker (A) PCR detection of mecA gene locus (A, 214-bp band). Lane 1 shows the DNA marker, lanes 1, positive control for mecA gene S. aureus strain ATCC 43300. Lane 7 negative control. Lanes 2, 4, 5, 6 were positive. Lane 3 negative case. (B) PCR detection of icaA gene locus (B, 189-bp band). Lane 2, positive control for icaA gene S. epidermidis reference strain (ATCC 25923). Lane 1, negative control for icaA gene S. epidermidis reference strain (ATCC 12228). Lanes 3, 5, 6, 7 were positive. Lane 4 negative case. Figure (C) PCR detection of icaD gene locus (C, 195-bp band). Lanes 2, 3, 6 positive cases. Lanes 4, 5 negative cases. Lane 1, positive control for icaD gene S. epidermidis reference strain (ATCC 25923). Lanes 7 negative control for icaD gene S. epidermidis reference strain (ATCC 12228).
Discussion
The current study was conducted on 170 neonates admitted in the NICUs of Beni-Suef University Hospital and Beni-Suef General Hospital, over a period of five months. Nasal swabs (340) were taken from neonates at the time of admission and after three days. Isolates that appeared only on the third day were included in the study as nosocomial colonization.
The present findings revealed that CoNS were the most prevalent isolates (50/100), a finding that is consistent with Ternes et al.,17 who found rates of colonization with CoNS of 55.9%.
Species identification is important for monitoring the reservoir and understanding the pathogenic potential of different CoNS species.18 The results revealed that S. haemolyticus (48%) and S. epidermidis (36%) were the most common species.
Similar results have been reported in other studies.19,20
The main risk factors for colonization in this study were the use of intravenous cannula and empirical antimicrobial therapy. These observations were supported by several studies.21,22 These findings could be explained by the fact that the exposure to a variety of procedures such as intubation, catheters, peripheral venous lines, and parenteral nutrition allow entry of microbes.18
A statistically significant association was reported between the clinically featured neonates with sepsis and blood culture positive for CoNS and the clinical status of patients regarding the clinical sepsis (p<0.001). Most of the neonates with clinically suspected sepsis proved by blood culture results were nasally colonized with CoNS (44/50, 88%). This finding was comparable to the work done by Shehab El-Din et al., who reported that the rate of neonatal sepsis confirmed to have bloodstream infection with CoNS was found to be significant (52.86%).20
Moreover, the prevalence of multidrug resistance was evident among CoNS isolates; that could be explained by the intensive selective pressure by antibiotic therapy administered to neonates in the NICU.23 In the current study, a high rate of resistance was detected to β-lactam antibiotics such as penicillin (98%), ceftriaxone (96%), oxacillin and cefoxitin (82%), which was confirmed by the presence of mecA gene in 82% of isolates. These findings paralleled the results obtained by Ternes et al.17
Biofilm is one of the most important virulence factors for CoNS that mediates resistance to antimicrobial therapy and guaranties chronicity of infection. CoNS isolates from the enrolled neonates (52%) were biofilm formers, using CRA medium, a finding that came in agreement with that of Nasr et al.,7 where 56% of CoNS isolates were biofilm producers.
It was found that co-expression of some genes such as icaA and icaD are necessary for phenotypic expression of biofilm in many isolates.24 icaA and icaD genes were detected by PCR as a reference of phenotypic method in 30 (60%) CoNS isolates in the present study. These results were consistent with Sharma et al.23 who found that 68.2% of the clinical isolates were positive for the ica genes. A clear relationship was present between biofilm formation and detection of icaA and icaD genes among the tested strains in this study, which was statistically significant with the icaA gene (p=0.025). A good correspondence between phenotypic characterization methods and the presence of ica genes was reported by Nasr et al.7
Unexpectedly, in the current study, 26.9% and 50% of the isolates were icaA and icaD negative and produced biofilm as detected by CRA methods, this concurs with the findings reported by Nasr et al.,7 and enlightened by the presence of genes inducing an alternative PIA-independent mechanism of biofilm formation in both S. aureus and S. epidermidis.25 However, lack of biofilm production by certain isolates despite their positivity for the ica genes like our findings, was elucidated by Post et al.26 They reported that although ica genes are responsible for biofilm formation, full phenotypic expression could be conditioned by additional genes (atlE, sarA, agrA and mecA) that have direct or indirect regulatory impact.
This study had the following limitations: follow-up of the neonates for colonization at several intervals and at discharge as well as following up the outcome of each case could have added more data to the present findings. However, due to lack of funding and materials available, we couldn't extend it any further.
Conclusions
Taken together, it can be concluded that nasal screening of neonates should be considered in NICUs to preclude nasal colonization that may predispose to neonatal sepsis. Prematurity, use of devices and abuse of antibiotics are serious risk factors for colonization with CoNS. Consequently, the quality of medical services in the NICU environment should be improved with constant training and education of healthcare providers for measures and procedures of infection control, over and above, the implementation of strict antibiotic policies.
Furthermore, the biofilm forming CoNS stains showed resistance to antibiotics potentially leading to treatment failure and development of more resistance. Hence, a better tool is recommended for screening of biofilm formation that might help to modify antibiotic therapy and averting hospital-acquired infections in NICUs. Similarly, anti-adhesive strategies could be applied to inhibit bacterial surface adhesion or to disrupt an already established biofilm, targeting the biofilm matrix. It is vital and imperative to continue investigating the environmental factors, which might contribute to increasing incidence of nasal colonization of neonates with resistant strains.
Acknowledgements
The authors would like to thank all participants and other healthcare workers who helped all through the process of the research.
Footnotes
Authors’ contributions statement: DMA designed the research plan and organized the study. SMAS carried out all laboratory tests, coordinated the data analysis, and had a major contribution in writing of the manuscript. MHM supervised all clinical issues of patients. NHI participated in the writing of the manuscript and had a major role in following-up all steps of the study. MAWAM participated in the main role of editing and revising the manuscript. All authors reviewed and approved the final version of the manuscript.
Conflicts of interest: All authors – none to declare.
Funding: None to declare.
References
- 1.Alfaleh KM. Incidence of late onset neonatal sepsis in very low birth weight infants in a tertiary hospital: an ongoing challenge. Sultan Qaboos Univ Med J. 2010;10:227–30. [PMC free article] [PubMed] [Google Scholar]
- 2.Becker K, Heilmann C, Peters G. Coagulase-negative staphylococci. Clin Microbiol Rev. 2014;27:870–926. doi: 10.1128/CMR.00109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walz JM, Memtsoudis SG, Heard SO. Prevention of central venous catheter bloodstream infections. J Intensive Care Med. 2010;25:131–8. doi: 10.1177/0885066609358952. [DOI] [PubMed] [Google Scholar]
- 4.Szczuka E, Jabłońska L, Kaznowski A. Coagulase-negative staphylococci: pathogenesis, occurrence of antibiotic resistance genes and in vitro effects of antimicrobial agents on biofilm-growing bacteria. J Med Microbiol. 2016;65:1405–13. doi: 10.1099/jmm.0.000372. [DOI] [PubMed] [Google Scholar]
- 5.Mathur T, Singhal S, Khan S, Upadhyay DJ, Fatma T, Rattan A. Detection of biofilm formation among the clinical isolates of Staphylococci: an evaluation of three different screening methods. Indian J Med Microbiol. 2006;24:25–9. doi: 10.4103/0255-0857.19890. [DOI] [PubMed] [Google Scholar]
- 6.Arciola CR, Baldassarri L, Montanaro L. Presence of icaA and icaD genes and slime production in a collection of staphylococcal strains from catheter-associated infections. J Clin Microbiol. 2001;39:2151–6. doi: 10.1128/JCM.39.6.2151-2156.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nasr RA, AbuShady HM, Hussein HS. Biofilm formation and presence of icaAD gene in clinical isolates of staphylococci Egypt. J Med Hum Genet. 2012;13:269–74. [Google Scholar]
- 8.Svensson K, Hellmark B, Söderquist B. Characterization of SCCmec elements in methicillin-resistant Staphylococcus epidermidis isolated from blood cultures from neonates during three decades. APMIS. 2011;119:885–93. doi: 10.1111/j.1600-0463.2011.02801.x. [DOI] [PubMed] [Google Scholar]
- 9.Miragaia M. Factors contributing to the evolution of mecA-mediated β-lactam resistance in staphylococci: Update and new insights from whole genome sequencing (WGS) Front Microbiol. 2018;9:2723. doi: 10.3389/fmicb.2018.02723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mermel LA, Farr BM, Sherertz RJ, et al. Guidelines for the management of intravascular catheter-related infections. Clin Infect Dis. 2001;32:1249–72. doi: 10.1086/320001. [DOI] [PubMed] [Google Scholar]
- 11.Clinical Laboratory Standards Institute antimicrobial susceptibility testing standards (CLSI) Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement CLSI document M100-S26. Wayne, PA, USA: Clinical and Laboratory Standards Institute; 2014. [Google Scholar]
- 12.Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 13.Freeman DJ, Falkiner FR, Keane CT. New method for detecting slime production by coagulase-negative staphylococci. J Clin Pathol. 1989;42:872–4. doi: 10.1136/jcp.42.8.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jain A, Agarwal A. Biofilm production, a marker of pathogenic potential of colonizing and commensal staphylococci. J Microbiol Methods. 2009;76:88–92. doi: 10.1016/j.mimet.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 15.Rigatti F, Tizotti MK, Hörner R, et al. Oxacillin-resistant coagulase-negative Staphylococci bacteremia at a teaching hospital in Santa Maria, State of Rio Grande do Sul, Brazil. Rev Soc Bras Med Trop. 2010;43:686–90. doi: 10.1590/s0037-86822010000600017. [DOI] [PubMed] [Google Scholar]
- 16.Nuryastuti T, Praseno P, Mustafa M. Preliminary study of biofilm formation properties and antibiotic susceptibility pattern of MRSA and MSSA isolates obtained in Yogyakarta, Indonesia. Malays J Microbiol. 2015;11:383–90. [Google Scholar]
- 17.Ternes YM, Lamaro-Cardoso J, ré MC, et al. Molecular epidemiology of coagulase-negative Staphylococcus carriage in neonates admitted to an intensive care unit in Brazil. BMC Infect Dis. 2013;13:572. doi: 10.1186/1471-2334-13-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kasim K, El Sadak A, Zayed K, Abdel-Wahed A, Mosaad M. Nosocomial infections in a neonatal intensive care unit. Middle East J Sci Res. 2014;19: 01–07. [Google Scholar]
- 19.Keim L, Torres-Filho, Silva PV, Teixeira LA. Prevalence, aetiology and antibiotic resistance profiles of coagulase negative staphylococci isolated in a teaching hospital. Braz J Microbiol. 2011;42:248–55. doi: 10.1590/S1517-83822011000100031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shehab El-Din EM, El-Sokkary MM, Bassiouny MR, Hassan R. Epidemiology of neonatal sepsis and implicated pathogens: A study from Egypt. Biomed Res Int. 2015;2015:509484. doi: 10.1155/2015/509484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang LR, Peng MJ, Li H, Pang Y. Pathogen distribution and risk factors of nosocomial infections in neonates in the neonatal intensive care unit. Zhongguo Dang Dai Er Ke Za Zhi. 2013;15:112–6. [PubMed] [Google Scholar]
- 22.Camacho-Gonzalez A, Spearman PW, Stoll BJ. Neonatal infectious diseases: evaluation of neonatal sepsis. Pediatr Clin North Am. 2013;60:367–89. doi: 10.1016/j.pcl.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma P, Lahiri KK, Kapila K. Conventional and molecular characterization of coagulase-negative Staphylococcus in hospital isolates. Indian J Pathol Microbiol. 2011;54:85–9. doi: 10.4103/0377-4929.77331. [DOI] [PubMed] [Google Scholar]
- 24.Gad GF, El-Feky MA, El-Rehewy MS, Hassan MA, Abolella H, El-Baky RM. Detection of icaA, icaD genes and biofilm production by Staphylococcus aureus and Staphylococcus epidermidis isolated from urinary tract catheterized patients. J Infect Dev Ctries. 2009;3:342–51. doi: 10.3855/jidc.241. [DOI] [PubMed] [Google Scholar]
- 25.Kogan G, Sadovskaya I, Chaignon P, Chokr A, Jabbouri S. Biofilms of clinical strains of Staphylococcus that do not contain polysaccharide intercellular adhesin. FEMS Microbiol Lett. 2006;255:11–6. doi: 10.1111/j.1574-6968.2005.00043.x. [DOI] [PubMed] [Google Scholar]
- 26.Post V, Harris LG, Morgenstern M, et al. Comparative genomics study of Staphylococcus epidermidis from orthopedic device-related infections correlated with patient outcome. J Clin Microbiol. 2017;55:3089–103. doi: 10.1128/JCM.00881-17. [DOI] [PMC free article] [PubMed] [Google Scholar]


