Abstract
More women are delaying child-birth. Thus, the diagnosis of pregnancy-associated breast cancer (PABC) will continue to increase. The aim of this study was to identify core candidate genes of PABC, and the relevance of the genes on the prognosis of PABC. GSE31192 and GSE53031 microarray profile datasets were downloaded from the Gene Expression Omnibus database and differentially expressed genes were analyzed using the R package and GEO2R tool. Then, Gene Ontology and Kyoto Encyclopedia of Gene and Genome pathway enrichment analyses were performed using the Database for Annotation, Visualization, and Integrated Discovery. Moreover, the Search Tool for the Retrieval of Interacting Genes and the Molecular Complex Detection Cytoscape software plug-in were utilized to visualize protein–protein interactions and to screen candidate genes. A total of 239 DEGs were identified in PABC, including 101 up-regulated genes mainly enriched in fatty acid activation and the fibroblast growth factor signaling pathway, while 138 down-regulated genes particularly involved in activation of DNA fragmentation factor and apoptosis-induced DNA fragmentation. Fourteen hub genes with a high degree of connectivity were selected, including CREB1, ARF3, UBA5, SIAH1, KLHL3, HECTD1, MMP9, TRIM69, MEX3C, ASB6, UBE2Q2, FBXO22, EIF4A3, and PXN. Overall survival (OS) analysis of core candidate genes was performed using the Gene Expression Profiling Interactive Analysis and UALCAN websites. High ASB6 expression was associated with worse OS of PABC patients. Molecular subtypes and menopause status were also associated with worse OS for PABC patients. In conclusion, ASB6 could be a potential predictor and therapeutic target in patient with PABC.
Keywords: bioinformatical analysis, differentially expressed genes, pregnancy-associated breast cancer, prognosis
Introduction
Pregnancy-associated breast cancer (PABC) refers to breast cancer that is diagnosed during gestation or within 1 year of delivery [1–3]. Pregnancy-related disease occurs in 0.2–3.8% of all breast cancer patients, comprising 1:10000 to 1:3000 pregnancies. The prevalence is much higher in pre-menopausal women, with 10–25% of breast cancers estimated to be pregnancy-associated [4–9].
Although breast cancer is the most frequently diagnosed malignancy among women and is increasing in incidence, PABC has remained a rare event in the last decade [5,8,10,11]. However, PABC has increased more recently as the government policy in China allows the birth of a second child and given the trend of women in postponing pregnancy [12–15]. While PABC formation and progression has been well-studied, the biology of PABC is still not clear. The poor prognosis of PABC has been attributed to a higher risk for nodal involvement, higher hormonal milieu, and more frequent delay in diagnosis [3,16–20]. However, several studies found no major differences in the expression of hormone receptors and a more common association with the high expression of potentially relevant cancer targets, such as insulin growth factor, Wnt/β-catenin and RANK ligand, and low prevalence of tumor-infiltrating lymphocytes [21–25]. There have been considerable efforts in managing breast cancer that develops during pregnancy. Yet, very little progress has been in understanding of the biology of this disease. This challenging scenario in PABC demands knowledge of the causes and potential molecular mechanisms of PABC, and molecular biomarkers for the early diagnosis and individualized therapy.
Gene microarray profiling is widely used in life sciences. This approach can quickly detect all the expressed genes within the same tissue and reveals differentially expressed genes (DEGs) [26]. The use of the gene chip can uncover thousands of DEGs associated with different biological processes, different signal pathways, or molecular functions of cancer. The popularity of gene microarray profiling has generated a huge amount of data that is available in public databases. These data can be scrutinized to integrate and re-analyze gene expression profiles, provide new methods, and obtain valuable clues for the disease of interest.
There are many reports that integrate gene expression profiles on breast cancer, however, hard to see it in PABC [27–29]. In this study, we screened two microarray profile datasets, GSE31192 [30] and GSE53031 [31], from the National Center for Biotechnology Information Gene Expression Omnibus database (NCBI-GEO). Then, we filtered DEGs using combined GEO2R and R package analyses, and performed Gene Ontology (GO) and Kyoto Encyclopedia of Gene and Genome (KEGG) pathway enrichment analyses of DEGs with the Database for Annotation, Visualization, and Integrated Discovery (DAVID). The protein–protein interaction (PPI) network of DEGs developed with the Search Tool for the Retrieval of Interacting Genes (STRING) and top modular analysis with Cytoscape software were used to identify core candidate genes in PABC. The association of the core candidate genes with overall survival (OS) was assessed using the Gene Expression Profiling Interactive Analysis (GEPIA) and UALCAN websites. The bioinformatics analyses revealed a number of DEGs. In particular, ASB6 (ankyrin repeat and suppressor of cytokine signaling (SOCS) box containing 6) could be potentially considered as a biomarker in PABC.
Materials and methods
Acquisition microarray datasets
Gene expression microarray data were obtained from the GSE31192 and GSE53031 microarray profile datasets in the NCBI-GEO database (http://www.ncbi.nlm.nih.gov/geo/), which is a public and freely available platform. GSE31192 is based on the GPL570 platform of the Affymetrix Human Genome U133 Plus 2.0 Array, which includes 20 PABC samples containing six cancer epithelial tissues, six cancer stroma tissues, three normal epithelial tissues, and five normal stroma tissues, and 13 non-PABC control samples that contained four cancer epithelial tissues, four cancer stroma tissues, three normal epithelial tissues, and two normal stroma tissues. GSE53031 is based on the GPL13667 platform of the Affymetrix Human Genome U219 Array, and contains 54 PABC tissues and 113 non-PABC control tissues.
Detection processing of DEGs
GEO2R (Online: https://www.ncbi.nlm.nih.gov/geo/geo2r/) is an online tool that can compare two or more groups of samples in the GEO series datasets. A P-value < 0.05 and |log FC| > 1 were set as the cut-off criteria. For the integrated analysis of the high-throughput functional genomic expression data, the raw data were processed in R according to the |log FC| > 1 and q-value < 0.05 criteria. The combination of GEO2R and R was used to select DEGs. The biological coefficient of variation, which was the square-root of the dispersions, was set to 0.01 following the suggestion in the R manual and Volcano plots was constructed [32]. Datasets that separated up-regulated and down-regulated DEGs were uploaded into FunRich version 3.1.3, a free software for functional enrichment analysis of gene sets that contains additional features [33]. Venn diagram of the analysis results plotted commonly changed DEGs using the P < 0.05 and |log FC| > 1 criteria.
GO and KEGG pathway analysis of DEGs
GO and KEGG pathway enrichment analyses were performed to detect the biological functions and potential pathway of DEGs. GO is a commonly used method to annotate genes and gene products, and identifies characteristic biological attributes for high-throughput genome or transcriptome data [34]. KEGG is a collection of databases dealing with genomes, biological pathways, diseases, drugs, and chemical substances [35,36]. GO and KEGG analyses were applied through the DAVID (Online: https://david.ncifcrf.gov/), a website bioinformatics resource that can provide tools for the gene annotation, visualization, and integrated discovery function [37]. The cut-off criterion was P < 0.05.
PPI network and module analyses
Search Tool for the Retrieval of Interacting Genes (STRING, https://string-db.org) is an online tool designed to evaluate PPIs [38]. The DEGs involved in the PPI network were identified using STRING, with confidence network edges and a medium confidence of 0.400 as the product criteria. The Tab Separated Values format document was downloaded as a simple tabular text output. Cytoscape_v3.6.0 was used to construct the PPI network and analyze the interactions of the candidate DEGs encoding proteins [39]. The Molecular Complex Detection (MCODE) software application was used to screen the top modules in the PPI network with a degree cut-off = 2, node score cut-off = 0.2, k-core = 2, and maximum depth = 100. The corresponding proteins in the central nodes and highly degree were potential core proteins encoded by key candidate genes that have important physiological regulatory functions.
OS analysis of core candidate genes in PABC
GEPIA (http://gepia.cancer-pku.cn/) is a web-based tool that rapidly delivers customizable functionalities based on TCGA and GTEx data. Patient survival analysis can be conducted [40]. UALCAN (Online: http://ualcan.path.uab.edu/index.html) is a user-friendly, interactive web resource used to analyze cancer transcriptome data. UALCAN allows users to identify up- and down-regulated expressed genes in individual cancer types, as well as to estimate the effect of gene expression level and clinicopathologic features on patient survival [41]. Presently, GEPIA was used to determine OS related to core candidate genes that had been identified. The effects of the expression of the core candidate genes on PABC OS according to molecular subtypes and menopause status were assessed by UALCAN and P-values were calculated.
Results
Identification of DEGs in PABC
NCBI-GEO is a free database of microarray profiles and next-generation sequencing data. The database was used to obtain PABC, non-PABC, and normal or adjacent stroma tissue gene expression profiles of the GSE31192 and GSE53031 datasets. A total of 1712 and 2642 DEGs were respectively up-regulated in these two datasets, while 2171 and 1388 DEGs were respectively down-regulated using P<0.05 and |logFC|>1 as the cut-off criteria (Figures 1 and 2). After integrated bioinformatic analysis, 239 common DEGs were identified from the two profile datasets. These included 101 up-regulated genes and 138 down-regulated genes in PABC, compared with non-PABC (Figure 2 and Table 1).
Figure 1. Analysis of the differentially expression genes between the PABC and non-PABC using Volcano plots.
Red and green points represent up-regulated and down-regulated genes. No differentially expressed genes in blue points. (A) DEGs based on GSE31192. (B) DEGs based on GSE53031.
Figure 2. Identification of 239 (101 up-regulated and 138 down-regulated) commonly changes differentially expression genes from GSE31192 and GSE53031 microarray profile datasets.
Different color areas stand for up-regulated and down-regulated genes, which were defined with P < 0.05 and |log FC| > 1 as the cut-off criterion.
Table 1. 239 differentially expressed genes were identified from two profile datasets, including 101 up-regulated genes and 138 down-regulated genes in PABC.
| DEGs | Genes name |
|---|---|
| Up-regulated | ASB6, GRIN2C, NOL8, TMEM97, TMEM132E, C11orf16, PDE11A, OR10A4, DIP2A, C15orf62, GRK5, RGS12, KRI1, SLC2A6, CD209, GGA1, CXCR5, VARS, PARVB, STAT5B, TBKBP1, UBASH3B, SOX6, RIN1, ATP6AP1L, BEST1, FOXRED2, ACSBG1, FCRLA, CPEB1, PLCG1, POLR2J4, GTDC1, MGAT4B, LSS, GRAPL, DISP1, MUC20, POU2F2, KCNJ16, BTN3A1, KIR2DL5A, PXN, PDE10A, LRCH3, DBF4B, AK2, PLEKHO2, PTPN7, PGBD5, ZDHHC3, FCRL2, TRIM69, RPS28, FMNL1, ADCY7, HLA-DOB, KCNIP2, CRYGS, LRRC28, GNL1, TOMM40L, AIF1, CCDC146, AP4B1, FGD2, FAM129C, SLCO3A1, DMRT3, IRF1, RAB7B, C8orf34, ADAMTSL1, LOC81691, SLC19A3, TNFRSF9, KLHL3, ACRC, ZNF93, AGFG2, CIDEC, TAP2, IL12RB1, SEZ6L, SGK494, CD22, FAM124A, VWCE, TBC1D1, THOC5, ITGB7, KCNJ10, APOBEC3G, SRGAP2, APOBEC3B, ACSL6, CENPN, CD274, TNFSF11, MMP9, CXCL13 |
| Down-regulated | FSIP1, RAB30, AR, RERG, COPB1, ACADSB, TPD52, DGUOK, ZNF33A, SYTL2, EPB41L5, WBP11, GLRB, ZNF33B, TRERF1, SLC25A15, AIMP1, AIM1, COX11, APLP2, RPRD1A, FBXO22, SLC19A2, CUL4A, SFXN2, FAM83H, ANO1, ZNF8, ZBTB38, ANG, ABHD12, MORF4L2, MICALCL, KDM4B, ZNF639, CCT8, ZNF562, ARF3, TBC1D12, SRPK2, ABI2, RAB14, BMPR1A, GFPT1, NDFIP2, G2E3, TIA1, DPY19L4, SCOC, NCOR1, TRUB1, SMARCA1, IFNAR1, PLEKHA1, ZADH2, USP14, TP53I11, KLHL8, VPS54, NUDT19, APOOL, ZNF551, PEX13, RPF1, AKT1S1, TSPYL1, DOCK1, TMEM106B, SS18, WDR61, YIPF6, REL, RAB11A, GNAQ, MIB1, SLC12A6, NOM1, SPCS2, ARMCX1, RMND5A, SEC23B, RNF103, TMEM33, EPS15L1, UBA5, PPP4R1, AKTIP, RALBP1, FBXO11, CEP290, ZNF664, FAR1, GTF2A2, ZRANB1, EIF4A3, RUFY2, ZNF302, RALGAPA1, HMGB1, KLF3, CTBS, ZNF3, EXOSC3, AKT2, GLUD1, MEX3C, UBE2Q2, KDELR2, CLIP1, SCARB2, SMARCE1, HIBADH, EIF1AX, STRN3, IER3IP1, HINT1, GFM1, SLK, PAN3, ZNF24, ZMYM2, SECISBP2, CREB1, PRPF39, NEK4, CDKN2B, NPTN, TWF1, KPNA1, OSBPL9, ZYG11B, SNRPB2, CCDC90B, HECTD1, MGST2, SIAH1, DNM1L, PICALM |
DEGs GO and KEGG pathway enrichment analyses
For a more in-depth understanding of the screened DEGs, we analyzed the GO and KEGG pathway enrichment of the identified DEGs through DAVID. All the DEGs were imported to DAVID and GO analysis comprised three functional groups: biological process (BP) group, molecular function (MF) group, and cellular component (CC) group. As shown in Figure 3A–C and Table 2, the GO BP group analysis indicated that the up-regulated genes were mainly enriched in the immune response, hemopoietic or lymphoid organ development, immune system development, antigen processing, and presentation and leukocyte differentiation. The down-regulated genes were mainly enriched in the macromolecule catabolic process, cellular macromolecule catabolic process, modification-dependent protein catabolic process, modification-dependent macromolecule catabolic process, and vesicle-mediated transport (Figure 3A). For the GO MF group, the up-regulated genes were mainly enriched in peptide binding, GTPase activator activity, 3′,5′-cyclic-GMP phosphodiesterase activity, antigen binding, and long-chain-fatty-acid-CoA ligase activity. The down-regulated genes were mainly enriched in small conjugating protein ligase activity, ubiquitin protein ligase activity, acid-amino acid ligase activity, ligase activity and formation of carbon-nitrogen bonds, and GTPase activity (Figure 3B). Concerning the GO CC group, the up-regulated genes were mainly enriched in plasma membrane, TAP complex, MHC class I peptide loading complex, plasma membrane, and integral to plasma membrane. The down-regulated genes were mainly enriched in nuclear lumen, Golgi apparatus, nucleolus, intracellular organelle lumen, and organelle lumen (Figure 3C).
Figure 3. Biology function and signaling pathway analysis of DEGs in PABC.
(A) GO significant enriched of biological process. (B) GO significant enriched of molecular function. (C) GO significant enriched of Cellular Component. (D) KEGG significant enriched of pathway.
Table 2. Genes ontology enrichment analysis of differentially expressed genes in PABC.
| DEGs | Category | Term | Count | Fold enrichment | P-value | FDR |
|---|---|---|---|---|---|---|
| Up-regulated | GOTERM_BP_DIRECT | GO:0006955∼immune response | 12 | 3.17 | 0.00 | 1.66 |
| GOTERM_BP_DIRECT | GO:0048534∼hemopoietic or lymphoid organ development | 7 | 4.92 | 0.00 | 4.10 | |
| GOTERM_BP_DIRECT | GO:0002520∼immune system development | 7 | 4.63 | 0.00 | 5.46 | |
| GOTERM_BP_DIRECT | GO:0019882∼antigen processing and presentation | 4 | 8.81 | 0.01 | 7.57 | |
| GOTERM_BP_DIRECT | GO:0002521∼leukocyte differentiation | 4 | 5.58 | 0.03 | 14.34 | |
| GOTERM_MF_DIRECT | GO:0042277∼peptide binding | 5 | 4.38 | 0.02 | 8.76 | |
| GOTERM_MF_DIRECT | GO:0005096∼GTPase activator activity | 5 | 4.04 | 0.03 | 5.52 | |
| GOTERM_MF_DIRECT | GO:0047555∼3′,5′-cyclic-GMP phosphodiesterase activity | 12 | 50.81 | 0.03 | 9.21 | |
| GOTERM_MF_DIRECT | GO:0003823∼antigen binding | 3 | 9.52 | 0.03 | 9.49 | |
| GOTERM_MF_DIRECT | GO:0004467∼long-chain-fatty-acid-CoA ligase activity | 2 | 35.56 | 0.04 | 10.89 | |
| GOTERM_CC_DIRECT | GO:0005886∼plasma membrane | 31 | 1.36 | 0.03 | 8.79 | |
| GOTERM_CC_DIRECT | GO:0042825∼TAP complex | 2 | 47.42 | 0.03 | 9.15 | |
| GOTERM_CC_DIRECT | GO:0042824∼MHC class I peptide loading complex | 2 | 36.88 | 0.04 | 7.21 | |
| GOTERM_CC_DIRECT | GO:0044459∼plasma membrane part | 20 | 1.50 | 0.04 | 8.84 | |
| GOTERM_CC_DIRECT | GO:0005887∼integral to plasma membrane | 12 | 1.67 | 0.04 | 8.27 | |
| Down-regulated | GOTERM_BP_DIRECT | GO:0009057∼macromolecule catabolic process | 15 | 2.18 | 0.00 | 12.00 |
| GOTERM_BP_DIRECT | GO:0044265∼cellular macromolecule catabolic process | 14 | 2.19 | 0.01 | 15.46 | |
| GOTERM_BP_DIRECT | GO:0019941∼modification-dependent protein catabolic process | 12 | 2.37 | 0.01 | 16.88 | |
| GOTERM_BP_DIRECT | GO:0043632∼modification-dependent macromolecule catabolic process | 12 | 2.37 | 0.01 | 16.88 | |
| GOTERM_BP_DIRECT | GO:0016192∼vesicle-mediated transport | 12 | 2.36 | 0.01 | 17.26 | |
| GOTERM_MF_DIRECT | GO:0019787∼small conjugating protein ligase activity | 9 | 6.12 | 1.05E-04 | 0.14 | |
| GOTERM_MF_DIRECT | GO:0004842∼ubiquitin-protein ligase activity | 8 | 6.14 | 3.10E-04 | 0.41 | |
| GOTERM_MF_DIRECT | GO:0016881∼acid-amino acid ligase activity | 9 | 5.05 | 3.89E-04 | 0.52 | |
| GOTERM_MF_DIRECT | GO:0016879∼ligase activity, forming carbon-nitrogen bonds | 9 | 4.39 | 9.70E-04 | 1.29 | |
| GOTERM_MF_DIRECT | GO:0003924∼GTPase activity | 8 | 4.28 | 0.00 | 3.40 | |
| GOTERM_CC_DIRECT | GO:0031981∼nuclear lumen | 21 | 1.85 | 0.00 | 8.61 | |
| GOTERM_CC_DIRECT | GO:0005794∼Golgi apparatus | 15 | 2.19 | 0.00 | 8.63 | |
| GOTERM_CC_DIRECT | GO:0005730∼nucleolus | 13 | 2.38 | 0.00 | 9.14 | |
| GOTERM_CC_DIRECT | GO:0070013∼intracellular organelle lumen | 24 | 1.72 | 0.00 | 9.95 | |
| GOTERM_CC_DIRECT | GO:0043233∼organelle lumen | 24 | 1.68 | 0.01 | 12.81 |
Abbreviations: BP, biological process; CC, cellular component; Count, the number of enriched genes in each term; FDR, false discovery rate; MF, molecular function.
In addition, functional and signaling pathway enrichment of the DEGs was conducted. The up-regulated genes were mainly enriched in fatty acid activation, fibroblast growth factor signaling pathway, lysophosphatidic acid receptor mediated events, cell-extracellular matrix interactions, and CXCR4-mediated signaling events. The down-regulated genes were mainly enriched in activation of DNA fragmentation factor, apoptosis-induced DNA fragmentation, valine degradation I, apoptotic execution phase, and Notch signaling pathway (Figure 3D and Table 3).
Table 3. Signaling pathway enrichment analysis of differentially expressed genes in PABC.
| DEGs | Biological pathway | Count | P-value | Mapped gene names |
|---|---|---|---|---|
| Up-regulated | Fatty acid activation | 2 | 0.00 | ACSBG1, SL6 |
| FGF signaling pathway | 3 | 0.00 | STAT5B, CG1, MP9 | |
| LPA receptor mediated events | 4 | 0.00 | PLCG1, XN, DCY7, MP9 | |
| Cell-extracellular matrix interactions | 2 | 0.00 | PARVB, XN | |
| CXCR4-mediated signaling events | 5 | 0.00 | STAT5B, LCG1, XN, TPN7, MP9 | |
| EGFR interacts with phospholipase C-gamma | 2 | 0.01 | PLCG1, DCY7 | |
| Signaling by EGFR | 3 | 0.02 | PLCG1, XN, DCY7 | |
| Chemokine receptors bind chemokines | 2 | 0.02 | CXCR5, XCL13 | |
| E-cadherin signaling in the nascent adherens junction | 5 | 0.02 | STAT5B, LCG1, DCY7, TGB7, MMP9 | |
| IL6-mediated signaling events | 2 | 0.03 | IRF1, TNFSF11 | |
| Down-regulated | Activation of DNA fragmentation factor | 2 | 0.00 | HMGB1, KPNA1 |
| Apoptosis induced DNA fragmentation | 2 | 0.00 | HMGB1, KPNA1 | |
| Valine degradation I | 2 | 0.00 | ACADSB, HIBADH | |
| Apoptotic execution phase | 3 | 0.00 | HMGB1, KPNA1, DNM1L | |
| Notch signaling pathway | 4 | 0.00 | AR, NCOR1, RAB11A, MIB1 | |
| Branched-chain amino acid catabolism | 2 | 0.00 | ACADSB, HIBADH | |
| PIP3 activates AKT signaling | 2 | 0.01 | AKT1S1, CREB1 | |
| Glutamate biosynthesis II | 1 | 0.01 | GLUD1 | |
| PI-3K cascade | 2 | 0.02 | AKT1S1, CREB1 | |
| AKT-mediated inactivation of FOXO1A | 1 | 0.02 | CREB1 |
Count: the number of enriched genes in each term.
DEGs PPI network and modular analysis
Using the STRING online database, a total of 139 DEGs (54 up-regulated and 85 down-regulated genes) of the 239 commonly altered DEGs were filtered into the PPI network complex. One hundred of the 239 DEGs did not fall into the PPI network. Based on the STRING output and Cytoscape software analysis, 54 nodes and 131 edges were identified in up-regulated DEGs and 85 nodes and 188 edges in down-regulated DEGs. The PPI network complex defaulter filter degree was between 1 and 20 inclusive (Figure 4A).
Figure 4. DEGs PPI network complex and the module analysis.
Up-regulated genes were marked in red, down-regulated genes were marked in light blue, and the lines show the interaction between the DEGs. A node degree is defined as the number of other nodes connected to the node. (A) DEGs were filtered into PPI network complex using STRING online database and Cytoscape software. (B) The module which the most highlighted circle areas. (C) High degree nodes value and genes name.
According to the degree of genes and the module was selected using the MCODE Cytoscape plug-in, the most significant fourteen node degree genes identified using a filtering degree >7 as the criterion were CREB1, ARF3, UBA5, SIAH1, KLHL3, HECTD1, MMP9, TRIM69, MEX3C, ASB6, UBE2Q2, FBXO22, EIF4A3, and PXN (Figure 4B,C). The proteins of these central nodes might be the core proteins and the genes could be key candidate genes with important roles in the progress of PABC and prognosis. They could be potential therapeutic targets.
Core candidate genes OS analysis
The prognostic information of the 14 core candidate genes is freely available in http://gepia.cancer-pku.cn. High expression of ASB6 (P < 0.01) was associated with worse OS of PABC patients. No difference on OS was evident with the high expression of KLHL3 (P = 0.64), MMP9 (P = 0.3), TRIM69 (P = 0.61), and PXN (P = 0.12), or with the low expression of CREB1 (P = 0.29), ARF3 (P = 0.053), UBA5 (P = 0.093), SIAH1 (P = 0.3), HECTD1 (P = 0.39), MEX3C (P = 0.72), UBE2Q2 (P = 0.85), FBXO22 (P = 0.43), and EIF4A3 (P = 0.21) (Figure 5A–N).
Figure 5. Overall survival values of core candidate genes in pregnancy-associated breast cancer.
(A) CREB1 (204314_s_at; P = 0.29). (B) ARF3 (200734_s_at; P = 0.053). (C) UBA5 (222579_at; P = 0.093). (D) SIAH1 (202981_x_at; P = 0.3). (E) KLHL3 (221221_s_at; P = 0.64). (F) HECTD1 (241683_at; P = 0.39). (G) MMP9 (203936_s_at; P = 0.3). (H) TRIM69 (229037_at; P = 0.61). (I) MEX3C (1556873_at; P = 0.72). (J) ASB6 (221657_s_at; P < 0.01). (K) UBE2Q2 (224747_at; P = 0.85). (L) FBXO22 (225736_at; P = 0.43). (M) EIF4A3 (201303_at; P = 0.21). (N) PXN (201087_at; P = 0.12).
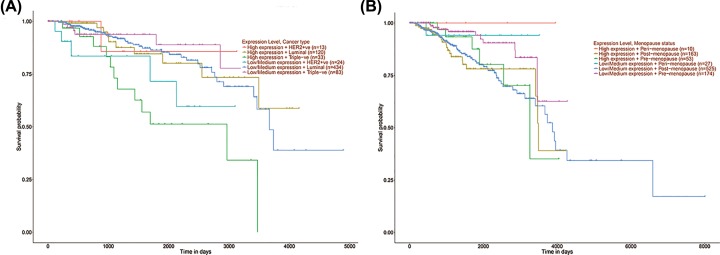
Of the 14 identified genes, the high expression of ASB6 was associated with worse OS for PABC patients according to molecular subtypes and menopause status. Compared with low expression of ASB6 in triple negative PABC, high expression of ASB6 was associated with worse OS for PABC patients, while the opposite result was obtained in the Her-2 overexpression subtype (Figure 6A). High expression of ASB6 was associated with poor OS in pre-menopausal PABC patients. By contrast, low expression of ASB6 was associated with a poor prognosis in peri-menopausal patients (Figure 6B). No statistical significance of ASB6 expression levels was evident according to luminal subtype and post-menopause status.
Figure 6. Effect of core candidate genes expression level in molecular subtypes and menopause status on pregnancy-associated breast cancer overall survival.
(A) ASB6 expression level effect OS of different subtypes on PABC (P < 0.01). (B) ASB6 expression level effect OS of different menopause status on PABC (P = 0.02).
Discussion
The prevalence of PABC is increasing, and the situation will become serious as the policy of planned birth has changed in China. Early diagnosis, prevention, and personalized therapy of PABC are vital and require more knowledge of the disease.
This study integrated two microarray profile datasets and utilized bioinformatics methods to identify 239 common DEGs. The genes comprised 101 up-regulated and 138 down-regulated genes. The 239 DEGs were divided into biological process, molecular function, and cellular component groups by GO and further clustered based on KEGG pathways with significant enrichment analysis. The up-regulated DEGs were mainly enriched in the immune response, peptide binding, plasma membrane, and fatty acid activation. The influences of fatty acids on cancer have been extensively studied, with variable influences apparent in different cancers [42–46]. The down-regulated DEGs were mainly enriched in the macromolecule catabolic process, small conjugating protein ligase activity, nuclear lumen, and activation of DNA fragmentation factor. Interestingly, the signaling pathway enrichment analysis indicated that epidermal growth factor receptor and protein kinase B, which is closely related to cell growth, proliferation, and apoptosis, play a critical role in cancer [47–52]. By reviewing the previous studies, Harvell et al. and Ma et al. found that PABC stroma tissue differentially expressed immune response genes, involved in angiogenesis and extracellular matrix deposition, associated with high-grade tumors [30,53]. Azim et al. and Muller et al. observed that CXCR4, belong to G protein-coupled receptor family of cell surface molecules, increase the metastatic potential of breast cancer cells during pregnancy [31,54]. These are consisted with our GO and KEGG enrichment results, will help us to further understand this cohort of patients.
Among the presently identified CREB1, ARF3, UBA5, SIAH1, KLHL3, HECTD1, MMP9, TRIM69, MEX3C, ASB6, UBE2Q2, FBXO22, EIF4A3, and PXN were identified as core genes that displayed a high degree of connectivity in the PPI network. These genes were expressed at higher or lower levels in PABC tissues. The up-regulation of ASB6 was significantly associated with worse OS of PABC patients. The same result was detected in triple negative molecular subtypes and pre-menopausal patients. Further analyses implicated ASB6 as having a central role in the biological process of PABC and in the prognosis. ASB6 might be a potentially valuable therapeutic molecular target.
ASB6 belongs to a family of ankyrin repeat proteins that contain a C-terminal SOCS box motif. ASB6 can couple to an adaptor protein containing PH and SH2 domains that interacts with insulin receptor to execute glucose transport [55,56]. Accumulating evidence suggests that the SOCS box acts as a bridge between specific substrate-binding domains and more generic proteins that comprise a large family of E3 ubiquitin protein ligases, which mediated the ubiquitination and subsequent proteasomal degradation of target proteins [57]. ASB6 up-regulation by Areca nut extracts has been identified in normal keratinocytes and oral cancer cells, and is highly associated with a worse prognosis of oral squamous cell carcinoma [58]. The authors also reported differential expression of ASB6 and its influence on prognosis, which are consistent with our findings. Whether these observations with ASB6 in two different types of cancer extend to other cancers remains to be determined.
In conclusion, this study identified DEGs through integrated bioinformatical analysis to find potential core candidate genes involved in PABC. ASB6 was identified as potentially having a pivotal role in the occurrence, development, and prognosis of PABC. ASB6 is thus implicated as a novel biomarker and potential therapeutic target.
Abbreviations
- ASB6
ankyrin repeat and suppressor of cytokine signaling box containing 6
- DAVID
Database for Annotation, Visualization, and Integrated Discovery
- DEG
differentially expressed gene
- GEPIA
gene expression profiling interactive analysis
- GO
gene ontology
- KEGG
Kyoto Encyclopedia of Gene and Genome
- MCODE
molecular complex detection
- OS
overall survival
- PABC
pregnancy-associated breast cancer
- PPI
protein–protein interaction
- SOCS
suppressor of cytokine signaling
- STRING
Search Tool for the Retrieval of Interacting Genes
Funding
This study was supported by the National Natural Science Foundation of China [grant numbers 81372843, 81502518, 81472472 and 81502300].
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Author Contribution
Jiao Zhang, Yan-Jun Zhou and Zhi-Hao Yu conceived and designed the study, performed the statistical analysis and wrote the manuscript; Ao-Xiang Chen and Yue Yu collected the data; Xin Wang and Xu-Chen Cao revised the manuscript. All authors read and approved the final manuscript.
References
- 1.Wallack M.K., Wolf J.A. Jr, Bedwinek J., Denes A.E., Glasgow G., Kumar B.. et al. (1983) Gestational carcinoma of the female breast. Curr. Probl. Cancer 7, 1–58 10.1016/S0147-0272(83)80006-3 [DOI] [PubMed] [Google Scholar]
- 2.Case A.S. (2016) Pregnancy-associated breast cancer. Clin. Obstet. Gynecol. 59, 779–788 10.1097/GRF.0000000000000235 [DOI] [PubMed] [Google Scholar]
- 3.Vinatier E., Merlot B., Poncelet E., Collinet P. and Vinatier D. (2009) Breast cancer during pregnancy. Eur. J. Obstet. Gynecol. Reprod. Biol. 147, 9–14 10.1016/j.ejogrb.2009.06.030 [DOI] [PubMed] [Google Scholar]
- 4.Andersson T.M., Johansson A.L., Hsieh C.C., Cnattingius S. and Lambe M. (2009) Increasing incidence of pregnancy-associated breast cancer in Sweden. Obstet. Gynecol. 114, 568–572 10.1097/AOG.0b013e3181b19154 [DOI] [PubMed] [Google Scholar]
- 5.Eibye S., Kjaer S.K. and Mellemkjaer L. (2013) Incidence of pregnancy-associated cancer in Denmark, 1977–2006. Obstet. Gynecol. 122, 608–617 10.1097/AOG.0b013e3182a057a2 [DOI] [PubMed] [Google Scholar]
- 6.Lee G.E., Mayer E.L. and Partridge A. (2017) Prognosis of pregnancy-associated breast cancer. Breast Cancer Res. Treat. 163, 417–421 10.1007/s10549-017-4224-6 [DOI] [PubMed] [Google Scholar]
- 7.Petrek J.A. (1994) Pregnancy safety after breast cancer. Cancer 74, 528–531 10.1002/cncr.2820741342 [DOI] [PubMed] [Google Scholar]
- 8.Rojas K.E., Bilbro N., Manasseh D.M. and Borgen P.I. (2018) A review of pregnancy-associated breast cancer: diagnosis, local and systemic treatment, and prognosis. J. Womens Health (Larchmt) 10.1089/jwh.2018.7264 [DOI] [PubMed] [Google Scholar]
- 9.Cottreau C.M., Dashevsky I., Andrade S.E., Li D.K., Nekhlyudov L., Raebel M.A.. et al. (2018) Pregnancy-associated cancer: a U.S. population-based study. J. Womens Health (Larchmt) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F.. et al. (2016) Cancer statistics in China, 2015. CA Cancer J. Clin. 66, 115–132 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 11.Siegel R.L., Miller K.D. and Jemal A. (2016) Cancer statistics, 2016. CA Cancer J. Clin. 66, 7–30 10.3322/caac.21332 [DOI] [PubMed] [Google Scholar]
- 12.Behboudi-Gandevani S., Ziaei S. and Khalajabadi-Farahani F. (2013) Delayed child bearing: new serious challenge in Iran. J. Res. Med. Sci. 18, 83. [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson J.A., Tough S. and Sogc Genetics C. (2012) Delayed child-bearing. J. Obstet. Gynaecol. Can. 34, 80–93 10.1016/S1701-2163(16)35138-6 [DOI] [PubMed] [Google Scholar]
- 14.Loibl S., Schmidt A., Gentilini O., Kaufman B., Kuhl C., Denkert C.. et al. (2015) Breast cancer diagnosed during pregnancy: adapting recent advances in breast cancer care for pregnant patients. JAMA Oncol. 1, 1145–1153 10.1001/jamaoncol.2015.2413 [DOI] [PubMed] [Google Scholar]
- 15.Merlo D.F., Ceppi M., Filiberti R., Bocchini V., Znaor A., Gamulin M.. et al. (2012) Breast cancer incidence trends in European women aged 20-39 years at diagnosis. Breast Cancer Res. Treat. 134, 363–370 10.1007/s10549-012-2031-7 [DOI] [PubMed] [Google Scholar]
- 16.Peccatori F.A., Lambertini M., Scarfone G., Del Pup L. and Codacci-Pisanelli G. (2018) Biology, staging, and treatment of breast cancer during pregnancy: reassessing the evidences. Cancer Biol. Med. 15, 6–13 10.20892/j.issn.2095-3941.2017.0146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schedin P. (2006) Pregnancy-associated breast cancer and metastasis. Nat. Rev. Cancer 6, 281–291 10.1038/nrc1839 [DOI] [PubMed] [Google Scholar]
- 18.Al-Amri A.M. (2015) Clinical presentation and causes of the delayed diagnosis of breast cancer in patients with pregnancy associated breast cancer. J. Family Community Med. 22, 96–100 10.4103/2230-8229.155383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johansson A.L.V., Andersson T.M., Hsieh C.C., Jirstrom K., Cnattingius S., Fredriksson I.. et al. (2018) Tumor characteristics and prognosis in women with pregnancy-associated breast cancer. Int. J. Cancer 142, 1343–1354 10.1002/ijc.31174 [DOI] [PubMed] [Google Scholar]
- 20.Basaran D., Turgal M., Beksac K., Ozyuncu O., Aran O. and Beksac M.S. (2014) Pregnancy-associated breast cancer: clinicopathological characteristics of 20 cases with a focus on identifiable causes of diagnostic delay. Breast Care (Basel) 9, 355–359 10.1159/000366436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amant F., von Minckwitz G., Han S.N., Bontenbal M., Ring A.E., Giermek J.. et al. (2013) Prognosis of women with primary breast cancer diagnosed during pregnancy: results from an international collaborative study. J. Clin. Oncol. 31, 2532–2539 10.1200/JCO.2012.45.6335 [DOI] [PubMed] [Google Scholar]
- 22.Azim H.A. Jr, Peccatori F.A., Brohee S., Branstetter D., Loi S., Viale G.. et al. (2015) RANK-ligand (RANKL) expression in young breast cancer patients and during pregnancy. Breast Cancer Res. 17, 24. 10.1186/s13058-015-0538-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azim H.A. Jr, Vingiani A., Peccatori F., Viale G., Loi S. and Pruneri G. (2015) Tumour infiltrating lymphocytes (TILs) in breast cancer during pregnancy. Breast 24, 290–293 10.1016/j.breast.2015.01.009 [DOI] [PubMed] [Google Scholar]
- 24.Hajiebrahimi M., Cnattingius S., Lambe M. and Bahmanyar S. (2016) Pregnancy history and risk of premenopausal breast cancer-a nested case-control study. Int. J. Epidemiol. 45, 816–824 10.1093/ije/dyv293 [DOI] [PubMed] [Google Scholar]
- 25.Kessous R., Walfisch A., Meirovitz M., Davidson E., Sergienko R. and Sheiner E. (2017) Preterm delivery and future maternal risk of female malignancies. Arch. Gynecol. Obstet. 295, 205–210 10.1007/s00404-016-4198-3 [DOI] [PubMed] [Google Scholar]
- 26.Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A. Jr and Kinzler K.W. (2013) Cancer genome landscapes. Science 339, 1546–1558 10.1126/science.1235122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang S., Wang Y., Gu Y., Zhu J., Ci C., Guo Z.. et al. (2018) Specific breast cancer prognosis-subtype distinctions based on DNA methylation patterns. Mol. Oncol. 12, 1047–1060 10.1002/1878-0261.12309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Z., Liang G., Tan L., Su A.N., Jiang W. and Gong C. (2017) High-efficient screening method for identification of key genes in breast cancer through microarray and bioinformatics. Anticancer Res. 37, 4329–4335 [DOI] [PubMed] [Google Scholar]
- 29.Wang Y., Zhang Y., Huang Q. and Li C. (2018) Integrated bioinformatics analysis reveals key candidate genes and pathways in breast cancer. Mol. Med. Rep. 17, 8091–8100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harvell D.M., Kim J., O’Brien J., Tan A.C., Borges V.F., Schedin P.. et al. (2013) Genomic signatures of pregnancy-associated breast cancer epithelia and stroma and their regulation by estrogens and progesterone. Horm. Cancer 4, 140–153 10.1007/s12672-013-0136-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azim H.A. Jr, Brohee S., Peccatori F.A., Desmedt C., Loi S., Lambrechts D.. et al. (2014) Biology of breast cancer during pregnancy using genomic profiling. Endocr. Relat. Cancer 21, 545–554 10.1530/ERC-14-0111 [DOI] [PubMed] [Google Scholar]
- 32.Robinson M.D., McCarthy D.J. and Smyth G.K. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pathan M., Keerthikumar S., Ang C.S., Gangoda L., Quek C.Y., Williamson N.A.. et al. (2015) FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics 15, 2597–2601 10.1002/pmic.201400515 [DOI] [PubMed] [Google Scholar]
- 34.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M.. et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du J., Yuan Z., Ma Z., Song J., Xie X. and Chen Y. (2014) KEGG-PATH: Kyoto encyclopedia of genes and genomes-based pathway analysis using a path analysis model. Mol. Biosyst. 10, 2441–2447 10.1039/C4MB00287C [DOI] [PubMed] [Google Scholar]
- 36.Kanehisa M. and Goto S. (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang da W., Sherman B.T. and Lempicki R.A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 38.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J.. et al. (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D.. et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang Z., Li C., Kang B., Gao G., Li C. and Zhang Z. (2017) GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, W98–W102 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chandrashekar D.S., Bashel B., Balasubramanya S.A.H., Creighton C.J., Ponce-Rodriguez I., Chakravarthi B.. et al. (2017) UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19, 649–658 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Currie E., Schulze A., Zechner R., Walther T.C. and Farese R.V. Jr (2013) Cellular fatty acid metabolism and cancer. Cell Metab. 18, 153–161 10.1016/j.cmet.2013.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hardy S., St-Onge G.G., Joly E., Langelier Y. and Prentki M. (2005) Oleate promotes the proliferation of breast cancer cells via the G protein-coupled receptor GPR40. J. Biol. Chem. 280, 13285–13291 10.1074/jbc.M410922200 [DOI] [PubMed] [Google Scholar]
- 44.Hopkins M.M., Zhang Z., Liu Z. and Meier K.E. (2016) Eicosopentaneoic acid and other free fatty acid receptor agonists inhibit lysophosphatidic acid- and epidermal growth factor-induced proliferation of human breast cancer cells. J. Clin. Med. 5, 10.3390/jcm5020016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu Q., Wang H., Zhao X., Shi Y., Jin M. and Wan B. (2013) Identification of G-protein-coupled receptor 120 as a tumor-promoting receptor that induces angiogenesis and migration in human colorectal carcinoma. Oncogene 32, 5541–5550 10.1038/onc.2013.264 [DOI] [PubMed] [Google Scholar]
- 46.Serafin D.S., Allyn B., Sassano M.F., Timoshchenko R.G., Mattox D., Brozowski J.M.. et al. (2018) Chemerin-activated functions of CMKLR1 are regulated by G protein-coupled receptor kinase 6 (GRK6) and beta-arrestin 2 in inflammatory macrophages. Mol. Immunol. 106, 12–21 10.1016/j.molimm.2018.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cidado J. and Park B.H. (2012) Targeting the PI3K/Akt/mTOR pathway for breast cancer therapy. J. Mammary Gland Biol. Neoplasia 17, 205–216 10.1007/s10911-012-9264-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grandis J.R. and Sok J.C. (2004) Signaling through the epidermal growth factor receptor during the development of malignancy. Pharmacol. Ther. 102, 37–46 10.1016/j.pharmthera.2004.01.002 [DOI] [PubMed] [Google Scholar]
- 49.Guerrero-Zotano A., Mayer I.A. and Arteaga C.L. (2016) PI3K/AKT/mTOR: role in breast cancer progression, drug resistance, and treatment. Cancer Metastasis Rev. 35, 515–524 10.1007/s10555-016-9637-x [DOI] [PubMed] [Google Scholar]
- 50.Liu Z.B., Hou Y.F., Zhu J., Hu D.L., Jin W., Ou Z.L.. et al. (2010) Inhibition of EGFR pathway signaling and the metastatic potential of breast cancer cells by PA-MSHA mediated by type 1 fimbriae via a mannose-dependent manner. Oncogene 29, 2996–3009 10.1038/onc.2010.70 [DOI] [PubMed] [Google Scholar]
- 51.Mohan C.D., Srinivasa V., Rangappa S., Mervin L., Mohan S., Paricharak S.. et al. (2016) Trisubstituted-imidazoles induce apoptosis in human breast cancer cells by targeting the oncogenic PI3K/Akt/mTOR signaling pathway. PLoS One 11, e0153155. 10.1371/journal.pone.0153155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang X., Gao D., Fang K., Guo Z. and Li L. (2018) Med19 is targeted by miR-101-3p/miR-422a and promotes breast cancer progression by regulating the EGFR/MEK/ERK signaling pathway. Cancer Lett. 444, 105–115 10.1016/j.canlet.2018.12.008 [DOI] [PubMed] [Google Scholar]
- 53.Ma X.J., Dahiya S., Richardson E., Erlander M. and Sgroi D.C. (2009) Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res. 11, R7. 10.1186/bcr2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muller A., Homey B., Soto H., Ge N., Catron D., Buchanan M.E.. et al. (2001) Involvement of chemokine receptors in breast cancer metastasis. Nature 410, 50–56 10.1038/35065016 [DOI] [PubMed] [Google Scholar]
- 55.Robic A., Faraut T., Liaubet L., Riquet J., Milan D. and Lobjois V. (2008) Characterization of porcine ASB6 gene and transcripts-comparison of mammalian transcripts. Anim. Biotechnol. 19, 138–143 10.1080/10495390801984721 [DOI] [PubMed] [Google Scholar]
- 56.Wilcox A., Katsanakis K.D., Bheda F. and Pillay T.S. (2004) Asb6, an adipocyte-specific ankyrin and SOCS box protein, interacts with APS to enable recruitment of elongins B and C to the insulin receptor signaling complex. J. Biol. Chem. 279, 38881–38888 10.1074/jbc.M406101200 [DOI] [PubMed] [Google Scholar]
- 57.Linossi E.M. and Nicholson S.E. (2012) The SOCS box-adapting proteins for ubiquitination and proteasomal degradation. IUBMB Life 64, 316–323 10.1002/iub.1011 [DOI] [PubMed] [Google Scholar]
- 58.Hung K.F., Lai K.C., Liu T.Y., Liu C.J., Lee T.C. and Lo J.F. (2009) Asb6 upregulation by Areca nut extracts is associated with betel quid-induced oral carcinogenesis. Oral. Oncol. 45, 543–548 10.1016/j.oraloncology.2008.10.004 [DOI] [PubMed] [Google Scholar]