Abstract
Background
Evidence suggests that high-level physical activity may potentially reduce cancer mortality through its immune enhancement effect. We therefore hypothesized that survival benefits associated with physical activity might be stronger in colorectal carcinomas with lower immune reaction at diagnosis.
Methods
Using molecular pathological epidemiology databases of 470 colon and rectal carcinoma cases in the Nurses’ Health Study and the Health Professionals Follow-up Study, we assessed the prognostic association of postdiagnosis physical activity in strata of densities of CD3+ cells, CD8+ cells, CD45RO (PTPRC)+ cells, or FOXP3+ cells in tumor tissue. Cox proportional hazards regression model was used to adjust for potential confounders, including microsatellite instability, CpG island methylator phenotype, long interspersed nucleotide element-1 methylation, KRAS, BRAF, and PIK3CA mutations, and expression of CTNNB1 (beta-catenin), PTGS2 (cyclooxygenase-2), and IRS1.
Results
The association of postdiagnosis physical activity with colorectal cancer-specific mortality differed by CD3+ cell density (Pinteraction < .001). Multivariable-adjusted colorectal cancer-specific mortality hazard ratios for a quartile-unit increase in physical activity were 0.56 (95% confidence interval = 0.38 to 0.83) among cases with the lowest quartile of CD3+ cell density compared with 1.14 (95% confidence interval = 0.79 to 1.65) in cases with the highest quartile. We observed no differential survival association of physical activity by densities of CD8+ cells, CD45RO+ cells, or FOXP3+ cells.
Conclusions
The association between postdiagnosis physical activity and colorectal cancer survival appeared stronger for carcinomas with lower T cell infiltrates, suggesting an interactive effect of exercise and immunity on colorectal cancer progression.
Innate and adaptive immunity play crucial roles in suppressing tumor progression (1,2). In colorectal carcinomas, high-level infiltrates of CD3+ cells, CD8+ cells, and CD45RO (PTPRC)+ cells in the tumor microenvironment have been associated with longer patient survival (3–7). Emerging evidence indicates that immunotherapies targeting immune checkpoint molecules such as PDCD1 (programmed cell death 1, PD-1) and CD274 (PDCD1 ligand 1, PD-L1) can be effective in treating several cancer types (8), including colorectal cancer with high-level microsatellite instability (MSI) (9,10). Elucidating the interplay between tumor cells and the immune system is of considerable importance to further improve the efficacy of immunoprevention and immunotherapy strategies for cancer (11–17).
High-level physical activity has been associated with lower incidence and mortality of colorectal cancer (18–28). Evidence suggests that physical activity may prolong colorectal cancer patient survival through decreasing chronic inflammation in the tumor microenvironment and enhancing the T cell-mediated antitumor immune response (29–32). Colorectal cancer consists of a heterogeneous group of neoplasms due to complex interactions with environmental factors, host immune cells, and transformed cells (1,33). We considered that tumors that had progressed despite the presence of higher T cell reaction might have developed mechanisms to escape immune surveillance; such tumors with higher immune reaction might exhibit refractoriness to immunomodulatory effects of physical activity after cancer diagnosis. We therefore hypothesized that survival benefits associated with increased physical activity levels might be stronger for tumors with lower T cell reaction than for tumors with a higher reaction.
To test our hypothesis, we used a molecular pathological epidemiology database derived from two large prospective cohort studies in the United States with data on physical activity levels after colorectal cancer diagnosis, tumor molecular and immune features, and survival outcomes. We examined the interactive prognostic association of postdiagnosis physical activity levels and tumor-infiltrating T cells.
Methods
Study Population
We examined data from two large prospective cohort studies in the United States: the Nurses’ Health Study (NHS, 121 701 women ages 30–55 years followed since 1976) and the Health Professionals Follow-up Study (HPFS, 51 529 men ages 40 to 75 years followed since 1986) (34). Study participants have been sent follow-up questionnaires biennially to update information on demographics, lifestyle factors, and medical history and to report newly diagnosed diseases including colorectal cancer. The follow-up rate has been more than 90% for each questionnaire cycle in both cohorts. The National Death Index was used to ascertain deaths of study participants and identify patients with unreported lethal colorectal cancer. Informed consent was obtained from all participants at study enrollment. This study was approved by the Human Subjects Committees at Harvard T.H. Chan School of Public Health and Brigham and Women’s Hospital (Boston, MA).
We included 470 colorectal cancer cases with available data on postdiagnosis physical activity levels and T cell densities in tumor tissue among participants diagnosed with colorectal cancer through 2008. We included both colon and rectal carcinoma cases based on the colorectal continuum model (35). Patients were followed until death or end of follow-up (June 30, 2014 for the NHS; January 1, 2014 for the HPFS), whichever came first. Study physicians, blinded to exposure data, reviewed medical records of patients diagnosed with colorectal cancer to collect data on tumor characteristics and to identify causes of death for deceased patients. We collected formalin-fixed paraffin-embedded tumor tissue samples from hospitals throughout the United States where participants with colorectal carcinoma underwent tumor resection. A single pathologist (SO), blinded to other data, conducted a central review of hematoxylin and eosin-stained tissue sections of all colorectal carcinoma cases and collected data on histopathological characteristics, including tumor differentiation and four lymphocytic reaction patterns (Crohn-like lymphoid reaction, peritumoral lymphocytic reaction, intratumoral periglandular reaction, and tumor-infiltrating lymphocytes) (36).
Assessment of Physical Activity Levels
In the NHS and HPFS, leisure-time physical activity has been assessed biennially since 1986 using a self-administered physical activity questionnaire, which was validated against physical activity diaries in which the participants documented the duration of time spent in activities as previously described (37). Participants reported the duration of physical activity for each component of physical activity. Based on this information, we calculated a metabolic equivalent task score (METS) (38), which was defined as the ratio of the metabolic rate of specific activities to the resting metabolic rate (39,40). We summed up the METS for each activity to obtain the total METS-hours/week. To avoid the period of active anti-cancer treatment, we used questionnaire data reported between 6 and 48 months after diagnosis of colorectal cancer. To minimize the bias arising from reduced physical activity due to the progression of disease, physical activity was evaluated at the earliest time period after the diagnosis of colorectal cancer. Taking into account sex differences in lifestyle and physical activity, we classified physical activity levels into sex-specific quartiles (39,40).
Immunohistochemistry
We constructed tissue microarrays from colorectal cancer tissue blocks (41) and conducted immunohistochemistry for CD3, CD8, CD45RO, and FOXP3, as previously described (7). We used an automated scanning microscope and the Ariol image analysis system (Genetix, CA) to measure T cell densities (cells/mm2) in tumor tissue. We assessed up to four tissue microarray cores from each tumor and calculated the average density of each tumor-infiltrating T cell subset. If applicable, we categorized T cell densities into low vs high by the median value.
Immunohistochemical analyses for CTNNB1 (beta-catenin) expression (42), PTGS2 (cyclooxygenase-2) expression (39), and IRS1 expression (40) were performed using a mouse anti-CTNNB1 antibody (BD Transduction Laboratories, CA), anti-PTGS2 antibody (Cayman Chemical, MI), and anti-IRS1 antibody (Millipore, Billerica, MA), respectively.
Analyses of Microsatellite Instability, DNA Methylation, and KRAS, BRAF, and PIK3CA Mutations
DNA was extracted from archival formalin-fixed, paraffin-embedded tumor tissue. We determined MSI status using 10 microsatellite markers (D2S123, D5S346, D17S250, BAT25, BAT26, BAT40, D18S55, D18S56, D18S67, and D18S487), as previously described (43). We defined MSI-high as the presence of instability in 30% or more of the markers, and non-MSI-high as instability in less than 30% of the markers. Using bisulfite-treated DNA, we quantified DNA methylation in eight CpG island methylator phenotype (CIMP)-specific promoters (CACNA1G, CDKN2A, CRABP1, IGF2, MLH1, NEUROG1, RUNX3, and SOCS1) and in long interspersed nucleotide element-1 (LINE-1) (44,45). We defined CIMP-high as the presence of six or more methylated promoters and CIMP-low/negative as five or fewer methylated promoters (45). We conducted polymerase chain reaction and pyrosequencing targeted for KRAS (codons 12, 13, 61, and 146) (46), BRAF (codon 600) (43), and PIK3CA (exons 9 and 20) (47).
Statistical Analysis
Detailed information on statistical methods is included in Supplementary Methods (available online). All statistical analyses were performed using SAS software (version 9.4, SAS Institute, Cary, NC), and all P values were two-sided. Our primary hypothesis testing was assessment of a statistical interaction (using the Wald test on the cross-product) between postdiagnosis physical activity levels (the median value of each decile category) and T cell densities in tumor tissue (the median value of each decile category) in the Cox proportional hazards regression model for colorectal cancer-specific mortality analysis. Variables for physical activity and T cell densities were treated as decile categorical variables to reduce the influential effect of a few arbitrary cutoff points. In our primary hypothesis testing on new discoveries, we used the α level of 0.005 (48). All other analyses including evaluations of stratum-specific hazard ratios (HRs) and survival curves represented secondary analyses. In our secondary and other exploratory analyses, we recognized multiple comparisons associated with those analyses and used the α level of 0.005. Outcome endpoints were colorectal cancer-specific mortality and overall mortality. Survival time was defined as the time since colorectal cancer diagnosis to death or the end of follow-up, whichever came first, and was left-truncated at the time of the first postdiagnosis questionnaire return.
To reduce bias due to the availability of postdiagnosis questionnaire data, the inverse probability weighting (IPW) method was used in all survival analyses (49–51). We estimated the probability of questionnaire return after colorectal cancer diagnosis using the multivariable logistic regression model as previously described (49) and used the inverse probability to weight each patient. When we performed sex-stratified IPW-adjusted Cox regression analyses without truncation of weight, the results remained consistent (data not shown). Multivariable sex-stratified Cox proportional hazards models initially included age at diagnosis, year of diagnosis, prediagnosis physical activity, postdiagnosis body mass index, history of colorectal cancer in any first-degree relatives, tumor location, tumor differentiation, disease stage, MSI status, CIMP, LINE-1 methylation level, BRAF mutation, KRAS mutation, PIK3CA mutation, nuclear CTNNB1 expression, PTGS2 expression, and IRS1 expression. A backward elimination was performed with a threshold of P equals .05 to select variables for the final models. We also estimated HRs for a quartile-unit increase of postdiagnosis physical activity levels in strata of levels of T cell densities using a re-parameterization of the interaction term in a single regression model (45). The cases with missing data were included in the majority category of a given categorical covariate to limit the degrees of freedom of the models. For cases with missing data on LINE-1 methylation level (2.1%) and IRS1 expression (12.0%), we assigned a separate indicator variable for each variable. We confirmed that excluding cases with missing information in any of the covariates did not alter our results substantially (data not shown). The proportionality of hazards assumption was evaluated using a time-dependent variable, which was the cross-product of the postdiagnosis physical activity variable and survival time (P > .05). Results of Cox regression analyses without IPW, which were similar to those with IPW, are shown in Supplementary Table 1 (available online). Survival probabilities were estimated using the IPW-adjusted Kaplan-Meier method and compared using the weighted log-rank test (52).
Results
We included 470 colorectal cancer cases with available data on postdiagnosis physical activity levels and T cell densities in tumor tissue. Table 1 summarizes the clinical, pathological, and molecular characteristics of colorectal cancer cases according to quartiles of postdiagnosis physical activity levels. During the median follow-up time of 17.3 years (interquartile range = 14.9 to 20.6 years) for all censored cases, there were 275 deaths from any cause, including 100 colorectal cancer-specific deaths. Postdiagnosis physical activity levels were associated with colorectal cancer-specific mortality overall (Table 2).
Table 1.
Clinical, pathological, and molecular characteristics of colorectal cancer cases according to postdiagnosis physical activity levels
| Postdiagnosis physical activity levels |
|||||
|---|---|---|---|---|---|
| Quartile 1 | Quartile 2 | Quartile 3 | Quartile 4 | ||
| All cases | (Lowest) | (Highest) | |||
| Characteristic* | (n = 470) | (n = 114) | (n = 121) | (n = 117) | (n = 118) |
| Postdiagnosis physical activity levels (METS-h/wk), median (range) | |||||
| Female (n = 252, NHS) | 7.3 (0–157.9) | 0.7 (0–2.2) | 3.7 (2.3–7.2) | 10.2 (7.3–16.9) | 36.0 (17.0–157.9) |
| Male (n = 218, HPFS) | 15.4 (0–155.9) | 1.0 (0–4.9) | 8.6 (5.0–15.3) | 23.0 (15.4–34.9) | 57.3 (35.0–155.9) |
| Mean age ± SD, y | 67.8 ± 8.0 | 69.4 ± 8.7 | 68.3 ± 7.9 | 67.0 ± 8.0 | 66.5 ± 7.2 |
| Year of diagnosis | |||||
| 1995 or before | 209 (44%) | 41 (36%) | 61 (50%) | 50 (43%) | 57 (48%) |
| 1996 to 2000 | 194 (41%) | 56 (49%) | 45 (37%) | 47 (40%) | 46 (39%) |
| 2001 to 2008 | 67 (14%) | 17 (15%) | 15 (12%) | 20 (17%) | 15 (13%) |
| Family history of colorectal cancer in first-degree relative(s) | |||||
| Absent | 369 (79%) | 89 (78%) | 89 (74%) | 100 (85%) | 91 (77%) |
| Present | 101 (21%) | 25 (22%) | 32 (26%) | 17 (15%) | 27 (23%) |
| Body mass index | |||||
| <25 kg/m2 | 209 (48%) | 41 (41%) | 55 (48%) | 58 (53%) | 55 (51%) |
| 25 to 29.9 kg/m2 | 156 (36%) | 35 (35%) | 42 (37%) | 37 (34%) | 42 (39%) |
| ≥30 kg/m2 | 68 (16%) | 25 (25%) | 17 (15%) | 15 (14%) | 11 (10%) |
| Tumor location | |||||
| Cecum | 89 (19%) | 27 (24%) | 21 (17%) | 19 (16%) | 22 (19%) |
| Ascending to transverse colon | 139 (30%) | 29 (25%) | 34 (28%) | 42 (36%) | 34 (29%) |
| Descending to sigmoid colon | 144 (31%) | 29 (25%) | 44 (36%) | 35 (30%) | 36 (31%) |
| Rectum | 97 (21%) | 29 (25%) | 22 (18%) | 20 (17%) | 26 (22%) |
| Tumor differentiation | |||||
| Well to moderate | 431 (92%) | 109 (96%) | 106 (88%) | 107 (92%) | 109 (93%) |
| Poor | 37 (7.9%) | 5 (4.4%) | 15 (12%) | 9 (7.8%) | 8 (6.8%) |
| AJCC disease stage | |||||
| I | 115 (26%) | 29 (27%) | 21 (19%) | 36 (33%) | 29 (26%) |
| II | 158 (36%) | 29 (27%) | 44 (40%) | 38 (35%) | 47 (42%) |
| III | 138 (31%) | 38 (35%) | 37 (34%) | 31 (28%) | 32 (29%) |
| IV | 29 (6.6%) | 13 (12%) | 8 (7.3%) | 5 (4.6%) | 3 (2.7%) |
| MSI status | |||||
| Non-MSI-high | 386 (83%) | 92 (81%) | 96 (81%) | 99 (85%) | 99 (84%) |
| MSI-high | 81 (17%) | 22 (19%) | 23 (19%) | 17 (15%) | 19 (16%) |
| CIMP status | |||||
| Low/negative | 384 (82%) | 87 (78%) | 99 (83%) | 101 (86%) | 97 (83%) |
| High | 82 (18%) | 25 (22%) | 21 (18%) | 16 (14%) | 20 (17%) |
| Mean LINE-1 methylation level ± SD (%) | 61.1 ± 9.6 | 60.5 ± 10.0 | 60.4 ± 9.3 | 60.8 ± 10.1 | 62.9 ± 9.0 |
| KRAS mutation | |||||
| Wild-type | 269 (58%) | 64 (56%) | 65 (55%) | 72 (63%) | 68 (58%) |
| Mutant | 195 (42%) | 50 (44%) | 53 (45%) | 43 (37%) | 49 (42%) |
| BRAF mutation | |||||
| Wild-type | 400 (86%) | 96 (85%) | 105 (88%) | 100 (88%) | 99 (85%) |
| Mutant | 64 (14%) | 17 (15%) | 15 (13%) | 14 (12%) | 18 (15%) |
| PIK3CA mutation | |||||
| Wild-type | 353 (82%) | 88 (87%) | 88 (81%) | 91 (83%) | 86 (77%) |
| Mutant | 77 (18%) | 13 (13%) | 20 (19%) | 19 (17%) | 25 (23%) |
| Nuclear CTNNB1 (beta-catenin) expression | |||||
| Negative | 238 (53%) | 62 (56%) | 60 (51%) | 59 (54%) | 57 (50%) |
| Positive | 213 (47%) | 48 (44%) | 58 (49%) | 51 (46%) | 56 (50%) |
| PTGS2 (cyclooxygenase-2) expression | |||||
| Negative | 179 (38%) | 48 (42%) | 44 (36%) | 45 (38%) | 42 (36%) |
| Positive | 291 (62%) | 66 (58%) | 77 (64%) | 72 (62%) | 76 (64%) |
| IRS1 expression | |||||
| Negative/low | 289 (70%) | 74 (70%) | 83 (76%) | 64 (65%) | 68 (67%) |
| High | 125 (30%) | 31 (30%) | 26 (24%) | 35 (35%) | 33 (33%) |
| CD3+ cell density | |||||
| Quartile 1 (lowest) | 111 (25%) | 32 (29%) | 28 (25%) | 29 (26%) | 22 (20%) |
| Quartile 2 | 111 (25%) | 32 (29%) | 18 (16%) | 32 (28%) | 29 (27%) |
| Quartile 3 | 112 (25%) | 17 (15%) | 37 (33%) | 26 (23%) | 32 (29%) |
| Quartile 4 (highest) | 111 (25%) | 29 (26%) | 30 (27%) | 26 (23%) | 26 (24%) |
| CD8+ cell density | |||||
| Quartile 1 (lowest) | 110 (25%) | 32 (30%) | 28 (25%) | 31 (28%) | 19 (18%) |
| Quartile 2 | 109 (25%) | 31 (29%) | 22 (20%) | 29 (26%) | 27 (25%) |
| Quartile 3 | 109 (25%) | 20 (19%) | 27 (24%) | 28 (25%) | 34 (32%) |
| Quartile 4 (highest) | 109 (25%) | 24 (22%) | 34 (31%) | 24 (21%) | 27 (25%) |
| CD45RO+ cell density | |||||
| Quartile 1 (lowest) | 113 (25%) | 28 (26%) | 27 (23%) | 35 (31%) | 23 (21%) |
| Quartile 2 | 113 (25%) | 32 (29%) | 33 (28%) | 24 (21%) | 24 (21%) |
| Quartile 3 | 113 (25%) | 27 (25%) | 29 (25%) | 26 (23%) | 31 (28%) |
| Quartile 4 (highest) | 112 (25%) | 22 (20%) | 28 (24%) | 28 (25%) | 34 (30%) |
| FOXP3+ cell density | |||||
| Quartile 1 (lowest) | 107 (25%) | 31 (30%) | 27 (24%) | 24 (24%) | 25 (24%) |
| Quartile 2 | 106 (25%) | 27 (26%) | 39 (34%) | 18 (18%) | 22 (21%) |
| Quartile 3 | 106 (25%) | 22 (21%) | 24 (21%) | 25 (25%) | 35 (33%) |
| Quartile 4 (highest) | 107 (25%) | 24 (23%) | 24 (21%) | 35 (34%) | 24 (23%) |
Percentage (%) indicates the proportion of cases with a specific clinical, pathological, or molecular characteristic of colorectal cancer cases in all cases or in strata of quartiles of postdiagnosis physical activity levels. AJCC = American Joint Committee on Cancer; CIMP = CpG island methylator phenotype; HPFS = Health Professionals Follow-up Study; LINE-1 = long interspersed nucleotide element-1; METS = metabolic equivalent task score; MSI = microsatellite instability; NHS = Nurses’ Health Study.
Table 2.
Colorectal cancer mortality according to postdiagnosis physical activity levels in all cases or in strata of quartiles of T cell densities
| Colorectal cancer-specific mortality HR for a quartile-unit increase of postdiagnosis physical activity levels |
Overall mortality HR for a quartile-unit increase of postdiagnosis physical activity levels |
||||||
|---|---|---|---|---|---|---|---|
| Characteristic | No. of cases | No. of events | Univariate HR (95% CI)* | Multivariable HR (95% CI)*† | No. of events | Univariate HR (95% CI)* | Multivariable HR (95% CI)*† |
| All colorectal cancer cases | 470 | 100 | 0.77 (0.64 to 0.92) | 0.78 (0.64 to 0.95) | 275 | 0.77 (0.69 to 0.86) | 0.83 (0.75 to 0.93) |
| CD3+ cell density | |||||||
| Quartile 1 (lowest) | 111 | 30 | 0.61 (0.42 to 0.88) | 0.56 (0.38 to 0.83) | 69 | 0.75 (0.59 to 0.95) | 0.76 (0.62 to 0.93) |
| Quartile 2 | 111 | 23 | 0.78 (0.53 to 1.15) | 0.80 (0.54 to 1.18) | 64 | 0.68 (0.55 to 0.84) | 0.72 (0.58 to 0.89) |
| Quartile 3 | 112 | 25 | 0.75 (0.49 to 1.14) | 0.73 (0.47 to 1.11) | 68 | 0.80 (0.63 to 1.02) | 0.83 (0.65 to 1.06) |
| Quartile 4 (highest) | 111 | 15 | 1.04 (0.73 to 1.49) | 1.14 (0.79 to 1.65) | 60 | 0.85 (0.67 to 1.08) | 0.96 (0.76 to 1.21) |
| Pinteraction‡ | .004 | <.001 | .35 | .17 | |||
| CD8+ cell density | |||||||
| Quartile 1 (lowest) | 110 | 34 | 0.67 (0.48 to 0.92) | 0.66 (0.47 to 0.94) | 66 | 0.72 (0.56 to 0.92) | 0.78 (0.59 to 1.02) |
| Quartile 2 | 109 | 23 | 0.84 (0.57 to 1.23) | 0.81 (0.56 to 1.18) | 65 | 0.81 (0.65 to 1.00) | 0.80 (0.65 to 0.99) |
| Quartile 3 | 109 | 18 | 0.58 (0.37 to 0.90) | 0.56 (0.33 to 0.95) | 59 | 0.62 (0.49 to 0.79) | 0.75 (0.59 to 0.95) |
| Quartile 4 (highest) | 109 | 19 | 1.03 (0.71 to 1.51) | 1.02 (0.67 to 1.56) | 64 | 0.87 (0.69 to 1.10) | 0.86 (0.70 to 1.05) |
| Pinteraction‡ | .060 | .14 | .22 | .45 | |||
| CD45RO+ cell density | |||||||
| Quartile 1 (lowest) | 113 | 33 | 0.95 (0.69 to 1.31) | 0.86 (0.62 to 1.19) | 73 | 0.73 (0.57 to 0.93) | 0.77 (0.62 to 0.95) |
| Quartile 2 | 113 | 34 | 0.70 (0.49 to 1.01) | 0.64 (0.44 to 0.95) | 69 | 0.77 (0.60 to 0.97) | 0.79 (0.62 to 0.99) |
| Quartile 3 | 113 | 19 | 0.61 (0.41 to 0.91) | 0.65 (0.42 to 1.02) | 63 | 0.74 (0.59 to 0.91) | 0.80 (0.65 to 1.00) |
| Quartile 4 (highest) | 112 | 10 | 0.87 (0.46 to 1.63) | 0.88 (0.44 to 1.72) | 60 | 0.82 (0.66 to 1.03) | 0.91 (0.74 to 1.13) |
| Pinteraction‡ | .81 | .84 | .76 | .80 | |||
| FOXP3+ cell density | |||||||
| Quartile 1 (lowest) | 107 | 32 | 0.71 (0.50 to 1.00) | 0.75 (0.51 to 1.10) | 76 | 0.67 (0.53 to 0.84) | 0.74 (0.58 to 0.94) |
| Quartile 2 | 106 | 23 | 0.69 (0.48 to 0.99) | 0.72 (0.48 to 1.08) | 67 | 0.71 (0.58 to 0.87) | 0.75 (0.61 to 0.92) |
| Quartile 3 | 106 | 20 | 0.94 (0.62 to 1.44) | 1.02 (0.68 to 1.51) | 56 | 0.98 (0.75 to 1.27) | 1.06 (0.85 to 1.33) |
| Quartile 4 (highest) | 107 | 13 | 0.66 (0.42 to 1.03) | 0.64 (0.40 to 1.03) | 49 | 0.73 (0.56 to 0.94) | 0.77 (0.61 to 0.98) |
| Pinteraction‡ | .46 | .33 | .73 | .31 | |||
IPW was applied to reduce a bias due to the availability of questionnaire data after cancer diagnosis (see Statistical Analysis subsection for details). CI = confidence interval; HR = hazard ratio; IPW = inverse probability weighting.
The multivariable sex-stratified IPW-adjusted Cox regression model initially included age, year of diagnosis, family history of colorectal cancer, body mass index, prediagnosis physical activity, tumor location, tumor differentiation, disease stage, microsatellite instability, CpG island methylator phenotype, long interspersed nucleotide element-1 methylation level, KRAS mutation, BRAF mutation, PIK3CA mutation, nuclear CTNNB1 (beta-catenin) expression, PTGS2 (cyclooxygenase-2) expression, and IRS1 expression. A backward elimination with a threshold of P equal to .05 was used to select variables for the final models. The variables that remained in the final models for analyses stratified by CD3+ cell density are described in Appendix Table A2.
P interaction was calculated using the Wald test for the cross-product of postdiagnosis physical activity levels (the median value of each decile category) and each T cell subset (the median value of each decile category) in the sex-stratified IPW-adjusted Cox regression model.
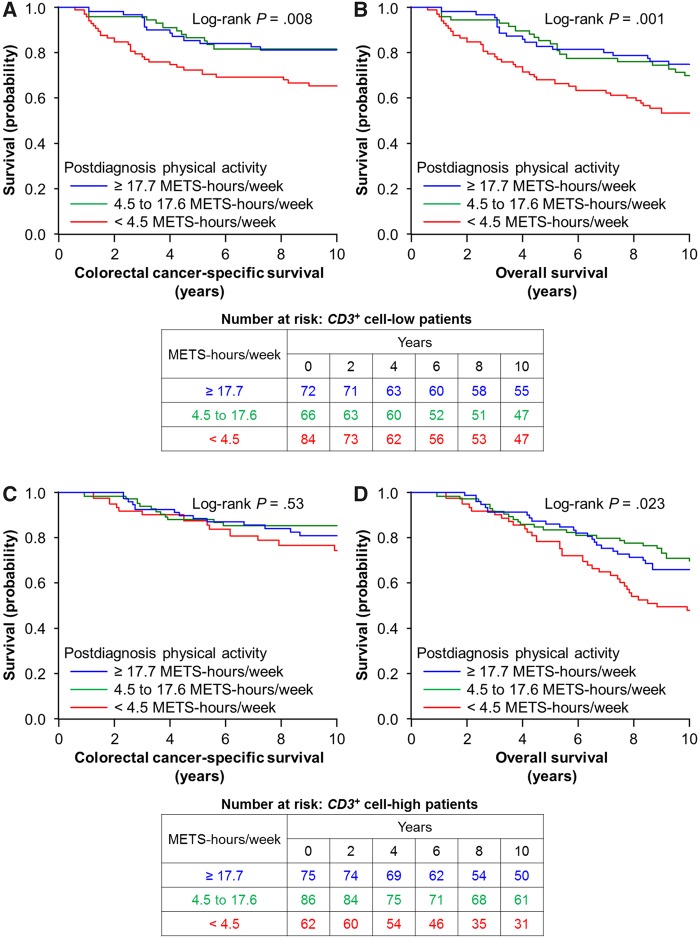
In our primary hypothesis testing, the association of postdiagnosis physical activity levels and colorectal cancer-specific mortality differed by CD3+ cell density (Pinteraction < .001; with the α level of 0.005; Table 2 and Supplementary Table 2, available online). The multivariable-adjusted HRs of colorectal cancer-specific mortality for a quartile-unit increase in postdiagnosis physical activity levels were 0.56 (95% confidence interval [CI] = 0.38 to 0.83) in the lowest quartile of CD3+ cell density and 1.14 (95% CI = 0.79 to 1.65) in the highest quartile of CD3+ cell density. The differential prognostic association was similarly observed in women and men, although statistical power was limited in each subgroup (Table 3). Figure 1 shows IPW-adjusted Kaplan-Meier survival curves of colorectal cancer-specific survival according to tertiles of postdiagnosis physical activity levels, and Table 4 shows HRs of cancer-specific mortality in each category of postdiagnosis physical activity levels. Taking into account the small number of events in each category, we used tertiles of postdiagnosis physical activity levels for these analyses. Considering the influence of arbitrary cutoff points, we entered postdiagnosis physical activity levels as a continuous variable into the models and obtained similar results (Supplementary Table 3, available online). When we excluded stage IV patients (Supplementary Table 4, available online) or patients who died within 6 months of the first postdiagnosis questionnaire return (n = 12, data not shown), we observed a similar interactive prognostic association of postdiagnosis physical activity levels and CD3+ cell density (Pinteraction < .001). We did not observe any statistically significant interaction of postdiagnosis physical activity levels with densities of CD8+ cells, CD45RO+ cells, or FOXP3+ cells (Pinteraction > .13; Table 2).
Table 3.
Colorectal cancer mortality according to postdiagnosis physical activity levels in all cases or in strata of quartiles of CD3+ cell density by sex
| Colorectal cancer-specific mortality HR for a quartile-unit increase of postdiagnosis physical activity levels |
Overall mortality HR for a quartile-unit increase of postdiagnosis physical activity levels |
||||||
|---|---|---|---|---|---|---|---|
| Characteristic | No. of cases | No. of events | Univariate HR (95% CI)* | Multivariable HR (95% CI)*† | No. of events | Univariate HR (95% CI)* | Multivariable HR (95% CI)*† |
| Female | |||||||
| All colorectal cancer cases | 252 | 55 | 0.72 (0.55 to 0.93) | 0.72 (0.55 to 0.94) | 134 | 0.68 (0.58 to 0.80) | 0.68 (0.57 to 0.80) |
| CD3+ cell density | |||||||
| Quartile 1 (lowest) | 56 | 15 | 0.49 (0.27 to 0.87) | 0.45 (0.25 to 0.82) | 27 | 0.52 (0.34 to 0.80) | 0.57 (0.35 to 0.91) |
| Quartile 2 | 61 | 14 | 0.78 (0.47 to 1.29) | 0.86 (0.50 to 1.48) | 30 | 0.56 (0.39 to 0.79) | 0.55 (0.39 to 0.77) |
| Quartile 3 | 65 | 15 | 0.90 (0.55 to 1.48) | 0.74 (0.44 to 1.25) | 40 | 0.92 (0.69 to 1.22) | 0.86 (0.65 to 1.15) |
| Quartile 4 (highest) | 53 | 6 | 0.55 (0.38 to 0.80) | 0.66 (0.44 to 1.00) | 29 | 0.67 (0.50 to 0.91) | 0.74 (0.53 to 1.02) |
| Pinteraction‡ | .071 | .11 | .60 | .37 | |||
| Male | |||||||
| All colorectal cancer cases | 218 | 45 | 0.83 (0.65 to 1.07) | 0.69 (0.50 to 0.96) | 141 | 0.87 (0.75 to 1.01) | 0.98 (0.85 to 1.13) |
| CD3+ cell density | |||||||
| Quartile 1 (lowest) | 55 | 15 | 0.73 (0.46 to 1.15) | 0.64 (0.39 to 1.05) | 42 | 0.90 (0.70 to 1.15) | 0.93 (0.74 to 1.17) |
| Quartile 2 | 50 | 9 | 0.77 (0.43 to 1.39) | 0.60 (0.32 to 1.13) | 34 | 0.79 (0.62 to 1.01) | 0.90 (0.70 to 1.15) |
| Quartile 3 | 47 | 10 | 0.50 (0.23 to 1.09) | 0.55 (0.26 to 1.20) | 28 | 0.63 (0.41 to 0.99) | 0.82 (0.53 to 1.26) |
| Quartile 4 (highest) | 58 | 9 | 1.58 (0.98 to 2.55) | 1.55 (0.86 to 2.80) | 31 | 1.08 (0.76 to 1.54) | 1.18 (0.84 to 1.65) |
| Pinteraction‡ | .001 | <.001 | .25 | .27 | |||
IPW was applied to reduce a bias due to the availability of questionnaire data after cancer diagnosis (see “Statistical Analysis” subsection for details). CI = confidence interval; HR = hazard ratio; IPW = inverse probability weighting.
The multivariable IPW-adjusted Cox regression model initially included age, year of diagnosis, family history of colorectal cancer, body mass index, prediagnosis physical activity, tumor location, tumor differentiation, disease stage, microsatellite instability, CpG island methylator phenotype, long interspersed nucleotide element-1 methylation level, KRAS mutation, BRAF mutation, PIK3CA mutation, nuclear CTNNB1 (beta-catenin) expression, PTGS2 (cyclooxygenase-2) expression, and IRS1 expression. A backward elimination with a threshold of P equal to .05 was used to select variables for the final models.
P interaction was calculated using the Wald test for the cross-product of postdiagnosis physical activity levels (the median value of each decile category) and each T cell subset (the median value of each decile category) in the IPW-adjusted Cox regression model.
Figure 1.
Inverse probability weighting-adjusted Kaplan-Meier curves of colorectal cancer-specific survival and overall survival according to tertiles of postdiagnosis physical activity levels (<4.5 vs 4.5 to 17.6 vs ≥17.7 METS-h/wk) in strata of CD3+ cell density. The P values were calculated using the weighted log-rank test (two-sided). A and B), CD3+ cell-low patients; C and D), CD3+ cell-high patients. METS = metabolic equivalent task score.
Table 4.
Colorectal cancer mortality according to tertiles (<4.5, 4.5 to 17.6, and ≥17.7 METS-h/wk) of postdiagnosis physical activity levels in strata of CD3+ cell density
| Colorectal cancer-specific mortality HR |
Overall mortality HR |
||||||
|---|---|---|---|---|---|---|---|
| Characteristic | No. of cases | No. of events | Univariate HR (95% CI)* | Multivariable HR (95% CI)*† | No. of events | Univariate HR (95% CI)* | Multivariable HR (95% CI)*† |
| CD3+ cell-low‡ | |||||||
| Postdiagnosis physical activity | |||||||
| <4.5 METS-h/wk | 84 | 29 | 1 (referent) | 1 (referent) | 59 | 1 (referent) | 1 (referent) |
| 4.5 to 17.6 METS-h/wk | 66 | 11 | 0.42 (0.21 to 0.84) | 0.37 (0.18 to 0.77) | 34 | 0.53 (0.34 to 0.81) | 0.54 (0.35 to 0.84) |
| ≥17.7 METS-h/wk | 72 | 13 | 0.44 (0.23 to 0.84) | 0.33 (0.16 to 0.67) | 40 | 0.53 (0.35 to 0.79) | 0.53 (0.35 to 0.79) |
| CD3+ cell-high‡ | |||||||
| Postdiagnosis physical activity | |||||||
| <4.5 METS-h/wk | 62 | 13 | 1 (referent) | 1 (referent) | 43 | 1 (referent) | 1 (referent) |
| 4.5 to 17.6 METS-h/wk | 86 | 13 | 0.62 (0.29 to 1.34) | 0.58 (0.27 to 1.24) | 43 | 0.58 (0.38 to 0.88) | 0.53 (0.35 to 0.81) |
| ≥17.7 METS-h/wk | 75 | 14 | 0.75 (0.36 to 1.57) | 0.62 (0.30 to 1.29) | 42 | 0.63 (0.41 to 0.96) | 0.72 (0.47 to 1.11) |
IPW was applied to reduce a bias due to the availability of questionnaire data after cancer diagnosis (see “Statistical Analysis” subsection for details). CI = confidence interval; HR = hazard ratio; IPW = inverse probability weighting; METS = metabolic equivalent task score.
The multivariable sex-stratified IPW-adjusted Cox regression model initially included age, year of diagnosis, family history of colorectal cancer, body mass index, prediagnosis physical activity, tumor location, tumor differentiation, disease stage, microsatellite instability, CpG island methylator phenotype, long interspersed nucleotide element-1 methylation level, KRAS mutation, BRAF mutation, PIK3CA mutation, nuclear CTNNB1 (beta-catenin) expression, PTGS2 (cyclooxygenase-2) expression, and IRS1 expression. A backward elimination with a threshold of P equal to .05 was used to select variables for the final models.
CD3+ cell density was categorized as low vs high by the median value.
Our previous studies have suggested that the association of postdiagnosis physical activity levels with colorectal cancer survival might differ by nuclear CTNNB1 (42), PTGS2 (39), or IRS1 expression status (40). Therefore, we performed secondary analyses stratified jointly by CD3+ cell density with nuclear CTNNB1, PTGS2, or IRS1 expression status (Table 5). Although statistical power was limited, there appeared to be a differential prognostic association by CD3+ cell density across nuclear CTNNB1 expression status and in PTGS2-positive or IRS1-negative cases.
Table 5.
Colorectal cancer mortality according to postdiagnosis physical activity levels in strata of combined CD3+ cell density and nuclear CTNNB1 (beta-catenin) expression, PTGS2 (cyclooxygenase-2) expression, or IRS1 expression status
| Colorectal cancer-specific mortality HR for a quartile-unit increase of postdiagnosis physical activity levels |
Overall mortality HR for a quartile-unit increase of postdiagnosis physical activity levels |
||||||
|---|---|---|---|---|---|---|---|
| Characteristic | No. of cases | No. of events | Univariate HR (95% CI)* | Multivariable HR (95% CI)*† | No. of events | Univariate HR (95% CI)* | Multivariable HR (95% CI)*† |
| Nuclear CTNNB1 (beta-catenin) expression | |||||||
| Negative | |||||||
| CD3+ cell-low‡ | 109 | 25 | 0.60 (0.39 to 0.91) | 0.57 (0.37 to 0.88) | 65 | 0.58 (0.46 to 0.74) | 0.61 (0.49 to 0.76) |
| CD3+ cell-high‡ | 117 | 22 | 0.82 (0.58 to 1.16) | 0.81 (0.57 to 1.14) | 67 | 0.79 (0.63 to 1.00) | 0.89 (0.72 to 1.10) |
| Positive | |||||||
| CD3+ cell-low‡ | 106 | 28 | 0.76 (0.55 to 1.06) | 0.74 (0.52 to 1.05) | 66 | 0.79 (0.64 to 0.98) | 0.87 (0.70 to 1.07) |
| CD3+ cell-high‡ | 97 | 16 | 0.93 (0.55 to 1.58) | 1.03 (0.62 to 1.73) | 55 | 0.91 (0.70 to 1.18) | 0.96 (0.75 to 1.24) |
| PTGS2 (cyclooxygenase-2) expression | |||||||
| Negative | |||||||
| CD3+ cell-low‡ | 74 | 14 | 0.62 (0.38 to 1.02) | 0.60 (0.37 to 0.99) | 43 | 0.71 (0.55 to 0.91) | 0.71 (0.54 to 0.94) |
| CD3+ cell-high‡ | 90 | 14 | 0.58 (0.36 to 0.96) | 0.55 (0.33 to 0.90) | 51 | 0.69 (0.53 to 0.89) | 0.78 (0.60 to 1.02) |
| Positive | |||||||
| CD3+ cell-low‡ | 148 | 39 | 0.71 (0.52 to 0.96) | 0.61 (0.44 to 0.86) | 90 | 0.71 (0.58 to 0.86) | 0.74 (0.61 to 0.89) |
| CD3+ cell-high‡ | 133 | 26 | 1.11 (0.78 to 1.58) | 1.06 (0.73 to 1.55) | 77 | 0.95 (0.77 to 1.18) | 1.01 (0.82 to 1.24) |
| IRS1 expression | |||||||
| Negative/low | |||||||
| CD3+ cell-low‡ | 136 | 38 | 0.62 (0.44 to 0.86) | 0.60 (0.42 to 0.86) | 89 | 0.68 (0.56 to 0.82) | 0.71 (0.58 to 0.86) |
| CD3+ cell-high‡ | 144 | 19 | 0.82 (0.53 to 1.26) | 0.76 (0.49 to 1.19) | 79 | 0.87 (0.71 to 1.08) | 0.91 (0.74 to 1.12) |
| High | |||||||
| CD3+ cell-low‡ | 57 | 7 | 1.73 (1.03 to 2.90) | 1.57 (0.93 to 2.64) | 30 | 0.98 (0.72 to 1.34) | 0.93 (0.72 to 1.20) |
| CD3+ cell-high‡ | 62 | 19 | 1.01 (0.69 to 1.46) | 1.18 (0.82 to 1.69) | 40 | 0.80 (0.61 to 1.07) | 0.89 (0.67 to 1.19) |
IPW was applied to reduce a bias due to the availability of questionnaire data after cancer diagnosis (see “Statistical Analysis” subsection for details). CI = confidence interval; HR = hazard ratio; IPW = inverse probability weighting.
The multivariable sex-stratified IPW-adjusted Cox regression model initially included age, year of diagnosis, family history of colorectal cancer, body mass index, prediagnosis physical activity, tumor location, tumor differentiation, disease stage, microsatellite instability, CpG island methylator phenotype, long interspersed nucleotide element-1 methylation level, KRAS mutation, BRAF mutation, PIK3CA mutation, nuclear CTNNB1 expression (except for CTNNB1-stratified analyses), PTGS2 expression (except for PTGS2-stratified analyses), and IRS1 expression (except for IRS1-stratified analyses). A backward elimination with a threshold of P equal to .05 was used to select variables for the final models.
CD3+ cell density was categorized as low vs high by the median value.
In our exploratory analysis, there were no statistically significant interactions between postdiagnosis physical activity levels and any of the lymphocytic reaction patterns examined (Supplementary Table 5, available online).
Discussion
Using two large prospective cohort studies, we tested the hypothesis that the association of postdiagnosis physical activity levels with colorectal cancer survival might differ by levels of tumor-infiltrating T cell subsets (CD3+ cells, CD8+ cells, CD45RO+ cells, or FOXP3+ cells). We found a stronger prognostic association of postdiagnosis physical activity levels for colorectal cancer accompanied by lower levels of CD3+ pan-T cells than for cancer accompanied by higher levels of CD3+ cells. Our results support an interactive effect of physical activity and immune status in the regulation of colorectal cancer progression. A future study is warranted to examine how the association between physical activity levels and colorectal cancer incidence may differ by tumor-infiltrating T cells.
Cancer immunotherapies such as immune checkpoint inhibitors have shown considerable promise with durable response (8,10). In colorectal carcinomas, clinical benefits from blockade therapies targeting the CD274 (PD-L1)-PDCD1 (PD-1) axis have shown to be greater for MSI-high tumors that are characterized by higher levels of mutation load and immunogenic neoantigens (9). However, a subset of MSI-high colorectal cancer responds poorly to immunotherapies. Intensity of the potential anti-tumor immune response is also a major determinant of the response to immunotherapies in colorectal cancer, which is affected by multiple endogenous and exogenous factors, including the gut microbiota (53–55). In this setting, there is an increasing need for integrative analyses of lifestyle factors, tumor features, and host immunity (6). A better understanding of the tumor-immune microenvironment would help us to optimize preventive and treatment strategies through immune modulation.
Evidence indicates the role of energy balance in carcinogenesis (56), and every tumor differs from other tumors (57). In the current study, we found a differential prognostic association of postdiagnosis physical activity levels by CD3+ cell density in colorectal carcinoma tissue. None of the more specific subsets of T cells statistically significantly modified the prognostic association of physical activity. This may indicate that, collectively, the density of pan-T cells (but not specific types of T cells) may indeed modify the prognostic effect of physical activity or that the measurements of the other specific markers of T cells may need further refinements. In addition, the density of CD3+ pan-T cells alone may be a potentially useful biomarker to identify the subpopulation that benefits more from postdiagnosis physical activity. To our knowledge, this is the first exploratory analysis on the interaction between postdiagnosis physical activity and immune response for colorectal cancer mortality. Therefore, our findings need to be validated in independent cohorts. Studies indicate that exercise can alter the number and function of circulating immune cells, including CD3+ cells, CD4+ cells, CD8+ cells, macrophages, and natural killer cells in healthy populations (29). Studies suggest that higher physical activity may increase plasma levels of ADIPOQ (adiponectin) (58–60). ADIPOQ can suppress inflammatory changes in the tumor microenvironment (61,62). Furthermore, higher levels of IL6 released from skeletal muscle during exercise may increase levels of cytokines (eg, IL1R1 and IL10) and cortisol, both of which exert anti-inflammatory properties (29,63–65). Preclinical studies suggest that exercise may increase circulating lymphocytes, promote the infiltration of natural killer cells to tumors, and increase apoptosis of cancer cells (29–31). In mouse models for colorectal cancer, exercise was associated with higher expression of cytotoxic T cell marker genes in intestinal mucosal tissue (32). Our population-based data support these mechanistic data, providing evidence for the immunomodulatory effects of physical activity in the regulation of colorectal cancer progression in humans. In the present study, there appeared to be no dose-response relationship between postdiagnosis physical activity levels and patient survival in CD3+ cell-low tumors. This may suggest a possibility of a threshold effect of physical activity for its immunomodulatory anti-tumor influence. However, we have been cautious in interpreting an individual HR estimate comparing each category of physical activity (to the referent category) in each stratum of patients according to T cell infiltrates, considering multiple hypothesis testing in such assessments. Further research is needed to assess potential dose-response effects of physical activity levels on colorectal cancer mortality in specific tumor types.
There are limitations in the current study. First, limited data on cancer treatments are available in our study populations. However, it was unlikely that treatment strategies were determined by levels of T cell densities, because attending physicians did not have access to such information. Second, the differential availability of questionnaire return for physical activity assessment after colorectal cancer diagnosis might have caused a bias. Thus, we used the IPW method in all survival analyses to reduce this potential bias. Third, available data on cancer recurrence were limited, but given the long follow-up duration of censored cases, colorectal cancer-specific mortality was considered as a reasonable surrogate for colorectal cancer-specific outcomes.
Strengths of the present study include the use of a molecular pathological epidemiology (66–69) database derived from two large prospective cohort studies, which included data on lifestyle factors and tumor molecular and immune characteristics. This integrated database allowed us to examine the interaction between postdiagnosis physical activity levels and tumor-infiltrating T cells while adjusting for a variety of potential confounders. In addition, this database enabled us to adjust for long-term physical activity levels before cancer diagnosis. Participants were enrolled at a large number of hospitals in diverse locations across the United States, which might improve the generalizability of our findings.
The current study suggests that the association of higher levels of postdiagnosis physical activity with better prognosis is stronger for colorectal carcinomas with lower CD3+ cell density than for carcinomas with higher CD3+ cell density. The present study provides evidence for a potential interaction between physical activity and anti-tumor immune response in suppressing colorectal cancer progression. Our findings suggest that the measurement of immune response based on densities of CD3+ in tumor tissue at diagnosis may help select patients who can gain the most benefit from exercise.
Funding
This work was supported by US National Institutes of Health (NIH) grants (P01 CA87969 to MJ Stampfer; UM1 CA186107 to MJ Stampfer; P01 CA55075 to WC Willett; UM1 CA167552 to WC Willett; U01 CA167552 to WC Willett and LA Mucci; P50 CA127003 to CSF; R01 CA118553 to CSF; R01 CA169141 to CSF; R01 CA137178 to ATC; K24 DK098311 to ATC; R35 CA197735 to SO; R01 CA151993 to SO; K07 CA190673 to RN; and K07 CA188126 to XZ); by Nodal Award (2016-02) from Dana-Farber Cancer Institute (SO); by the Stand Up To Cancer Colorectal Cancer Dream Team Translational Research Grant (grant number SU2C-AACR-DT22-17 administered by the American Association for Cancer Research, scientific partner of SU2C to MGi and CSF); and by grants from the Project P Fund, The Friends of the Dana-Farber Cancer Institute, Bennett Family Fund, and the Entertainment Industry Foundation through National Colorectal Cancer Research Alliance. LL was supported by a scholarship grant from Chinese Scholarship Council and a fellowship grant from Huazhong University of Science and Technology. KK was supported by grants from Overseas Research Fellowship (JP2017-775). ATC is a Stuart and Suzanne Steele MGH Research Scholar. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Notes
Affiliations of authors: Department of Oncologic Pathology (HK, TH, LL, AdS, TT, YM, KK, YS, MGu, WaLi, CD, YCh, WeLi, HL, CL, RN, SO) and Department of Medical Oncology (MGi, JAM), Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA; Department of Nutrition (MS, LL, KW, AH, RN), Department of Epidemiology (KW, RN, SO), Department of Immunology and Infectious Diseases (ATC), and Department of Biostatistics, (RN), Harvard T.H. Chan School of Public Health, Boston, MA; Clinical and Translational Epidemiology Unit, Massachusetts General Hospital, Harvard Medical School, Boston, MA (MS, ATC); Division of Gastroenterology, Massachusetts General Hospital and Harvard Medical School, Boston, MA (MS, ATC); Division of Public Health Sciences, Department of Surgery, Washington University School of Medicine, St Louis, MO (YCa); Program in MPE Molecular Pathological Epidemiology, Department of Pathology, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA (JAN, RN, SO); Laboratory of Human Carcinogenesis, National Cancer Institute, National Institutes of Health, Bethesda, MD (SAK); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA (KW, XZ, ATC); Broad Institute of MIT and Harvard, Cambridge, MA (MGi, ATC, SO); Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA (MGi); Yale Cancer Center, New Haven, CT (CSF); Department of Medicine, Yale School of Medicine, New Haven, CT (CSF); Smilow Cancer Hospital, New Haven, CT (CSF); Department of Hematology, Graduate School of Medicine, Osaka City University, Osaka, Japan (HK); Department of Epidemiology and Biostatistics, and the Ministry of Education Key Lab of Environment and Health, School of Public Health, Huazhong University of Science and Technology, Wuhan, P.R. China (LL); Department of Pathology, Graduate School of Medicine, The University of Tokyo, Tokyo, Japan (TM); Department of Medical Oncology, Chinese PLA General Hospital, Beijing, P.R. China (YS); College of Pharmacy, Zhejiang Chinese Medical University, Zhejiang, P.R. China (MGu); Department of Gastroenterology, Rheumatology, and Clinical Immunology, Sapporo Medical University School of Medicine, Sapporo, Japan (KN); Division of Pathology, The Cancer Institute, Japanese Foundation for Cancer Research, Tokyo, Japan (KI); Department of Ophthalmology, Keio University School of Medicine, Tokyo, Japan (AH) .
ATC previously served as a consultant for Bayer Pharma AG and Pfizer Inc. CSF has been a consultant for Eli Lilly and Co; Entrinsic Health Solutions, Inc; Pfizer Inc; Merck & Co., Inc; Sanofi S.A.; F. Hoffmann-La Roche Ltd; Genentech. Inc; Merrimack Pharmaceuticals, Inc; Dicerna Pharmaceuticals, Inc; Bayer Pharma AG; Agios Pharmaceuticals, Inc; Gilead Sciences, Inc; Five Prime Therapeutics, Inc; and Taiho Pharmaceutical Co, Ltd. This study was not funded by all these companies. No other conflicts of interest exist. The other authors declare that they have no conflicts of interest.
We thank the participants and staff of the NHS and the HPFS for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, and WY. The authors assume full responsibility for analyses and interpretation of these data.
Use of standardized official symbols: We use HUGO (Human Genome Organisation)-approved official symbols (or root symbols) for genes and gene products, including ADIPOQ, BRAF, CACNA1G, CD3, CD4, CD8, CD274, CDKN2A, CRABP1, CTNNB1, FOXP3, IGF2, IL1R1, IL6, IL10, IRS1, KRAS, MLH1, NEUROG1, PDCD1, PIK3CA, PTGS2, PTPRC, RUNX3, and SOCS1, all of which are described at www.genenames.org. The official symbols are italicized to differentiate from nonitalicized colloquial names that are used along with the official symbols. This format enables readers to familiarize themselves with the official symbols for genes and gene products together with common colloquial names.
Supplementary Material
References
- 1. Ogino S, Nowak J, Hamada T, et al. Integrative analysis of exogenous, endogenous, tumour, and immune factors for precision medicine. Gut. 2018;67(6):1168–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fridman WH, Zitvogel L, Sautès-Fridman C, Kroemer G.. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol. 2017;14(12):717–734. [DOI] [PubMed] [Google Scholar]
- 3. Prizment AE, Vierkant RA, Smyrk TC, et al. Cytotoxic T cells and granzyme B associated with improved colorectal cancer survival in a prospective cohort of older women. Cancer Epidemiol Biomarkers Prev. 2017;26(4):622–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Governa V, Trella E, Mele V, et al. The interplay between neutrophils and CD8(+) T cells improves survival in human colorectal cancer. Clin Cancer Res. 2017;23(14):3847–3858. [DOI] [PubMed] [Google Scholar]
- 5. Rozek LS, Schmit SL, Greenson JK, et al. Tumor-infiltrating lymphocytes, Crohn's-like lymphoid reaction, and survival from colorectal cancer. J Natl Cancer Inst. 2016;108(8):djw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mlecnik B, Bindea G, Angell HK, et al. Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity. 2016;44(3):698–711. [DOI] [PubMed] [Google Scholar]
- 7. Nosho K, Baba Y, Tanaka N, et al. Tumour-infiltrating T-cell subsets, molecular changes in colorectal cancer, and prognosis: cohort study and literature review. J Pathol. 2010;222(4):350–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Basile D, Garattini SK, Bonotto M, et al. Immunotherapy for colorectal cancer: where are we heading? Expert Opin Biol Ther. 2017;17(6):709–721. [DOI] [PubMed] [Google Scholar]
- 9. Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bever KM, Le DT.. An expanding role for immunotherapy in colorectal cancer. J Natl Compr Canc Netw. 2017;15(3):401–410. [DOI] [PubMed] [Google Scholar]
- 11. Pages F, Mlecnik B, Marliot F, et al. International validation of the consensus immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet. 2018;391(10135):2128–2139. [DOI] [PubMed] [Google Scholar]
- 12. Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24(5):541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ogino S, Giannakis M.. Immunoscore for (colorectal) cancer precision medicine. Lancet. 2018;391(10135):2084–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grizzi F, Basso G, Borroni EM, et al. Evolving notions on immune response in colorectal cancer and their implications for biomarker development. Inflamm Res. 2018;67(5):375–389. [DOI] [PubMed] [Google Scholar]
- 15. Fletcher R, Wang YJ, Schoen RE, et al. Colorectal cancer prevention: immune modulation taking the stage. Biochim Biophys Acta Rev Cancer. 2018;1869(2):138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Giannakis M, Mu XJ, Shukla SA, et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 2016;15(4):857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Galon J, Mlecnik B, Bindea G, et al. Towards the introduction of the ‘immunoscore’ in the classification of malignant tumours. J Pathol. 2014;232(2):199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jayasekara H, English DR, Haydon A, et al. Associations of alcohol intake, smoking, physical activity and obesity with survival following colorectal cancer diagnosis by stage, anatomic site and tumor molecular subtype. Int J Cancer. 2018;142(2):238–250. [DOI] [PubMed] [Google Scholar]
- 19. Phipps AI, Shi Q, Zemla TJ, et al. Physical activity and outcomes in patients with stage III colon cancer: a correlative analysis of phase III trial NCCTG N0147 (Alliance). Cancer Epidemiol Biomarkers Prev. 2018;27(6):696–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Walter V, Jansen L, Knebel P, et al. Physical activity and survival of colorectal cancer patients: population-based study from Germany. Int J Cancer. 2017;140(9):1985–1997. [DOI] [PubMed] [Google Scholar]
- 21. Moore SC, Lee IM, Weiderpass E, et al. Association of leisure-time physical activity with risk of 26 types of cancer in 1.44 million adults. JAMA Intern Med. 2016;176(6):816–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kyu HH, Bachman VF, Alexander LT, et al. Physical activity and risk of breast cancer, colon cancer, diabetes, ischemic heart disease, and ischemic stroke events: systematic review and dose-response meta-analysis for the Global Burden of Disease Study 2013. BMJ. 2016;354:i3857.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Keum N, Bao Y, Smith-Warner SA, et al. Association of physical activity by type and intensity with digestive system cancer risk. JAMA Oncol. 2016;2(9):1146–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Friedenreich CM, Neilson HK, Farris MS, Courneya KS.. Physical activity and cancer outcomes: a precision medicine approach. Clin Cancer Res. 2016;22(19):4766–4775. [DOI] [PubMed] [Google Scholar]
- 25. Campbell PT, Patel AV, Newton CC, Jacobs EJ, Gapstur SM.. Associations of recreational physical activity and leisure time spent sitting with colorectal cancer survival. J Clin Oncol. 2013;31(7):876–885. [DOI] [PubMed] [Google Scholar]
- 26. Vrieling A, Kampman E.. The role of body mass index, physical activity, and diet in colorectal cancer recurrence and survival: a review of the literature. Am J Clin Nutr. 2010;92(3):471–490. [DOI] [PubMed] [Google Scholar]
- 27. Meyerhardt JA, Giovannucci EL, Ogino S, et al. Physical activity and male colorectal cancer survival. Arch Intern Med. 2009;169(22):2102–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meyerhardt JA, Giovannucci EL, Holmes MD, et al. Physical activity and survival after colorectal cancer diagnosis. J Clin Oncol. 2006;24(22):3527–3534. [DOI] [PubMed] [Google Scholar]
- 29. Koelwyn GJ, Quail DF, Zhang X, White RM, Jones LW.. Exercise-dependent regulation of the tumour microenvironment. Nat Rev Cancer. 2017;17(10):620–632. [DOI] [PubMed] [Google Scholar]
- 30. Pedersen L, Idorn M, Olofsson GH, et al. Voluntary running suppresses tumor growth through epinephrine- and IL-6-dependent NK cell mobilization and redistribution. Cell Metab. 2016;23(3):554–562. [DOI] [PubMed] [Google Scholar]
- 31. Ashcraft KA, Peace RM, Betof AS, Dewhirst MW, Jones LW.. Efficacy and mechanisms of aerobic exercise on cancer initiation, progression, and metastasis: a critical systematic review of in vivo preclinical data. Cancer Res. 2016;76(14):4032–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McClellan JL, Steiner JL, Day SD, et al. Exercise effects on polyp burden and immune markers in the ApcMin/+ mouse model of intestinal tumorigenesis. Int J Oncol. 2014;45(2):861–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ogino S, Nowak JA, Hamada T, Milner DA Jr, Nishihara R.. Insights into pathogenic interactions among environment, host, and tumor at the crossroads of molecular pathology and epidemiology [published online ahead of print August 20, 2018]. Annu Rev Pathol. doi:10.1146/annurev-pathmechdis-012418-012818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nishihara R, Wu K, Lochhead P, et al. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N Engl J Med. 2013;369(12):1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yamauchi M, Morikawa T, Kuchiba A, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012;61(6):847–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ogino S, Nosho K, Irahara N, et al. Lymphocytic reaction to colorectal cancer is associated with longer survival, independent of lymph node count, microsatellite instability, and CpG island methylator phenotype. Clin Cancer Res. 2009;15(20):6412–6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chasan-Taber S, Rimm EB, Stampfer MJ, et al. Reproducibility and validity of a self-administered physical activity questionnaire for male health professionals. Epidemiology. 1996;7(1):81–86. [DOI] [PubMed] [Google Scholar]
- 38. Ainsworth BE, Haskell WL, Leon AS, et al. Compendium of physical activities: classification of energy costs of human physical activities. Med Sci Sports Exerc. 1993;25(1):71–80. [DOI] [PubMed] [Google Scholar]
- 39. Yamauchi M, Lochhead P, Imamura Y, et al. Physical activity, tumor PTGS2 expression, and survival in patients with colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2013;22(6):1142–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hanyuda A, Kim SA, Martinez-Fernandez A, et al. Survival benefit of exercise differs by tumor IRS1 expression status in colorectal cancer. Ann Surg Oncol. 2016;23(3):908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chan AT, Ogino S, Fuchs CS.. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. N Engl J Med. 2007;356(21):2131–2142. [DOI] [PubMed] [Google Scholar]
- 42. Morikawa T, Kuchiba A, Yamauchi M, et al. Association of CTNNB1 (beta-catenin) alterations, body mass index, and physical activity with survival in patients with colorectal cancer. JAMA. 2011;305(16):1685–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58(1):90–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Irahara N, Nosho K, Baba Y, et al. Precision of pyrosequencing assay to measure LINE-1 methylation in colon cancer, normal colonic mucosa, and peripheral blood cells. J Mol Diagn. 2010;12(2):177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nosho K, Irahara N, Shima K, et al. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS One. 2008;3(11):e3698.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Imamura Y, Lochhead P, Yamauchi M, et al. Analyses of clinicopathological, molecular, and prognostic associations of KRAS codon 61 and codon 146 mutations in colorectal cancer: cohort study and literature review. Mol Cancer. 2014;13(1):135.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liao X, Lochhead P, Nishihara R, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med. 2012;367(17):1596–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Benjamin D, Berger J, Johannesson M, et al. Redefine statistical significance. Nat Hum Behav. 2018;2(1):6–10. [DOI] [PubMed] [Google Scholar]
- 49. Hamada T, Cao Y, Qian ZR, et al. Aspirin use and colorectal cancer survival according to tumor CD274 (Programmed Cell Death 1 Ligand 1) expression status. J Clin Oncol. 2017;35(16):1836–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu L, Nevo D, Nishihara R, et al. Utility of inverse probability weighting in molecular pathological epidemiology. Eur J Epidemiol. 2018;33(4):381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Seaman SR, White IR.. Review of inverse probability weighting for dealing with missing data. Stat Methods Med Res. 2013;22(3):278–295. [DOI] [PubMed] [Google Scholar]
- 52. Xie J, Liu C.. Adjusted Kaplan-Meier estimator and log-rank test with inverse probability of treatment weighting for survival data. Stat Med. 2005;24(20):3089–3110. [DOI] [PubMed] [Google Scholar]
- 53. Rajpoot M, Sharma AK, Sharma A, Gupta GK.. Understanding the microbiome: emerging biomarkers for exploiting the microbiota for personalized medicine against cancer. Semin Cancer Biol. 2018;52(pt 1):1–8. [DOI] [PubMed] [Google Scholar]
- 54. Tilg H, Adolph TE, Gerner RR, Moschen AR.. The intestinal microbiota in colorectal cancer. Cancer Cell. 2018;33(6):954–964. [DOI] [PubMed] [Google Scholar]
- 55. Inamura K. Colorectal cancers: an update on their molecular pathology. Cancers (Basel). 2018;10(1):26.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Martinez-Useros J, García-Foncillas J.. Obesity and colorectal cancer: molecular features of adipose tissue. J Transl Med. 2016;14:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ogino S, Fuchs CS, Giovannucci E.. How many molecular subtypes? Implications of the unique tumor principle in personalized medicine. Expert Rev Mol Diagn. 2012;12(6):621–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Voss SC, Nikolovski Z, Bourdon PC, Alsayrafi M, Schumacher YO.. The effect of cumulative endurance exercise on leptin and adiponectin and their role as markers to monitor training load. Biol Sport. 2016;33(1):23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang X, You T, Murphy K, Lyles MF, Nicklas BJ.. Addition of exercise increases plasma adiponectin and release from adipose tissue. Med Sci Sports Exerc. 2015;47(11):2450–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee S, Kwak HB.. Effects of interventions on adiponectin and adiponectin receptors. J Exerc Rehabil. 2014;10(2):60–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Murphy N, Jenab M, Gunter MJ.. Adiposity and gastrointestinal cancers: epidemiology, mechanisms and future directions. Nat Rev Gastroenterol Hepatol. 2018;15(11):659–670. [DOI] [PubMed] [Google Scholar]
- 62. Deng T, Lyon CJ, Bergin S, Caligiuri MA, Hsueh WA.. Obesity, inflammation, and cancer. Annu Rev Pathol. 2016;11:421–449. [DOI] [PubMed] [Google Scholar]
- 63. Hojman P, Gehl J, Christensen JF, Pedersen BK.. Molecular mechanisms linking exercise to cancer prevention and treatment. Cell Metab. 2018;27(1):10–21. [DOI] [PubMed] [Google Scholar]
- 64. Knudsen JG, Gudiksen A, Bertholdt L, et al. Skeletal muscle IL-6 regulates muscle substrate utilization and adipose tissue metabolism during recovery from an acute bout of exercise. PLoS One. 2017;12(12):e0189301.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Steensberg A, Fischer CP, Keller C, Moller K, Pedersen BK.. IL-6 enhances plasma IL-1ra, IL-10, and cortisol in humans. Am J Physiol Endocrinol Metab. 2003;285(2):E433–E437. [DOI] [PubMed] [Google Scholar]
- 66. Ogino S, Stampfer M.. Lifestyle factors and microsatellite instability in colorectal cancer: the evolving field of molecular pathological epidemiology. J Natl Cancer Inst. 2010;102(6):365–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ogino S, Chan AT, Fuchs CS, Giovannucci E.. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60(3):397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rescigno T, Micolucci L, Tecce MF, Capasso A.. Bioactive nutrients and nutrigenomics in age-related diseases. Molecules. 2017;22(1):105.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hughes LAE, Simons C, van den Brandt PA, van Engeland M, Weijenberg MP.. Lifestyle, diet, and colorectal cancer risk according to (epi)genetic instability: current evidence and future directions of molecular pathological epidemiology. Curr Colorectal Cancer Rep. 2017;13(6):455–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.