MIC testing using the Bactec mycobacteria growth indicator tube system 960 of 70 phylogenetically diverse, isoniazid-resistant clinical strains of Mycobacterium tuberculosis revealed a complex pattern of overlapping MIC distributions. Whole-genome sequencing explained most of the levels of resistance observed.
KEYWORDS: MGIT 960, MIC, Mycobacterium tuberculosis, isoniazid resistance, whole-genome sequencing
ABSTRACT
MIC testing using the Bactec mycobacteria growth indicator tube system 960 of 70 phylogenetically diverse, isoniazid-resistant clinical strains of Mycobacterium tuberculosis revealed a complex pattern of overlapping MIC distributions. Whole-genome sequencing explained most of the levels of resistance observed. The MIC distribution of strains with only inhA promoter mutations was split by the current concentration endorsed by the Clinical and Laboratory Standards Institute to detect low-level resistance to isoniazid and is, consequently, likely not optimally set.
TEXT
In light of the continued selection and spread of drug-resistant tuberculosis, coupled with the dearth of novel antibiotics, the question of whether low-level resistance can be overcome by increasing the dose of a drug has become increasingly urgent (1). In 2018, the World Health Organization (WHO) formally endorsed this possibility for moxifloxacin, whereby a dose of 800 mg/day can be used to treat low-level resistance to this fluoroquinolone, although the corresponding clinical breakpoint has not been recognized by the Clinical and Laboratory Standards Institute (CLSI) (2–5). Conversely, for at least 15 years, the position of CLSI has been to stratify resistance to the core drug isoniazid (INH) into low- and high-level resistance by testing two concentrations of this drug, whereas WHO has not endorsed this concept to date (6–8). Specifically, the CLSI recommendation is to include the following statement in the antimicrobial susceptibility testing (AST) reports of strains that are only low-level resistant (i.e., are resistant to INH at the critical concentration [CC] of 0.1 μg/ml but not the higher clinical breakpoint of 0.4 μg/ml): “A specialist in the treatment of [multidrug-resistant tuberculosis] should be consulted concerning the appropriate therapeutic regimen and dosages” (3). However, WHO is in the process of reviewing its recommendation for INH and, in its most recent manual for AST, has begun to stratify INH resistance on the genotypic level but has not yet set corresponding clinical breakpoints to align the phenotype (7). We, therefore, set out to compare the phenotypic definitions of low- and high-level resistance of CLSI with the genotypic stratification proposed by WHO.
To this end, we used the BC Bactec mycobacteria growth indicator tube (MGIT) 960 system to conduct comprehensive MIC testing of a select set of phylogenetically diverse strains (70 INH-resistant and 5 INH-susceptible isolates), along with Mycobacterium tuberculosis H37Rv ATCC 27294 as the control strain. Four serial 2-fold dilutions were prepared from an INH stock solution to provide a final test range of 0.016 to 0.25 μg/ml for susceptible strains and H37Rv, 0.25 to 4 μg/ml for inhA promoter mutant isolates with or without a concurrent inhA coding mutation, 1 to 16 μg/ml for S315T/N mutant isolates, and 4 to 64 μg/ml for isolates with double mutations in katG and the inhA promoter. Whole-genome sequencing (WGS) was carried out with the Nextera-XT DNA kit to prepare paired-end libraries of 150-bp read lengths for Illumina sequencing. Data analysis and single-nucleotide polymorphism calling were performed using the MTBseq pipeline (9) (for additional details, see Supplementary methods in the supplemental material).
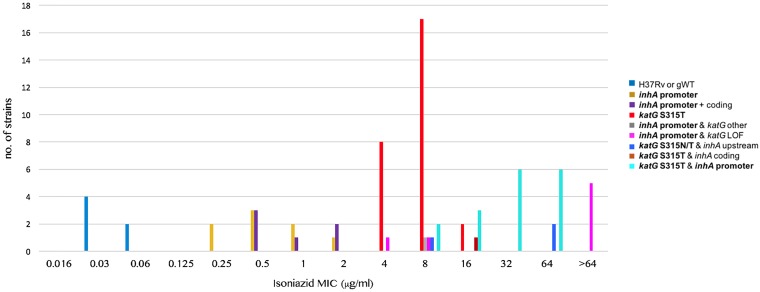
The six susceptible controls had INH MICs of 0.03 to 0.06 μg/ml (Fig. 1 and Table S2 in the supplemental material). In contrast, resistant strains displayed a series of overlapping MIC distributions. Strains that had only mutations that are interrogated by the WHO-endorsed genotypic AST assays (i.e., the Hain GenoType MTBDRplus version 2 and Nipro NTM+MDRTB version 2) resulted in three MIC distributions (10), i.e., strains that had only an inhA promoter change or a mutation at codon 315 of katG had nonoverlapping MIC distributions of 0.25 to 2 and 4 to 16 μg/ml, respectively, and strains with both mutations displayed MICs of 8 to 64 μg/ml. The variation in these distributions was likely largely due to the normal variation in MIC testing (i.e., even in the same laboratory, a variation of plus or minus one dilution is inevitable, which is further exacerbated by the variation in testing between laboratories).
FIG 1.
Isoniazid MIC results stratified by known or likely resistance mutations in the coding region of katG or inhA or mutations that result in the overexpression of inhA. All of the latter mutations are upstream of inhA, but “promoter” is used to highlight mutations in the −16 to −8 region upstream of the transcriptional start site of the fabG1-inhA operon, which can be detected by the WHO-endorsed Hain GenoType MTBDRplus version 2 and Nipro NTM+MDRTB version 2 assays (all mutations interrogated by these assays are shown in bold in the key of the plot [25]). gWT, genotypically wild-type strain (i.e., strain without known resistance mutations); LOF, loss-of-function mutation (i.e., insertion, deletion, or nonsense mutation).
The precise level of resistance cannot be predicted by using the Hain and Nipro assays alone because mutations that are not interrogated by these assays can increase the MICs. To some extent, this can be overcome by using WGS data, provided that known mutations with predictable effects are identified (i.e., the level of resistance could not be fully explained even with WGS). For example, some but not all strains with loss-of-function mutations in katG had MICs of >64 μg/ml (Fig. 1). Moreover, a C deletion 34 nucleotides upstream of the main transcriptional start site of inhA likely accounted for the unusually high MIC of 64 μg/ml for the katG S315N mutant (11). In contrast, another mutation upstream of inhA at codon 203 of fabG1, which is known to result in the overexpression of inhA by creating an alternative promoter, did not appear to increase the MIC above the level explained by the katG S315T mutation in the strain in question (i.e., 8 μg/ml) (12). Similarly, there was an almost complete overlap between the MIC distributions of strains that harbored only inhA promoter mutations and those that had additional inhA coding mutations at codon 21, 94, or 194 (i.e., 0.25 to 2 μg/ml versus 0.5 to 2 μg/ml).
This study was conducted in a single center, so we could not quantify the effect of laboratory-to-laboratory variation. Data from additional laboratories are needed to define robust quality-control ranges/targets to evaluate whether the current CC of 0.1 μg/ml corresponds to the epidemiological cutoff and to define the lower and upper ends of the various resistance mechanisms more accurately (3, 7, 13). Moreover, breakpoints cannot be set based on MIC data alone (5, 14, 15). Nevertheless, the MIC distributions in this study have implications for defining low-level resistance. The current clinical breakpoint of CLSI to define low-level resistance (i.e., 0.4 μg/ml, which corresponds to 0.5 μg/ml using our dilution series) does not correspond to the upper end of the MIC distribution of inhA promoter mutants. Instead, the upper end of the MIC distribution of a mechanism has to be considered when assessing whether it is treatable with either the standard or an elevated dose of INH (16–18). For strains with only inhA promoter mutations, this target concentration would be 1 or 2 μg/ml (i.e., at least 10 times higher than the current CC) (3, 7). Should pharmacokinetic/pharmacodynamic, drug penetration, and clinical outcome data confirm that this target is achievable, 1 or 2 μg/ml may be adopted instead of 0.4 μg/ml (14, 15). This would avoid splitting the MIC distribution of inhA promoter mutants and would, consequently, reduce or eliminate their misclassification as high-level resistant because of the technical variation in AST, as is the case with the current clinical breakpoint of CLSI.
One argument against setting a clinical breakpoint at 1 or 2 μg/ml might be that it would result in the misclassification of strains with both inhA promoter and coding mutations as low-level resistant, as stressed in a previous consensus statement (7, 19). However, two aspects should be borne in mind. First, only 3% (95% confidence interval, 1% to 6%) of strains with inhA promoter mutations in the −16 to −8 region that do not have katG mutations have additional inhA coding mutations based on recent WHO population-level surveillance data from seven countries (20). Therefore, in most settings, misclassifications of double mutants would be rare compared with the increased ability to detect inhA promoter mutants with a higher clinical breakpoint. Second, the effect of these coding mutations on the INH MIC and thus clinical outcome is likely modest at worst, but more MIC data are needed for the mutations at different inhA codons (21–24). Nonetheless, it might be advisable for countries that conduct routine WGS to err on the side of caution by classifying these double mutants as high-level resistant until clinical data to the contrary are available, although, in practice, it would be challenging to conduct a sufficiently powered study to address this question because these mutations are so rare.
Supplementary Material
ACKNOWLEDGMENTS
S.N. received support by the German Center for Infection Research, the Deutsche Forschungsgemeinschaft (German Research Foundation) under Germany’s Excellence Strategy (EXC 22167-390884018), and the Leibniz Science Campus EvoLUNG (Evolutionary Medicine of the Lung).
The funders had no role in study design, data collection, interpretation, or the decision to submit the work for publication.
C.U.K. is a consultant for the WHO Regional Office for Europe, QuantuMDx Group Ltd., and the Foundation for Innovative New Diagnostics, which involves work for Cepheid Inc., Hain Lifescience, and WHO. C.U.K. is an advisor to GenoScreen. The Bill & Melinda Gates Foundation, Janssen Pharmaceutica, and PerkinElmer covered C.U.K.’s travel and accommodation to present at meetings. The Global Alliance for TB Drug Development Inc. and Otsuka Novel Products GmbH have supplied C.U.K. with antibiotics for in vitro research. C.U.K. is collaborating with YD Diagnostics.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00092-19.
REFERENCES
- 1.World Health Organization. 2018. Global tuberculosis report. http://apps.who.int/iris/bitstream/handle/10665/274453/9789241565646-eng.pdf. Accessed 6 December 2018.
- 2.World Health Organization. Technical report on critical concentrations for drug susceptibility testing of medicines used in the treatment of drug-resistant tuberculosis [WHO/CDS/TB/2018.5]. http://apps.who.int/iris/bitstream/10665/260470/1/WHO-CDS-TB-2018.5-eng.pdf. Accessed 19 December 2018.
- 3.Clinical and Laboratory Standards Institute. 2018. Susceptibility testing of susceptibility testing of mycobacteria, Nocardia spp., and other aerobic actinomycetes—3rd ed CLSI standard M24. Clinical and Laboratory Standards Institute, Wayne, PA. [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2018. Performance standards for susceptibility testing of mycobacteria, Nocardia spp., and other aerobic actinomycetes—1st ed CLSI supplement M62. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 5.Köser CU, Maurer FP, Kranzer K. 2019. 'Those who cannot remember the past are condemned to repeat it': drug-susceptibility testing for bedaquiline and delamanid. Int J Infect Dis 80:S32–S35. doi: 10.1016/j.ijid.2019.02.027. [DOI] [PubMed] [Google Scholar]
- 6.NCCLS. 2003. Susceptibility testing of mycobacteria, Nocardiae, and other aerobic actinomycetes; approved standard. NCCLS document M24-A. NCCLS, Wayne, PA. [PubMed] [Google Scholar]
- 7.World Health Organization. 2018. Technical manual for drug susceptibility testing of medicines used in the treatment of tuberculosis [WHO/CDS/TB/2018.24]. World Health Organization, Geneva, Switzerland: http://www.who.int/tb/publications/2018/WHO_technical_drug_susceptibility_testing/en/. Accessed 17 November 2018. [Google Scholar]
- 8.Van Deun A, Decroo T, Piubello A, de Jong BC, Lynen L, Rieder HL. 2018. Principles for constructing a tuberculosis treatment regimen: the role and definition of core and companion drugs. Int J Tuber Lung Dis 22:239–245. doi: 10.5588/ijtld.17.0660. [DOI] [PubMed] [Google Scholar]
- 9.Kohl TA, Utpatel C, Schleusener V, De Filippo MR, Beckert P, Cirillo DM, Niemann S. 2018. MTBseq: a comprehensive pipeline for whole genome sequence analysis of Mycobacterium tuberculosis complex isolates. PeerJ 6:e5895. doi: 10.7717/peerj.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Health Organization. 2016. The use of molecular line probe assays for the detection of resistance to isoniazid and rifampicin. Policy update [WHO/HTM/TB/2016.12]. World Health Organization, Geneva, Switzerland: https://www.who.int/tb/publications/molecular-test-resistance/en/. Accessed 17 December 2018. [Google Scholar]
- 11.Karunaratne G, Wijesundera SS, Vidanagama D, Adikaram CP, Perera J. 2018. Significance of coexisting mutations on determination of the degree of isoniazid resistance in Mycobacterium tuberculosis strains. Microb Drug Resist 24:844–851. doi: 10.1089/mdr.2017.0330. [DOI] [PubMed] [Google Scholar]
- 12.Ando H, Miyoshi-Akiyama T, Watanabe S, Kirikae T. 2014. A silent mutation in mabA confers isoniazid resistance on Mycobacterium tuberculosis. Mol Microbiol 91:538–547. doi: 10.1111/mmi.12476. [DOI] [PubMed] [Google Scholar]
- 13.Schön T, Matuschek E, Mohamed S, Utukuri M, Heysell S, Alffenaar JW, Shin S, Martinez E, Sintchenko V, Maurer FP, Keller PM, Kahlmeter G, Köser CU. 2019. Standards for MIC testing that apply to the majority of bacterial pathogens should also be enforced for Mycobacterium tuberculosis complex. Clin Microbiol Infect 25:403–405. doi: 10.1016/j.cmi.2019.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kahlmeter G. 2015. The 2014 Garrod Lecture: EUCAST - are we heading towards international agreement? J Antimicrob Chemother 70:2427–2439. doi: 10.1093/jac/dkv145. [DOI] [PubMed] [Google Scholar]
- 15.Chesov D, Ciobanu N, Lange C, Schön T, Heyckendorf J, Crudu V. 2017. Lack of evidence of isoniazid efficacy for the treatment of MDR/XDR-TB in the presence of the katG 315T mutation. Eur Respir J 50:1701752. doi: 10.1183/13993003.01752-2017. [DOI] [PubMed] [Google Scholar]
- 16.Mouton JW, Muller AE, Canton R, Giske CG, Kahlmeter G, Turnidge J. 2018. MIC-based dose adjustment: facts and fables. J Antimicrob Chemother 73:564–568. doi: 10.1093/jac/dkx427. [DOI] [PubMed] [Google Scholar]
- 17.Goutelle S, Genestet C, Ader F, Lina G, Dumitrescu O. 2018. Comment on: MIC-based dose adjustment: facts and fables. J Antimicrob Chemother 73:2584–2585. doi: 10.1093/jac/dky131. [DOI] [PubMed] [Google Scholar]
- 18.Mouton JW, Muller AE, Canton R, Christian GG, Kahlmeter G, Turnidge J. 2018. MIC-based dose adjustment: facts and fables: authors' response. J Antimicrob Chemother 73:2585–2586. doi: 10.1093/jac/dky195. [DOI] [PubMed] [Google Scholar]
- 19.Domínguez J, Boettger EC, Cirillo D, Cobelens F, Eisenach KD, Gagneux S, Hillemann D, Horsburgh R, Molina-Moya B, Niemann S, Tortoli E, Whitelaw A, Lange C, TBNET, RESIST-TB networks. 2016. Clinical implications of molecular drug resistance testing for Mycobacterium tuberculosis: a TBNET/RESIST-TB consensus statement. Int J Tuber Lung Dis 20:24–42. doi: 10.5588/ijtld.15.0221. [DOI] [PubMed] [Google Scholar]
- 20.Zignol M, Cabibbe AM, Dean AS, Glaziou P, Alikhanova N, Ama C, Andres S, Barbova A, Borbe-Reyes A, Chin DP, Cirillo DM, Colvin C, Dadu A, Dreyer A, Driesen M, Gilpin C, Hasan R, Hasan Z, Hoffner S, Hussain A, Ismail N, Kamal SMM, Khanzada FM, Kimerling M, Kohl TA, Mansjö M, Miotto P, Mukadi YD, Mvusi L, Niemann S, Omar SV, Rigouts L, Schito M, Sela I, Seyfaddinova M, Skenders G, Skrahina A, Tahseen S, Wells WA, Zhurilo A, Weyer K, Floyd K, Raviglione MC. 2018. Genetic sequencing for surveillance of drug resistance in tuberculosis in highly endemic countries: a multi-country population-based surveillance study. Lancet Infect Dis 18:675–683. doi: 10.1016/S1473-3099(18)30073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vilchèze C, Wang F, Arai M, Hazbon MH, Colangeli R, Kremer L, Weisbrod TR, Alland D, Sacchettini JC, Jacobs WR Jr. 2006. Transfer of a point mutation in Mycobacterium tuberculosis inhA resolves the target of isoniazid. Nat Med 12:1027–1029. doi: 10.1038/nm1466. [DOI] [PubMed] [Google Scholar]
- 22.Machado D, Perdigão J, Ramos J, Couto I, Portugal I, Ritter C, Boettger EC, Viveiros M. 2013. High-level resistance to isoniazid and ethionamide in multidrug-resistant Mycobacterium tuberculosis of the Lisboa family is associated with inhA double mutations. J Antimicrob Chemother 68:1728–1732. doi: 10.1093/jac/dkt090. [DOI] [PubMed] [Google Scholar]
- 23.Heyckendorf J, Andres S, Köser CU, Olaru ID, Schön T, Sturegård E, Beckert P, Schleusener V, Kohl TA, Hillemann D, Moradigaravand D, Parkhill J, Peacock SJ, Niemann S, Lange C, Merker M. 2018. What is resistance? Impact of phenotypic versus molecular drug resistance testing on therapy for multi- and extensively drug-resistant tuberculosis. Antimicrob Agents Chemother 62:e01550-17. doi: 10.1128/AAC.01550-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lempens P, Meehan CJ, Vandelannoote K, Fissette K, de Rijk P, Van Deun A, Rigouts L, de Jong BC. 2018. Isoniazid resistance levels of Mycobacterium tuberculosis can largely be predicted by high-confidence resistance-conferring mutations. Sci Rep 8:3246. doi: 10.1038/s41598-018-21378-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miotto P, Tessema B, Tagliani E, Chindelevitch L, Starks AM, Emerson C, Hanna D, Kim PS, Liwski R, Zignol M, Gilpin C, Niemann S, Denkinger CM, Fleming J, Warren RM, Crook D, Posey J, Gagneux S, Hoffner S, Rodrigues C, Comas I, Engelthaler DM, Murray M, Alland D, Rigouts L, Lange C, Dheda K, Hasan R, Ranganathan UDK, McNerney R, Ezewudo M, Cirillo DM, Schito M, Köser CU, Rodwell TC. 2017. A standardised method for interpreting the association between mutations and phenotypic drug resistance in Mycobacterium tuberculosis. Eur Respir J 50:1701354. doi: 10.1183/13993003.01354-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.