LETTER
Extended-spectrum β-lactamases (ESBL) are spread worldwide in the order Enterobacterales (1, 2) but are less common in Pseudomonas aeruginosa; consequently, little is known regarding the genetic environment and plasmid-carrying blaESBL genes in this species (3). The predominant ESBL enzymes are those in the CTX-M family (1). The GES family is a less common group of ESBL enzymes comprising 40 members, which have been found in various Gram-negative bacilli (4).
One P. aeruginosa strain, clinical strain 1206/13 (here called Pa1206/13), isolated from cerebrospinal fluid at a hospital in São Paulo State, Brazil, from 2007 to 2014 and resistant to third- and fourth-generation cephalosporins, aztreonam, or carbapenems, was studied. The antimicrobial resistance genes were investigated by PCR (5–9). Plasmid incompatibility groups were investigated by the PCR-based replicon typing (PBRT) (10, 11) and Acinetobacter baumannii PBRT (AB-PBRT) (12) methods. Pa1206/13, displaying an extensively drug-resistant (XDR) phenotype (13) (Table 1), carried blaCTX-M-2 and blaGES-1 genes. S1 and I-Ceu-I nuclease digestion followed by pulsed-field gel electrophoresis (PFGE) and Southern blot hybridization with specific probes was performed to determine the locations of the bla genes. Based on S1-PFGE, Pa1206/13 possessed a single ∼340-kb plasmid (p1206/13), which was nontypeable by PBRT, IncU, IncR, or AB-PBRT. Although these methodologies are not optimized for the typing of Pseudomonas aeruginosa plasmids, they are the most commonly used plasmid-typing methodologies. Southern blotting followed by hybridization with blaCTX-M-2- and blaGES-1-specific probes revealed that both bla genes were carried by p1206/13. Hybridization with probes for a Pseudomonas sp. 16S rRNA gene and the two bla genes after I-Ceu-I-PFGE further excluded a chromosomal location. Whole-genome sequencing of Pa1206/13 was then performed using Illumina NextSeq 250-bp paired-end sequencing. De novo assembly was carried out using CLC Genomics Workbench, version 8.0 (CLC bio, Aarhus, Denmark), and generated 565 contigs, with a contig N50 of 125,375 bp, an average coverage of 84×, and an assembled genome of approximately 7.1 Mb (draft sequence). Gene prediction was performed for the draft sequence using the RAST server (http://rast.nmpdr.org/).
TABLE 1.
In vitro evaluation of activities of antimicrobial drugs against P. aeruginosa 1206/13
| Druga | Susceptibility profileb | MIC (μg/ml)c |
|---|---|---|
| TZP | I | |
| TIM | R | |
| CZA | S | |
| C/T | R | |
| CAZ | R | ≥256 (R) |
| CPM | R | ≥256 (R) |
| ATM | R | 16 (I) |
| IPM | R | ≥32 (R) |
| MER | R | ≥32 (R) |
| GEN | R | |
| TOB | R | |
| AMK | R | |
| CIP | R | |
| LVX | R |
TZP, piperacillin-tazobactam; TIM, ticarcillin-clavulanate; CZA, ceftazidime-avibactam; C/T, ceftolozane-tazobactam; CAZ, ceftazidime; CPM, cefepime; ATM, aztreonam; IPM, imipenem; MER, meropenem; GEN, gentamicin; TOB, tobramycin; AMK, amikacin; CIP, ciprofloxacin; LVX, levofloxacin.
S, susceptible; I, intermediate; R, resistant.
MIC testing was performed by Etest (bioMérieux). MIC breakpoints were evaluated according to CLSI guidelines (19).
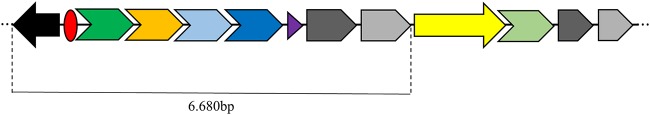
According to multilocus sequence typing (http://pubmlst.org/paeruginosa/), Pa1206/13 belongs to sequence type 1602 (ST1602), which was recently characterized in two P. aeruginosa clinical isolates from Brazil (14), and Pa1206/13 seems to be the first reported ST1602 isolate producing ESBL. The sequencing data revealed blaGES-1 as a gene cassette on a previously unreported class 1 integron, In1600 (http://integrall.bio.ua.pt/) (Fig. 1). Furthermore, blaCTX-M-2 was found downstream of ISCR1 associated with In1600, resulting in a complex class 1 integron of ∼11,680 bp (15). Additional antimicrobial resistance genes were predicted using ResFinder, version 3.1 (https://cge.cbs.dtu.dk/services/ResFinder/), which showed a resistome consisting of 15 resistance genes [aadA2, aphA-6, aph(3′)-IIb, aacA4, blaOXA-395, blaCTX-M-2, blaPAO, blaOXA-2, blaGES-1, crpP, fosA, cmlA4, catB7, sul1, dfrB5]. PlasmidFinder was also used to determine the type of plasmid and, again, confirmed it as nontypeable. In silico analysis of the draft sequence showed that the plasmid was closely related to IncP2 plasmids (GenBank accession numbers KC543497.1 and KY494864.1). IncP2 plasmids have been found in environmental bacteria and have been observed carrying a tellurite resistance determinant (16). p1206/13 possessed conjugation (tra family; TraV, TraB, TraG) and partitioning (par family; ParA and ParB) genes, showing that in vivo conjugation may occur. Furthermore, p1206/13 carried diverse virulence determinants, including pil proteins (PilT and PilG), which govern twitching motility, as well as type IV pili and biofilm formation, and the che operon, which is known to be essential for flagellum chemotaxis in P. aeruginosa (17). These virulence factors have also been detected in other IncP2 plasmids from P. aeruginosa (pBJ37 [18] and pOZ176 [16]). However, the mer operon present in those plasmids was not detected in p1206/13.
FIG 1.
Schematic representation of the complex class 1 integron characterized in the GES-1- and CTX-M-2-producing Pseudomonas aeruginosa 1206/13 isolate. Arrows indicate the gene orientations. The black arrow represents intI (the class I integron integrase gene); the red circle, attI (the integron-associated recombination site). The four cassette genes/proteins that follow are blaGES-1/β-lactamase (dark green arrow), aacA4/aminoglycoside-modifying enzyme (orange arrow), cmlA4/chloramphenicol exporter (light blue arrow), and aadA2/aminoglycoside-modifying enzyme (dark blue arrow). The purple triangle represents attC (the cassette-associated recombination sites). The 3′ conserved segment consists of fused genes for disinfectant and sulfonamide resistance (qacEΔ [dark gray arrow] and sul1 [light gray arrow], respectively). Downstream of sul1 are ISCR1 (yellow arrow) associated with blaCTX-M-2 (light green arrow) and duplicate qacEΔ/sul1 genes.
blaCTX-M-2 inserted into the P. aeruginosa chromosome has been described previously; however, this is the first report of an IncP2 plasmid coharboring two ESBL genes, blaCTX-M-2 and blaGES-1, in P. aeruginosa.
Accession number(s).
This sequence has been deposited in the DDBJ/ENA/GenBank database under BioSample accession number SAMN08384001.
ACKNOWLEDGMENTS
We thank the São Paulo Research Foundation (FAPESP) and the National Council for Scientific and Technological Development (CNPq, Brazil) for their constant support for our research. We also thank Dr. Vaughn Cooper for his assistance with whole-genome sequencing.
This work was supported by FAPESP (grant 2014/14494-8). The efforts of Y.D. were supported by research grants from the National Institutes of Health (grants R21AI123747, R21AI135522, and R01AI104895). A.S.B. was supported by a Ph.D. fellowship (grant 2015/23484-9). R.G. was supported by a postdoctoral fellowship from FAPESP (grant 2015/11728-0). Also, this study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES) (Finance Code 001).
We have no conflicts of interest to declare.
REFERENCES
- 1.Cantón R, González-Alba JM, Galán JC. 2012. CTX-M enzymes: origin and diffusion. Front Microbiol 3:110. doi: 10.3389/fmicb.2012.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adeolu M, Alnajar S, Naushad S, Gupta RS. 2016. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int J Syst Evol Microbiol 66:5575–5599. doi: 10.1099/ijsem.0.001485. [DOI] [PubMed] [Google Scholar]
- 3.Galetti R, Andrade LN, da Costa Darini AL. 2015. Pseudomonas aeruginosa carrying blaCTX-M-2 in Brazil: the occurrence of “high-risk clones”? J Glob Antimicrob Resist 3:153–154. doi: 10.1016/j.jgar.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Cuzon G, Bogaerts P, Bauraing C, Huang TD, Bonnin RA, Glupczynski Y, Naas T. 2016. Spread of plasmids carrying multiple GES variants. Antimicrob Agents Chemother 60:5040–5043. doi: 10.1128/AAC.00360-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrade LN, Minarini LAR, Pitondo-Silva A, Clímaco EC, Palazzo ICV, Medeiros MIC, Darini ALC. 2010. Determinants of β-lactam resistance in meningitis-causing Enterobacteriaceae in Brazil. Can J Microbiol 56:399–407. doi: 10.1139/W10-020. [DOI] [PubMed] [Google Scholar]
- 6.Taylor E, Sriskandan S, Woodford N, Hopkins KL. 2018. High prevalence of 16S rRNA methyltransferases among carbapenemase-producing Enterobacteriaceae in the UK and Ireland. Int J Antimicrob Agents 52:278–282. doi: 10.1016/j.ijantimicag.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 7.Bogaerts P, de Castro RR, de Mendonça R, Huang TD, Denis O, Glupczynski Y. 2013. Validation of carbapenemase and extended-spectrum β-lactamase multiplex endpoint PCR assays according to ISO 15189. J Antimicrob Chemother 68:1576–1582. doi: 10.1093/jac/dkt065. [DOI] [PubMed] [Google Scholar]
- 8.Cattoir V, Poirel L, Rotimi V, Soussy CJ, Nordmann P. 2007. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J Antimicrob Chemother 60:394–397. doi: 10.1093/jac/dkm204. [DOI] [PubMed] [Google Scholar]
- 9.Ellington MJ, Kistler J, Livermore DM, Woodford N. 2007. Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J Antimicrob Chemother 59:321–322. doi: 10.1093/jac/dkl481. [DOI] [PubMed] [Google Scholar]
- 10.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 11.García-Fernández A, Fortini D, Veldman K, Mevius D, Carattoli A. 2009. Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J Antimicrob Chemother 63:274–281. doi: 10.1093/jac/dkn470. [DOI] [PubMed] [Google Scholar]
- 12.Bertini A, Poirel L, Mugnier PD, Villa L, Nordmann P, Carattoli A. 2010. Characterization and PCR-based replicon typing of resistance plasmids in Acinetobacter baumannii. Antimicrob Agents Chemother 54:4168–4177. doi: 10.1128/AAC.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magiorakos A-P, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 14.Cacci LC, Chuster SG, Martins N, do Carmo PR, Girão VBDC, Nouér SA, de Freitas WV, de Matos JA, Magalhães ACDG, Ferreira ALP, Picão RC, Moreira BM. 2016. Mechanisms of carbapenem resistance in endemic Pseudomonas aeruginosa isolates after an SPM-1 metallo-β-lactamase producing strain subsided in an intensive care unit of a teaching hospital in Brazil. Mem Inst Oswaldo Cruz 111:551–558. doi: 10.1590/0074-02760160116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Partridge SR, Kwong SM, Firth N, Jensen SO. 2018. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev 31:e00088-17. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong J, Alexander DC, Ma JH, Déraspe M, Low DE, Jamieson FB, Roy PH. 2013. Complete sequence of pOZ176, a 500-kilobase IncP-2 plasmid encoding IMP-9-mediated carbapenem resistance, from outbreak isolate Pseudomonas aeruginosa 96. Antimicrob Agents Chemother 57:3775–3782. doi: 10.1128/AAC.00423-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burrows LL. 2012. Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu Rev Microbiol 66:493–520. doi: 10.1146/annurev-micro-092611-150055. [DOI] [PubMed] [Google Scholar]
- 18.Botelho J, Grosso F, Quinteira S, Mabrouk A, Peixe L. 2017. The complete nucleotide sequence of an IncP-2 megaplasmid unveils a mosaic architecture comprising a putative novel blaVIM-2-harbouring transposon in Pseudomonas aeruginosa. J Antimicrob Chemother 72:2225–2229. doi: 10.1093/jac/dkx143. [DOI] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial susceptibility testing, 28th ed, M100-S28E. CLSI, Wayne, PA. [Google Scholar]