Clinical phenotypic fluoroquinolone susceptibility testing of Mycobacterium tuberculosis is currently based on M. tuberculosis growth at a single critical concentration, which provides limited information for a nuanced clinical response. We propose using specific resistance-conferring M. tuberculosis mutations in gyrA together with population pharmacokinetic and pharmacodynamic modeling as a novel tool to better inform fluoroquinolone treatment decisions.
KEYWORDS: fluoroquinolone, pharmacodynamics, pharmacokinetics, treatment, tuberculosis
ABSTRACT
Clinical phenotypic fluoroquinolone susceptibility testing of Mycobacterium tuberculosis is currently based on M. tuberculosis growth at a single critical concentration, which provides limited information for a nuanced clinical response. We propose using specific resistance-conferring M. tuberculosis mutations in gyrA together with population pharmacokinetic and pharmacodynamic modeling as a novel tool to better inform fluoroquinolone treatment decisions. We sequenced the gyrA resistance-determining region of 138 clinical M. tuberculosis isolates collected from India, Moldova, Philippines, and South Africa and then determined each strain’s MIC against ofloxacin, moxifloxacin, levofloxacin, and gatifloxacin. Strains with specific gyrA single-nucleotide polymorphisms (SNPs) were grouped into high or low drug-specific resistance categories based on their empirically measured MICs. Published population pharmacokinetic models were then used to explore the pharmacokinetics and pharmacodynamics of each fluoroquinolone relative to the empirical MIC distribution for each resistance category to make predictions about the likelihood of patients achieving defined therapeutic targets. In patients infected with M. tuberculosis isolates containing SNPs associated with a fluoroquinolone-specific low-level increase in MIC, models suggest increased fluoroquinolone dosing improved the probability of achieving therapeutic targets for gatifloxacin and moxifloxacin but not for levofloxacin and ofloxacin. In contrast, among patients with isolates harboring SNPs associated with a high-level increase in MIC, increased dosing of levofloxacin, moxifloxacin, gatifloxacin, or ofloxacin did not meaningfully improve the probability of therapeutic target attainment. We demonstrated that quantifiable fluoroquinolone drug resistance phenotypes could be predicted from rapidly detectable gyrA SNPs and used to support dosing decisions based on the likelihood of patients reaching therapeutic targets. Our findings provide further supporting evidence for the moxifloxacin clinical breakpoint recently established by the World Health Organization.
INTRODUCTION
Prior to the advent of antimicrobials, the 10-year case fatality rate (CFR) for individuals with smear-positive tuberculosis (TB) was estimated to be 70% (1). In contrast, the current CFR for all TB cases reported to the World Health Organization (WHO) for 2017 was only 16% (2). This reduction in mortality is impressive; however, universal treatment success remains elusive and is particularly dismal among individuals who harbor drug-resistant Mycobacterium tuberculosis. According to the WHO, among individuals with multidrug-resistant TB (MDR-TB), the treatment success rate is currently 55% (2016 cohort), and among those with extensively drug-resistant TB (XDR-TB), the treatment success rate is only 34% (2015 cohort) (2). If M/XDR-TB treatment success rates are to be significantly improved, a more individualized approach to treatment is needed. We propose that available molecular diagnostics combined with simple probabilistic pharmacokinetic and pharmacodynamic (PKPD) modeling would provide an additional tool for clinicians that could rapidly determine optimal treatment and dosing regimens that better align with the infecting pathogen’s unique resistance profile and ultimately result in better treatment outcomes (3).
Currently, if a patient is considered at risk for drug resistance or does not respond to standard treatment as anticipated, drug susceptibility testing (DST) is used as a guide to help physicians design an appropriate TB treatment regimen. Both phenotypic and genotypic DST are used to classify clinical M. tuberculosis strains as either “resistant” or “susceptible” to specific drugs which are then included or excluded from a treatment plan. Routine phenotypic DST has historically been based on M. tuberculosis resistance in vitro at a single WHO-recommended “critical concentration” of drug (4). Genotypic DST, on the other hand, classifies resistance based on the detection of single-nucleotide polymorphisms (SNPs) documented to confer phenotypic resistance at the critical concentration. However, both these overly simplistic dichotomous classifications can potentially limit treatment options and exclude the use of critical drugs that might still be effective against strains considered resistant (4–6).
The MIC of a drug required to achieve antimycobacterial activity provides a more nuanced approach to resistance classification and can resolve some phenotypic/genotypic DST discordance for isolates with MICs that near the critical concentration. However, phenotypic characterization of each patient’s M. tuberculosis MICs for multiple drugs is resource intensive and unrealistic in low-resource settings given the biosafety hazards and required specialized laboratory skills. Additionally, the slow growth of M. tuberculosis adds weeks to the diagnostic process. We propose that single, rapidly detected resistance-conferring SNPs can be used as clinically reliable proxies for phenotypic MIC. The uptake and recent proliferation of molecular-based DSTs would allow for sufficiently accurate approximations of MICs without significant changes in current diagnostic practices or laboratory workflows (7).
Optimized dosing that results in serum concentrations that maximize the probability of successfully killing or sterilizing M. tuberculosis in individual patients, while minimizing potential drug toxicity effects, is based both on individual patient pharmacokinetic parameters and drug-specific MICs (3, 8). However, as with phenotypic MIC estimation, empirical measurements of patient-specific drug levels are invasive, technically challenging, and costly (9, 10). Probabilistic pharmacokinetic and pharmacodynamic (PKPD) modeling uses population PKPD parameters to describe the relationship between dose and probable serum drug concentration over time (0 to 24 h) or area under the concentration curve (AUC), providing an alternative method to estimate likely AUC/MIC ratios.
The goal of this study was 2-fold. First, using a small clinical M. tuberculosis isolate set, we demonstrated that resistance-conferring mutations for gyrA SNPs can contribute to a more nuanced understanding of resistance for gatifloxacin (GFX), levofloxacin (LFX), moxifloxacin (MFX), and ofloxacin (OFX). Second, using gyrA SNPs as proxies for either a low- or high-level increase in fluoroquinolone-specific MIC above the critical concentration, we used probabilistic population PKPD modeling to estimate what proportion of a patient population would achieve therapeutic AUC/MIC targets based on the likely fluoroquinolone-specific MICs of M. tuberculosis strains with specific gyrA SNPs. Given the increasing availability of molecular-based DSTs, we are proposing that individual SNPs detected by molecular-based tests be used as predictors of MIC or “level of resistance” and that treatment response be predicted based on identified gyrA SNPs.
RESULTS
SNP classification.
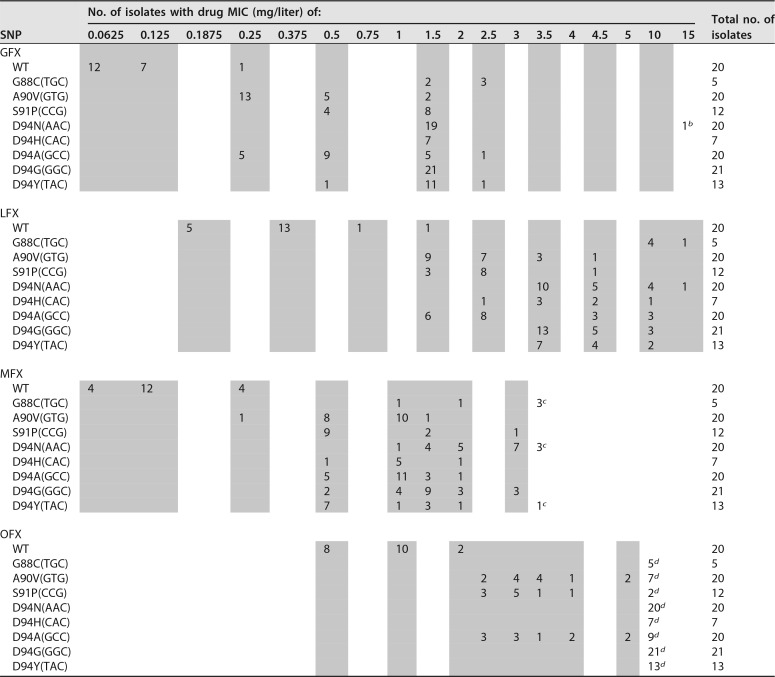
One-hundred thirty-eight isolates were selected from the UCSD archive and successfully revived from frozen MicroBank beads. Isolate cultures were then used to inoculate MGIT tubes at approximate serial dilutions for GFX, LFX, MFX, and OFX (Table 1). MIC results for each isolate were aggregated by SNP. MIC distributions varied between fluoroquinolones. GFX and MFX MICs were consistently lower than that for LFX. OFX MICs were greater than the highest dilution tested for a majority of the isolates.
TABLE 1.
Frequencies of GFX, LFX, MFX, and OFX MGIT MICs by SNP among 138 M. tuberculosis isolatesa
Shaded columns indicate MICs tested.
bMIC of >10 mg/liter.
cMIC of >3 mg/liter.
dMIC of >5 mg/liter.
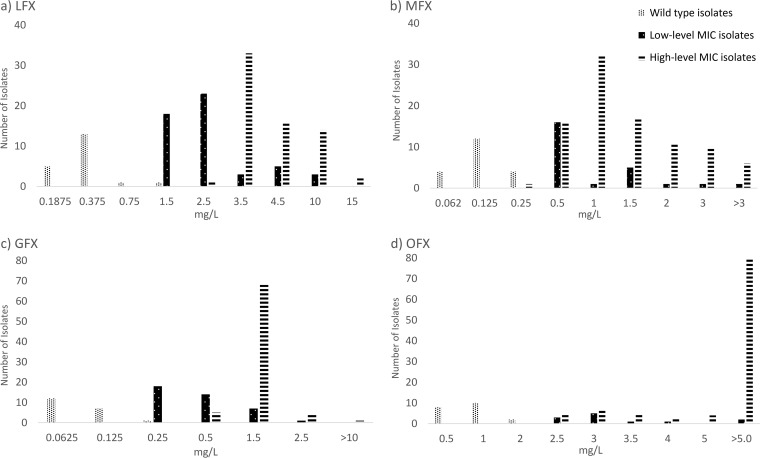
Isolates with specific gyrA SNPs were grouped by their MIC mode into high-level and low-level MIC categories (Table 2). Overall, SNP categorizations were similar between all fluoroquinolones with a few exceptions. In our isolate set, SNP A90V(GTG) was consistently categorized as a low-level increase MIC mutation across all fluoroquinolones tested, and SNPs G88C(TGC), D94N(AAC), and D94G(GGC) were consistently categorized as high-level increase MIC mutations. Specific MICs (on a dilution scale) were graphed by classification categories in Fig. 1. Kruskal-Wallis and Dunn’s tests indicated that there was a statistically significant difference (P < 0.001) in MIC distributions between wild-type isolates, isolates harboring SNPs categorized as low-level MICs, and isolates harboring SNPs categorized as high-level MICs for all fluoroquinolones. A majority of OFX resistance-conferring SNPs had MICs greater than the highest dilution measured and were categorized using the highest dilution tested.
TABLE 2.
Fluoroquinolones by MIC category, SNP, corresponding amino acid, and MTBDRsl probe
| Fluoroquinolone | MIC (mg/liter) [mode (median)] | SNP | AAa | Corresponding Hain LPA probe |
|---|---|---|---|---|
| Gatifloxacin | 0.0625 (0.0625) | Wild type | ||
| Low-level MIC | 0.25 (0.5) | 90GTG | A90V (Val) | gyrAMUT1 |
| 94GCC | D94A (Ala) | gyrAMUT3A | ||
| High-level MIC | 1.5 (1.5) | 88TGC | G88C (Cys) | |
| 91CCG | S91P (Pro) | gyrAMUT2 | ||
| 94TAC | D94Y (Tyr) | gyrAMUT3B | ||
| 94AAC | D94N (Asn) | gyrAMUT3B | ||
| 94GGC | D94G (Gly) | gyrAMUT3C | ||
| 94CAC | D94H (His) | gyrAMUT3D | ||
| Levofloxacin | 0.375 (0.375) | Wild type | ||
| Low-level MIC | 2.5 (2.5) | 90GTG | A90V (Val) | gyrAMUT1 |
| 91CCG | S91P (Pro) | gyrAMUT2 | ||
| 94GCC | D94A (Ala) | gyrAMUT3A | ||
| High-level MIC | 3.5 (3.5) | 88TGC | G88C (Cys) | |
| 94AAC | D94N (Asn) | gyrAMUT3B | ||
| 94TAC | D94Y (Tyr) | gyrAMUT3B | ||
| 94GGC | D94G (Gly) | gyrAMUT3C | ||
| 94CAC | D94H (His) | gyrAMUT3D | ||
| Moxifloxacin | 0.125 (0.125) | Wild type | ||
| Low-level MIC | 0.5 (0.5) | 91CCG | S91P (Pro) | gyrAMUT2 |
| 94TAC | D94Y (Tyr) | gyrAMUT3B | ||
| High-level MIC | 1.0 (1.0) | 88TGC | G88C (Cys) | |
| 90GTG | A90V (Val) | gyrAMUT1 | ||
| 94GCC | D94A (Ala) | gyrAMUT3A | ||
| 94AAC | D94N (Asn) | gyrAMUT3B | ||
| 94GGC | D94G (Gly) | gyrAMUT3C | ||
| 94CAC | D94H (His) | gyrAMUT3D | ||
| Ofloxacin | 1.0 (1.0) | Wild type | ||
| Low-level MIC | 3.0 (3.0) | 91CCG | S91P (Pro) | gyrAMUT2 |
| High-level MIC | 5.0 (5.0) | 88TGC | G88C (Cys) | |
| 90GTG | A90V (Val) | gyrAMUT1 | ||
| 94GCC | D94A (Ala) | gyrAMUT3A | ||
| 94TAC | D94Y (Tyr) | gyrAMUT3B | ||
| 94AAC | D94N (Asn) | gyrAMUT3B | ||
| 94GGC | D94G (Gly) | gyrAMUT3C | ||
| 94CAC | D94H (His) | gyrAMUT3D |
AA, amino acid.
FIG 1.
Bar graphs of MICs for wild-type, low-level MIC, and high-level MIC categories by fluoroquinolone. (a) LFX; (b) MFX; (c) GFX; (d) OFX.
PKPD modeling.
Monte Carlo simulations were used to investigate the probability of AUC/MIC target attainment for each fluoroquinolone using the distribution of empirically derived MICs from the study isolates for each MIC category. Population PKPD models were used to assess the probability of therapeutic target attainment for each fluoroquinolone by MIC category and were plotted for each drug dose.
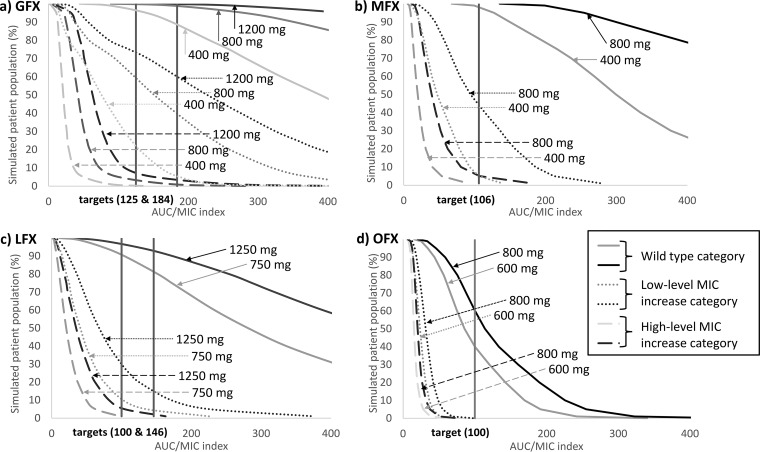
Among patients with low-level increase GFX MIC mutations, dosing of 400 mg/day of GFX resulted in only 4% of the theoretical patient population achieving a target AUC/MIC of 184; in contrast, increased dosing of 800 and 1,200 mg/day of GFX resulted in 40% and 60% of patients, respectively, achieving the target AUC/MIC (Fig. 2a). Among patients with high-level increase GFX MIC mutations, the highest dosing modeled (1,200 mg/day) resulted in only 3% of the patient population achieving therapeutic targets. Among patients with wild-type M. tuberculosis, dosing of 400, 800, and 1,200 mg/day of GFX resulted in 89%, 99%, and 100% of the patient population, respectively, achieving the target AUC/MIC of 184.
FIG 2.
Graphs of proportions of population achieving therapeutic target (AUC/MIC) by fluoroquinolone dose and SNP category. (a) GFX; (b) MFX; (c) LFX; (d) OFX.
MFX treatment of 400 mg/day among patients with low-level increase MFX MIC mutations resulted in 5% of the patient population achieving a target AUC/MIC of 106. Increased MFX dosing to 800 mg/day increased the proportion of the patient population attaining the target AUC/MIC to 44%. Among patients with high-level increase MFX MIC mutations, increased dosing resulted in only 5% of the population achieving the target AUC/MIC. Models indicated that 98% of patients with wild-type M. tuberculosis would achieve AUC/MIC targets with standard 400-mg/day dosing and 100% would achieve targets with 800-mg/day dosing (Fig. 2b).
Among patients with low-level increase LFX MIC mutations, increased dosing of LFX from 750 mg/day to 1,250 mg/day only increased the proportion of the patient population likely to achieve a target AUC/MIC of 146 from 4% to 14%. Our models indicated that among patients with high-level LFX MIC mutations, only 2% would achieve a target AUC/MIC of 146 at 750 mg/day in comparison to less than 1% with standard dosing. Among patients with wild-type M. tuberculosis, model estimates demonstrated that 82% of the patient population would achieve therapeutic targets with standard 750-mg/day dosing, while increased dosing of LFX to 1,250 mg/day would result in 93% of the patient population achieving therapeutic targets (Fig. 2c).
Among patients with either low-level or high-level OFX MIC mutations, OFX dosing of 600 or 800 mg/day resulted in 0% of patients achieving a target AUC/MIC of 100. Even among patients with wild-type M. tuberculosis, dosing of 600 and 800 mg/day resulted in only 40% and 60% of patients, respectively, achieving therapeutic targets (Fig. 2d).
DISCUSSION
We demonstrated that SNPs in the gyrA gene of M. tuberculosis can be used to rapidly predict approximate and statistically differentiable MICs to specific fluoroquinolones, allowing for the potentially clinically relevant categorization of M. tuberculosis strains based on gyrA SNPs. This MIC categorization, together with PKPD simulations, can be used to predict the probability a patient will achieve therapeutic targets for individual fluoroquinolones.
SNPs predict level of resistance.
Previous investigators have aggregated the distributions of wild-type M. tuberculosis fluoroquinolone MICs; however, only with the recent release of the WHO technical report on critical concentrations for drug susceptibility testing of medicines used in the treatment of drug-resistant tuberculosis have the MICs associated with individual resistance-associated gyrA SNPs been exhaustively aggregated (4, 11, 12). We used our limited isolate set to demonstrate the proof of principal that specific SNPs can be used to predict statistically distinct high- or low-level increase fluoroquinolone-specific MIC categories. Additionally, differences in our SNP categorization between fluoroquinolones illustrate the complexity of fluoroquinolone resistance and the importance of individual fluoroquinolone resistance determination given the incomplete cross-resistance between fluoroquinolones used for TB treatment (13).
Our categorization of SNPs for individual fluoroquinolones was relatively consistent with previous studies, which classified D94N(AAC) and D94G(GGC) as SNPs associated with a high-level increase in MFX MIC and D94Y(TAC) as a SNP associated with a low-level increase in MFX MIC (14, 15). OFX MIC classifications were also consistent and identified D94G(GGC), D94N(AAC), D94Y(TAC), and D94A(GCC) as high-level OFX MIC SNPs (16). Some classification discrepancies were apparent for SNPs with MICs that demonstrated midlevel resistance, but were likely due to differing classification cutoffs.
Population PKPD modeling.
Our models demonstrated that among simulated patients harboring M. tuberculosis isolates with SNPs associated with low-level increase in GFX MIC, only 5% treated with 400 mg/day of GFX would achieve an AUC/MIC target of 184. However, if dosing were increased to 800 or 1,200 mg/day, approximately 40% or 60% of patients, respectively, would attain the therapeutic target, suggesting increased GFX dosing may play a useful role in the treatment of M. tuberculosis strains with gyrA SNPs associated with low-level increase in MIC. If assessing the AUC/MIC target of 125, increased dosing to 800 or 1,200 mg/day would result in 59% or 73% of the patient population, respectively, attaining the therapeutic target for GFX. Among simulated patients with SNPs associated with low-level increase in MFX MIC, only 5% would achieve a target AUC/MIC of 106 with standard dosing of 400 mg/day of MFX. Increased MFX dosing to 800 mg/day would result in 44% of the patient population attaining therapeutic targets, which suggests high-dose MFX may still be worth considering as part of a multiple drug regimen in patients with strains with SNPs associated with low-level increase in MIC in settings where alternative options are limited. This provides further supporting evidence for the moxifloxacin clinical breakpoint recently established by the World Health Organization (4). In contrast, among patients harboring low-level LFX or OFX MICs, increased dosing of either drug did not meaningfully increase the proportion of patients likely to attain therapeutic targets, emphasizing the limited value of older-generation fluoroquinolones in drug-resistant TB treatment. Additionally, among patients with isolates harboring SNPs associated with high-level fluoroquinolone resistance, increased dosing of GFX, LFX, MFX, or OFX did not increase the probability of therapeutic target attainment, suggesting that fluoroquinolones have a limited role to play in M. tuberculosis strains with these mutations.
Based on our OFX modeling, it appears that among patients harboring wild-type M. tuberculosis, only 40% and 60% of would achieve the generic AUC/MIC therapeutic target of 100 at 600 and 800 mg/day, respectively, providing a possible explanation of poorer treatment results among patients with OFX-containing treatment regimens than with MFX-containing regimens.
Our study results appear to confirm results from Gumbo et al, who demonstrated that based on the variability in distribution and clearance of MFX in human subjects as well as the variability of the MICs, the currently recommended dose of MFX of 400 mg/day is likely to suppress drug resistance in ∼60% of patients. Approximately 93% of patients would achieve this goal with 800 mg/day (17). Monte Carlo simulations by Smythe et al. predicted that 90% of patients would achieve an AUC/MIC of >125 only when M. tuberculosis MIC was <0.125 mg/liter with a GFX dose of 800 mg/day (18). This compares favorably to our results for wild-type M. tuberculosis (MIC mode of 0.0625 mg/liter), in which Monte Carlo simulations predict that 99% of patients harboring wild-type M. tuberculosis would achieve an AUC/MIC target of 184.
Given the importance of fluoroquinolones in the recently published WHO guidelines for all-oral regimens to treat MDR-TB, it is critical that we use these drugs effectively and sustainably (19). In fact, ignoring these considerations may have contributed to the lack of success observed in some of the most recent phase three trials of short-course fluoroquinolone treatments (20, 21). While several new drugs have recently been added to the anti-TB arsenal, their use is still limited, making stewardship and optimization of current anti-TB drugs imperative if a return to the preantimicrobial era is to be avoided (22).
Limitations.
This study was based on limited set of isolates and only considered mutations in the gyrA gene. Larger aggregated data sets and including mutations in the gyrB gene would have likely provided a more finely calibrated distribution of MICs based on specific SNPs; however, setting conclusive MIC distributions for each gyrA SNP along with any synergistic effects of gyrB SNPs was beyond the scope of this study (6).
Lacking in the TB research literature are well-defined fluoroquinolone-specific AUC/MIC index targets associated with clinical outcomes and long-term treatment success. We included both generic fluoroquinolone AUC/MIC index targets and fluoroquinolone-specific targets associated with either microbial kill or resistance suppression. Additional trials are needed to confirm clinical relevance of fluoroquinolone-specific AUC/MIC targets.
Conclusions.
In conclusion, we demonstrated using a limited data set that SNPs in the gyrA gene of M. tuberculosis can be used to rapidly predict statistically distinct approximate MICs of individual fluoroquinolones, allowing for potentially clinically relevant MIC categorization of M. tuberculosis strains. This MIC categorization, together with Monte Carlo simulations, can be used to predict the probability a patient will achieve therapeutic targets for individual fluoroquinolones based on the presence of a specific gyrA SNP. This novel approach to interpreting molecular-based DSTs could potentially provide a template for evaluation of antituberculosis drugs in real time using currently available tools and move closer toward a precision medicine approach for treatment of drug-resistant TB.
MATERIALS AND METHODS
This study was determined by the University of California Human Subject Research Protections Program to be exempt from review.
Specimen archive.
M. tuberculosis isolates were selected from our laboratory archive containing approximately 800 clinical M. tuberculosis specimens collected under diverse enrollment and specimen selection criteria as previously described (23, 24). In brief, isolates include retrospectively collected samples from TB hospital or research facility repositories in India, Moldova, Philippines, and South Africa. To ensure genotypic diversity, strains from these repositories were selected to maximize the diversity of local phenotypic resistance patterns, and only one isolate was selected per TB patient. Prospectively acquired isolates were collected as part of a diagnostic evaluation cohort study conducted in India, Moldova, and South Africa from 2012 and 2013. Specimens were collected from patients presenting with presumed drug-resistant TB, and only one isolate from each patient was included to ensure clinical independence of samples. All archive isolates were subjected to standard DST using the MGIT960 method (25). Isolates were sequenced at the gyrA gene from codon 88 to 94 using one or more of the following methods: Pyrosequencing using a PyroMark Q96 ID system, Sanger sequencing, or PacBio sequencing. A detailed description of sequencing methods was previously published (23, 26).
Isolate selection.
Approximately 20 isolates were randomly selected from the archive specimen set for MIC analysis for each of the following SNPs: G88C(TGC), A90V(GTG), S91P(CCG), D94G(GGC), D94A(GCC), D94N(AAC), D94Y(TAC), and D94H(CAC) (27). If fewer than 20 isolates in the archive set harbored a desired SNP, all isolates containing the desired mutation were selected for analysis. For comparison, an additional 20 phenotypically OFX-susceptible isolates harboring no SNPs between gyrA codon 88 and 94 were randomly selected from the archived isolates to represent wild-type strains and were also subjected to MIC analysis. Of the 20 OFX-susceptible isolates, 15 were phenotypically pan-susceptible for isoniazid, rifampin, capreomycin, kanamycin, and amikacin and 5 were resistant to isoniazid and rifampin. M. tuberculosis H37Rv (ATCC 27294) was included as a control strain.
Isolate culturing and MIC analysis.
Selected isolates were revived from frozen MicroBank beads using the semiautomated Bactec MGIT960 (Becton, Dickinson, Sparks, MD). One frozen bead from a Microbank bead cryovial was added to a 7-ml MGIT tube containing 800 μl MGIT supplement and incubated in the MGIT960 until flagged positive by the instrument. OFX (O8757; Sigma), LFX (28266; Sigma), and GFX (G7298; Sigma) were dissolved in 0.1 N NaOH and MFX (047903; Matrix Scientific) was dissolved in UltraPure water (10977015; Invitrogen) to create concentrated stock solutions. Stock solutions were filter sterilized using a 0.22-μm membrane microfilter (SLGS033SS; Millex) and diluted in UltraPure water to create the desired working concentrations. A positive M. tuberculosis culture was inoculated simultaneously in a growth control MGIT tube with multiple dilutions of GFX, MFX, LFX, and OFX. Drug concentrations initially included five concentrations above and three dilutions below the critical concentration for a total of nine dilutions; critical concentrations were 0.5, 0.25, 1.5, and 2.0 mg/liter for GFX, MFX, LFX, and OFX, respectively. If no growth was detected at the lowest concentration, isolates were regrown at two additional dilutions. The MIC DST setups were monitored by the MGIT960 linked to the BD EpiCenter software equipped with the TB eXiST module for extended susceptibility testing.
SNP classification.
Isolates with specific gyrA SNPs were visually grouped by MIC mode and categorized as low-level or high-level increase category MIC for each fluoroquinolone. Differences in MIC distributions between MIC categories were assessed using the nonparametric Kruskal-Wallis test, and pairwise differences between categories were assessed post hoc using the Dunn’s test. For context with existing reference standard molecular diagnostics, we also identified corresponding probes from the GenoType MTBDRsl assay (Hain Lifescience, Germany) to each of the SNPs assessed.
PKPD modeling.
Using NONMEM version 7.3 (ICON, Dublin, Ireland), virtual PK parameters for 10,000 simulated subjects were linked (randomly) to the distribution of observed MICs for each MIC category. AUCs were calculated using standard PK equations (AUC = bioavailability × dose/clearance) and based on previously published PKPD models for GFX, LFX, MFX, and OFX (18, 20, 28, 29). The proportion of subjects achieving optimal AUC/MIC thresholds was simulated using SAS 9.4 software (Cary, NC).
Optimal AUC/MIC thresholds were selected based on previously published targets associated with clinical response or with in vitro resistance acquisition suppression, and if optimal TB AUC/MIC targets were not well defined in the literature for M. tuberculosis, thresholds were selected based on generic fluoroquinolone targets or specific targets associated with either microbial kill or resistance suppression in various models. The AUC/MIC target ratio(s) for MFX was 106, for LFX was 100 and 146, for OFX was 100, and for GFX was 125 and 184 (17, 18, 30–32). Target achievement was assessed for each drug using a combination of published standard dosing recommendations and investigational dosing for each drug (33). We used dosing of 750 and 1,250 mg/day for LFX, 400 and 800 mg/day for MFX, 400, 800, and 1,200 mg/day for GFX, and 600 and 800 mg/day for OFX to assess the impact of increased dosing on target achievement.
ACKNOWLEDGMENTS
We thank Mark Pettigrove for his laboratory support for this study.
This work was supported by the National Institute of Allergy and Infectious Diseases at the National Institutes of Health (grant numbers P30 AI036214-20 and R01AI111435) and by the National Hearth Lung and Blood Institute at the National Institutes of Health (grant number T32HL134632).
REFERENCES
- 1.Tiemersma EW, van der Werf MJ, Borgdorff MW, Williams BG, Nagelkerke NJ. 2011. Natural history of tuberculosis: duration and fatality of untreated pulmonary tuberculosis in HIV negative patients: a systematic review. PLoS One 6:e17601. doi: 10.1371/journal.pone.0017601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. 2018. Global Tuberculosis Report 2018. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.Aljayyoussi G, Jenkins VA, Sharma R, Ardrey A, Donnellan S, Ward SA, Biagini GA. 2017. Pharmacokinetic-pharmacodynamic modelling of intracellular Mycobacterium tuberculosis growth and kill rates is predictive of clinical treatment duration. Sci Rep 7:502. doi: 10.1038/s41598-017-00529-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO. 2018. Technical report on critical concentrations for drug susceptibility testing of medicines used in the treatment of drug-resistant tuberculosis. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 5.Zheng X, Zheng R, Hu Y, Werngren J, Forsman LD, Mansjo M, Xu B, Hoffner S. 2016. Determination of MIC breakpoints for second-line drugs associated with clinical outcomes in multidrug-resistant tuberculosis treatment in China. Antimicrob Agents Chemother 60:4786–4792. doi: 10.1128/AAC.03008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farhat MR, Jacobson KR, Franke MF, Kaur D, Sloutsky A, Mitnick CD, Murray M. 2016. Gyrase mutations are associated with variable levels of fluoroquinolone resistance in Mycobacterium tuberculosis. J Clin Microbiol 54:727–733. doi: 10.1128/JCM.02775-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghimire S, Van't Boveneind-Vrubleuskaya N, Akkerman OW, de Lange WCM, van Soolingen D, Kosterink JGW, van der Werf TS, Wilffert B, Touw DJ, Alffenaar J-W. 2016. Pharmacokinetic/pharmacodynamic-based optimization of levofloxacin administration in the treatment of MDR-TB. J Antimicrob Chemother 71:2691–2703. doi: 10.1093/jac/dkw164. [DOI] [PubMed] [Google Scholar]
- 8.Pasipanodya JG, McIlleron H, Burger A, Wash PA, Smith P, Gumbo T. 2013. Serum drug concentrations predictive of pulmonary tuberculosis outcomes. J Infect Dis 208:1464–1473. doi: 10.1093/infdis/jit352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alsultan A, Peloquin CA. 2014. Therapeutic drug monitoring in the treatment of tuberculosis: an update. Drugs 74:839–854. doi: 10.1007/s40265-014-0222-8. [DOI] [PubMed] [Google Scholar]
- 10.WHO. 2018. Technical report on the pharmacokinetics and pharmacodynamics (PK/PD) of medicines used in the treatment of drug-resistant tuberculosis. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 11.Angeby K, Giske CG, Jureen P, Schon T, Pasipanodya JG, Gumbo T. 2011. Wild-type MIC distributions must be considered to set clinically meaningful susceptibility testing breakpoints for all bacterial pathogens, including Mycobacterium tuberculosis. Antimicrob Agents Chemother 55:4492–4493. doi: 10.1128/AAC.00232-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu X, Wang G, Chen S, Wei G, Shang Y, Dong L, Schon T, Moradigaravand D, Parkhill J, Peacock SJ, Koser CU, Huang H. 2016. Wild-type and non-wild-type Mycobacterium tuberculosis MIC distributions for the novel fluoroquinolone antofloxacin compared with those for ofloxacin, levofloxacin, and moxifloxacin. Antimicrob Agents Chemother 60:5232–5237. doi: 10.1128/AAC.00393-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farhat MR, Mitnick CD, Franke MF, Kaur D, Sloutsky A, Murray M, Jacobson KR. 2015. Concordance of Mycobacterium tuberculosis fluoroquinolone resistance testing: implications for treatment. Int J Tuberc Lung Dis 19:339–341. doi: 10.5588/ijtld.14.0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willby M, Sikes RD, Malik S, Metchock B, Posey JE. 2015. Correlation between GyrA substitutions and ofloxacin, levofloxacin, and moxifloxacin cross-resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 59:5427–5434. doi: 10.1128/AAC.00662-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kambli P, Ajbani K, Sadani M, Nikam C, Shetty A, Udwadia Z, Rodwell TC, Catanzaro A, Rodrigues C. 2015. Correlating minimum inhibitory concentrations of ofloxacin and moxifloxacin with gyrA mutations using the genotype MTBDRsl assay. Tuberculosis (Edinb) 95:137–141. doi: 10.1016/j.tube.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Gao X, Luo T, Wu J, Sun G, Liu Q, Jiang Y, Zhang Y, Mei J, Gao Q. 2014. Association of gyrA/B mutations and resistance levels to fluoroquinolones in clinical isolates of Mycobacterium tuberculosis. Emerg Microbes Infect 3:e19. doi: 10.1038/emi.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gumbo T, Louie A, Deziel MR, Parsons LM, Salfinger M, Drusano GL. 2004. Selection of a moxifloxacin dose that suppresses drug resistance in Mycobacterium tuberculosis, by use of an in vitro pharmacodynamic infection model and mathematical modeling. J Infect Dis 190:1642–1651. doi: 10.1086/424849. [DOI] [PubMed] [Google Scholar]
- 18.Smythe W, Merle CS, Rustomjee R, Gninafon M, Lo MB, Bah-Sow O, Olliaro PL, Lienhardt C, Horton J, Smith P, McIlleron H, Simonsson US. 2013. Evaluation of initial and steady-state gatifloxacin pharmacokinetics and dose in pulmonary tuberculosis patients by using Monte Carlo simulations. Antimicrob Agents Chemother 57:4164–4171. doi: 10.1128/AAC.00479-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.WHO. 2018. WHO treatment guidelines for multidrug- and rifampicin-resistant tuberculosis, 2018 update. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 20.Peloquin CA, Hadad DJ, Molino LP, Palaci M, Boom WH, Dietze R, Johnson JL. 2008. Population pharmacokinetics of levofloxacin, gatifloxacin, and moxifloxacin in adults with pulmonary tuberculosis. Antimicrob Agents Chemother 52:852–857. doi: 10.1128/AAC.01036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanoix JP, Chaisson RE, Nuermberger EL. 2016. Shortening tuberculosis treatment with fluoroquinolones: lost in translation? Clin Infect Dis 62:484–490. doi: 10.1093/cid/civ911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pai M, Behr MA, Dowdy D, Dheda K, Divangahi M, Boehme CC, Ginsberg A, Swaminathan S, Spigelman M, Getahun H, Menzies D, Raviglione M. 2016. Tuberculosis. Nat Rev Dis Primers 2:16076. doi: 10.1038/nrdp.2016.76. [DOI] [PubMed] [Google Scholar]
- 23.Rodwell TC, Valafar F, Douglas J, Qian L, Garfein RS, Chawla A, Torres J, Zadorozhny V, Kim MS, Hoshide M, Catanzaro D, Jackson L, Lin G, Desmond E, Rodrigues C, Eisenach K, Victor TC, Ismail N, Crudu V, Gler MT, Catanzaro A. 2014. Predicting extensively drug-resistant Mycobacterium tuberculosis phenotypes with genetic mutations. J Clin Microbiol 52:781–789. doi: 10.1128/JCM.02701-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hillery N, Groessl EJ, Trollip A, Catanzaro D, Jackson L, Rodwell TC, Garfein RS, Lin SY, Eisenach K, Ganiats TG, Park D, Valafar F, Rodrigues C, Crudu V, Victor TC, Catanzaro A. 2014. The Global Consortium for Drug-resistant Tuberculosis Diagnostics (GCDD): design of a multi-site, head-to-head study of three rapid tests to detect extensively drug-resistant tuberculosis. Trials 15:434. doi: 10.1186/1745-6215-15-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siddiqi SH, Rusch-Gerdes S. 2006. MGIT procedure manual for BACTEC MGIT 960 TB System (FIND). http://www.finddx.org/wp-content/uploads/2016/02/mgit_manual_nov2006.pdf. Accessed 16 June 2016.
- 26.Georghiou SB, Seifert M, Lin SY, Catanzaro D, Garfein RS, Jackson RL, Crudu V, Rodrigues C, Victor TC, Catanzaro A, Rodwell TC. 2016. Shedding light on the performance of a pyrosequencing assay for drug-resistant tuberculosis diagnosis. BMC Infect Dis 16:458. doi: 10.1186/s12879-016-1781-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miotto P, Tessema B, Tagliani E, Chindelevitch L, Starks AM, Emerson C, Hanna D, Kim PS, Liwski R, Zignol M, Gilpin C, Niemann S, Denkinger CM, Fleming J, Warren RM, Crook D, Posey J, Gagneux S, Hoffner S, Rodrigues C, Comas I, Engelthaler DM, Murray M, Alland D, Rigouts L, Lange C, Dheda K, Hasan R, Ranganathan UDK, McNerney R, Ezewudo M, Cirillo DM, Schito M, Koser CU, Rodwell TC. 2017. A standardised method for interpreting the association between mutations and phenotypic drug resistance in Mycobacterium tuberculosis. Eur Respir J 50:1701354. doi: 10.1183/13993003.01354-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zvada SP, Denti P, Sirgel FA, Chigutsa E, Hatherill M, Charalambous S, Mungofa S, Wiesner L, Simonsson US, Jindani A, Harrison T, McIlleron HM. 2014. Moxifloxacin population pharmacokinetics and model-based comparison of efficacy between moxifloxacin and ofloxacin in African patients. Antimicrob Agents Chemother 58:503–510. doi: 10.1128/AAC.01478-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chigutsa E, Meredith S, Wiesner L, Padayatchi N, Harding J, Moodley P, Mac Kenzie WR, Weiner M, McIlleron H, Kirkpatrick CM. 2012. Population pharmacokinetics and pharmacodynamics of ofloxacin in South African patients with multidrug-resistant tuberculosis. Antimicrob Agents Chemother 56:3857–3863. doi: 10.1128/AAC.00048-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deshpande D, Pasipanodya JG, Mpagama SG, Bendet P, Srivastava S, Koeuth T, Lee PS, Bhavnani SM, Ambrose PG, Thwaites G, Heysell SK, Gumbo T. 2018. Levofloxacin pharmacokinetics/pharmacodynamics, dosing, susceptibility breakpoints, and artificial intelligence in the treatment of multidrug-resistant tuberculosis. Clin Infect Dis 67:S293–S302. doi: 10.1093/cid/ciy611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deshpande D, Pasipanodya JG, Srivastava S, Bendet P, Koeuth T, Bhavnani SM, Ambrose PG, Smythe W, McIlleron H, Thwaites G, Gumusboga M, Van Deun A, Gumbo T. 2018. Gatifloxacin pharmacokinetics/pharmacodynamics-based optimal dosing for pulmonary and meningeal multidrug-resistant tuberculosis. Clin Infect Dis 67:S274–S283. doi: 10.1093/cid/ciy618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forrest A, Nix DE, Ballow CH, Goss TF, Birmingham MC, Schentag JJ. 1993. Pharmacodynamics of intravenous ciprofloxacin in seriously ill patients. Antimicrob Agents Chemother 37:1073–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.WHO. 2014. Companion handbook to the WHO guidelines for the programmatic management of drug-resistant tuberculosis. World Health Organization, Geneva, Switzerland. [PubMed] [Google Scholar]