The purpose of this study was to evaluate the clinical impacts of ampicillin-susceptible but penicillin-resistant (ASPR) phenotypes of Enterococcus faecalis on clinical outcomes in patients with bloodstream infection (BSI). A total of 295 patients with an E. faecalis BSI from six sentinel hospitals during a 2-year period (from May 2016 to April 2018) were enrolled in this study.
KEYWORDS: CC28, Enterococcus faecalis, ampicillin, clinical outcome, penicillin
ABSTRACT
The purpose of this study was to evaluate the clinical impacts of ampicillin-susceptible but penicillin-resistant (ASPR) phenotypes of Enterococcus faecalis on clinical outcomes in patients with bloodstream infection (BSI). A total of 295 patients with an E. faecalis BSI from six sentinel hospitals during a 2-year period (from May 2016 to April 2018) were enrolled in this study. Putative risk factors, including host-, treatment-, and pathogen-related variables, were assessed to determine the associations with the 30-day mortality rate of patients with an E. faecalis BSI. The proportion of ASPR E. faecalis isolates was 22.7% (67/295). ASPR isolates (adjusted odds ratio, 2.27; 95% confidence interval, 1.01 to 5.02) exhibited a significant association with an increased 30-day mortality rate, and a significant difference in survival was identified in a group of patients treated with ampicillin- and/or piperacillin-based regimens who were stratified according to the penicillin susceptibility of the causative pathogen (P = 0.011 by a log rank test). ASPR E. faecalis BSIs resulted in a >2-fold-higher 30-day mortality rate (26.9%; 18/67) than for the BSIs caused by penicillin-susceptible strains (12.3%; 28/228). The differences in mortality rates of patients stratified by penicillin susceptibility were likely due to the treatment failures of ampicillin and/or piperacillin in patients with an ASPR E. faecalis BSI.
INTRODUCTION
Enterococci are common Gram-positive pathogens that cause opportunistic infections, including bloodstream infections (BSIs), particularly in immunocompromised patients. Enterococcus faecalis was previously the dominant human pathogen among enterococci; however, Enterococcus faecium has recently become a more frequent human pathogen than E. faecalis, possibly due to the differing abilities of these species to acquire antimicrobial resistance determinants (1). The mortality rate of patients with an enterococcal BSI has been reported to range from 20% to 50%, and comorbidities of the patients and the antimicrobial resistance of the causative pathogens are considered major risk factors for early mortality (2–4).
Antimicrobial regimens that inhibit bacterial cell wall synthesis required for the treatment of enterococcal infections are determined according to in vitro susceptibilities of infection-causing pathogens to ampicillin and vancomycin (5). Ampicillin resistance in enterococci is associated with two different mechanisms: (i) penicillinase production, which is rarely observed in E. faecalis (6), and (ii) increased production of penicillin-binding proteins (PBPs) with a low affinity for beta-lactams, which is predominantly observed in E. faecium (7). Vancomycin resistance is mainly associated with the acquisition of the vanA or vanB gene cluster (8). For the antimicrobial treatment of patients with an E. faecalis BSI, the administration of ampicillin either alone or in combination with aminoglycosides is recommended because the species rarely acquires resistance to ampicillin and/or vancomycin (1).
Recently, ampicillin-susceptible but penicillin-resistant (ASPR) E. faecalis strains have been reported in many countries around the world, including Greece, Denmark, and Brazil (9–11). The resistance phenotype is associated with mutations in the pbp4 gene and its promoter region, resulting in the overproduction of altered PBP4 with a low affinity for beta-lactam antimicrobials (12, 13). However, the clinical impacts of ASPR E. faecalis infections in humans have never been reported, and thus, a consensus regarding the proper antimicrobial treatment options for these infections is unavailable. Therefore, we conducted a comprehensive analysis by performing a prospective, multicenter, observational study to investigate the risk factors affecting the early mortality rate in patients with an E. faecalis BSI, focusing on the penicillin resistance of the pathogen.
RESULTS
Description of the patients with E. faecalis BSI.
Among the 296 patients with an E. faecalis BSI, one patient was excluded due to death on the day of the initial blood culture. Ultimately, 295 patients were enrolled in this study (Table 1). The median age of the patients was 70.0 years (interquartile range [IQR], 58.0 to 79.0 years), and 61.7% (182/295) were male. Malignancy (28.8%; n = 85) was the most common underlying disease, followed by diabetes mellitus (23.1%; n = 68), end-stage renal diseases (19.0%; n = 56), cardiovascular diseases (18.0%; n = 53), cerebrovascular diseases (16.3%; n = 48), and liver cirrhosis (8.8%; n = 26). The median Charlson comorbidity index value was 2.0 (IQR, 1.0 to 3.0). More than half (59.3%, n = 175) of the patients acquired a hospital-originated (HO) infection, and 33.9% (n = 100) of the patients were hospitalized in intensive care units (ICUs). The median sequential organ failure assessment (SOFA) score was 4.0 (IQR, 1.0 to 6.0). Secondary BSI was identified by culture in 49 cases (16.6%), including 33 (11.2%) that originated from the urinary tract. Central line-associated BSI was identified in 5 cases (1.7%), and the primary site of infection was not identified 241 cases (81.7%). Only six patients (2.0%) were associated with infective endocarditis. Appropriate antimicrobial regimens were administered to 43.4% (n = 128) of patients as empirical treatment and to 62.4% (n = 184) of patients as definitive treatment. The 30-day all-cause mortality rate of patients with an E. faecalis BSI was 15.6% (46/295); however, all six patients associated with infective endocarditis survived during the 30-day follow-up.
TABLE 1.
Patient-, treatment-, and pathogen-related variables associated with 30-day mortality in patients with E. faecalis BSIsc
| Variable | Value for group |
Univariable analysisa |
Multivariable analysisa |
||||
|---|---|---|---|---|---|---|---|
| Total (n = 295; 100%) | Nonsurvivors (n = 46; 15.6%) | Survivors (n = 249; 84.4%) | OR (95% CI) | P value | aOR (95% CI) | P value | |
| Patients | |||||||
| Mean age (yr) (range) | 70.0 (58.0–79.0) | 76.0 (62.0–81.0) | 69.0 (57.0–79.0) | 1.02 (0.99–1.04) | 0.083 | ||
| No. (%) of males | 182 (61.7) | 27 (58.7) | 155 (62.2) | 0.85 (0.45–1.63) | 0.622 | ||
| No. (%) with comorbidity | |||||||
| Malignancy | 85 (28.8) | 18 (39.1) | 67 (26.9) | 1.69 (0.87–3.23) | 0.117 | ||
| Diabetes mellitus | 68 (23.1) | 18 (39.1) | 50 (20.1) | 2.48 (1.26–4.82) | 0.008 | ||
| Cardiovascular disease | 53 (18.0) | 16 (34.8) | 37 (14.9) | 2.97 (1.45–5.94) | 0.002 | 2.98 (1.29–6.83) | 0.010 |
| Cerebrovascular disease | 48 (16.3) | 7 (15.2) | 41 (16.5) | 0.88 (0.34–2.01) | 0.782 | ||
| Liver cirrhosis | 26 (8.8) | 7 (15.2) | 19 (7.6) | 2.12 (0.78–5.18) | 0.115 | ||
| End-stage renal disease | 56 (19.0) | 13 (28.3) | 43 (17.3) | 1.89 (0.89–3.82) | 0.086 | ||
| Mean Charlson comorbidity index (IQR) | 2.0 (1.0–3.0) | 3.0 (2.0–4.0) | 2.0 (1.0–3.0) | 1.26 (1.10–1.44) | 0.001 | 1.21 (1.02–1.43) | 0.025 |
| No. (%) with host factor | |||||||
| ICU admission | 100 (33.9) | 25 (54.3) | 75 (30.1) | 2.77 (1.46–5.31) | 0.002 | ||
| Hospital-originated infection | 175 (59.3) | 31 (67.4) | 144 (57.8) | 1.47 (0.77–2.93) | 0.258 | ||
| Polymicrobial infection | 67 (22.7) | 15 (32.6) | 52 (20.9) | 1.78 (0.87–3.50 | 0.102 | ||
| Primary site of infection of: | |||||||
| Urinary tract | 33 (11.2) | 5 (10.9) | 28 (11.2) | 0.94 (0.30–2.38) | 0.898 | ||
| Central line | 5 (1.7) | 0 (0) | 5 (2.0) | ||||
| Others | 16 (5.4) | 3 (6.5) | 13 (5.2) | ||||
| Unknown | 241 (81.7) | 38 (82.6) | 203 (81.5) | ||||
| Concurrent infective endocarditis | 6 (2.0) | 0 (0) | 6 (2.4) | ||||
| Mean SOFA score (IQR) | 4.0 (1.0–6.0) | 7.0 (5.0–12.0) | 3.0 (1.0–5.0) | 1.34 (1.23–1.47) | <0.001 | 1.33 (1.21–1.47) | <0.001 |
| No. (%) of patients with adequate antimicrobial treatment | |||||||
| Empirical | 128 (43.4) | 28 (60.9) | 100 (40.2) | 2.32 (1.23–4.48) | 0.011 | 1.84 (0.87–3.94) | 0.111 |
| Definitive | 184 (62.4) | 29 (63.0) | 155 (62.2) | 1.03 (0.54–2.02) | 0.919 | ||
| Pathogens | |||||||
| No. (%) of isolates of strain type | |||||||
| CC16 | 97 (32.9) | 15 (32.6) | 82 (32.0) | 0.99 (0.49–1.90) | 0.967 | ||
| CC28b | 67 (22.7) | 17 (37.0) | 50 (20.1) | 2.38 (1.19–4.66) | 0.012 | ||
| CC507 | 23 (7.8) | 2 (4.3) | 21 (8.4) | 0.48 (0.08–1.72) | 0.334 | ||
| Other STs | 108 (36.6) | 12 (26.1) | 96 (38.6) | 0.56 (0.27–1.11) | 0.107 | ||
| No (%) of isolates nonsusceptible to: | |||||||
| Ampicillin | 0 (0) | 0 (0) | 0 (0) | ||||
| Penicillin | 67 (22.7) | 18 (39.1) | 49 (19.7) | 2.68 (1.35–5.23) | 0.004 | 2.27 (1.01–5.02) | 0.045 |
| Imipenemb | 66 (22.4) | 17 (37.0) | 49 (19.7) | 2.44 (1.22–4.78) | 0.010 | ||
| Ciprofloxacin | 174 (59.0) | 31 (67.4) | 143 (57.4) | 1.50 (0.78–2.98) | 0.237 | ||
| High-level gentamicin | 116 (39.3) | 21 (45.7) | 95 (38.2) | 1.35 (0.71–2.56) | 0.349 | ||
| High-level streptomycin | 44 (14.9) | 9 (19.6) | 35 (14.1) | 1.50 (0.63–3.27) | 0.333 | ||
| Vancomycin | 1 (0.3) | 0 (0) | 1 (0.4) | ||||
| Teicoplanin | 1 (0.3) | 0 (0) | 1 (0.4) | ||||
| Tetracycline | 217 (73.6) | 38 (82.6) | 179 (71.9) | 1.85 (0.86–4.44) | 0.139 | ||
| Linezolid | 19 (6.4) | 3 (6.5) | 16 (6.4) | ||||
| Tigecycline | 6 (2.0) | 0 (0) | 6 (2.4) | ||||
| No. (%) of isolates with MDR phenotype | 92 (31.2) | 21 (45.7) | 71 (28.5) | 2.12 (1.11–4.03) | 0.022 | ||
Six patients censored before 30 days were excluded from the univariate and multivariate logistic regression analyses.
CC28 (VIF = 15.1) and nonsusceptibility to imipenem (VIF = 6,496,558) were excluded from multivariate analysis due to multicollinearity with nonsusceptibility to penicillin.
Boldface type indicates variables significantly associated with 30-day mortality in the multivariable analysis. Abbreviations: CC, clonal complex; CI, confidence interval; ICU, intensive care unit; IQR, interquartile range; MDR, multidrug resistant; OR, odds ratio; aOR, adjusted odds ratio; SOFA, sequential organ failure assessment; ST, sequence type.
Antimicrobial susceptibilities of E. faecalis blood isolates.
All 295 E. faecalis blood isolates were susceptible to ampicillin, while 22.4% (n = 66) of them were not susceptible to either penicillin or imipenem, and one isolate was resistant to penicillin but susceptible to imipenem; i.e., 22.7% (n = 67) of E. faecalis blood isolates exhibited ASPR phenotypes. Beta-lactamase was not produced in any of the ASPR isolates, according to the Cefinase test. High-level aminoglycoside resistance was observed in 39.3% (n = 116) of the isolates for gentamicin and 14.9% (n = 44) for streptomycin. Only one isolate exhibiting resistance to both vancomycin and teicoplanin harbored the vanA gene. Antimicrobial susceptibilities of the isolates stratified according to strain type are summarized in Table S1 in the supplemental material.
Risk factors for 30-day mortality.
Among the 295 patients enrolled in this study, 6 patients were censored before 30 days and were excluded from the logistic regression analysis. In the univariable analyses stratified by host factor variables (Table 1), an increased Charlson comorbidity index (odds ratio [OR], 1.26; 95% confidence interval [CI], 1.10 to 1.44), diabetes mellitus (OR, 2.48; 95% CI, 1.26 to 4.82), cardiovascular diseases (OR, 2.97; 95% CI, 1.45 to 5.94), admission to ICUs (OR, 2.77; 95% CI, 1.46 to 5.31), and an increased SOFA score (OR, 1.34; 95% CI, 1.23 to 1.47) were significantly correlated with an increased 30-day mortality rate in patients with an E. faecalis BSI. Nonsusceptibilities of the causative pathogens to penicillin (OR, 2.68; 95% CI, 1.35 to 5.23) and imipenem (OR, 2.44; 95% CI, 1.22 to 4.78) were associated with an increased 30-day mortality rate. When isolates were stratified by strain type, clonal complex 28 (CC28) (OR, 2.38; 95% CI, 1.19 to 4.66) was significantly associated with an increased 30-day mortality rate compared to non-CC28 isolates. Both the variables nonsusceptibility to imipenem (variance inflation factor [VIF] = 6,496,558) and CC28 (VIF = 15.1) were excluded from the multivariate analyses due to multicollinearity with nonsusceptibility to penicillin.
The forward stepwise multivariate logistic regression analyses revealed that three host factor variables, an increased Charlson comorbidity index (adjusted OR [aOR], 1.21; 95% CI, 1.02 to 1.43), cardiovascular diseases (aOR, 2.98; 95% CI, 1.29 to 6.83), and an increased SOFA score (aOR, 1.33; 95% CI, 1.21 to 1.47), were risk factors for an increased 30-day mortality rate. In addition, a pathogenic factor variable, nonsusceptibility to penicillin (aOR, 2.27; 95% CI, 1.01 to 5.02), was also an independent risk factor.
ASPR E. faecalis BSI.
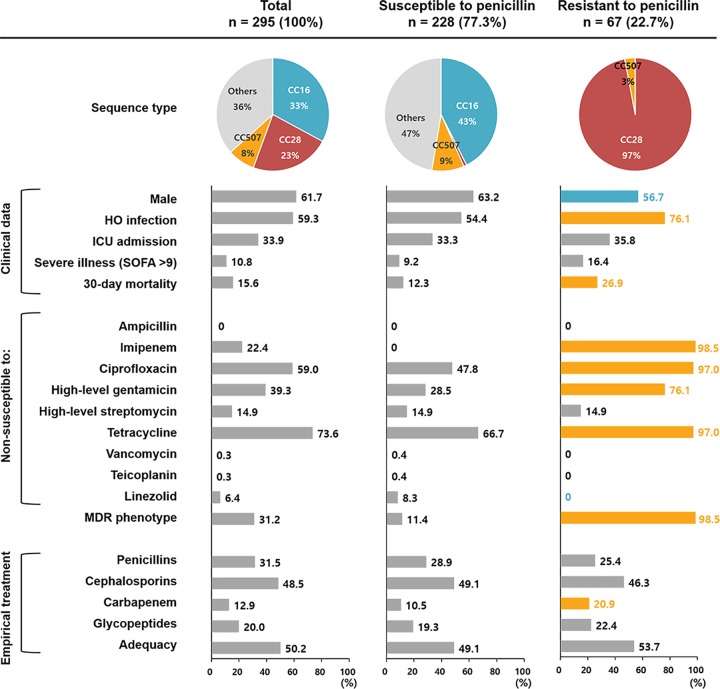
The majority (97.0%; 65/67) of ASPR E. faecalis isolates were identified as belonging to CC28, including 62 isolates of sequence type 28 (ST28) (gdh-gyd-pstS-gki-aroE-xpt-yqiL, 4-4-8-3-8-1-3) and 1 isolate each of ST207 (4-4-46-3-2-1-3), ST867 (4-4-8-3-8-7-3), and ST882 (4-4-8-99-8-1-3). The ASPR E. faecalis isolates mainly (98.5%; 66/67) exhibited multidrug-resistant (MDR) phenotypes (Fig. 1). ASPR E. faecalis BSIs resulted in a >2-fold-higher 30-day mortality rate (26.9%; 18/67) than BSIs caused by penicillin-susceptible strains (12.3%; 28/228). A log rank test showed a significant difference in survival (P = 0.002) in patients stratified according to the penicillin susceptibility of the causative E. faecalis pathogens (Fig. 2A). The impacts of the penicillin susceptibility of BSI-causing E. faecalis on patients’ clinical outcomes were evaluated in groups stratified by the antimicrobial treatment regimen administered. A statistically significant difference in survival was observed in patients stratified according to the penicillin susceptibility of BSI-causative pathogens using the log rank test in a group of patients treated with ampicillin- and/or piperacillin-based regimens (P = 0.011) (Fig. 2B) but not in another group of patients treated with glycopeptide-based regimens (P = 0.290) (Fig. 2C).
FIG 1.
Comparison of characteristics of patients with E. faecalis BSIs and their causative pathogens stratified according to penicillin susceptibility. The pie charts indicate the proportions of major sequence types, and the proportions of each variable are indicated in the bar charts. Statistical significance (P < 0.05), assessed using the chi-square test, is presented for the penicillin-resistant group with colored bars and numbers (orange, more prevalent; blue, less prevalent).
FIG 2.
Survival analysis. Kaplan-Meier curves were constructed for three groups stratified by penicillin susceptibility: all patients (A), patients treated with an ampicillin- and/or a piperacillin-based regimen as a definitive treatment (B), and patients treated with a glycopeptide-based regimen as a definitive treatment (C). Statistical significance was determined using the log rank test.
The effects of the penicillin, ampicillin, and imipenem MICs for causative E. faecalis isolates on the 30-day mortality rate of BSI patients treated with an ampicillin- and/or a piperacillin-based regimen as a definitive treatment were evaluated using Pearson’s correlation coefficients. The penicillin MIC exhibited a strong and positive linear correlation (correlation coefficients [r], 0.960) with the 30-day mortality rate: at an MIC of ≤4 μg/ml, the 30-day mortality rate was 13.0% (6/46); at an MIC of 8 μg/ml, the rate was 20.0% (4/20); and at an MIC of ≥16 μg/ml, the rate was 41.2% (7/17) (Fig. 3A). Furthermore, MICs of both imipenem (r = 0.902) and ampicillin (r = 0.988) also showed strong positive linear correlations with the 30-day mortality rate (Fig. 3B and C).
FIG 3.
Thirty-day mortality rates in patients treated with an ampicillin- and/or a piperacillin-based regimen stratified according to the MICs of penicillin, imipenem, and ampicillin. Bar charts indicate numbers of patients stratified by the MIC of each antimicrobial. Red rhombi indicate 30-day mortality rates. As MICs of each antimicrobial increase, trends in the 30-day mortality rates are plotted with dotted lines obtained from Pearson’s correlation coefficients.
Characteristics of ASPR isolates.
Penicillin MICs for the 295 E. faecalis blood isolates ranged from ≤1 to 64 μg/ml (MIC50 = 4 μg/ml; MIC90 = 32 μg/ml), and those for the 67 ASPR isolates ranged from 16 to 64 μg/ml (MIC50 = 32 μg/ml; MIC90 = 32 μg/ml) (Table 2). All ASPR isolates were identified as belonging to CC28 (n = 65), except for two isolates identified as belonging to CC507. Nucleotide deletion of a single A residue in a string of seven A residues upstream of a putative −35 region for the pbp4 gene, a variant identical to the one described previously by Rice et al., was identified in all the 65 ASPR isolates of CC28 but not in other isolates (13). Eight amino acid substitutions in PBP4, including seven in the penicillin-binding domain (PBD) (A369V, T418A, L475M, F499I, P520S, M652L, and D666P) and one in the non-penicillin-binding domain (nPBD) (T50I), were identified compared to the reference E. faecalis strain ATCC 29212. All 67 CC28 isolates, including 65 ASPR isolates and 2 penicillin-susceptible isolates, shared both substitutions T50I in the nPBD and A369V in the PBD, and the 65 ASPR isolates of CC28 carried 2 to 4 additional substitutions in the PBD along with a nucleotide deletion in the promoter region of the pbp4 gene. All 23 CC507 isolates shared the A369V and P520S substitutions in the PBD; however, two CC507 isolates exhibiting ASPR phenotypes did not show any additional substitution in PBP4, nor did they show a nucleotide deletion in the promoter region of the pbp4 gene. Other amino acid substitutions in PBP4 observed in penicillin-resistant isolates in previous studies, i.e., L218N and V231I in the nPBD and D573E, A617T, and D632E in the PBD, were not identified in this study (12, 13).
TABLE 2.
Amino acid substitutions in PBP4 and a nucleotide deletion in the promoter region of the pbp4 gene compared with MICs of penicillin, imipenem, and ampicillind
aDeletion of a single A residue in a string of seven A residues upstream of a putative −35 region for the pbp4 gene described previously by Rice et al. (13).
bThe amino acid sequences of PBP4 of E. faecalis isolates were compared with that of the reference E. faecalis strain ATCC 29212.
cMICs designating intermediate resistance and resistance are indicated by boldface type and gray-shaded areas, respectively.
“O” indicates that the specified sequence variant was identified. Abbreviations: CC, clonal complex; nPBD, non-penicillin-binding domain; NE, not evaluated; PBD, penicillin-binding domain.
DISCUSSION
Clinical isolates of E. faecalis are mostly susceptible to ampicillin in vitro. MICs of penicillin and ampicillin show good concordance, and most E. faecalis clinical isolates are regularly cross-susceptible to penicillin and imipenem (14). Therefore, antimicrobial susceptibility testing for either ampicillin or penicillin has been recommended as a critical assessment to determine an antimicrobial regimen for bacterial cell wall inhibition in patients with an enterococcal BSI (5). Recently, ASPR E. faecalis clinical strains have been identified worldwide (9–11); however, to the best of our knowledge, the adequacy of the cell wall-inhibiting beta-lactam regimens as a treatment for ASPR E. faecalis BSIs has not yet been evaluated.
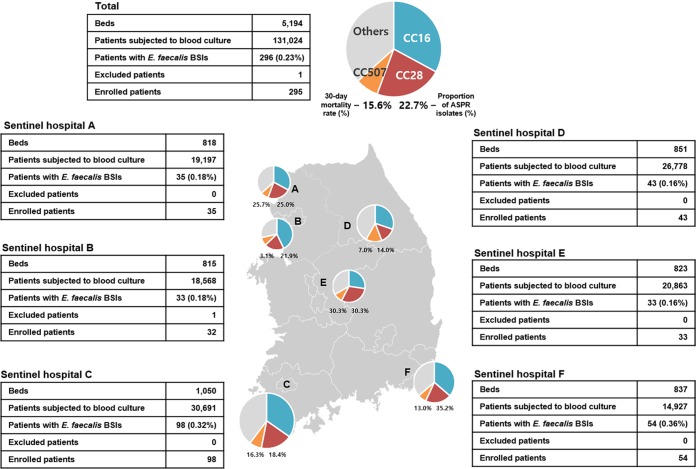
ASPR phenotypes were identified in a considerable proportion (22.7%) of E. faecalis blood isolates collected in this study, ranging from 14.0% to 35.2% by sentinel hospital (Fig. 4). The rate of resistance to penicillin in E. faecalis clinical isolates has rarely been reported so far, although a high frequency of ASPR E. faecalis isolates, at 30% of the total, was described in Brazil in 2006 (11). Multivariable analyses using putative risk factors, including host-, treatment-, and pathogen-related variables, showed that the penicillin resistance of the causative pathogen was an independent risk factor for 30-day mortality. In addition, a significant difference in survival was observed in a group of patients treated with ampicillin and/or piperacillin who were stratified according to the penicillin susceptibility of the causative pathogen. Based on these observations, this study provides the first clinical evidence that the administration of ampicillin or piperacillin to patients with an ASPR E. faecalis BSI might lead to a treatment failure, resulting in a high early mortality rate for patients even though they exhibited in vitro susceptibility to ampicillin.
FIG 4.
Geographic distribution of sentinel hospitals (hospitals A to F) in the Korean peninsula. The numbers of beds and E. faecalis bloodstream infection cases in each hospital are indicated in the tables. The pie charts indicate the proportions of major sequence types of causative E. faecalis strains, and the percentages below the pie charts indicate the 30-day mortality rates for patients with E. faecalis BSIs and the proportions of ASPR strains. The relative sizes of the pie charts indicate the number of E. faecalis cases in each hospital.
The Clinical and Laboratory Standards Institute (CLSI) recommends clinical breakpoints of resistance determination as MICs of ≥16 μg/ml for both ampicillin and penicillin for enterococci (15). However, all isolates in the present study had 1- to 3-fold-lower MICs for ampicillin than for penicillin (MIC50 of 2 μg/ml and MIC90 of 8 μg/ml for ampicillin; MIC50 of 4 μg/ml and MIC90 of 32 μg/ml for penicillin), consistent with data from previous reports (12, 14). The emergence of ASPR E. faecalis might be due to the breakpoint. From this perspective, penicillin is a better surrogate marker than ampicillin in tests of the antimicrobial susceptibilities of E. faecalis clinical isolates to antienterococcal beta-lactam agents to avoid reporting substantial very major errors in the results. Furthermore, additional MIC determinations for penicillin should be performed for E. faecalis isolates exhibiting MICs of 4 or 8 μg/ml for ampicillin to exclude the ASPR phenotypes.
Among the 8 amino acid substitutions in PBP4 identified in this study, both T50I and A369V were reported to be irrelevant to the beta-lactam resistance of bacterial hosts, consistent with our findings (16). P520S was suggested to be a key substitution required for the development of high-level resistance to beta-lactams in E. faecalis isolates (16); however, this substitution was identified in both penicillin-susceptible and -resistant isolates in this study, indicating that P520S might be irrelevant to the beta-lactam resistance of bacterial hosts. The remaining 5 amino acid substitutions in PBP4, T418A, L475M, F499I, M652L, and D666P, were novel, and they were exclusively identified in 65 ASPR E. faecalis isolates of CC28. Additional studies are needed to investigate the roles of these substitutions in elevating the MICs of penicillin for bacterial hosts. In addition, the nucleotide deletion of a single A residue in the promoter region of the pbp4 gene, identical to that described previously by Rice et al., was also identified in these 65 ASPR isolates, which might result in overexpression of the altered PBP4 protein (13). However, two ASPR E. faecalis isolates of CC507 did not display any relevant amino acid substitutions in PBP4, nor did they show any nucleotide deletions in the promoter region, suggesting that the resistance phenotypes might be related to alterations in another PBP.
A limitation of the present study is that the sentinel hospitals are geographically restricted to a single country, South Korea. Therefore, ethnic or racial diversities were not considered in this study, and the high proportion of ASPR E. faecalis isolates observed in this study might be limited to South Korea. However, previous studies have reported the emergence of ASPR strains in other countries (9–11). In addition, ASPR E. faecalis isolates were identified in all sentinel hospitals in this study, although they are located in different provinces. The lack of a functional evaluation of amino acid substitutions in PBP4 represents another potential limitation of the study. Further studies should be performed to confirm the possible changes in the beta-lactam affinities of PBP4 induced by the amino acid substitutions.
In conclusion, in the present prospective, multicenter, observational study, ASPR phenotypes in E. faecalis resulted in an inadequate choice of antimicrobial treatment regimens for patients with BSIs, ultimately resulting in fatal clinical outcomes. ASPR E. faecalis should be considered clinically resistant to all antienterococcal beta-lactams, including ampicillin and piperacillin, when choosing antimicrobial regimens for the definitive treatment of BSIs.
MATERIALS AND METHODS
Study design.
This prospective observational study was performed with all patients with an E. faecalis BSI during the 2-year period between May 2016 and April 2018 in six general hospitals participating in the Global Antimicrobial Surveillance System (GLASS) in South Korea (Kor-GLASS) (17). The six hospitals are located in different districts of the Korean peninsula, and the number of beds ranges from 815 to 1,050 per hospital (Fig. 4). A total of 131,024 patients were subjected to blood culture for suspected BSIs, and 296 patients (0.2%) were diagnosed with an E. faecalis BSI. Clinical information about the demographic conditions, underlying diseases, and antimicrobial treatments were retrieved from the electronic medical records of each hospital. The Charlson comorbidity index and the sequential organ failure assessment (SOFA) score were assessed at the time of the initial blood culture (18, 19). Clinical outcomes were assessed by calculating the 30-day mortality rate. Only the first isolate from each patient was collected for microbiological evaluation, and sequential isolates were discarded. All the isolates collected at each hospital were transferred to the analysis center. The requirement for informed consent from the participants was waived by all local ethical committees of the six sentinel hospitals.
Definition.
A hospital-originated (HO) infection was defined when an initial blood culture was performed after ≥2 calendar days of hospitalization. The primary infection site was determined when a site-specific infection with an E. faecalis isolate was identified at another body site by bacterial culture (20). The multidrug-resistant (MDR) phenotype of the causative pathogen was determined as an in vitro lack of susceptibility to ≥3 drug classes, as described previously by Magiorakos et al. (21). Empirical antimicrobial treatment was defined as an initial blind antimicrobial treatment without in vitro antimicrobial susceptibility information for the responsible pathogen, and definitive treatment was defined as a revised antimicrobial treatment based on the in vitro susceptibility of the responsible pathogen within 72 h from the initial blood culture. An ampicillin- or piperacillin-based regimen was defined as an administration of ampicillin or piperacillin with or without any combination with drugs of other classes except glycopeptides, and a glycopeptide-based regimen was defined as the administration of vancomycin or teicoplanin with or without any combination with drugs of other classes. An appropriate antimicrobial treatment was defined as the use of in vitro-susceptible cell wall-inhibiting antimicrobials, irrespective of their combination with drugs of other classes.
Microbiological analysis.
Bacterial species were identified with a MALDI Biotyper (Bruker Daltonik GmbH, Bremen, Germany) and using 16S rRNA gene sequencing. MICs of ampicillin, penicillin, imipenem, vancomycin, teicoplanin, gentamicin (high level), and streptomycin (high level) were determined with the broth microdilution test using Mueller-Hinton broth (Difco Laboratories, Detroit, MI) according to CLSI guidelines (15). In addition, antimicrobial susceptibilities to ciprofloxacin and tetracycline were tested using the disc diffusion method on cation-adjusted Mueller-Hinton agar (Difco Laboratories). For penicillin-nonsusceptible isolates, beta-lactamase production was assessed using the nitrocefin disc test. Nucleotide sequences of the pbp4 gene and its promoter region for all the E. faecalis isolates were determined by PCR and direct sequencing using primer sets, as previously described (12, 16), and were compared to those of the E. faecalis reference strain ATCC 29212. The vanA, vanB, and vanM genes were detected in vancomycin-nonsusceptible isolates using PCR (22). For strain typing, multilocus sequence typing (MLST) was performed by comparing partial sequences of seven housekeeping genes, gdh, gyd, pstS, gki, aroE, xpt, and yqiL, to the E. faecalis MLST database (https://pubmlst.org/efaecalis/) to determine the allelic types and the sequence types (STs) (23).
Statistical analysis.
Analyses were performed using R software version 3.4.3 (R Development Core Team 2017 [http://www.R-project.org/]). Differences between groups were analyzed using the Mann-Whitney U test and Fisher’s exact test for continuous variables and categorical variables, respectively. A univariable logistic regression analysis was performed to calculate odds ratio (OR) of each variable to 30-day mortality. Adjusted ORs (aORs) were calculated for variables with P values of <0.05 using a forward stepwise multivariable logistic regression analysis. Kaplan-Meier curves were constructed, and log rank tests were performed to compare the mortality dynamics between groups. The results of the statistical analyses were considered significant when the P values were <0.05.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kor-GLASS participants for their contribution to the program.
This study was supported by the research program funded by the Korea Centers for Disease Control and Prevention (number 2017E4400101). The funder of the study had no role in study design, data collection, data interpretation, or writing of the report.
We do not have conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00291-19.
REFERENCES
- 1.Weiner LM, Webb AK, Limbago B, Dudeck MA, Patel J, Kallen AJ, Edwards JR, Sievert DM. 2016. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect Control Hosp Epidemiol 37:1288–1301. doi: 10.1017/ice.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Billington EO, Phang SH, Gregson DB, Pitout JD, Ross T, Church DL, Laupland KB, Parkins MD. 2014. Incidence, risk factors, and outcomes for Enterococcus spp. blood stream infections: a population-based study. Int J Infect Dis 26:76–82. doi: 10.1016/j.ijid.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 3.DiazGranados CA, Zimmer SM, Klein M, Jernigan JA. 2005. Comparison of mortality associated with vancomycin-resistant and vancomycin-susceptible enterococcal bloodstream infections: a meta-analysis. Clin Infect Dis 41:327–333. doi: 10.1086/430909. [DOI] [PubMed] [Google Scholar]
- 4.Noskin GA, Peterson LR, Warren JR. 1995. Enterococcus faecium and Enterococcus faecalis bacteremia: acquisition and outcome. Clin Infect Dis 20:296–301. doi: 10.1093/clinids/20.2.296. [DOI] [PubMed] [Google Scholar]
- 5.Murray BE. 2018. Treatment of enterococcal infections In Sexton DJ. (ed), UpToDate. UpToDate, Waltham, MA: https://www.uptodate.com/contents/treatment-of-enterococcal-infections. [Google Scholar]
- 6.Murray BE. 1992. Beta-lactamase-producing enterococci. Antimicrob Agents Chemother 36:2355–2359. doi: 10.1128/AAC.36.11.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fontana R, Aldegheri M, Ligozzi M, Lopez H, Sucari A, Satta G. 1994. Overproduction of a low-affinity penicillin-binding protein and high-level ampicillin resistance in Enterococcus faecium. Antimicrob Agents Chemother 38:1980–1983. doi: 10.1128/AAC.38.9.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cetinkaya Y, Falk P, Mayhall CG. 2000. Vancomycin-resistant enterococci. Clin Microbiol Rev 13:686–707. doi: 10.1128/CMR.13.4.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conceição N, de Oliveira CDCHB, da Silva PR, Ávila BGM, de Oliveira AG. 2011. Trends in antimicrobial resistance among clinical isolates of enterococci in a Brazilian tertiary hospital: a 4-year study. Rev Soc Bras Med Trop 44:177–181. doi: 10.1590/S0037-86822011005000009. [DOI] [PubMed] [Google Scholar]
- 10.Guardabassi L, Larsen J, Skov R, Schonheyder HC. 2010. Gentamicin-resistant Enterococcus faecalis sequence type 6 with reduced penicillin susceptibility: diagnostic and therapeutic implications. J Clin Microbiol 48:3820–3821. doi: 10.1128/JCM.01252-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metzidie E, Manolis EN, Pournaras S, Sofianou D, Tsakris A. 2006. Spread of an unusual penicillin- and imipenem-resistant but ampicillin-susceptible phenotype among Enterococcus faecalis clinical isolates. J Antimicrob Chemother 57:158–160. doi: 10.1093/jac/dki427. [DOI] [PubMed] [Google Scholar]
- 12.Conceição N, da Silva LE, Darini AL, Pitondo-Silva A, de Oliveira AG. 2014. Penicillin-resistant, ampicillin-susceptible Enterococcus faecalis of hospital origin: pbp4 gene polymorphism and genetic diversity. Infect Genet Evol 28:289–295. doi: 10.1016/j.meegid.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 13.Rice LB, Desbonnet C, Tait-Kamradt A, Garcia-Solache M, Lonks J, Moon TM, D’Andréa ÉD, Page R, Peti W. 2018. Structural and regulatory changes in PBP4 trigger decreased beta-lactam susceptibility in Enterococcus faecalis. mBio 9:e00361-18. doi: 10.1128/mBio.00361-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinstein MP. 2001. Comparative evaluation of penicillin, ampicillin, and imipenem MICs and susceptibility breakpoints for vancomycin-susceptible and vancomycin-resistant Enterococcus faecalis and Enterococcus faecium. J Clin Microbiol 39:2729–2731. doi: 10.1128/JCM.39.7.2729-2731.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clinical and Laboratory Standards Institute. 2016. Performance standards for antimicrobial susceptibility testing: twenty-sixth informational supplement. CLSI document M100-S26 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 16.Ono S, Muratani T, Matsumoto T. 2005. Mechanisms of resistance to imipenem and ampicillin in Enterococcus faecalis. Antimicrob Agents Chemother 49:2954–2958. doi: 10.1128/AAC.49.7.2954-2958.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee H, Yoon EJ, Kim D, Jeong SH, Shin JH, Shin JH, Shin KS, Kim YA, Uh Y, Park C, Lee KJ. 2018. Establishment of the South Korean national antimicrobial resistance surveillance system, Kor-GLASS, in 2016. Euro Surveill 23(42):1700734. doi: 10.2807/1560-7917.ES.2018.23.42.1700734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charlson ME, Pompei P, Ales KL, MacKenzie CR. 1987. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 40:373–383. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 19.Vincent JL, Moreno R, Takala J, Willatts S, De Mendonça A, Bruining H, Reinhart CK, Suter PM, Thijs LG. 1996. The SOFA (sepsis-related organ failure assessment) score to describe organ dysfunction/failure. On behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. Intensive Care Med 22:707–710. doi: 10.1007/BF01709751. [DOI] [PubMed] [Google Scholar]
- 20.Horan TC, Andrus M, Dudeck MA. 2008. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control 36:309–332. doi: 10.1016/j.ajic.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 21.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 22.Xu X, Lin D, Yan G, Ye X, Wu S, Guo Y, Zhu D, Hu F, Zhang Y, Wang F, Jacoby GA, Wang M. 2010. vanM, a new glycopeptide resistance gene cluster found in Enterococcus faecium. Antimicrob Agents Chemother 54:4643–4647. doi: 10.1128/AAC.01710-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruiz-Garbajosa P, Bonten MJ, Robinson DA, Top J, Nallapareddy SR, Torres C, Coque TM, Cantón R, Baquero F, Murray BE, del Campo R, Willems RJ. 2006. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J Clin Microbiol 44:2220–2228. doi: 10.1128/JCM.02596-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.