We report on a carbapenemase-producing hypervirulent Klebsiella pneumoniae (CP-hvKP) isolate collected from a U.S. patient at an outpatient clinic.
KEYWORDS: Klebsiella, hypervirulence, plasmid-mediated resistance
ABSTRACT
We report on a carbapenemase-producing hypervirulent Klebsiella pneumoniae (CP-hvKP) isolate collected from a U.S. patient at an outpatient clinic. The isolate was identified as K. pneumoniae serotype K1 sequence type 23 and included both a hypervirulence (with rmpA, rmpA2 iroBCDN, peg-344, and iucABCD-iutA genes) and a carbapenemase-encoding (blaKPC-2) plasmid. The emergence of CP-hvKP underscores the importance of clinical awareness of this pathotype and the need for continued monitoring of CP-hvKP in the United States.
INTRODUCTION
Carbapenemase-producing Klebsiella pneumoniae (CP-KP) isolates exhibit broad resistance to most β-lactams, including the carbapenems ertapenem, imipenem, meropenem, and doripenem, which often serve as last-line therapeutic options for infections caused by highly resistant Enterobacteriaceae (1). Resistance to β-lactams in CP-KP is frequently mediated by K. pneumoniae carbapenemase (KPC), a gene that commonly resides on a mobile genetic element, allowing the gene to spread horizontally between different bacteria (2, 3). Carbapenem-resistant Enterobacteriaceae (CRE) strains, such as CP-KP, can cause a number of serious infections, including intra-abdominal infections, pneumonia, urinary tract infections, and device-associated infections (1). The U.S. Centers for Disease Control and Prevention (CDC) estimates that >9,000 health care-associated infections are caused by carbapenem-resistant Escherichia coli and Klebsiella species each year in the United States (4). Importantly, infections caused by CRE are associated with mortality rates as high as 47% (5, 6).
Over the past decade, hypervirulent variants of K. pneumoniae (hvKP) have emerged. These isolates, which often display a mucoid or hypermucoviscous phenotype, are concerning because they are associated with severe infections, such as pyogenic liver abscesses and osteomyelitis, in immunocompetent healthy individuals (7). Although CP-KP and hvKP have both been identified across the globe, they typically occupy nonoverlapping clonal groups and strain types (8). However, recent reports of carbapenemase-producing hvKP (CP-hvKP) in China (9, 10) and Argentina (11) have signaled the concerning convergence of CP-KP and hvKP, with the potential for increased pathogenicity and mortality.
Aware of the public health threat presented by CP-hvKP, we searched for five virulence markers (peg-344, iroB, iucA, rmpA, and rmpA2) associated with hvKP (12), using whole-genome sequence data from 600 K. pneumoniae isolates collected through CDC surveillance and reference activities from March 2015 to May 2018.
A K. pneumoniae isolate was cultured in 2016 from a urine specimen collected from a patient >65 years of age at an outpatient clinic. The patient had recently traveled to South America but was not hospitalized while traveling. No signs or symptoms of disease were reported, indicating probable asymptomatic bacteriuria. The isolate was submitted to the CDC as part of the Emerging Infections Program Multi-site Gram-negative Surveillance Initiative (MuGSI), (13), which has conducted active population- and laboratory-based surveillance for carbapenem-resistant pathogens since 2012.
Isolate DHQP1701672, identified as a K. pneumoniae isolate by using matrix-assisted laser desorption ionization–time of flight mass spectrometry, underwent antimicrobial susceptibility testing using reference broth microdilution according to Clinical and Laboratory Standards Institute (CLSI) guidelines (14). DHQP1701672 displayed resistance to ampicillin, ceftriaxone, cefotaxime, ertapenem, and piperacillin-tazobactam but remained susceptible or intermediate to all other drugs tested (Table 1). Carbapenemase activity was confirmed using the modified carbapenem inactivation method (15), and a blaKPC gene was detected using multiplex real-time PCR (16). Colonies grown from the isolate were not hypermucoviscous, as the string test was negative (viscous strings were <5 mm) (17). Whole-genome sequencing of DHQP1701672 was performed using both the Illumina MiSeq (San Diego, CA) and Oxford Nanopore Technologies MinION (Oxford, UK) platforms.
TABLE 1.
Antimicrobial susceptibility test results for K. pneumoniae isolate DHQP1701672
| Antimicrobial agent | Minimum inhibitory concentration (μg/ml) | Interpretationa |
|---|---|---|
| Amikacin | ≤1 | S |
| Ampicillin | >32 | R |
| Aztreonam | 8 | I |
| Cefotaxime | 4 | R |
| Ceftriaxone | >32 | R |
| Ceftazidime | ≤1 | S |
| Ceftazidime-avibactam | ≤0.5 | S |
| Cefepime | 2 | S |
| Ciprofloxacin | ≤0.25 | S |
| Colistin | ≤0.25 | NWT |
| Doripenem | 1 | S |
| Ertapenem | 2 | R |
| Gentamicin | ≤0.25 | S |
| Imipenem | 2 | I |
| Levofloxacin | ≤0.125 | S |
| Meropenem | 2 | I |
| Piperacillin-tazobactam | 128/4 | R |
| Tetracycline | ≤2 | S |
| Tigecycline | ≤0.5 | S |
| Trimethoprim-sulfamethoxazole | ≤0.5 | S |
Interpretative criteria were applied according to CLSI document M100 (14), except for tigecycline, for which criteria established by the U.S. Food and Drug Administration were used. NWT, non-wild type; R, resistant; S, susceptible.
DHQP1701672 was identified as K. pneumoniae serotype K1 sequence type 23 (ST23, per multilocus sequence typing definitions from http://bigsdb.pasteur.fr/klebsiella/klebsiella.html), which belongs to the globally disseminated hypervirulent clonal complex 23 (18) and is associated with life-threatening liver abscesses (19–21). The chromosome of DHQP1701672 was 5,393,085 bp and included multiple virulence factors associated with ST23 isolates (22), such as allS (allatonin metabolism), kfu (iron uptake), magA (mucoviscosity), mrkD (type 3 fimbrial adhesion), wcaG (capsule biosynthesis), and ybtS (yersiniabactin siderophore). Four antimicrobial resistance genes were detected on the chromosome: the β-lactamase blaSHV-36, the fosfomycin resistance gene fosA, and the multidrug efflux pump genes oqxA and oqxB.
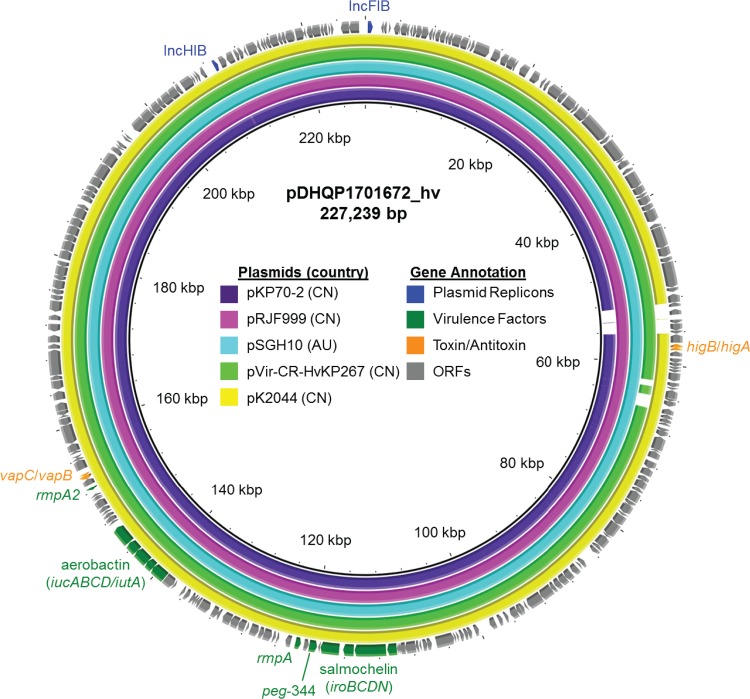
The genome of DHQP1701672 included two plasmids. The larger plasmid (pDHQP1701672_hv) was 227,239 bp and had IncFIB and IncHI1B replicons (Fig. 1). Among the 259 open reading frames were multiple virulence factors, including the mucoid phenotype transcription factors rmpA and rmpA2 (truncated at residue 99), the salmochelin siderophore iroBCDN, the metabolite transporter peg-344, and the ferric aerobactin and receptor iucABCD-iutA. The plasmid carried two type II toxin-antitoxin systems, higB-higA and vapC-vapB (23). A BLAST search revealed that pDHQP1701672_hv shared >98% sequence identity and coverage with previously reported hypervirulent plasmids from China and Australia (24–26). Like pDHQP1701672_hv, two of these plasmids originated from CP-hvKP isolates; pKP70-2 (GenBank accession no. MF398271.1) was from an ST23 isolate and carried a mobile element that included blaKPC-2, making it both a carbapenemase-producing and a hypervirulent plasmid (24), and pVir-CR-hvKP267 (GenBank accession no. MG053312.1) was from an ST11 isolate with blaKPC-2 on a separate plasmid (26).
FIG 1.
Sequence alignment of the 227-kb pDHQP1701672_hv hypervirulence plasmid to the five most similar plasmids from an NCBI BLAST search. pVir-CR-HvKP267 was found in a K. pneumoniae ST11 isolate, and the other plasmids were from K. pneumoniae ST23. Similar plasmids were found in isolates from China (CN) and Australia (AU). Open reading frames (ORFs) of pDHQP1701672_hv are shown as the outermost ring, with plasmid replicons, virulence factors, and toxin-antitoxin systems highlighted.
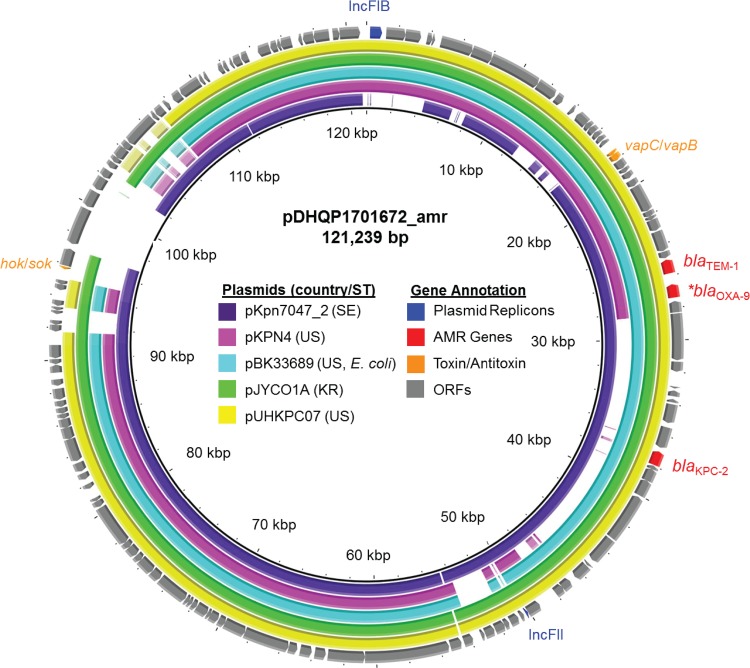
The smaller plasmid from DHQP1701672 (pDHQP1701672_amr) was 121,239 bp long and included IncFIB and IncFII replicons (Fig. 2). The plasmid had 135 open reading frames and two antimicrobial resistance genes, blaKPC-2 and blaTEM-1A, as well as a copy of blaOXA-9 truncated at residue 118 by a nonsense mutation. Carbapenemase gene blaKPC-2 was found on a Tn4401 transposon, the mobile genetic element most commonly associated with blaKPC genes (27). Two toxin-antitoxin systems were also found on the plasmid, the type I hok-sok and the type II vapC-vapB systems. A BLAST search revealed that the plasmid shared >99% sequence identity with 69% to 95% coverage of multiple pKpQIL-like plasmids (28), which have been largely responsible for the spread of blaKPC genes among Enterobacteriaceae in the United States and elsewhere (29, 30).
FIG 2.
Sequence alignment of the 121-kb pDHQP1701672_amr carbapenemase-encoding plasmid to the five most similar plasmids from an NCBI BLAST search. All of the plasmids were from CRE, four from K. pneumoniae and one (pBK33689) from E. coli isolates. Similar plasmids were found in isolates from Sweden (SE), the United States (US), and South Korea (KR). Open reading frames (ORFs) of pDHQP1701672_amr are shown as the outermost ring, with plasmid replicons, antimicrobial resistance (AMR) genes (*, including truncated blaOXA-9), and toxin-antitoxin systems highlighted.
Previous studies identified blaKPC-2 in ST23 isolates from Argentina (11), China (10, 31), and Poland (32). In contrast to DHQP1701672, these isolates displayed resistance to all β-lactams tested, including carbapenems. The low-level carbapenem MICs associated with DHQP1701672 may be the result of numerous genetic factors affecting the level of KPC production (e.g., copy number, level of expression), as well as the absence of other mechanisms (e.g., porin mutations) contributing to a carbapenem-resistant phenotype (33). Similar to DHQP1701672 and one of the isolates from China (24), the ST23 isolate from Argentina was positive for the virulence gene rmpA (as determined by PCR) (11), suggesting that it may have also harbored a hypervirulence and an antimicrobial resistance plasmid.
Although DHQP1701672 did not have a positive string test, it did carry five biomarkers (peg-344, iroB, iucA, rmpA, and rmpA2) described as more accurate predictors of hypervirulence in K. pneumoniae than the string test (12). All of these markers were located on the same IncFIB/IncHI1B plasmid (pDHQP1701672_hv), suggesting that horizontal acquisition of this virulence gene cluster is a possibility. The presence of toxin-antitoxin systems on both plasmids further increases the likelihood that these will be maintained across generations.
Other reports indicated different distributions of virulence factors in Klebsiella isolates associated with invasive infections (34). Thus, further studies are needed to fully understand the elements underlying hypervirulence and how they are related to a hypermucoviscous phenotype (34). To what extent DHQP1701672 is associated with enhanced in vivo virulence remains to be determined.
Although infections due to hvKP have been reported in the United States (7), this report represents the first documented CP-hvKP isolate. Together, the global distribution of ST23 (18) and the fact that plasmids carrying blaKPC genes are ubiquitous in many U.S. health care settings (35) represent potential stages on which additional CP-hvKP strains may emerge in the United States. Such emergence of CP-hvKP strains underscores the importance of clinical awareness of this pathotype and the need for continued monitoring of CP-hvKP in the United States and abroad. Given the patient’s history of travel to South America and that CP-hvKP ST23 has been reported from Argentina, it is possible that this isolate was imported. However, with high rates of human mobility, the recognition that these CP-hvKP strains are being imported and potentially circulating domestically is critical for containing their spread, especially given their potential for increased pathogenicity and mortality.
The present CP-hvKP isolate was identified through population-based surveillance conducted by the Emerging Infections Program, a network of 10 state health departments and academic partners working with the CDC to track the incidence and describe the epidemiology of infections caused by resistant or health care-associated pathogens. To enhance the ability to rapidly detect and contain emerging antimicrobial-resistant organisms, the CDC recently established the Antibiotic Resistance Laboratory Network (AR Lab Network), which supports infrastructure in 56 state and local public health laboratories to detect emerging antimicrobial-resistant pathogens, including carbapenemase-producing Enterobacteriaceae (36). Thus, the AR Lab Network allows for identification and characterization of CRE nationwide and complements the Emerging Infections Program’s intensive epidemiological efforts. At the CDC, whole-genome sequence analysis is routinely applied to surveillance isolates and a subset of CRE collected through the AR Lab Network. This report demonstrates the broad utility of whole-genome sequencing and emphasizes its role as a tool for detecting emerging pathotypes of public health importance, such as hypervirulence.
Accession number(s).
All whole-genome sequencing data are deposited in the NCBI database under BioSample accession number SAMN11054834.
ACKNOWLEDGMENTS
We thank staff at the Minnesota Department of Public Health for collecting and submitting clinical isolates to the CDC through the Multi-site Gram-negative Surveillance Initiative. We also thank Ruth Lynfield, Paula Vagnone-Snippes, Brittany VonBank, Isaac See, Shelley Magill, and L. Clifford McDonald for critical reading of the manuscript.
The findings and conclusions in this article are those of the authors and do not necessarily represent the official position of the U.S. Centers for Disease Control and Prevention.
REFERENCES
- 1.Morrill HJ, Pogue JM, Kaye KS, LaPlante KL. 2015. Treatment options for carbapenem-resistant Enterobacteriaceae infections. Open Forum Infect Dis 2:ofv050. doi: 10.1093/ofid/ofv050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC. 2001. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yigit H, Queenan AM, Rasheed JK, Biddle JW, Domenech-Sanchez A, Alberti S, Bush K, Tenover FC. 2003. Carbapenem-resistant strain of Klebsiella oxytoca harboring carbapenem-hydrolyzing beta-lactamase KPC-2. Antimicrob Agents Chemother 47:3881–3889. doi: 10.1128/AAC.47.12.3881-3889.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tamma PD, Goodman KE, Harris AD, Tekle T, Roberts A, Taiwo A, Simner PJ. 2017. Comparing the outcomes of patients with carbapenemase-producing and non-carbapenemase-producing carbapenem-resistant Enterobacteriaceae bacteremia. Clin Infect Dis 64:257–264. doi: 10.1093/cid/ciw741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akova M, Daikos GL, Tzouvelekis L, Carmeli Y. 2012. Interventional strategies and current clinical experience with carbapenemase-producing Gram-negative bacteria. Clin Microbiol Infect 18:439–448. doi: 10.1111/j.1469-0691.2012.03823.x. [DOI] [PubMed] [Google Scholar]
- 6.Bratu S, Landman D, Haag R, Recco R, Eramo A, Alam M, Quale J. 2005. Rapid spread of carbapenem-resistant Klebsiella pneumoniae in New York City: a new threat to our antibiotic armamentarium. Arch Intern Med 165:1430–1435. doi: 10.1001/archinte.165.12.1430. [DOI] [PubMed] [Google Scholar]
- 7.Lederman ER, Crum NF. 2005. Pyogenic liver abscess with a focus on Klebsiella pneumoniae as a primary pathogen: an emerging disease with unique clinical characteristics. Am J Gastroenterol 100:322–331. doi: 10.1111/j.1572-0241.2005.40310.x. [DOI] [PubMed] [Google Scholar]
- 8.Bialek-Davenet S, Criscuolo A, Ailloud F, Passet V, Jones L, Delannoy-Vieillard A-S, Garin B, Le Hello S, Arlet G, Nicolas-Chanoine M-H, Decré D, Brisse S. 2014. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis 20:1812–1820. doi: 10.3201/eid2011.140206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, Chan EW, Shu L, Yu J, Zhang R, Chen S. 2018. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis 18:37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 10.Zhang R, Lin D, Chan EW, Gu D, Chen GX, Chen S. 2016. Emergence of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae strains in China. Antimicrob Agents Chemother 60:709–711. doi: 10.1128/AAC.02173-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cejas D, Fernandez Canigia L, Rincon Cruz G, Elena AX, Maldonado I, Gutkind GO, Radice MA. 2014. First isolate of KPC-2-producing Klebsiella pneumoniae sequence type 23 from the Americas. J Clin Microbiol 52:3483–3485. doi: 10.1128/JCM.00726-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russo TA, Olson R, Fang CT, Stoesser N, Miller M, MacDonald U, Hutson A, Barker JH, La Hoz RM, Johnson JR. 2018. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol 56:e00776-18. doi: 10.1128/JCM.00776-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magill SS, Dumyati G, Ray SM, Fridkin SK. 2015. Evaluating epidemiology and improving surveillance of infections associated with health care, United States. Emerg Infect Dis 21:1537–1542. doi: 10.3201/eid2109.150508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.CLSI. 2018. Performance standards for antimicrobial susceptibility testing—28th ed CLSI supplement M100 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 15.Pierce VM, Simner PJ, Lonsway DR, Roe-Carpenter DE, Johnson JK, Brasso WB, Bobenchik AM, Lockett ZC, Charnot-Katsikas A, Ferraro MJ, Thomson RB Jr, Jenkins SG, Limbago BM, Das S. 2017. Modified carbapenem inactivation method for phenotypic detection of carbapenemase production among Enterobacteriaceae. J Clin Microbiol 55:2321–2333. doi: 10.1128/JCM.00193-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasheed JK, Kitchel B, Zhu W, Anderson KF, Clark NC, Ferraro MJ, Savard P, Humphries RM, Kallen AJ, Limbago BM. 2013. New Delhi metallo-beta-lactamase-producing Enterobacteriaceae, United States. Emerg Infect Dis 19:870–878. doi: 10.3201/eid1906.121515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagiya H, Watanabe N, Maki M, Murase T, Otsuka F. 2014. Clinical utility of string test as a screening method for hypermucoviscosity-phenotype Klebsiella pneumoniae. Acute Med Surg 1:245–246. doi: 10.1002/ams2.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Struve C, Roe CC, Stegger M, Stahlhut SG, Hansen DS, Engelthaler DM, Andersen PS, Driebe EM, Keim P, Krogfelt KA. 2015. Mapping the evolution of hypervirulent Klebsiella pneumoniae. mBio 6:e00630. doi: 10.1128/mBio.00630-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coutinho RL, Visconde MF, Descio FJ, Nicoletti AG, Pinto FC, Silva AC, Rodrigues-Costa F, Gales AC, Furtado GH. 2014. Community-acquired invasive liver abscess syndrome caused by a K1 serotype Klebsiella pneumoniae isolate in Brazil: a case report of hypervirulent ST23. Mem Inst Oswaldo Cruz 109:970–971. doi: 10.1590/0074-0276140196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Decre D, Verdet C, Emirian A, Le Gourrierec T, Petit JC, Offenstadt G, Maury E, Brisse S, Arlet G. 2011. Emerging severe and fatal infections due to Klebsiella pneumoniae in two university hospitals in France. J Clin Microbiol 49:3012–3014. doi: 10.1128/JCM.00676-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gundestrup S, Struve C, Stahlhut SG, Hansen DS. 2014. First case of liver abscess in Scandinavia due to the international hypervirulent Klebsiella pneumoniae clone ST23. Open Microbiol J 8:22–24. doi: 10.2174/1874285801408010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Compain F, Babosan A, Brisse S, Genel N, Audo J, Ailloud F, Kassis-Chikhani N, Arlet G, Decré D. 2014. Multiplex PCR for detection of seven virulence factors and K1/K2 capsular serotypes of Klebsiella pneumoniae. J Clin Microbiol 52:4377–4380. doi: 10.1128/JCM.02316-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Unterholzner SJ, Poppenberger B, Rozhon W. 2013. Toxin-antitoxin systems: biology, identification, and application. Mob Genet Elements 3:e26219. doi: 10.4161/mge.26219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong N, Lin D, Zhang R, Chan EW, Chen S. 2018. Carriage of blaKPC-2 by a virulence plasmid in hypervirulent Klebsiella pneumoniae. J Antimicrob Chemother 73:3317–3321. doi: 10.1093/jac/dky358. [DOI] [PubMed] [Google Scholar]
- 25.Lam MMC, Wyres KL, Duchene S, Wick RR, Judd LM, Gan YH, Hoh CH, Archuleta S, Molton JS, Kalimuddin S, Koh TH, Passet V, Brisse S, Holt KE. 2018. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat Commun 9:2703. doi: 10.1038/s41467-018-05114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao H, Qin S, Chen S, Shen J, Du XD. 2018. Emergence of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Lancet Infect Dis 18:25. doi: 10.1016/S1473-3099(17)30628-X. [DOI] [PubMed] [Google Scholar]
- 27.Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. 2014. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol 22:686–696. doi: 10.1016/j.tim.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leavitt A, Chmelnitsky I, Carmeli Y, Navon-Venezia S. 2010. Complete nucleotide sequence of KPC-3-encoding plasmid pKpQIL in the epidemic Klebsiella pneumoniae sequence type 258. Antimicrob Agents Chemother 54:4493–4496. doi: 10.1128/AAC.00175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doumith M, Findlay J, Hirani H, Hopkins KL, Livermore DM, Dodgson A, Woodford N. 2017. Major role of pKpQIL-like plasmids in the early dissemination of KPC-type carbapenemases in the UK. J Antimicrob Chemother 72:2241–2248. doi: 10.1093/jac/dkx141. [DOI] [PubMed] [Google Scholar]
- 30.Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Carmeli Y, Brolund A, Giske CG. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother 53:3365–3370. doi: 10.1128/AAC.00126-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi Y, Wei Z, Li L, Ji S, Du X, Shen P, Yu Y. 2010. Detection of a common plasmid carrying blaKPC-2 in Enterobacteriaceae isolates from distinct cities in China. Microb Drug Resist 16:297–301. doi: 10.1089/mdr.2010.0023. [DOI] [PubMed] [Google Scholar]
- 32.Baraniak A, Grabowska A, Izdebski R, Fiett J, Herda M, Bojarska K, Zabicka D, Kania-Pudlo M, Mlynarczyk G, Zak-Pulawska Z, Hryniewicz W, Gniadkowski M, KPC-PL Study Group. 2011. Molecular characteristics of KPC-producing Enterobacteriaceae at the early stage of their dissemination in Poland, 2008–2009. Antimicrob Agents Chemother 55:5493–5499. doi: 10.1128/AAC.05118-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitchel B, Rasheed JK, Endimiani A, Hujer AM, Anderson KF, Bonomo RA, Patel JB. 2010. Genetic factors associated with elevated carbapenem resistance in KPC-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 54:4201–4207. doi: 10.1128/AAC.00008-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Catalan-Najera JC, Garza-Ramos U, Barrios-Camacho H. 2017. Hypervirulence and hypermucoviscosity: two different but complementary Klebsiella spp. phenotypes? Virulence 8:1111–1123. doi: 10.1080/21505594.2017.1317412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woodworth KR, Walters MS, Weiner LM, Edwards J, Brown AC, Huang JY, Malik S, Slayton RB, Paul P, Capers C, Kainer MA, Wilde N, Shugart A, Mahon G, Kallen AJ, Patel J, McDonald LC, Srinivasan A, Craig M, Cardo DM. 2018. Vital signs: containment of novel multidrug-resistant organisms and resistance mechanisms - United States, 2006–2017. MMWR Morb Mortal Wkly Rep 67:396–401. doi: 10.15585/mmwr.mm6713e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Centers for Disease Control and Prevention. 2018. Lab capacity: Antibiotic Resistance Laboratory Network (AR Lab Network). https://www.cdc.gov/drugresistance/solutions-initiative/ar-lab-network.html. Accessed 30 October 2018.