Plasmid-mediated colistin resistance of the mobile colistin resistance (MCR) type is a growing concern in Enterobacteriaceae since it has been described worldwide in humans and animals. Here, we identified a series of MCR-producing Escherichia coli isolates corresponding to two different clones (represented by isolates PS1 and PS8b) producing MCR-1 and MCR-5, respectively, obtained from pig fecal samples in France.
KEYWORDS: MCR, MITE, colistin, mobilization, pig, resistance, transposition
ABSTRACT
Plasmid-mediated colistin resistance of the mobile colistin resistance (MCR) type is a growing concern in Enterobacteriaceae since it has been described worldwide in humans and animals. Here, we identified a series of MCR-producing Escherichia coli isolates corresponding to two different clones (represented by isolates PS1 and PS8b) producing MCR-1 and MCR-5, respectively, obtained from pig fecal samples in France. Plasmid analysis showed that the plasmid carrying the mcr-1 gene (pPS1) possesses an IncHI2 backbone, whereas the mcr-5 gene was carried onto a 6,268-bp nontypeable non-self-conjugative plasmid (pPS8b). Detailed analysis of plasmid pPS8b revealed a 3,803-bp-long cassette containing the mcr-5 gene that was bracketed by two inverted-repeat (IR) sequences with 5-bp-long direct repeats at each extremity, similarly to an insertion sequence, but with the exception that no transposase gene was identified within this cassette. By performing in vitro transposition experiments, we showed that the mcr-5 cassette could be mobilized by the TnAs1 transposase provided in trans, displaying a mobilization mechanism similar to that of miniature inverted-repeat transposable elements (MITEs).
INTRODUCTION
Mobile genetic elements represent a key feature in Gram-negative bacteria to adapt to their environment, and particularly, to selective pressures. Acquisition of antibiotic resistance may be either due to mutations in chromosomal genes being intrinsic to the species or to the acquisition of foreign DNA through horizontal gene transfer. Acquisition and horizontal transfer of antimicrobial resistance genes are mainly mediated by transposons, integrons, and plasmids (1, 2). Altogether, those genetic structures contribute to the occurrence of multidrug-resistant isolates and to the large spread of resistance determinants, not only on a geographical point of view, but also within bacterial species.
Plasmid-mediated colistin resistance genes have been recently widely identified among enterobacterial isolates recovered from humans and animals (3). Since the identification of the first colistin resistance determinant, namely, mobile colistin resistance 1 (MCR-1), seven other different MCR-type enzymes have been characterized (4–11). Those enzymes are phosphoethanolamine transferases being responsible for adding phosphoethanolamine to the lipid A moiety of the lipopolysaccharide (LPS) (the target of polymyxins) (3). Colistin resistance is therefore explained by a lack of interaction between polymyxins and a modified LPS structure. The progenitors of the different mcr genes have been shown to correspond to a variety of Gram-negative bacterial species, namely, Moraxella spp. for mcr-1 and mcr-2, Aeromonas spp. for mcr-3, and Shewanella spp. for mcr-4 (6, 7, 12, 13). The process of acquisition of mcr-like genes from their progenitors to their respective plasmid supports remains poorly understood so far, with the exception of mcr-1 that was shown to be, in some cases, associated with an ISApl1-made functional composite transposon (2, 14, 15). Other mcr-like genes were often identified into structures comprising insertion sequences (IS), but the role of these IS in the acquisition of the resistance genes was not elucidated. The mcr-5 gene was first described in a Salmonella enterica isolate, and in silico analysis identified an association with a Tn3-like transposon sharing 90% nucleotide identity with ISPa38 (8).
It is speculated that the use of colistin in veterinary medicine worldwide has contributed to the selection of colistin-resistant Enterobacteriaceae and thereby of MCR-producing isolates in animals (16). A recent prospective study performed in Portugal evidenced a strong link between the high use of MCR-producing Escherichia coli and Klebsiella pneumoniae strains and the use of colistin as a prophylactic treatment in pigs (17). Here, we aimed to characterize the genetic features of colistin resistance acquisition among colistin-resistant E. coli isolates recovered from pig farms in France.
RESULTS
Identification of colistin-resistant E. coli isolates.
Among the 147 collected samples, 35 samples showed a positive culture for a colistin-resistant E. coli isolate. PCR experiments showed that out of 35 colistin-resistant E. coli isolates recovered, 15 isolates were positive for the mcr-1 gene and 8 isolates were positive for the mcr-5 genes. No mcr-like genes were identified among the other colistin-resistant isolates. They likely possessed mutations in chromosomally encoded proteins involved in lipid A modifications, as recently reported, and are under investigation (18). All MCR-positive isolates were identified from a single pig farm, being the only one where colistin is frequently given for treating piglet diarrhea. Pulsed-field gel electrophoresis (PFGE) analysis showed that all MCR-1 and all MCR-5 producers belonged to two distinct clones (data not shown). A single MCR-1-producing isolate (PS1) and a single MCR-5-producing isolate (PS8b) were retained for further characterization.
Both isolates had an MIC value for colistin of 8 μg/ml. Multilocus sequence typing results identified the PS1 and PS8b isolates as belonging to sequence type 5409 (ST5409) and ST5786, respectively. Both isolates belonged to the phylogroup B1, suggesting a commensal origin. Isolate PS1 showed resistance to penicillins, sulfamethoxazole-trimethoprim, chloramphenicol, and tetracycline, although isolate PS8b showed resistance only to narrow-spectrum penicillins and sulfamethoxazole-trimethoprim.
Plasmid analysis.
Mating-out assays followed by plasmid analysis showed that the plasmid carrying the mcr-1 gene in isolate PS1 (pPS1) was a conjugative IncHI2 plasmid. However, no E. coli transconjugant could be obtained using isolate PS8b as a donor, but electrotransformation experiments using a plasmid extract from isolate PS8b allowed us to obtain an E. coli transformant carrying a plasmid (pPS8b) on which the mcr-5 gene was located. Plasmid pPS8b was nontypeable by PCR-based replicon typing (PBRT). Susceptibility testing showed that the E. coli transconjugant carrying plasmid pPS1 showed coresistance to sulfonamides and tetracycline, whereas no other resistance determinant was provided by plasmid pPS8b.
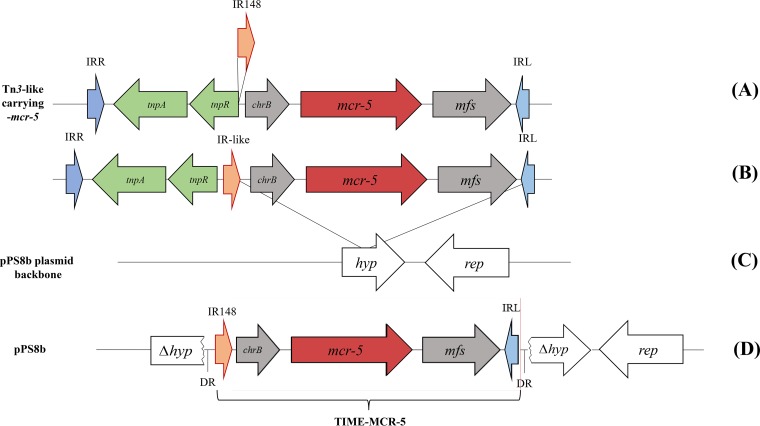
Sequencing of plasmid pPS8b (GenBank accession number MH674200.1) revealed a size of 6,268 bp and a sequence identical to that of the recently identified mcr-5-bearing plasmid pEC0674 (GenBank accession number MF684783.1) (19). By analyzing the sequence of plasmid pPS8b, we found that mcr-5 was part of a 3,810-bp-long inserted cassette. This cassette also contained a chromate resistance gene, chrB, and the inverted repeat left (IRL) of the Tn3-like structure, as previously identified in the original study that identified the mcr-5 gene (8) (Fig. 1). Noteworthy, a 5-bp-long direct repeat (AATTA) was identified at each extremity of the cassette, suggesting that this cassette had been acquired through a transposition mechanism, even though no transposase gene was identified on plasmid pPS8b. A 148-bp-long sequence sharing 85% identity with the IRL of a Tn3-like transposon was identified at the 5′ extremity of the mcr-5 gene (Fig. 2).
FIG 1.
(A and B) Schematic representation of the mcr-5 cassette inserted into the pPS8b plasmid (A) and comparison with a similar structure previously described (B). The direct repeats (DRs) being signature of transposition events are in bold and underlined. IRL corresponds to a Tn3-like left inverted-repeat sequence.
FIG 2.
Alignment of the 40 first nucleotides of the IR148 with the original IR identified on the mcr-5 cassette.
In silico analysis showed that this 148-bp-long IR-like structure was actually present in similar genetic contexts. Indeed, this exact same sequence, later named IR148, was identified in the near vicinity of Tn3-like transposons that were lacking their cognate transposase gene. Hence, IR148, together with the original IR of a truncated Tn3-like structure, likely corresponded to the boundaries of the whole cassette, being bracketed by direct repeats (Fig. 1). This suggested that IR148 could be involved in the mobilization of the mcr-5 cassette that likely occurred through a nonautonomous transposition mechanism. Those features resemble those described as miniature inverted-repeat transposable elements (MITEs), being reported as mobile elements possessing left and right IRs and lacking a transposase gene. Some studies even confirmed that they may be mobilized by the in trans activity of specific transposases recognizing those IRs (20).
Mobilization of the mcr-5 cassette by the TnAs1 transposase.
By using the BLAST tool of the ISfinder webpage (https://isfinder.biotoul.fr/blast.php) (21), we found that the sequence of IR148 shared the highest nucleotide identity with the IRL of the TnAs1 transposase (90%). In addition, the IR right (IRR) of the TnAs1 shared 80% nucleotide identity with the Tn3-like IRL of the mcr-5 transposon. Hence, we speculated that the TnAs1 transposase might be able to mobilize the mcr-5-containing structure in trans and therefore performed corresponding in vitro transposition experiments.
Initial attempts to mobilize the mcr-5 cassette by using the transposase identified in the original Tn3 transposon found in association with the mcr-5 gene (8) were unsuccessful, despite the high level of nucleotide identity between its IRs and those of the mcr-5 cassette. Therefore, transposition experiments were done using the E. coli RZ211 recombinant strain containing the pOX38 conjugative plasmid, together with the pACYC-mcr-5 and the pBAD-TnAs1.1 recombinant plasmids. That recombinant strain was therefore used as the host of putative transposition events, speculating that the mcr-5 cassette could be mobilized onto plasmid pOX38 by transposition through the in trans activity of the TnAs1.1 transposase. In order to evidence such a putative transposition event, mating-out assays were performed using the azide-resistant J53 strain as a recipient, and 10 E. coli transconjugants were analyzed. Susceptibility testing confirmed that those transconjugants were resistant to gentamicin, sodium azide, and colistin but remained susceptible to chloramphenicol and ampicillin, which was in agreement with a transfer of plasmid pOX38 carrying the mcr-5 cassette. PCR experiments confirmed that the mcr-5 cassette had indeed transposed onto the pOX38 conjugative plasmid.
Sequence analysis identified 10 different insertion sites of this transposed structure that were systematically bracketed by a 5-bp direct repeat (DR) sequence (Fig. 3A and B). Analysis of the GC content of the close genetic environment of the insertion site revealed an AT-rich sequence whose consensus was found to be A/T, A/T, A/T, B, A/T (Fig. 3C). The genetic environment surrounding the different target sites did not display a particular feature.
FIG 3.
Target site analysis of the mcr-5 cassette. (A) Schematic representation of the positions of the integration sites of the MITE-like structure onto plasmid pOX38. (B) Sequence alignment of 10 transposition events identified onto plasmid pOX38. The IR148 (IR-like) and inverted repeat left (IRL) sequences of this MITE structure are boxed. The 5-bp duplicated direct repeat (DR) sequences are highlighted in bold and underlined. (C) Pictogram showing the relative frequencies of A, T, C, and G nucleotides at the target site and their vicinities 10 bp downstream and upstream.
DISCUSSION
We first identified here MCR-5-producing E. coli isolates in France. Sequence analysis of plasmid pPS8b revealed a particular genetic structure displaying an mcr-5 cassette surrounded by two imperfect inverted-repeat sequences but with no transposase-encoding gene, suggesting that the mcr-5 cassette could correspond to a nonautonomous mobile genetic structure, similarly to MITEs. MITEs are defined as small genetic elements bracketed by two IRs that can be recognized by a transposase acting in trans (21). Such transposase activity can therefore mobilize the MITE element by recognizing the two IR sequences. In the case that such a mobilized genetic element actually contains an antibiotic resistance gene, such a MITE therefore constitutes a genetic structure being an original source of acquired resistance. This has been shown for instance with the GES-5 carbapenemase-encoding gene (22) or with the QnrS2 quinolone resistance-encoding gene (23). Here, we showed that the TnAs1 transposase was able to recognize the extremities of this mcr-5 cassette leading to its mobilization through a transposition mechanism. The TnAs1 transposase belongs to the Tn3 family, and its inverted-repeat sequences shared high nucleotide identities with the IR148 and IRL of the mcr-5 cassette. In silico analysis showed that TnAs1 is not found in Aeromonas spp. A particular class of MITEs whose mobilization is mediated by Tn3 elements but which does not carry any putative open reading frame, accordingly designated TIME (standing for Tn3-derived inverted-repeat miniature element), has been previously reported (24). Here, we may consider our structure bearing the mcr-5 cassette to be part of this class of transposable elements, and we therefore named it TIME–MCR-5.
This finding constitutes the first evidence of a MITE-mediated mobilization of an mcr-like gene. In addition, the mcr-5-bearing cassette was identified on a small nonconjugative plasmid that cannot be transferred by conjugation; however, it can be mobilized by another plasmid, therefore prompting it to be horizontally transferred and to disseminate among different strains.
Noteworthy, in silico analysis revealed that this same mcr-5 cassette was identified in E. coli and Aeromonas sp. isolates, respectively (19, 25), suggesting that it constituted an effective transposable element. According to our experimental data, the chronology of the genetic events might be the following: the IR148 sequence was inserted into the original Tn3 transposon carrying the mcr-5 gene (Fig. 4A and B). Then, the IR-like structure and the IRL of Tn3 were recognized by transposase activity provided in trans that was responsible for the excision of the new cassette and of its insertion into the pPS8b plasmid backbone sequence (Fig. 4C), accordingly generating DRs (Fig. 4D). Further experiments will be necessary in order to investigate the mechanism responsible for the initial step corresponding to insertion of IR148 into transposon Tn3. Interestingly, in silico investigations showed that IR148 sequences have been found in similar genetic structures, likely resulting from a similar MITE-like mobilization mechanism (Fig. 5). This IR-like structure was found to be associated with other IRs, all sharing high identity with Tn3-like IRs. This suggests that the IR148 element may be responsible for the mobilization of numerous genes and represent an efficient mobilization unit.
FIG 4.
Proposed model of the chronology of acquisition of the mcr-5 cassette onto the pPS8b plasmid backbone.
FIG 5.
Examples of genetic contexts in which the IR-like structure in different genetic contexts forming a MITE-like structure in the genomes of V. parahaemolyticus and A. salmonicida. A 5-nucleotide (nt) direct repeat sequence was identified in silico in every MITE-like structure. The direct repeats (DRs) being signature of transposition events are in bold and underlined.
This is the first study identifying a MITE-like element at the origin of acquisition of a colistin resistance determinant. Our study highlights the diversity of mobile elements associated with resistance determinants and the remarkable genetic engineering of bacterial genomes for adaptive resistance toward antimicrobial agents.
MATERIALS AND METHODS
Design of the study.
A collection of 147 rectal samples recovered from pigs and from 4 different farms were collected in the Bourgogne region in France in 2016. All samples were cultured overnight in Luria-Bertani (LB) broth supplemented with colistin (1 μg/ml). After 16 h of incubation, 10 μl of culture was inoculated onto Superpolymyxin agar plates (ELITech, Signes, France) and was incubated at 37°C for another 16 h. Colistin resistance of the recovered isolates was confirmed by the broth microdilution method using cation-adjusted Mueller-Hinton (MH) broth (Bio-Rad, Cressier, Switzerland), as recommended by the CLSI (26).
The presence of mcr-like genes (mcr-1 to -8) was investigated by PCR analysis using specific primers, and all positive signals were sequenced using the Sanger method (Microsynth, Balgach, Switzerland) (Table 1). Antimicrobial susceptibility testing was performed according to the standard disk diffusion method with MH agar plates (http://www.eucast.org/).
TABLE 1.
Oligonucleotides used in this work
| Primer | Direction | Sequence (5′ to 3′) | Reference |
|---|---|---|---|
| mcr-1 | Forward | ATGCCAGTTTCTTTCGCGTG | 29 |
| Reverse | TCGGCAAATTGCGCTTTTGGC | ||
| mcr-2 | Forward | AAGGCTGACACCCCATGTCAT | 29 |
| Reverse | GATGGCGGTCTATCCTGTAT | ||
| mcr-3 | Forward | ACCAGTAAATCTGGTGGCGT | 29 |
| Reverse | AGGACAACCTCGTCATAGCA | ||
| mcr-4 | Forward | TTGCAGACGCCCATGGAATA | 29 |
| Reverse | GCCCGCATGAGCTAGTATCGT | ||
| mcr-5 | Forward | GGACGCGACTCCCTAACTTC | 29 |
| Reverse | ACAACCAGTACGAGAGCACG | ||
| mcr-7 | Forward | GGTGAATTTGTTGCTGGTGC | This study |
| Reverse | GGCACTGGCTGAAAATATCG | ||
| mcr-8 | Forward | TCGGCAACATAGCACTTTGG | This study |
| Reverse | TGTGTTTGTTCATTGGGGGC |
Clonality evaluation was performed by pulsed-field gel electrophoresis (PFGE). Briefly, total DNA of all MCR producers was digested by using the XbaI enzyme (New England BioLabs, Ipswich, MA, USA). The generated fragments were separated by PFGE using a CHEF-DR III system (Bio-Rad, Cressier, Switzerland). Multilocus sequence type was obtained using the Center for Genomic Epidemiology server (MLST 1.8). Phylogroup determination was performed using the PCR-based Clermont method (27).
Plasmid analysis.
Plasmid carrying the mcr-5 gene was extracted using the Zyppy miniprep kit (Zymo Research, Irvine, CA, USA) and was sequenced by the Sanger method (Microsynth, Balgach, Switzerland) using a PCR walking method. PBRT was used to determine the incompatibility group of the plasmids carrying mcr genes (28).
Conjugation experiments were performed using the azide-resistant E. coli J53 recipient strain. Both donor and recipient strains were cultured in exponential phase and then mixed on solid LB agar using filters at a 1:10 donor/recipient ratio. After 5 h of incubation, filters were resuspended in 0.85% NaCl, and the bacterial mixture was plated onto agar plates supplemented with colistin (1 μg/ml) and sodium azide (100 μg/ml).
Plasmid constructs for the transposition assay.
Two different recombinant plasmids were obtained for the transposition experiments. pACYC-mcr-5 was obtained by cloning the mcr-5 cassette (containing the mcr-5 gene bracketed by the two identified IRs) into the tetracycline resistance gene of the low-copy-number cloning vector pACYC184 carrying both tetracycline and chloramphenicol resistance genes. The pBAD-TnAs1.1 plasmid was obtained by cloning the resolvase and transposase genes of transposon TnAs1 into the l-arabinose-inducible pBADb plasmid carrying an ampicillin resistance determinant. The pBAD-Tn3-like plasmid was obtained by cloning the resolvase and transposase of the original Tn3 transposon containing the mcr-5 cassette in the pBADb plasmid (14).
Transposition experiments.
Both pACYC-mcr-5 and pBAD-TnAs1.1 were transformed into the E. coli RZ211 strain carrying the conjugative pOX38 plasmid used as a DNA receptor for transposition events and carrying the gentamicin resistance determinant. The RZ211 E. coli strain containing the three plasmids was incubated overnight at 37°C in LB broth supplemented in l-arabinose (0.1%), ampicillin (100 μg/ml), gentamicin (8 μg/ml), chloramphenicol (25 μg/ml), and colistin (1 μg/ml). After mating-out assays with the J53 strain, the putative transposants were selected on LB agar supplemented with azide (100 μg/ml), gentamicin (8 μg/ml), and colistin (1 μg/ml). Each colony obtained was checked for chloramphenicol and ampicillin susceptibility in order to eliminate strains in which whole-plasmid integration events might have occurred.
Determination of the location of the transposition events was performed by PCR, followed by sequencing using one outward primer located at the 5′ extremity of the mcr-5 cassette and a second primer further designed after sequencing of this 5′ extremity and targeting the adjacent sequences on plasmid pOX38.
ACKNOWLEDGMENT
This work has been funded by the University of Fribourg and by the Swiss National Science Foundation (project FNS-407240_177381).
REFERENCES
- 1.Toleman MA, Walsh TR. 2011. Combinatorial events of insertion sequences and ICE in Gram-negative bacteria. FEMS Microbiol Rev 35:912–935. doi: 10.1111/j.1574-6976.2011.00294.x. [DOI] [PubMed] [Google Scholar]
- 2.Partridge SR, Kwong SM, Firth N, Jensen SO. 2018. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev 31:e00088-17. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poirel L, Jayol A, Nordmann P. 2017. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev 30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistinresistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 5.Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, Malhotra-Kumar S. 2016. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill 21:pii=30280. doi: 10.2807/1560-7917.ES.2016.21.27.30280. [DOI] [PubMed] [Google Scholar]
- 6.Yin W, Li H, Shen Y, Liu Z, Wang S, Shen Z, Zhang R, Walsh TR, Shen J, Wang Y. 2017. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio 8:e00543-17. doi: 10.1128/mBio.00543-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carattoli A, Villa L, Feudi C, Curcio L, Orsini S, Luppi A, Pezzotti G, Magistrali CF. 2017. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill 22:pii=30589. doi: 10.2807/1560-7917.ES.2017.22.31.30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borowiak M, Fischer J, Hammerl JA, Hendriksen RS, Szabo I, Malorny B. 2017. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother 72:3317–3324. doi: 10.1093/jac/dkx327. [DOI] [PubMed] [Google Scholar]
- 9.AbuOun M, Stubberfield EJ, Duggett NA, Kirchner M, Dormer L, Nunez-Garcia J, Randall LP, Lemma F, Crook DW, Teale C, Smith RP, Anjum MF. 2018. mcr-1 and mcr-2 (mcr-6.1) variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015. J Antimicrob Chemother 73:2904. doi: 10.1093/jac/dky272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang YQ, Li YX, Lei CW, Zhang AY, Wang HN. 2018. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J Antimicrob Chemother 73:1791–1795. doi: 10.1093/jac/dky111. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Wang Y, Zhou Y, Li J, Yin W, Wang S, Zhang S, Shen J, Shen Z, Wang Y. 2018. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect 7:122. doi: 10.1038/s41426-018-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kieffer N, Nordmann P, Poirel L. 2017. Moraxella species as potential sources of MCR-like polymyxin resistance determinants. Antimicrob Agents Chemother 61:e00129-17. doi: 10.1128/AAC.00129-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poirel L, Kieffer N, Fernandez-Garayzabal JF, Vela AI, Larpin Y, Nordmann P. 2017. MCR-2-mediated plasmid-borne polymyxin resistance most likely originates from Moraxella pluranimalium. J Antimicrob Chemother 72:2947–2949. doi: 10.1093/jac/dkx225. [DOI] [PubMed] [Google Scholar]
- 14.Poirel L, Kieffer N, Nordmann P. 2017. In vitro study of ISApl1-mediated mobilization of the colistin resistance gene mcr-1. Antimicrob Agents Chemother 61:e00127-17. doi: 10.1128/AAC.00127-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Partridge SR. 2017. mcr-2 in the IncX4 plasmid pKP37-BE is flanked by directly oriented copies of ISEc69. J Antimicrob Chemother 72:1533–1535. doi: 10.1093/jac/dkw575. [DOI] [PubMed] [Google Scholar]
- 16.Poirel L, Nordmann P. 2016. Emerging plasmid-encoded colistin resistance: the animal world as the culprit? J Antimicrob Chemother 71:2326–2327. doi: 10.1093/jac/dkw074. [DOI] [PubMed] [Google Scholar]
- 17.Kieffer N, Aires-de-Sousa M, Nordmann P, Poirel L. 2017. High rate of MCR-1-producing Escherichia coli and Klebsiella pneumoniae among pigs, Portugal. Emerg Infect Dis 23:2023–2029. doi: 10.3201/eid2312.170883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bourrel AS, Poirel L, Royer G, Darty M, Vuillemin X, Kieffer N, Clermont O, Denamur E, Nordmann P, Decousser JW, IAME Resistance Group. 2019. Colistin resistance in Parisian inpatient faecal Escherichia coli as the result of two distinct evolutionary pathways. J Antimicrob Chemother, in press. [DOI] [PubMed] [Google Scholar]
- 19.Hammerl JA, Borowiak M, Schmoger S, Shamoun D, Grobbel M, Malorny B, Tenhagen BA, Käsbohrer A. 2018. mcr-5 and a novel mcr-5.2 variant in Escherichia coli isolates from food and food-producing animals, Germany, 2010 to 2017. J Antimicrob Chemother 73:1433–1435. doi: 10.1093/jac/dky020. [DOI] [PubMed] [Google Scholar]
- 20.Fattash I, Rooke R, Wong A, Hui C, Luu T, Bhardwaj P, Yang G. 2013. Miniature inverted-repeat transposable elements: discovery, distribution, and activity. Genome 56:475–486. doi: 10.1139/gen-2012-0174. [DOI] [PubMed] [Google Scholar]
- 21.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poirel L, Carrër A, Pitout JD, Nordmann P. 2009. Integron mobilization unit as a source of mobility of antibiotic resistance genes. Antimicrob Agents Chemother 53:2492–2498. doi: 10.1128/AAC.00033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cattoir V, Poirel L, Aubert C, Soussy CJ, Nordmann P. 2008. Unexpected occurrence of plasmid-mediated quinolone resistance determinants in environmental Aeromonas spp. Emerg Infect Dis 14:231–237. doi: 10.3201/eid1402.070677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szuplewska M, Ludwiczak M, Lyzwa K, Czarnecki J, Bartosik D. 2014. Mobility and generation of mosaic non-autonomous transposons by Tn3-derived inverted-repeat miniature elements (TIMEs). PLoS One 9:e105010. doi: 10.1371/journal.pone.0105010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma S, Sun C, Hulth A, Li J, Nilsson LE, Zhou Y, Börjesson S, Bi Z, Bi Z, Sun Q, Wang Y. 2018. Mobile colistin resistance gene mcr-5 in porcine Aeromonas hydrophila. J Antimicrob Chemother 73:1777–1780. doi: 10.1093/jac/dky110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial susceptibility testing; 28th informational supplement. CLSI document M100. Clinical and Laborratory Standards Institute, Wayne, PA, USA. [Google Scholar]
- 27.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66:4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 29.Lescat M, Poirel L, Nordmann P. 2018. Rapid multiplex polymerase chain reaction for detection of mcr-1 to mcr-5 genes. Diagn Microbiol Infect Dis 92:267–269. doi: 10.1016/j.diagmicrobio.2018.04.010. [DOI] [PubMed] [Google Scholar]