ABSTRACT
Chickens are the reservoir host of Salmonella Enteritidis. Salmonella Enteritidis colonizes the gastro-intestinal tract of chickens and replicates within macrophages without causing clinically discernable illness. Persistence of S. Enteritidis in the hostile environments of intestinal tract and macrophages allows it to disseminate extra-intestinally to liver, spleen, and reproductive tract. Extra-intestinal dissemination into reproductive tract leads to contamination of internal contents of eggs, which is a major risk factor for human infection. Understanding the genes that contribute to S. Enteritidis persistence in the chicken host is central to elucidate the genetic basis of the unique pathobiology of this public health pathogen. The aim of this study was to identify a succinct set of genes associated with infection-relevant in vitro environments to provide a rational foundation for subsequent biologically-relevant research. We used in silico prediction of gene expression and RNA-seq technology to identify a core set of 73 S. Enteritidis genes that are consistently highly expressed in multiple S. Enteritidis strains cultured at avian physiologic temperature under conditions that represent intestinal and intracellular environments. These common highly expressed (CHX) genes encode proteins involved in bacterial metabolism, protein synthesis, cell-envelope biogenesis, stress response, and a few proteins with uncharacterized functions. Further studies are needed to dissect the contribution of these CHX genes to the pathobiology of S. Enteritidis in the avian host. Several of the CHX genes could serve as promising targets for studies towards the development of immunoprophylactic and novel therapeutic strategies to prevent colonization of chickens and their environment with S. Enteritidis.
Keywords: Salmonella, transcriptome, common highly expressed genes, chicken, environment
INTRODUCTION
Salmonella enterica sub sp. enterica serovar Enteritidis (S. Enteritidis) is a leading foodborne pathogen worldwide. S. Enteritidis causes persistent infection in chickens without inducing clinically discernible illness and has a unique pathobiology that allows it to contaminate internal contents of eggs (Denagamage et al., 2017; Gast et al., 2017). S. Enteritidis infection in chickens is relevant for public health because human infections are associated with consumption of contaminated eggs and meat [(Shah et al., 2017b; Goncalves-Tenorio et al., 2018) and reviewed in Chousalkar et al. (2018)]. Incidence of S. Enteritidis has increased steadily since the early 2000s, in both humans and chickens, according to laboratory-confirmed human cases (FoodNet and CDC Laboratory-based Enteric Diseases Surveillance or LEDS) and broiler chicken rinsate surveys (VetNet). In the last decade, at least 2 large multistate outbreaks reported in the US were attributed to shell eggs contaminated with S. Enteritidis, resulting in approximately 3,578 human illnesses and a nationwide voluntary egg recall from suspected suppliers (Centers for Disease Control and Prevention, 2010; Chai et al., 2012; Boore et al., 2015). Thus, S. Enteritidis poses a major challenge to poultry production, food-safety, and public health.
S. Enteritidis infects and persists in poultry largely because of its ability to survive and replicate in the harsh environments encountered in the gastrointestinal tract and systemic sites of the avian host (Shah et al., 2017a). Interestingly, wild-type strains of S. Enteritidis exhibit extensive phenotypic diversity, including dissimilar ability to infect chickens (Yim et al., 2010; Shah et al., 2011a; Gast et al., 2018). For instance, S. Enteritidis strains better able to tolerate acid and oxidative stress are also better able to colonize the avian gastrointestinal tract, internal organs and survive within the internal content of eggs (Shah et al., 2012; Baron et al., 2017). Phenotypic factors such as presence of high-molecular-mass LPS, high cell density growth and motility also correlate with the ability of certain S. Enteritidis strains to infect chickens, contaminate eggs, and cause human infection (Guard-Petter, 2001; Yim et al., 2011; Shah et al., 2011b). Studies using negative selection screens of mutant libraries and in vivo expression technology have identified genes that contribute to intestinal colonization and systemic dissemination in chickens and survival of S. Enteritidis within the internal contents of eggs (Silva et al., 2012; Raspoet et al., 2014a; Raspoet et al., 2014b). The genes identified by these studies, however, are generally present in all the S. Enteritidis strains due to the extensive within serovar genetic homogeneity at the gene content level (Shah et al., 2012). Moreover, epidemiologically distinct strains of S. Enteritidis show similar gene content that contrasts their phenotypic diversity and differential pathobiology in chickens (Allard et al., 2013). This poses significant challenges in defining gene function in the context of infection. Newer approaches are, therefore, needed to dissect the genetic mechanisms underlying phenotypic diversity and differential ability of S. Enteritidis strains to cause infection in chickens.
Recent studies show that the differential pathogenicity of genetically related S. Enteritidis strains might be driven at the transcriptional level. For instance, S. Enteritidis strains with high colonizing ability in chickens display distinct transcriptional profiles relative to the strains that are not as efficient in their colonization capabilities (Shah, 2014). This raises the possibility that strains with high colonizing ability may exhibit common transcriptional signatures that enable them to persist in the avian host. In this context, our hypothesis is that S. Enteritidis strains that successfully persist in the avian host consistently express a common set of genes that may play an important role in persistence of this organism. Consequently, we aimed to identify such common set of genes that are consistently highly expressed among multiple pathogenic S. Enteritidis strains, independently of the surrounding host microenvironment. To accomplish this, we coupled in silico prediction of gene expression (Karlin and Mrazek, 2000) with global in vitro transcriptome analysis of 3 highly pathogenic S. Enteritidis strains cultured under conditions that resemble the gastrointestinal and intra-macrophage compartments within the avian host. With this dual approach, we show that the genes predicted to be highly expressed in silico correlate with the experimental global transcriptomes generated from these pathogenic S. Enteritidis strains. Our results also show that a relatively small set of genes (n = 73) are consistently highly expressed in S. Enteritidis independently of strain or surrounding microenvironment. These genes encode proteins that are involved in protein synthesis, stress response, cell-envelope, or membrane biogenesis, bacterial metabolism, and a few proteins with poorly characterized functions. We discuss the functions of these genes in Salmonella or other related organisms and show that several of these genes likely play an important role in the pathobiology of S. Enteritidis. This study provides foundation for further investigations needed to dissect the contributions of the highly expressed genes in biologically-relevant systems and their potential application in the development of new immunoprophylactic and therapeutic measures to control this public health pathogen.
MATERIALS AND METHODS
In Silico Prediction of Gene Expression Levels
The complete genome sequence of S. Enteritidis str. P125109 (UK, phage type 4, NCBI accession number NC_011294) was analyzed in silico using the software package GEMBASSY-gphx (Itaya et al., 2013). For comparison, complete genome sequences of the genetically related S. Typhimurium LT2 (NC_003197) and other Gram-negative bacterial strains such as E. coli K12, C. jejuni NCTC 11168 (NC_002163), Deinococcus radiodurans, and Gram-positive bacterium such as B. subtilis were also analyzed (Table 1). GEMBASSY-gphx predicts highly expressed (PHX) genes based on their predicted general expression level [E(g)] (Karlin and Mrazek, 2000; Karlin et al., 2003). Prediction of expression level takes into account the difference in codon usage (B) between each specific protein-coding gene of interest (g) relative to all other protein-coding genes within the genome (C). The difference in codon usage is also calculated between the gene of interest (g) and three reference gene classes known to be highly expressed across different microorganisms. These gene classes include (i) RP = ribosomal protein genes, (ii) CH = chaperone genes, and (iii) TF = translation/transcription associated genes. The predicted expression level of a gene of interest relative to each of the reference family classes is calculated as follows:
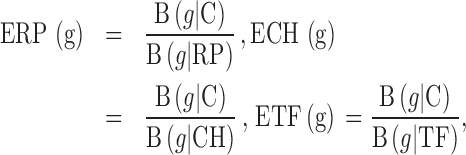 |
E = Expression relative to RP, CH, or TF. The codon usage difference between each test gene (g) and reference gene classes (RP, CH, or TF) is denoted by B (g ∣ Reference Gene Class). A predicted highly expressed (PHX) gene displays a high ratio difference relative to the rest of protein-coding genes [B(g∣C)] and a low ratio difference relative to the 3 representative gene classes [B(g/RP) or (g/CH) or (g/TF)]. Finally, the predicted general expression level [E(g)] is calculated by combining these 3 ratios, as follows:
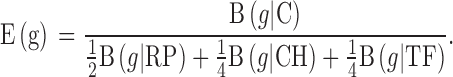 |
The following 2 conditions must be met for a gene to be categorized as PHX: (i) the predicted general expression level [E(g)] must be ≥1; and (ii) at least 2 of the 3 values (ERP, ECH, or ETF) must exceed 1.05 (Karlin and Mrazek, 2000; Karlin et al., 2003).
Table 1.
General statistics for in silico predicted highly expressed (PHX) genes in selected bacterial genomes.
| Genome | Genome size (kb) | Total CDS1 | Total number of PHX2 genes | % PHX genes from total CDS | CDS ≥ 100 codons3 | PHX genes from CDS ≥ 100 codons | % PHX genes (CDS ≥ 100 codons) | Max E(g)4 | Reference | NCBI accession |
|---|---|---|---|---|---|---|---|---|---|---|
| S. Enteritidis P125109 | 4,685 | 4206 | 161 | 3.8 | 3849 | 124 | 3.2 | 3.53 | This study | NC_011294 |
| S. Typhimurium LT2 | 4,857 | 4423 | 171 | 3.9 | 4009 | 130 | 3.2 | 3.75 | This study | NC_003197 |
| B. subtilis str.168 | 4,215 | 4174 | 145 | 3.5 | 3615(3612) | 96(148) | 2.7(4) | 4.54 (2.34) | This study and (23) | NC_000964 |
| E.coli K-12 substr.MG1655 | 4,641 | 4144 | 248 | 6.0 | 3800(3898) | 204(306) | 5.4(8) | 3.71 (2.66) | This study and (23) | NC_000913 |
| C. jejuni (NCTC 11168) | 1,641 | 1622 | 129 | 8.0 | 1452 | 100 | 6.9 | 1.69 | This study | NC_002163 |
| D. radiodurans R1 (BAA-816) | 3,284 | 2997 | 325 | 10.8 | 2828(2629) | 310(362) | 11(14) | 2.94 (1.66) | This study and (37) | NC_001263 NC_001264 |
1CDS, coding sequences.
2PHX, Predicted Highly Expressed.
3Values in parenthesis have been reported previously by the studies cited in the Reference column.
4E(g), predicted general expression level.
Transcriptome Analysis
S. Enteritidis UK (phage type 4), G1 (phage type 4), and BC8 (phage type 8) were cultured in LB-salt (300 mM NaCl) and LPM broth (5 mM KCl, 7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 38 mM glycerol, 0.1% casamino acids, 8μM MgCl2, 337 μM PO3−4, pH 5.8) (Coombes et al., 2004) at avian physiologic temperature (42°C) until reaching exponential phase with constant agitation (200 RPM) as described previously (Shah, 2014). These conditions resemble the salt rich environment within the avian intestinal lumen (LB-salt) (Razdan et al., 1997; Huang et al., 2014) and the low pH, low-magnesium, low-phosphate conditions encountered within macrophages (LPM) (Coombes et al., 2004; Gibbons et al., 2005; Niemann et al., 2011; Heithoff et al., 2012). RNA extraction, mRNA enrichment, and RNA-Seq were performed as described previously (Shah, 2014). For comparison, we also included RNA-Seq data obtained from strains grown under nutrient rich conditions (LB) from a previously published report (Shah, 2014). Paired-end reads (75 bp) were trimmed and aligned against the S. Enteritidis str. P125109 reference genome using Geneious v 11.15 (Biomatters Ltd, New Zealand). Read counts were adjusted to Reads per kilobase per million (RPKM). Transcripts with RPKM values below 10 were removed from analysis and the remaining data was transformed to Log2 units for further analysis. The transcriptome data have been deposited in NCBI’s gene expression omnibus (Edgar et al., 2002) under the GEO series accession number GSE122177. In addition, data compiled for LB, LB-salt, and LPM media is included in Supplementary files 1, 2, and 3, respectively. The Pearson correlation between the transcriptome data (RPKM-log2 transformed values) obtained from the 3 strains grown under each condition and in silico prediction of gene expression (Eg) was determined using NCSS 2007 version 07.1.19 (NCSS, USA). The most abundant transcripts from the experimental transcriptome analysis (top 10%) in all strains and all conditions that were concurrently identified as PHX were designated as common highly expressed (CHX) genes.
In Silico Determination of Binding Affinity Between Shine-Dalgarno (SD) Region and the Anti-Shine-Dalgarno Sequence (aSD)
The SD region was defined as 20 nucleotides upstream of the start codon of each gene (Nakagawa et al., 2017). The SD region was extracted from all coding sequences (CDs) annotated in S. Enteritidis P125109 genome (NC_011294). Binding affinity (kcal/mol) between the SD region and the extended aSD (5′-CCUCCUUA-3′) (Wei et al., 2017) was calculated in silico using RNAcofold v2.4.9 with default parameters (Lorenz et al., 2011). Protein localization prediction was performed with PSORTb v3.0 (Yu et al., 2010).
RESULTS AND DISCUSSION
Identification of S. Enteritidis PHX Genes Using GEMBASSY-gphx
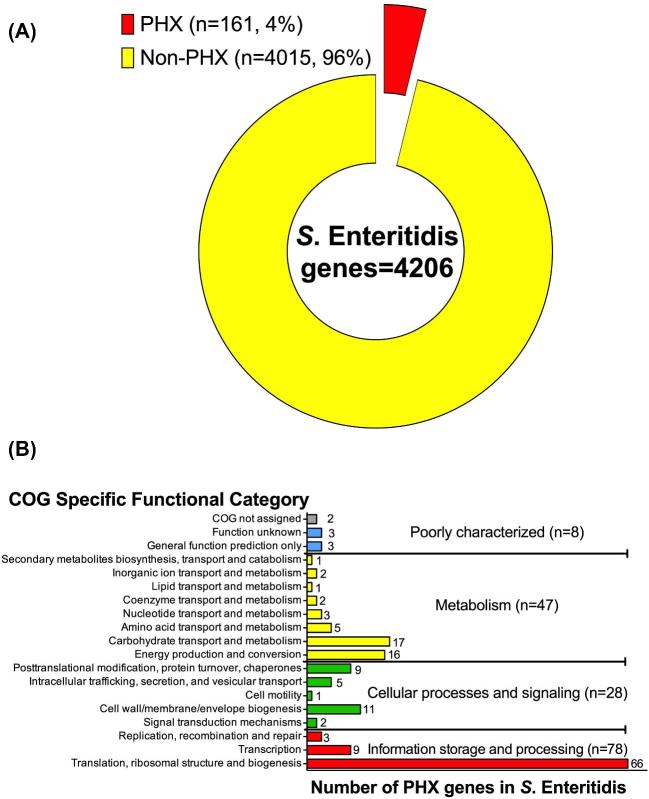
In this study, we first identified predicted highly expressed genes (PHX) in silico in the S. Enteritidis genome using GEMBASSY-gphx software tool. This in silico prediction is based on the difference in codon usage between a test gene and 3 different gene classes (RP, CH, and TF) (Karlin and Mrazek, 2000) known to be highly expressed as they encode highly abundant proteins relevant to bacterial physiology (VanBogelen et al., 1990; Ishihama et al., 2008; Maier et al., 2011). This in silico analysis of 4206 genes within the S. Enteritidis genome predicted a total of 161 PHX genes (3.8%) (Figure 1a), whose codon usage is similar to the reference highly expressed genes. The remaining 4015 genes were classified as non-PHX and will not be discussed in this study. The proportion and identity of PHX genes in the S. Typhimurium LT2 genome was similar to S. Enteritidis (Table 1). The proportion of PHX genes differed in other organisms including the Gram-positive B. subtilis (3.5%), the Gram-negatives E. coli K12 (6%) (Karlin and Mrazek, 2000), Campylobacter jejuni (8%), and the radiation-resistant Deinococcus radiodurans (10.8%) (Na Gao et al., 2009) (Table 1). The difference in proportion of PHX genes among these diverse bacterial genomes is expected in lieu of their individual genome content, niche-specificity and divergent evolution.
Figure 1.
(A) Classification of S. Enteritidis str. P125109 genome into predicted highly expressed (PHX) genes according to GEMBASSY-gphx. (B) Classification of S. Enteritidis str. P125109 PHX genes according to cluster of orthologous groups (COG) specific functional categories.
Functional Classes of S. Enteritidis PHX Genes Identified Using GEMBASSY-gphx
Classification of 161 PHX genes into functional categories based on cluster of orthologous genes (COG) revealed that the majority of PHX genes contribute to information storage and processing (78/161, 48%) (Figure 1b). Of these 78 genes, 66 genes encode proteins involved in translation, ribosomal structure and biogenesis. The second most abundant functional category of PHX genes is related to bacterial metabolism (47/161 = 29%), wherein the majority of genes are involved in carbohydrate transport and metabolism (17/47, 36%). Identification of these functional classes is consistent with previous reports showing strong association between functional groups J (translation, ribosome structure, and biogenesis) and C (energy production and conversion) with highly expressed genes in bacteria (Ma et al., 2002; Rollenhagen and Bumann, 2006). Such association is not surprising given that genes within these functional categories are highly conserved and play critical roles in bacterial physiology (Nei and Kumar, 2000; Rocha and Danchin, 2004; Drummond et al., 2005). Indeed, the functional relevance and evolutionary selection pressure are so central for these PHX genes that their proteins are less prone to mutations that lead to amino acid substitutions (Drummond et al., 2005), allowing their efficient recognition by the most abundant tRNAs and translation from multiple tRNA isoacceptors to ensure appropriate protein synthesis rate (Abernathy et al., 2013). The third most abundant functional category of PHX genes is related to cellular processes and signaling functions (28/161 = 17%), where the majority of genes contribute to cell-envelope or membrane biogenesis (11/28). Finally, a small proportion of PHX genes (8/161 = 5%) with unknown or poorly characterized functions were also identified in this study (Figure 1b).
Identification of Highly Expressed Genes in pathogenic S. Enteritidis Strains Using RNA-Seq
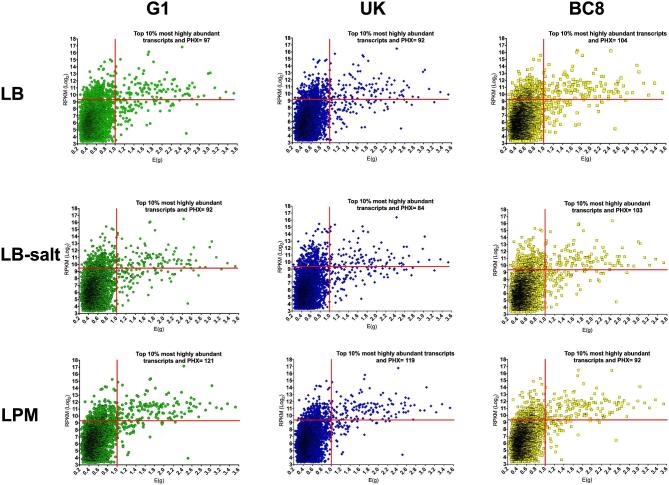
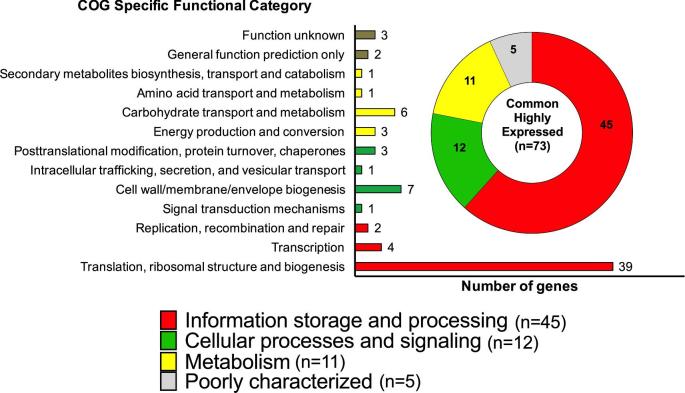
We used RNA-Seq to determine transcriptomic profiles of 3 S. Enteritidis strains (UK, G1 and BC8) cultured at avian physiologic temperature under 2 different growth conditions that resemble the intestinal (LB-salt medium) and intramacrophage (LPM medium) microenvironments encountered in the chicken host. For comparison, we also included the transcriptomes of these S. Enteritidis strains cultured in nutrient rich LB medium from a previously published report (Shah, 2014). In general, Pearson correlation between RPKM-Log2 values from RNA-seq analysis and in silico predicted general expression level (Eg) ranged between 0.42 and 0.52, with limited variation between strains cultured in each microenvironment (Table 2). In a previous study, the correlation between protein abundance and in silico prediction of expression levels in E. coli strain MC4100 grown to exponential phase at 37°C was 0.52, indicating that our results are in agreement with this report (Ishihama et al., 2008). Next, we selected genes that were concurrently predicted as PHX and that also produced the most abundant transcripts (top 10%) in each strain and under each culture condition (Figure 2). The number of such highly expressed genes between the test strains ranged from 92 to 104 (LB), 84 to 103 (LB-salt), and 92 to 121 (LPM) (Figure 2). Subsequently, we identified a core set of 73 highly expressed genes that were common across all strains and all microenvironments tested (Figure 3). We refer to these genes as common highly expressed genes (CHX) for the rest of this manuscript. The broad and specific functional classification of these genes according to cluster of orthologous genes (COG) is shown in Figure 3.
Table 2.
Pearson correlation between RPKM-log2 adjusted in vitro transcriptomic data and in silico predicted general expression levels E(g).
| S. Enteritidis strain | |||
|---|---|---|---|
| Growth medium | G1 | UK | BC8 |
| LB1 | 0.44 | 0.45 | 0.47 |
| LB-salt2 | 0.42 | 0.42 | 0.46 |
| LPM3 | 0.51 | 0.50 | 0.52 |
1LB, Luria Bertani.
2LB-salt, Luria Bertani supplemented with 300 mM of sodium chloride.
3LPM, Low phosphate-low magnesium media.
Figure 2.
In silico predicted levels of gene expression (Eg) and in vitro transcript abundance in RPKM-Log2 adjusted values in S. Enteritidis strains G1, UK, and BC8 cultured in nutrient rich (LB), LB-salt, and LPM media at avian physiologic temperature (42°C). The top right quadrant of each chart corresponds to genes simultaneously predicted to be highly expressed (PHX) and among the top 10% most abundant transcripts. The genes common to these quadrants are considered as common highly expressed (CHX) genes.
Figure 3.
Specific functional classification of common highly expressed (CHX) genes in S. Enteritidis according to cluster of orthologous genes (COG). The inset corresponds to broad functional classification according to COG.
CHX Genes Involved in Information Storage and Processing
The majority of CHX genes (n = 45) identified in this study are involved in information storage and processing (Figure 3). Of these, 39 genes participate in translation, ribosomal structure, and biogenesis by encoding ribosomal proteins part of the large (50S) and small (30S) ribosomal subunits (Suppl. file 4). Most of these genes are identified as essential because of their indispensable role in assembly of the protein synthesis machinery and will not be discussed in detail here [reviewed in Kaczanowska and Ryden-Aulin (2007) and Shajani et al. (2011)]. In the following section, we focus our discussion on non-essential CHX genes that have been reported as dispensable in genetic screening studies conducted in either E. coli, S. Typhimurium, S. Typhi, or S. Enteritidis and, therefore, genetic manipulation is technically feasible in S. Enteritidis for follow-up investigations (Baba et al., 2006; Santiviago et al., 2009; Barquist et al., 2013; Porwollik et al., 2014). The 4 non-essential genes in this functional category include SEN_RS19340 encoding transcription termination factor Rho and cspA, cspC, and cspE genes encoding cold-shock proteins (Baba et al., 2006; Barquist et al., 2013; Porwollik et al., 2014) (Table 3). Termination factor Rho plays a general regulatory function in bacterial transcription (Kriner and Groisman, 2017). Cold-shock proteins are known to function as transcription antiterminators to regulate expression of stress-responsive and virulence-associated genes in E. coli (Bae et al., 2000; Phadtare and Inouye, 2004) and in S. Typhimurium during intra-macrophage infection (Michaux et al., 2017). The CHX genes hupA and hupB encode the proteins HupAB, which bind to DNA and regulate various genes involved in ATP synthesis, glutathione metabolism, peptidoglycan biosynthesis and protein export. HupAB also controls expression of virulence related genes involved in motility (motA and motB), flagellar structure, and assembly (flgE, fliA, fljK) and SPI-2 genes (Guo and Adhya, 2007; Mangan et al., 2011). Salmonella Typhimurium hupA and hupB mutants are attenuated in chickens, pigs, and cattle (Chaudhuri et al., 2013). Taken together, these data suggest that CHX genes belonging to this functional group are involved in controlling or regulating expression of genes relevant for bacterial physiology and virulence. Therefore, it can be expected that the absence of these genes may negatively impact S. Enteritidis infection and persistence in chickens.
Table 3.
Non-ribosomal common highly expressed (CHX) genes in three pathogenic S. Enteritidis strains cultured under distinct environmental conditions (n = 34).
| Locus1 | Gene | Product | COG2 specific functional category | Localization3 | E(g)4 | Hybridization energy (ΔG) in kcal/mol5 | Protein abundance in S. Typhimurium6 |
|---|---|---|---|---|---|---|---|
| SEN_RS19340 | rho | Transcription termination factor Rho | Transcription | Cytoplasmic | 1.42 | -1.56 | Top 10 to 25% |
| SEN_RS18060 | cspA | Cold-shock protein | Transcription | Cytoplasmic | 1.72 | -3.34 | Top 30 to 40% |
| SEN_RS03055 | cspE | Cold-shock protein | Transcription | Cytoplasmic | 1.70 | -3.50 | Top 5 to 10% |
| SEN_RS06220 | cspC | Cold-shock protein CspC | Transcription | Cytoplasmic | 1.82 | -6.00 | Top 5 to 10% |
| SEN_RS20610 | hupA | DNA-binding protein HU-alpha CDS | Replication, recombination, and repair | Cytoplasmic | 2.05 | -5.70 | Top 5% |
| SEN_RS02220 | hupB | DNA-binding protein HU-beta | Replication, recombination, and repair | Cytoplasmic | 1.45 | -6.88 | Top 5% |
| SEN_RS21340 | groEL | Molecular chaperone GroEL | Post-translational modification, protein turnover, and chaperones | Cytoplasmic | 2.38 | -6.07 | Top 5% |
| SEN_RS21335 | groES | molecular chaperone groES | Post-translational modification, protein turnover, and chaperones | Cytoplasmic | 1.65 | -5.12 | Top 5% |
| SEN_RS01975 | Peroxiredoxin | Peroxiredoxin | Post-translational modification, protein turnover, and chaperones | Cytoplasmic | 1.67 | -7.45 | Top 5 to 10% |
| SEN_RS17750 | uspA | Universal stress protein A | Signal transduction mechanisms | Cytoplasmic | 1.54 | -7.89 | Top 5% |
| SEN_RS03070 | tatE | tatE | Intracellular trafficking, secretion, and vesicular transport | Cytoplasmic membrane | 1.12 | -4.30 | 25 to 75% |
| SEN_RS04830 | ompA | Outer membrane protein A | Cell wall/membrane/envelope biogenesis | Outer membrane | 3.03 | -5.59 | Top 5% |
| SEN_RS11705 | ompC | Phosphoporin PhoE | Cell wall/membrane/envelope biogenesis | Outer membrane | 1.63 | -5.44 | Top 5% |
| SEN_RS07685 | nmpC | Phosphoporin PhoE (ompD) | Cell wall/membrane/envelope biogenesis | Outer membrane | 2.99 | -6.09 | Top 5 to 25% |
| SEN_RS04025 | ompX | ompX | Cell wall/membrane/envelope biogenesis | Outer membrane | 2.65 | -5.24 | Top 10% |
| SEN_RS08655 | lpp | Major outer membrane lipoprotein | Cell wall/membrane/envelope biogenesis | Outer membrane | 2.43 | -5.81 | Top 5% |
| SEN_RS03605 | pal | Peptidoglycan-associated lipoprotein Pal | Cell wall/membrane/envelope biogenesis | Outer membrane | 2.49 | -5.71 | Top 5 to 10% |
| SEN_RS01165 | hlpA | Chaperone protein Skp | Cell wall/membrane/envelope biogenesis | Periplasmic | 1.58 | -8.42 | Top 5% |
| SEN_RS09130 | gapA | Aldehyde dehydrogenase | Carbohydrate transport and metabolism | Cytoplasmic | 2.58 | -4.83 | Top 5% |
| SEN_RS14530 | eno | Enolase | Carbohydrate transport and metabolism | Cytoplasmic | 2.57 | -5.07 | Top 5% |
| SEN_RS15150 | fba | Class II fructose-bisphosphate aldolase | Carbohydrate transport and metabolism | Cytoplasmic | 2.89 | -4.13 | Top 5 to 10% |
| SEN_RS12575 | ptsI | Phosphoenolpyruvate–protein phosphotransferase | Carbohydrate transport and metabolism | Cytoplasmic | 1.25 | -8.63 | Top 10 to 25% |
| SEN_RS12570 | ptsH | Phosphocarrier protein HPr | Carbohydrate transport and metabolism | Cytoplasmic | 1.29 | -5.81 | Top 5% |
| SEN_RS12580 | crr | Glucose-specific phosphotransferase enzyme IIA component | Carbohydrate transport and metabolism | Cytoplasmic | 1.94 | -6.87 | Top 5% |
| SEN_RS19120 | atpE | ATP synthase subunit C | Energy production and conversion | Cytoplasmic membrane | 1.61 | -6.07 | NA7 |
| SEN_RS19115 | atpF | ATP synthase subunit B | Energy production and conversion | Cytoplasmic membrane | 1.45 | -3.44 | Top 5 to 10% |
| SEN_RS04545 | pflB | Formate C-acetyltransferase | Energy production and conversion | Cytoplasmic | 3.36 | -7.11 | Top 10% |
| SEN_RS21315 | aspA | Aspartate ammonia-lyase | Amino acid transport and metabolism | Cytoplasmic | 1.61 | -3.26 | Top 10% |
| SEN_RS09630 | acpP | Acyl carrier protein | Secondary metabolites biosynthesis, transport and catabolism | Cytoplasmic | 1.60 | -4.26 | Top 10 to 25% |
| SEN_RS10710 | yeeX | DUF496 domain-containing protein | Unknown | Cytoplasmic | 1.27 | -4.56 | Top 5% |
| SEN_RS20875 | yjbJ | CsbD family protein—stress response | Unknown | Unknown | 1.50 | -6.09 | Top 5% |
| SEN_RS21370 | ecnB | Entericidin B lipoprotein | Unknown | Cytoplasmic membrane | 1.31 | -6.00 | Top 10 to 25% |
| SEN_RS06655 | hns | DNA-binding protein H-NS | General function prediction only | Cytoplasmic | 1.42 | -2.73 | Top 5% |
| SEN_RS13375 | yfiD | Autonomous glycil radical cofactor GrcA | General function prediction only | Cytoplasmic | 1.49 | -3.76 | Top 5 to 10% |
1Locus in reference sequence S. Enteritidis P125109 (NCBI Accession NC_011294).
2COG, cluster of orthologous groups.
3Localization according to PSORTb v3.0 (Yu et al., 2010).
4E(g), predicted general expression level ≥ 1.05 indicates predicted highly expressed gene (PHX) based on codon usage similar to reference highly expressed gene classes.
5Hybridization energy (ΔG) according to RNAcofold V2.4.9 with default parameters (Lorenz et al., 2011).
6Protein abundance averaged by PaxDb protein abundance database version 4.1 (https://pax-db.org/) based on S. Typhimurium strain LT2 cultured under nutrient rich media (LB) at log and stationary phase, and low-magnesium/low-pH media (Adkins et al., 2006; Wang et al., 2015).
7NA, Data Not Available.
CHX Genes Involved in Cellular Processes and Signaling
A total of 12 CHX genes are involved in cellular processing and signaling (Figure 3). Among these, groES, groEL, and SEN_RS01975 are involved in post-translational modification, protein turnover, and chaperone function (Table 3). Whereas, groES is considered an essential gene, the essentiality of groEL appears ambiguous (Baba et al., 2006; Barquist et al., 2013). These genes encode the protein complex GroES-GroEL that prevents aggregation of misfolded proteins in the bacterial cytoplasm and thus contribute to overall bacterial health [reviewed by Hayer-Hartl et al. (2016)]. The CHX gene SEN_RS01975 encodes peroxiredoxin, or alkyl hydroperoxide reductase, a member of the AhpC-TSA superfamily that reduces reactive oxygen species to alcohols to protect bacteria against oxidative stress. In S. Typhimurium, tsa is highly expressed in murine macrophages and contributes to improve survival during oxidative stress in the absence of other catalases (Hebrard et al., 2009). TSA is also an immunodominant antigen in H. pylory, inducing early humoral responses in patients infected with this microorganism (Kimmel et al., 2000; Nurgalieva et al., 2005). The CHX gene uspA encodes the universal stress protein A, whose transcription is induced in S. Typhimurium upon exposure to oxidative, nutritional, or temperature stress (Liu et al., 2007; Karatzas et al., 2008), disinfectants (Karatzas et al., 2008), and bile salts (Hernandez et al., 2012). Upregulation of uspA in presence of bile salts may contribute to adaptation to bile and aid colonization and persistence in liver (Hernandez et al., 2012). S. Typhimurium uspA mutants are attenuated in orally infected mice (Liu et al., 2007), however uspA mutants do not colonize oviduct tissues efficiently and expression of uspA is induced in S. Enteritidis exposed to egg white, suggesting that uspA likely supports invasion, colonization, or persistence of S. Enteritidis in egg contents (Gantois et al., 2008; Raspoet et al., 2011). The CHX gene tatE encodes TatE, an active component of the twin-arginine translocation system that exports fully folded proteins across the bacterial inner membrane (Patel et al., 2014; Eimer et al., 2015). Although the function of tatE is not well characterized, mutations in Tat components induce failure to translocate Tat substrates resulting in cell-envelope defects, sensitivity to bile salts and virulence attenuation in S. Typhimurium (Reynolds et al., 2011; Craig et al., 2013). This potential role in cell-envelope fitness may be related to the survival of S. Enteritidis in egg contents as a recent proteomic survey shows that TatE abundance increases in S. Enteritidis exposed to egg white (Qin et al., 2018).
The CHX genes ompA, ompC, ompD (nmpC), ompX, lpp, and pal encode outer membrane proteins (OMPs), whereas hlpA (skp in E.coli) encodes a periplasmic chaperone (Table 3). OmpA is one of the most abundant structural proteins of the bacterial OM (reviewed in Silhavy et al. (2010)). Expression of ompA increases upon exposure to antimicrobial compounds such as fluoroquinolones (Coldham et al., 2006) and chlorine in S. Typhimurium and S. Enteritidis (Wang et al., 2010). OmpA protein levels decrease after treatment with common disinfectants in S. Typhimurium (Karatzas et al., 2008) and organic acids in S. Enteritidis (de Almeida et al., 2017). Mutants of ompA in S. Enteritidis do not colonize avian tissues efficiently (Zhou et al., 2016), whereas OmpA abundance is increased in S. Enteritidis strains hypersensitive to human serum (Dudek et al., 2016), suggesting that OmpA is critical for cell-membrane homeostasis during infection in vivo. The porins OmpC and OmpD allow passive diffusion of small nutrients across the OM and adjust the OM permeability in response to stress such as high osmolarity, low pH, and toxic compounds (Santiviago et al., 2002; Hernandez et al., 2012; van der Heijden et al., 2016). Reduced expression or mutation of ompC and ompD favors resistance against cephalosporins (Hu et al., 2005) and beta-lactamases in S. Typhimurium (Sun et al., 2009). Moreover, S. Typhimurium ompC mutants are attenuated in orally infected mice, chickens, and pigs (Chatfield et al., 1991; Chaudhuri et al., 2013) and ompC and ompX are required for growth of S. Typhi in presence of bile salts (Langridge et al., 2009). Vaccination and passive transfer of antibodies against OmpD confer protection against S. Paratyphi A in mice (Yang et al., 2012). The extracellular L3 loop of OmpX binds to host complement system and cell membrane proteins, whereas its L1 loop contains an antibody binding domain (Vogt and Schulz, 1999). Taken together, CHX genes encoding these OMPs appear to play important functions in maintenance, response, and adaptation of the OM to the host environment and thereby may contribute to pathogenicity and antigenicity of S. Enteritidis in the chicken host.
The genes lpp, pal, and hlpA encode proteins central to biogenesis and stabilization of the OM. Lpp or Braun lipoprotein links the OM to muropeptides in the adjacent peptidoglycan layer (Braun, 1975). Deletion of lpp induces virulence attenuation and triggers protective immune responses in mice challenged with S. Typhimurium (Fadl et al., 2005; Erova et al., 2016). Reduced levels of Lpp in response to bile salts leads to cell-envelope reorganization in S. Typhimurium (Hernandez et al., 2015), a phenotype that is likely associated with reduced pathogenicity and altered antigenicity in Salmonella. Peptidoglycan associated lipoprotein Pal exists in complex with TolB and facilitates its interaction with OmpA and Lpp to maintain OM integrity (Lloubes et al., 2001). Indeed, pal mutants in S. Typhimurium display cell-envelope aberrations and are impaired in their survival within murine macrophages and colonization of extra-intestinal tissues in mice (Masilamani et al., 2018). The hlpA gene encodes the periplasmic chaperone HlpA (a.k.a. OmpH), a member of the Skp family that prevents non-specific auto aggregation of OM proteins (Burmann et al., 2013). Moreover, hlpA mutants in S. Typhimurium display competitive disadvantage in colonization of extra-intestinal tissues in mice (Rowley et al., 2011). Expression of hlpA is also downregulated in piglets orally infected with Shigella dysenteriae (Kuntumalla et al., 2011). It appears that CHX genes involved in cellular processes and signaling participate in cell-envelope fitness which is intimately coupled to stress response in bacteria. Functions encoded by these CHX genes may enable S. Enteritidis to withstand hazardous antimicrobial compounds and harsh environments encountered in the chicken host with repercussions in pathogenicity and immunogenicity of S. Enteritidis.
CHX Genes Involved in Salmonella Metabolism
A total of 11 CHX genes hold functions in metabolism (Figure 3). Six of these genes (gapA, eno, fba, ptsI, ptsH, and crr) are involved in carbohydrate transport and metabolism (Table 3). The genes gapA, eno, and fba are potentially essential, or at a minimum their essentiality is ambiguous in Salmonella (Hartman et al., 2014; Porwollik et al., 2014). The gapA gene encodes glyceraldehyde-3-phosphate dehydrogenase (GAPDH) that catalyzes oxidation and phosphorylation of glyceraldehyde-3-phosphate (GAP) to 1,3-biphosphoglycerate in the glycolysis pathway. GAPDH from Lactobacillus plantarum can bind to mucin, a major component of the extracellular matrix protecting the intestinal epithelium layer (Kinoshita et al., 2008), whereas GAPDH from E. coli can bind directly to the surface of human enteric epithelial cells (Egea et al., 2007). It is likely that GAPDH aids S. Enteritidis in gaining a foothold in the chicken intestinal tract via direct interactions with chicken enteric epithelial cells. The CHX gene eno encodes Eno protein that catalyzes the conversion of 2-phosphoglycerate into phosphoenolpyruvate in the glycolysis pathway. As a part of the RNaseE/degradosome complex, Eno also influences bacterial morphology during anaerobiosis (Murashko and Lin-Chao, 2017). The CHX gene fba encodes fructose 1,6-biphosphate aldolase, which performs reversible aldol condensation of dihydroxyacetone with GAP during glycolysis (reviewed in (Shams et al., 2014)). Although its function in Salmonella is not fully understood, Fba contributes indirectly to virulence via its role in glycolysis and gluconeogenesis and is important for intracellular growth and survival of microbes such as Mycobacterium tuberculosis (Puckett et al., 2014), Toxoplasma gondii (Blume et al., 2015), and Francisella novicida (Ziveri et al., 2017). Fba is also a promising target to develop antimicrobial compounds that can interrupt central glycolysis (Puckett et al., 2014). The genes ptsH, ptsI, and crr are located in the ptsHI-crr operon that encodes the phosphotransferase HPr, phosphotransferase enzyme I, and the glucose-specific phosphotransferase enzyme IIA, respectively (De Reuse and Danchin, 1988). These genes participate in uptake and phosphorylation of glucose and catabolite repression via the PTSGlc system for carbon metabolism (De Reuse and Danchin, 1988). In S. Typhimurium, ptsH, ptsI, and crr mutants do not replicate efficiently within cultured murine macrophages (Kok et al., 2003; Bowden et al., 2014) and ptsI mutants are attenuated in intraperitoneally challenged mice (Kok et al., 2003). Failure in transport and metabolism of glucose within the Salmonella containing vacuole may contribute to these phenotypes (Kok et al., 2003).
The CHX genes atpE and atpF encode ATP synthase subunits that belong to the ubiquitous F-type (F1F0) ATPase that produces ATP from ADP in a proton gradient mechanism [reviewed by Yoshida et al. (2001) and Ruhle and Leister (2015)]. Interestingly, atpE and atpF mutants of S. Typhimurium are attenuated in orally infected chicken, pigs, and cattle (Chaudhuri et al., 2013). The CHX gene pflB encodes formate-C-acetyltransferase-1 that converts pyruvate and Coenzyme A into formate and acetyl-CoA, and regulates anaerobic glucose metabolism in bacteria (Nnyepi et al., 2007). In E. coli, expression of pflB is induced by oxidative and osmotic stress after exposure to sodium salicylate (Pomposiello et al., 2001) and benzalkonium chloride (Moen et al., 2012), although a specific role in stress response has not been determined. PflB protein levels increase in S. Enteritidis exposed to egg white (Qin et al., 2018), suggesting that pflB may contribute to colonization and survival of S. Enteritidis in egg contents. The CHX gene aspA encodes aspartate ammonia-lyase that catalyzes conversion of aspartate to fumarate and is also involved in catabolism of the amino acids aspartate, glutamine, glutamate, proline, and asparagine as carbon sources. In C. jejuni, aspA mutants showed defects in caecal colonization in orally infected chickens (Guccione et al., 2008), and in Y. pseudotuberculosis, aspA contributes to acid stress tolerance by producing ammonia as byproduct of aspartate catabolism (Hu et al., 2010). The CHX gene acpP is an essential gene that encodes an acyl carrier protein involved in fatty acid biosynthesis and lipid metabolism (Baba et al., 2006; De Lay and Cronan, 2006; Byers and Gong, 2007; Porwollik et al., 2014). The antibiotic class pantothenamide inactivates AcpP by covalent modification, leading to accumulation of inactive AcpP and inhibition of fatty acid synthesis (Zhang et al., 2004). Several CHX genes encoding proteins involved in metabolic activities also have “moonlighting” abilities to perform additional roles in Salmonella. Their abundance is likely advantageous for acquisition of various carbon sources in nutrient-deprived microcompartments in the chicken host, thereby facilitating survival, persistence, and pathogenic potential of S. Enteritidis.
CHX Genes with Poorly Characterized or Unknown Functions
A total of 5 CHX genes (yeeX, yjbJ, ecnB, hns, and yfiD) identified in this study encode poorly characterized proteins (Table 3). Expression of the CHX gene yeeX (SEN_RS10710) is upregulated in avian pathogenic E. coli (APEC) exposed to chicken serum (Li et al., 2011), suggesting that this gene is likely important for cecal colonization of S. Enteritidis in chickens. The CHX gene yjbJ is controlled by rpoS (Weber et al., 2005) and its expression is upregulated in response to cadmium (Worden et al., 2009) and osmotic stress in E. coli (Weber et al., 2006) and S. Typhimurium (Kroger et al., 2013). Thus, yjbJ potentially contributes to stress response in hostile environments such as intestinal lumen, host macrophages, reproductive tract and eggs of chickens infected with S. Enteritidis. The CHX gene ecnB encodes entericidin B membrane lipoprotein, which is the toxin component of the antitoxin/toxin pair ecnAB in E. coli (Bishop et al., 1998). Expression of ecnAB is also regulated by rpoS in response to osmoregulatory and starvation stress in E. coli to modulate competing bacterial populations (Bishop et al., 1998). EcnB promotes death of competing microbes by destabilizing cell membranes via a porin forming mechanism (Bishop et al., 1998). EcnB from E. coli and Enterobacter C6–6 inhibits in vitro growth of Flavobacterium psychrophilum, the causative agent of cold water disease in rainbow trout, implying potential antimicrobial applications (Schubiger et al., 2014). In S. Typhimurium, encB expression is upregulated in the presence of deoxicolate, suggesting that encB may play a role in adaptation to bile and potentially contribute to colonization of liver (Hernandez et al., 2012). Although the role of encB has not been investigated in S. Enteritidis, it would be interesting to determine if ecnB plays a role in inhibition of competing microorganisms during S. Enteritidis colonization of intestinal tissue in chickens.
The CHX gene hns encodes Hns protein, a master regulator of at least 60 different genes, including carbohydrate metabolism and osmotic regulation genes (Dorman, 2007). Hns binds to promoter regions and represses transcription of virulence genes from pathogenicity islands in Shigella flexeneri, enteroinvasive E. coli (Beloin and Dorman, 2003), Vibrio cholerae (Stonehouse et al., 2011), and S. Typhimurium (Dorman, 2007). Hns-deficient mutants in E. coli display increased sensitivity to low pH, high osmolarity, and bile (Erol et al., 2006), conditions that S. Enteritidis encounters in the chicken gut, macrophages, and liver. Hns also silences SPI-1 genes by repressing hilA transcription as well as various SPI-2, SPI-3, and SPI-5 genes in S. Typhimurium (Schechter et al., 2003; Navarre et al., 2006). In S. Enteritidis, expression of hns is downregulated when exposed to chlorine (Wang et al., 2010) and in biofilms exposed to benzalkonium chloride (Mangalappalli-Illathu and Korber, 2006). The CHX gene yfiD encodes the autonomous glycil radical cofactor GrcA, which promotes protection from oxidative stress by reactivating the oxygen sensitive PFL enzyme (pflB, see previous section) (Wagner et al., 2001). In E. coli, yfiD is strongly induced under aerobic and anaerobic acidic conditions and upon exposure to sublethal doses of benzalkonium chloride (Blankenhorn et al., 1999; Wyborn et al., 2002; Moen et al., 2012). Although the functions of these CHX genes in S. Enteritidis remain poorly characterized, the potential role of these genes appears centered around stress responses or associated with expression of Salmonella pathogenicity genes. Therefore, it is of interest to define the roles of these genes and their products in pathogenicity and persistence of S. Enteritidis in the chicken host.
Translation Initiation of CHX Genes is Energetically Favorable
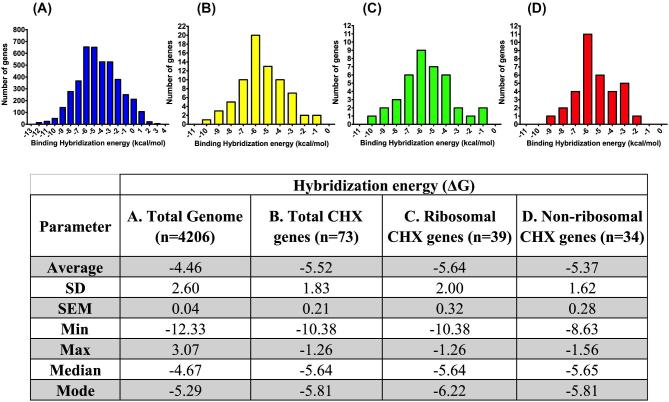
The presence of SD sequences in the 5′ UTR of bacterial mRNA promotes expression of highly expressed genes. The SD sequence interacts with the aSD sequence in the 16S rRNA during recruitment of bacterial mRNA to the small ribosomal subunit 30S (Shine and Dalgarno, 1974). This interaction facilitates translation initiation and efficiency by aligning the ribosome and the mRNA start codon (Shine and Dalgarno, 1974; Vimberg et al., 2007). This SD:aSD pairing relies on complementarity between both sequences (Shine and Dalgarno, 1974) and inherent characteristics such as reduced secondary structures at the 5′UTR of mRNA (Keller et al., 2012; Scharff et al., 2017). The strength of this RNA-RNA pairing or binding can be determined in silico based on hybridization energy (ΔG) (Bernhart et al., 2006; Lorenz et al., 2011; de Almeida et al., 2017). The more negative ΔG values indicate more favorable SD:aSD binding affinity due to small energy requirements (Bernhart et al., 2006; Lorenz et al., 2011). We hypothesized that the CHX genes identified in this study display more favorable SD:aSD binding affinities relative to the entire S. Enteritidis genome. In silico ΔG analysis revealed that the entire protein-coding genome of S. Enteritidis displayed a relatively higher average ΔG (-4.46±0.04 kcal/mol) than the 73 CHX genes identified in this study (ΔG = -5.52±0.21 kcal/mol) (Figure 4). Among the CHX genes, the ΔG of ribosomal (-5.64±0.32 kcal/mol) and non-ribosomal genes (-5.37±0.28 kcal/mol) was lower than the average ΔG for the entire genome (Figure 4). Because ΔG for CHX genes is lower, the SD:aSD interaction is more energetically favorable for CHX genes. These CHX genes bear 2 key characteristics of highly expressed genes: (1) codon usage similar to highly expressed reference genes that results in predicted Eg ≥1.05, and (2) energetically favorable SD:aSD interactions (Table 3). Both key features can drive translation initiation and protein synthesis from CHX genes more efficiently, resulting in higher abundance of proteins encoded by CHX genes. Although no data is available for S. Enteritidis, comprehensive global proteomic analysis of the closely related serovar S. Typhimurium reveals that proteins encoded by CHX genes are highly abundant across various conditions including growth in nutrient rich media (logarithmic and stationary phase) and low magnesium/low pH media (Adkins et al., 2006; Wang et al., 2015). The functional abundance of these CHX-encoded proteins appears confined to the top 5 to 10% of the total proteome, suggesting that they are highly abundant regardless of the condition (Table 3). These findings partly corroborate the relationship between trancriptomic data, SD:aSD binding affinities and protein abundance previously demonstrated in the taxonomically related E.coli (Wei et al., 2017). Collectively, these data suggest that S. Enteritidis CHX genes identified in this study are not only highly expressed at the transcript level, but the proteins encoded by these CHX genes can also be expected to be highly abundant as well. The CHX genes identified in this study likely play important roles during S. Enteritidis infection and persistence in chickens and possibly the environment. Therefore, follow-up functional studies are warranted in biologically-relevant systems.
Figure 4.
Hybridization energy (kcal/mol) between the Shine-Dalgarno (SD) region of S. Enteritidis mRNA and the anti-Shine-Dalgarno (aSD) region in S. Enteritidis 16S rRNA in (A) the entire protein-coding genome, (B) total common highly expressed (CHX) genes, (C) ribosomal CHX genes, and (D) non-ribosomal CHX genes. Hybridization energies were calculated using the software package RNAcofold v2.4.9 with default parameters.
CONCLUSIONS
By combining in silico prediction of gene expression and in vitro transcriptomic analysis of pathogenic S. Enteritidis at chicken physiologic temperature (42°C) under microenvironments resembling those encountered in the chicken host, we identified a core of 73 common highly expressed genes (CHX). We show that a subset (n = 34) of these CHX genes encode proteins that participate in cell-envelope fitness, stress response, nutritional and metabolic fitness in S. Enteritidis. The genes encoding outer membrane proteins OmpA, OmpC, OmpD, and OmpX are particularly relevant for modulation of cell-envelope structure in response to stress, antimicrobials, disinfectants, and also induce immunogenic responses in the chicken host. The CHX genes ptsH, ptsI, and crr are of interest due to their role in metabolism of glucose, which serves as the source of energy in the nutrient-limited Salmonella containing vacuole within the host macrophage (Kok et al., 2003). CHX genes including yjbJ and yfiD are also of interest due to their role in stress responses, whereas hns is directly involved in control of Salmonella pathogenicity genes. The potential role of encB in promoting S. Enteritidis colonization of the chicken gastro-intestinal tract by inhibiting competing microbes is also intriguing. Given the potential role of these genes in infection, the current in vitro study provides a strong foundation to perform biologically-relevant follow-up investigations to clarify the role of this concise set of genes in S. Enteritidis pathobiology in the chicken host. Moreover, our in silico analysis shows that CHX genes display at least 2 features that cooperatively optimize their translation to favor protein abundance. First, the CHX genes display codon usage similar to reference highly expressed genes, resulting in high Eg (≥ 1.05) (Table 3). Second, CHX gene transcripts have energetically favorable SD:aSD binding affinities, which facilitates translation initiation (Table 3 and Figure 4). Therefore, proteins produced from CHX gene transcripts are expected to be abundant in S. Enteritidis irrespective of the surrounding environment. Indeed, the proteins encoded by all CHX genes identified in this study are among the most abundant proteins identified in S. Typhimurium in culture conditions similar to ours (Table 3) (Adkins et al., 2006). Finally, our transcriptomic compendiums obtained from distinct S. Enteritidis strains cultured under multiple growth conditions expand the number of publicly available tools to study this pathogen. Because such transcriptomic compendiums are already available for S. Typhimurium (Kroger et al., 2013; Srikumar et al., 2015; Li et al., 2018), the current study also allows meaningful comparisons to dissect gene function and relevance specific to S. Enteritidis infection in chickens. These efforts will aid in the development of new immunoprophylactic and therapeutic strategies to prevent S. Enteritidis infection in chickens, improve food safety, and prevent human infection with this pathogen.
SUPPLEMENTARY DATA
Supplementary File 4. Ribosomal Common highly expressed (CHX) genes in three S. Enteritidis strains under distinct environmental conditions (n=39).
ACKNOWLEDGMENTS
This project was funded in part by the Agricultural Animal Health Program, College of Veterinary Medicine, Washington State University. Kim Lam Chiok was supported in part by the National Institutes of Health (NIH) Biotechnology Training Program at Washington State University and by the Agricultural Animal Health and Research Program at Washington State University, Pullman, WA.
REFERENCES
- Abernathy J., Corkill C., Hinojosa C., Li X., Zhou H.. 2013. Deletions in the pyruvate pathway of Salmonella typhimurium alter SPI1-mediated gene expression and infectivity. J. Anim. Sci. Biotechnol. 4:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adkins J. N., Mottaz H. M., Norbeck A. D., Gustin J. K., Rue J., Clauss T. R., Purvine S. O., Rodland K. D., Heffron F., Smith R. D.. 2006. Analysis of the Salmonella typhimurium proteome through environmental response toward infectious conditions. Mol. Cell. Proteomics 5:1450–1461. [DOI] [PubMed] [Google Scholar]
- Allard M. W., Luo Y., Strain E., Pettengill J., Timme R., Wang C., Li C., Keys C. E., Zheng J., Stones R., Wilson M. R., Musser S. M., Brown E. W.. 2013. On the evolutionary history, population genetics and diversity among isolates of Salmonella Enteritidis PFGE pattern JEGX01.0004. PLoS One 8:e55254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., Mori H.. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae W., Xia B., Inouye M., Severinov K.. 2000. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc. Natl. Acad. Sci. 97:7784–7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron F., Bonnassie S., Alabdeh M., Cochet M. F., Nau F., Guerin-Dubiard C., Gautier M., Andrews S. C., Jan S. 2017. Global gene-expression analysis of the response of Salmonella Enteritidis to egg white exposure reveals multiple egg white-imposed stress responses. Front Microbiol. 8:829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barquist L., Langridge G. C., Turner D. J., Phan M. D., Turner A. K., Bateman A., Parkhill J., Wain J., Gardner P. P.. 2013. A comparison of dense transposon insertion libraries in the Salmonella serovars Typhi and Typhimurium. Nucleic Acids Res. 41:4549–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloin C., Dorman C. J.. 2003. An extended role for the nucleoid structuring protein H-NS in the virulence gene regulatory cascade of Shigella flexneri. Mol. Microbiol. 47:825–838. [DOI] [PubMed] [Google Scholar]
- Bernhart S. H., Tafer H., Muckstein U., Flamm C., Stadler P. F., Hofacker I. L.. 2006. Partition function and base pairing probabilities of RNA heterodimers. Algorithms Mol. Biol. 1:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop R. E., Leskiw B. K., Hodges R. S., Kay C. M., Weiner J. H.. 1998. The entericidin locus of Escherichia coli and its implications for programmed bacterial cell death. J. Mol. Biol. 280:583–596. [DOI] [PubMed] [Google Scholar]
- Blankenhorn D., Phillips J., Slonczewski J. L.. 1999. Acid- and base-induced proteins during aerobic and anaerobic growth of Escherichia coli revealed by two-dimensional gel electrophoresis. J. Bacteriol. 181:2209–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume M., Nitzsche R., Sternberg U., Gerlic M., Masters S. L., Gupta N., McConville M. J.. 2015. A Toxoplasma gondii Gluconeogenic enzyme contributes to robust central carbon metabolism and is essential for replication and virulence. Cell Host Microbe 18:210–220. [DOI] [PubMed] [Google Scholar]
- Boore A. L., Hoekstra R. M., Iwamoto M., Fields P. I., Bishop R. D., Swerdlow D. L.. 2015. Salmonella enterica infections in the United States and assessment of coefficients of variation: a novel approach to identify epidemiologic characteristics of individual serotypes, 1996–2011. PLoS One 10:e0145416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden S. D., Hopper-Chidlaw A. C., Rice C. J., Ramachandran V. K., Kelly D. J., Thompson A.. 2014. Nutritional and metabolic requirements for the infection of HeLa cells by Salmonella enterica serovar Typhimurium. PLoS One 9:e96266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V. 1975. Covalent lipoprotein from the outer membrane of Escherichia coli. Biochim. Biophys. Acta Rev. Biomembr. 415:335–377. [DOI] [PubMed] [Google Scholar]
- Burmann B. M., Wang C., Hiller S.. 2013. Conformation and dynamics of the periplasmic membrane-protein–chaperone complexes OmpX–Skp and tOmpA–Skp. Nat. Struct. Mol. Biol. 20:1265–1272. [DOI] [PubMed] [Google Scholar]
- Byers D. M., Gong H.. 2007. Acyl carrier protein: structure–function relationships in a conserved multifunctional protein family. Biochem. Cell Biol. 85:649–662. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention 2010. Multistate Outbreak of Human Salmonella Enteritidis Infections Associated with Shell Eggs. Accessed 2 Oct. 2018. https://www.cdc.gov/salmonella/2010/shell-eggs-12-2-10.html. [Google Scholar]
- Chai S. J., White P. L., Lathrop S. L., Solghan S. M., Medus C., McGlinchey B. M., Tobin-D’Angelo M., Marcus R., Mahon B. E.. 2012. Salmonella enterica serotype Enteritidis: increasing incidence of domestically acquired infections. Clin. Infect. Dis. 54 Suppl5:S488–S497. [DOI] [PubMed] [Google Scholar]
- Chatfield S. N., Dorman C. J., Hayward C., Dougan G.. 1991. Role of ompR-dependent genes in Salmonella Typhimurium virulence: mutants deficient in both ompC and ompF are attenuated in vivo. Infect. Immun. 59:449–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri R. R., Morgan E., Peters S. E., Pleasance S. J., Hudson D. L., Davies H. M., Wang J., van Diemen P. M., Buckley A. M., Bowen A. J., Pullinger G. D., Turner D. J., Langridge G. C., Turner A. K., Parkhill J., Charles I. G., Maskell D. J., Stevens M. P.. 2013. Comprehensive assignment of roles for Salmonella typhimurium genes in intestinal colonization of food-producing animals. PLoS Genet 9:e1003456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chousalkar K., Gast R., Martelli F., Pande V.. 2018. Review of egg-related salmonellosis and reduction strategies in United States, Australia, United Kingdom and New Zealand. Crit. Rev. Microbiol. 44:290–303. [DOI] [PubMed] [Google Scholar]
- Coldham N. G., Randall L. P., Piddock L. J., Woodward M. J.. 2006. Effect of fluoroquinolone exposure on the proteome of Salmonella enterica serovar Typhimurium. J. Antimicrob. Chemother. 58:1145–1153. [DOI] [PubMed] [Google Scholar]
- Coombes B. K., Brown N. F., Valdez Y., Brumell J. H., Finlay B. B.. 2004. Expression and secretion of Salmonella Pathogenicity Island-2 Virulence genes in response to acidification exhibit differential requirements of a functional type iii secretion apparatus and SsaL. J. Biol. Chem. 279:49804–49815. [DOI] [PubMed] [Google Scholar]
- Craig M., Sadik A. Y., Golubeva Y. A., Tidhar A., Slauch J. M.. 2013. Twin-arginine translocation system (tat) mutants of Salmonella are attenuated due to envelope defects, not respiratory defects. Mol. Microbiol. 89:887–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida F. A., Pimentel-Filho N. J., Carrijo L. C., Bento C. B. P., Baracat-Pereira M. C., Pinto U. M., de Oliveira L. L., Vanetti M. C. D.. 2017. Acyl homoserine lactone changes the abundance of proteins and the levels of organic acids associated with stationary phase in Salmonella Enteritidis. Microb. Pathog. 102:148–159. [DOI] [PubMed] [Google Scholar]
- De Lay N. R., Cronan J. E.. 2006. Gene-specific random mutagenesis of Escherichia coli in vivo: isolation of temperature-sensitive mutations in the acyl carrier protein of fatty acid synthesis. J. Bacteriol. 188:287–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Reuse H., Danchin A.. 1988. The ptsH, ptsI, and crr genes of the Escherichia coli phosphoenolpyruvate-dependent phosphotransferase system: a complex operon with several modes of transcription. J. Bacteriol. 170:3827–3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denagamage T. N., Jayarao B. M., Wallner-Pendleton E., Patterson P. H., Kariyawasam S.. 2017. A retrospective study of Salmonella Enteritidis isolated from commercial layer flocks. Avian Dis. 61:330–334. [DOI] [PubMed] [Google Scholar]
- Dorman C. J. 2007. H-NS, the genome sentinel. Nat. Rev. Micro. 5:157–161. [DOI] [PubMed] [Google Scholar]
- Drummond D. A., Bloom J. D., Adami C., Wilke C. O., Arnold F. H.. 2005. Why highly expressed proteins evolve slowly. Proc. Natl. Acad. Sci. 102:14338–14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudek B., Krzyzewska E., Kapczynska K., Rybka J., Pawlak A., Korzekwa K., Klausa E., Bugla-Ploskonska G.. 2016. Proteomic analysis of outer membrane proteins from Salmonella enteritidis strains with different sensitivity to human serum. PLoS One 11:e0164069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A. E.. 2002. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea L., Aguilera L., Gimenez R., Sorolla M. A., Aguilar J., Badia J., Baldoma L.. 2007. Role of secreted glyceraldehyde-3-phosphate dehydrogenase in the infection mechanism of enterohemorrhagic and enteropathogenic Escherichia coli: interaction of the extracellular enzyme with human plasminogen and fibrinogen. Int. J. Biochem. Cell Biol. 39:1190–1203. [DOI] [PubMed] [Google Scholar]
- Eimer E., Frobel J., Blummel A. S., Muller M.. 2015. TatE as a Regular constituent of bacterial Twin-arginine protein translocases. J. Biol. Chem. 290:29281–29289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erol I., Jeong K. C., Baumler D. J., Vykhodets B., Choi S. H., Kaspar C. W.. 2006. H-NS controls metabolism and stress tolerance in Escherichia coli O157:H7 that influence mouse passage. BMC Microbiol 6:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erova T. E., Kirtley M. L., Fitts E. C., Ponnusamy D., Baze W. B., Andersson J. A., Cong Y., Tiner B. L., Sha J., Chopra A. K.. 2016. Protective immunity elicited by oral immunization of mice with Salmonella enterica serovar Typhimurium Braun lipoprotein (Lpp) and acetyltransferase (MsbB) mutants. Front Cell Infect. Microbiol. 6:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadl A. A., Sha J., Klimpel G. R., Olano J. P., Niesel D. W., Chopra A. K.. 2005. Murein lipoprotein is a critical outer membrane component involved in Salmonella enterica serovar Typhimurium systemic infection. Infect. Immun. 73:1081–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantois I., Ducatelle R., Pasmans F., Haesebrouck F., Van Immerseel F.. 2008. Salmonella enterica serovar Enteritidis genes induced during oviduct colonization and egg contamination in laying hens. Appl. Environ. Microbiol. 74:6616–6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gast R. K., Guard J., Guraya R., Locatelli A.. 2018. Multiplication in egg yolk and survival in egg albumen of genetically and phenotypically characterized Salmonella enteritidis strains. J. Food Prot. 81:876–880. [DOI] [PubMed] [Google Scholar]
- Gast R. K., Guraya R., Jones D. R., Anderson K. E., Karcher D. M.. 2017. Frequency and duration of fecal shedding of Salmonella enteritidis by experimentally infected laying hens housed in enriched colony cages at different stocking densities. Front Vet. Sci. 4:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons H. S., Kalb S. R., Cotter R. J., Raetz C. R.. 2005. Role of Mg2+ and pH in the modification of Salmonella lipid A after endocytosis by macrophage tumour cells. Mol. Microbiol. 55:425–440. [DOI] [PubMed] [Google Scholar]
- Goncalves-Tenorio A., Silva B. N., Rodrigues V., Cadavez V., Gonzales-Barron U.. 2018. Prevalence of pathogens in poultry meat: a meta-analysis of european published surveys. Foods 7:pii E69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guard-Petter J. 2001. The chicken, the egg and Salmonella enteritidis. Environ. Microbiol. 3:421–430. [DOI] [PubMed] [Google Scholar]
- Guccione E., Leon-Kempis Mdel R., Pearson B. M., Hitchin E., Mulholland F., van Diemen P. M., Stevens M. P., Kelly D. J.. 2008. Amino acid-dependent growth of Campylobacter jejuni: key roles for aspartase (AspA) under microaerobic and oxygen-limited conditions and identification of AspB (Cj0762), essential for growth on glutamate. Mol. Microbiol. 69:77–93. [DOI] [PubMed] [Google Scholar]
- Guo F., Adhya S.. 2007. Spiral structure of Escherichia coli HU provides foundation for DNA supercoiling. Proc. Natl. Acad. Sci. 104:4309–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman H. B., Fell D. A., Rossell S., Jensen P. R., Woodward M. J., Thorndahl L., Jelsbak L., Olsen J. E., Raghunathan A., Daefler S., Poolman M. G.. 2014. Identification of potential drug targets in Salmonella enterica sv. Typhimurium using metabolic modelling and experimental validation. Microbiology 160:1252–1266. [DOI] [PubMed] [Google Scholar]
- Hayer-Hartl M., Bracher A., Hartl F. U.. 2016. The GroEL–GroES chaperonin machine: a nano-cage for protein folding. Trends Biochem. Sci. 41:62–76. [DOI] [PubMed] [Google Scholar]
- Hebrard M., Viala J. P., Meresse S., Barras F., Aussel L.. 2009. Redundant hydrogen peroxide scavengers contribute to Salmonella virulence and oxidative stress resistance. J. Bacteriol. 191:4605–4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heithoff D. M., Shimp W. R., House J. K., Xie Y., Weimer B. C., Sinsheimer R. L., Mahan M. J.. 2012. Intraspecies variation in the emergence of hyperinfectious bacterial strains in nature. PLoS Pathog. 8:e1002647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez S. B., Cava F., Pucciarelli M. G., Garcia-Del Portillo F., de Pedro M. A., Casadesus J.. 2015. Bile-induced peptidoglycan remodelling in Salmonella enterica. Environ. Microbiol. 17:1081–1089. [DOI] [PubMed] [Google Scholar]
- Hernandez S. B., Cota I., Ducret A., Aussel L., Casadesus J.. 2012. Adaptation and preadaptation of Salmonella enterica to bile. PLoS Genet 8:e1002459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W. S., Li P. C., Cheng C. Y.. 2005. Correlation between ceftriaxone resistance of Salmonella enterica serovar Typhimurium and expression of outer membrane proteins OmpW and Ail/OmpX-like protein, which are regulated by BaeR of a two-component system. Antimicrob. Agents Chemother. 49:3955–3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Lu P., Zhang Y., Li L., Chen S.. 2010. Characterization of an aspartate-dependent acid survival system in Yersinia pseudotuberculosis. FEBS Lett. 584:2311–2314. [DOI] [PubMed] [Google Scholar]
- Huang C., Guo Y., Yuan J.. 2014. Dietary taurine impairs intestinal growth and mucosal structure of broiler chickens by increasing toxic bile acid concentrations in the intestine. Poult Sci 93:1475–1483. [DOI] [PubMed] [Google Scholar]
- Ishihama Y., Schmidt T., Rappsilber J., Mann M., Hartl F. U., Kerner M. J., Frishman D.. 2008. Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics 9:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itaya H., Oshita K., Arakawa K., Tomita M.. 2013. GEMBASSY: an EMBOSS associated software package for comprehensive genome analyses. Source Code Biol. Med. 8:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczanowska M., Ryden-Aulin M.. 2007. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol. Mol. Biol. Rev. 71:477–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karatzas K. A., Randall L. P., Webber M., Piddock L. J., Humphrey T. J., Woodward M. J., Coldham N. G.. 2008. Phenotypic and proteomic characterization of multiply antibiotic-resistant variants of Salmonella enterica serovar Typhimurium selected following exposure to disinfectants. Appl. Environ. Microbiol. 74:1508–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S., Barnett M. J., Campbell A. M., Fisher R. F., Mrazek J.. 2003. Predicting gene expression levels from codon biases in -proteobacterial genomes. Proc. Natl. Acad. Sci. 100:7313–7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S., Mrazek J.. 2000. Predicted highly expressed genes of diverse prokaryotic genomes. J. Bacteriol. 182:5238–5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller T. E., Mis S. D., Jia K. E., Wilke C. O.. 2012. Reduced mRNA secondary-structure stability near the start codon indicates functional genes in prokaryotes. Genome Biol. Evol. 4:80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel B., Bosserhoff A., Frank R., Gross R., Goebel W., Beier D.. 2000. Identification of immunodominant antigens from Helicobacter pylori and evaluation of their reactivities with sera from patients with different gastroduodenal pathologies. Infect. Immun. 68:915–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita H., Uchida H., Kawai Y., Kawasaki T., Wakahara N., Matsuo H., Watanabe M., Kitazawa H., Ohnuma S., Miura K., Horii A., Saito T.. 2008. Cell surface Lactobacillus plantarum LA 318 glyceraldehyde-3-phosphate dehydrogenase (GAPDH) adheres to human colonic mucin. J. Appl. Microbiol. 104:1667–1674. [DOI] [PubMed] [Google Scholar]
- Kok M., Bron G., Erni B., Mukhija S.. 2003. Effect of enzyme I of the bacterial phosphoenolpyruvate : sugar phosphotransferase system (PTS) on virulence in a murine model. Microbiology 149:2645–2652. [DOI] [PubMed] [Google Scholar]
- Kriner M. A., Groisman E. A.. 2017. RNA secondary structures regulate three steps of Rho-dependent transcription termination within a bacterial mRNA leader. Nucleic Acids Res. 45:631–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroger C., Colgan A., Srikumar S., Handler K., Sivasankaran S. K., Hammarlof D. L., Canals R., Grissom J. E., Conway T., Hokamp K., Hinton J. C.. 2013. An infection-relevant transcriptomic compendium for Salmonella enterica Serovar Typhimurium. Cell Host Microbe 14:683–695. [DOI] [PubMed] [Google Scholar]
- Kuntumalla S., Zhang Q., Braisted J. C., Fleischmann R. D., Peterson S. N., Donohue-Rolfe A., Tzipori S., Pieper R.. 2011. In vivo versus in vitro protein abundance analysis of Shigella dysenteriae type 1 reveals changes in the expression of proteins involved in virulence, stress and energy metabolism. BMC Microbiol. 11:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge G. C., Phan M. D., Turner D. J., Perkins T. T., Parts L., Haase J., Charles I., Maskell D. J., Peters S. E., Dougan G., Wain J., Parkhill J., Turner A. K.. 2009. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res. 19:2308–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Tivendale K. A., Liu P., Feng Y., Wannemuehler Y., Cai W., Mangiamele P., Johnson T. J., Constantinidou C., Penn C. W., Nolan L. K.. 2011. Transcriptome analysis of avian pathogenic Escherichia coli O1 in chicken serum reveals adaptive responses to systemic infection. Infect. Immun. 79:1951–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Li X., Lv R., Jiang X., Cao H., Du Y., Jiang L., Liu B.. 2018. Global regulatory function of the low oxygen-induced transcriptional regulator LoiA in Salmonella Typhimurium revealed by RNA sequencing. Biochem. Biophys. Res. Commun. 503:2022–2027. [DOI] [PubMed] [Google Scholar]
- Liu W. T., Karavolos M. H., Bulmer D. M., Allaoui A., Hormaeche R. D., Lee J. J., Khan C. M.. 2007. Role of the universal stress protein UspA of Salmonella in growth arrest, stress and virulence. Microb. Pathog. 42:2–10. [DOI] [PubMed] [Google Scholar]
- Lloubes R., Cascales E., Walburger A., Bouveret E., Lazdunski C., Bernadac A., Journet L.. 2001. The Tol-Pal proteins of the Escherichia coli cell envelope: an energized system required for outer membrane integrity? Res. Microbiol. 152:523–529. [DOI] [PubMed] [Google Scholar]
- Lorenz R., Bernhart S. H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P. F., Hofacker I. L.. 2011. ViennaRNA package 2.0. Algorithms Mol. Biol. 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Campbell A., Karlin S.. 2002. Correlations between Shine-Dalgarno sequences and gene features such as predicted expression levels and operon structures. J. Bacteriol. 184:5733–5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier T., Schmidt A., Guell M., Kuhner S., Gavin A. C., Aebersold R., Serrano L.. 2014. Quantification of mRNA and protein and integration with protein turnover in a bacterium. Mol. Syst. Biol. 7:511–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangalappalli-Illathu A. K., Korber D. R.. 2006. Adaptive resistance and differential protein expression of Salmonella enterica serovar Enteritidis biofilms exposed to benzalkonium chloride. Antimicrob. Agents Chemother. 50:3588–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan M. W., Lucchini S., Ó Cróinín T., Fitzgerald S., Hinton J. C., Dorman C. J.. 2011. Nucleoid-associated protein HU controls three regulons that coordinate virulence, response to stress and general physiology in Salmonella enterica serovar Typhimurium. Microbiology 157:1075–1087. [DOI] [PubMed] [Google Scholar]
- Masilamani R., Cian M. B., Dalebroux Z. D.. 2018. Salmonella Tol-Pal Reduces outer membrane glycerophospholipid levels for envelope homeostasis and survival during bacteremia. Infect. Immun. 86:e00173–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaux C., Holmqvist E., Vasicek E., Sharan M., Barquist L., Westermann A. J., Gunn J. S., Vogel J.. 2017. RNA target profiles direct the discovery of virulence functions for the cold-shock proteins CspC and CspE. Proc. Natl. Acad. Sci. USA 114:6824–6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moen B., Rudi K., Bore E., Langsrud S.. 2012. Subminimal inhibitory concentrations of the disinfectant benzalkonium chloride select for a tolerant subpopulation of Escherichia coli with inheritable characteristics. Int. J. Mol. Sci. 13:4101–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashko O. N., Lin-Chao S.. 2017. Escherichia coli responds to environmental changes using enolasic degradosomes and stabilized DicF sRNA to alter cellular morphology. Proc. Natl. Acad. Sci. USA 114:E8025–E8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na Gao B.-G. M., Zhang Yu-Sheng, Song Qin, Chen Ling-Ling, Zhang Hong-Yu. 2009. Gene expression analysis of four radiation-resistant bacteria. 2:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S., Niimura Y., Gojobori T.. 2017. Comparative genomic analysis of translation initiation mechanisms for genes lacking the Shine-Dalgarno sequence in prokaryotes. Nucleic Acids Res. 45:3922–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre W. W., Porwollik S., Wang Y., McClelland M., Rosen H., Libby S. J., Fang F. C.. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313:236–238. [DOI] [PubMed] [Google Scholar]
- Nei M., Kumar S.. 2000. Molecular Evolution and Phylogenetics. Oxford University Press, Oxford, New York. [Google Scholar]
- Niemann G. S., Brown R. N., Gustin J. K., Stufkens A., Shaikh-Kidwai A. S., Li J., McDermott J. E., Brewer H. M., Schepmoes A., Smith R. D., Adkins J. N., Heffron F.. 2011. Discovery of novel secreted virulence factors from Salmonella enterica serovar Typhimurium by proteomic analysis of culture supernatants. Infect. Immun. 79:33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nnyepi M. R., Peng Y., Broderick J. B.. 2007. Inactivation of E. coli pyruvate formate-lyase: role of AdhE and small molecules. Arch. Biochem. Biophys. 459:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurgalieva Z. Z., Conner M. E., Opekun A. R., Zheng C. Q., Elliott S. N., Ernst P. B., Osato M., Estes M. K., Graham D. Y.. 2005. B-cell and T-cell immune responses to experimental Helicobacter pylori infection in humans. Infect. Immun. 73:2999–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R., Vasilev C., Beck D., Monteferrante C. G., van Dijl J. M., Hunter C. N., Smith C., Robinson C.. 2014. A mutation leading to super-assembly of twin-arginine translocase (Tat) protein complexes. Biochim. Biophys. Acta – Mol. Cell Res. 1843:1978–1986. [DOI] [PubMed] [Google Scholar]
- Phadtare S., Inouye M.. 2004. Genome-wide transcriptional analysis of the cold shock response in wild-type and cold-sensitive, quadruple-csp-deletion strains of Escherichia coli. J. Bacteriol. 186:7007–7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomposiello P. J., Bennik M. H., Demple B.. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183:3890–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porwollik S., Santiviago C. A., Cheng P., Long F., Desai P., Fredlund J., Srikumar S., Silva C. A., Chu W., Chen X., Canals R., Reynolds M. M., Bogomolnaya L., Shields C., Cui P., Guo J., Zheng Y., Endicott-Yazdani T., Yang H. J., Maple A., Ragoza Y., Blondel C. J., Valenzuela C., Andrews-Polymenis H., McClelland M.. 2014. Defined single-gene and multi-gene deletion mutant collections in Salmonella enterica sv Typhimurium. PLoS One 9:e99820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puckett S., Trujillo C., Eoh H., Marrero J., Spencer J., Jackson M., Schnappinger D., Rhee K., Ehrt S.. 2014. Inactivation of fructose-1,6-bisphosphate aldolase prevents optimal co-catabolism of glycolytic and gluconeogenic carbon substrates in Mycobacterium tuberculosis. PLoS Pathog. 10:e1004144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X., He S., Zhou X., Cheng X., Huang X., Wang Y., Wang S., Cui Y., Shi C., Shi X.. 2019. Quantitative proteomics reveals the crucial role of YbgC for Salmonella enterica serovar Enteritidis survival in egg white. Int. J. Food Microbiol. 289:115–126. [DOI] [PubMed] [Google Scholar]
- Raspoet R., Appia-Ayme C., Shearer N., Martel A., Pasmans F., Haesebrouck F., Ducatelle R., Thompson A., Van Immerseel F.. 2014. Microarray-based detection of Salmonella enterica serovar Enteritidis genes involved in chicken reproductive tract colonization. Appl. Environ. Microbiol. 80:7710–7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raspoet R., Gantois I., Devloo R., Martel A., Haesebrouck F., Pasmans F., Ducatelle R., Van Immerseel F.. 2011. Salmonella enteritidis universal stress protein (usp) gene expression is stimulated by egg white and supports oviduct colonization and egg contamination in laying hens. Vet. Microbiol. 153:186–190. [DOI] [PubMed] [Google Scholar]
- Raspoet R., Shearer N., Appia-Ayme C., Haesebrouck F., Ducatelle R., Thompson A., Van Immerseel F.. 2014. A genome-wide screen identifies Salmonella Enteritidis lipopolysaccharide biosynthesis and the HtrA heat shock protein as crucial factors involved in egg white persistence at chicken body temperature. Poult. Sci. 93:1263–1269. [DOI] [PubMed] [Google Scholar]
- Razdan A., Pettersson D., Pettersson J.. 1997. Broiler chicken body weights, feed intakes, plasma lipid and small-intestinal bile acid concentrations in response to feeding of chitosan and pectin. Br. J. Nutr. 78:283–291. [DOI] [PubMed] [Google Scholar]
- Reynolds M. M., Bogomolnaya L., Guo J., Aldrich L., Bokhari D., Santiviago C. A., McClelland M., Andrews-Polymenis H.. 2011. Abrogation of the twin arginine transport system in Salmonella enterica serovar Typhimurium leads to colonization defects during infection. PLoS One 6:e15800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha E. P., Danchin A.. 2004. An analysis of determinants of amino acids substitution rates in bacterial proteins. Mol. Biol. Evol. 21:108–116. [DOI] [PubMed] [Google Scholar]
- Rollenhagen C., Bumann D.. 2006. Salmonella enterica highly expressed genes are disease specific. Infect. Immun. 74:1649–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley G., Skovierova H., Stevenson A., Rezuchova B., Homerova D., Lewis C., Sherry A., Kormanec J., Roberts M.. 2011. The periplasmic chaperone Skp is required for successful Salmonella Typhimurium infection in a murine typhoid model. Microbiology 157:848–858. [DOI] [PubMed] [Google Scholar]
- Ruhle T., Leister D.. 2015. Assembly of F1F0-ATP synthases. Biochim. Biophys. Acta– Bioenerg. 1847:849–860. [DOI] [PubMed] [Google Scholar]
- Santiviago C. A., Fuentes J. A., Bueno S. M., Trombert A. N., Hildago A. A., Socias L. T., Youderian P., Mora G. C.. 2002. The Salmonella enterica sv. Typhimurium smvA, yddG and ompD (porin) genes are required for the efficient efflux of methyl viologen. Mol. Microbiol. 46:687–698. [DOI] [PubMed] [Google Scholar]
- Santiviago C. A., Reynolds M. M., Porwollik S., Choi S. H., Long F., Andrews-Polymenis H. L., McClelland M.. 2009. Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice. PLoS Pathog. 5:e1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharff L. B., Ehrnthaler M., Janowski M., Childs L. H., Hasse C., Gremmels J., Ruf S., Zoschke R., Bock R.. 2017. Shine-Dalgarno sequences play an essential role in the translation of plastid mRNAs in Tobacco. Plant Cell 29:3085–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter L. M., Jain S., Akbar S., Lee C. A.. 2003. The small nucleoid-binding proteins H-NS, HU, and Fis affect hilA expression in Salmonella enterica serovar Typhimurium. Infect. Immun. 71:5432–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubiger C. B., Orfe L. H., Ponnerassery S. S., Cain K. D., Shah D. H., Call D. R.. 2014. Entericidin is requisite for a probiotic treatment (Enterobacter C6-6) to protect trout from cold-water disease challenge. Appl. Environ. Microbiol. 81:658–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah D. H. 2014. RNA sequencing reveals differences between the global transcriptomes of Salmonella enterica serovar enteritidis strains with high and low pathogenicities. Appl. Environ. Microbiol. 80:896–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah D. H., Casavant C., Hawley Q., Addwebi T., Call D. R., Guard J.. 2012. Salmonella enteritidis strains from poultry exhibit differential responses to acid stress, oxidative stress, and survival in the egg albumen. Foodborne Path. Dis. 9:258–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah D. H., Elder J. R., Chiok K. L., Paul N. C.. 2017a. Chapter 10 - genetic basis of Salmonella enteritidis pathogenesis in chickens. Pages 187–208 in Producing Safe Eggs. Ricke S. C., Gast R. K. eds. Academic Press, San Diego. [Google Scholar]
- Shah D. H., Paul N. C., Sischo W. C., Crespo R., Guard J.. 2017b. Population dynamics and antimicrobial resistance of the most prevalent poultry-associated Salmonella serotypes. Poult. Sci. 96:687–702. [DOI] [PubMed] [Google Scholar]
- Shah D. H., Zhou X., Addwebi T., Davis M. A., Call D. R.. 2011. In vitro and in vivo pathogenicity of Salmonella enteritidis clinical strains isolated from North America. Arch. Microbiol. 193:811–821. [DOI] [PubMed] [Google Scholar]
- Shah D. H., Zhou X., Addwebi T., Davis M. A., Orfe L., Call D. R., Guard J., Besser T. E.. 2011. Cell invasion of poultry-associated Salmonella enterica serovar Enteritidis isolates is associated with pathogenicity, motility and proteins secreted by the type III secretion system. Microbiology 157:1428–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajani Z., Sykes M. T., Williamson J. R.. 2011. Assembly of bacterial ribosomes. Annu. Rev. Biochem. 80:501–526. [DOI] [PubMed] [Google Scholar]
- Shams F., Oldfield N. J., Wooldridge K. G., Turner D. P.. 2014. Fructose-1,6-bisphosphate aldolase (FBA)–a conserved glycolytic enzyme with virulence functions in bacteria: ‘ill met by moonlight’. Biochim. Soc. Trans. 42:1792–1795. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L.. 1974. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. 71:1342–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy T. J., Kahne D., Walker S.. 2010. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2:a000414–a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva C. A., Blondel C. J., Quezada C. P., Porwollik S., Andrews-Polymenis H. L., Toro C. S., Zaldivar M., Contreras I., McClelland M., Santiviago C. A.. 2012. Infection of mice by Salmonella enterica serovar Enteritidis involves additional genes that are absent in the genome of serovar Typhimurium. Infect. Immun. 80:839–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikumar S., Kroger C., Hebrard M., Colgan A., Owen S. V., Sivasankaran S. K., Cameron A. D., Hokamp K., Hinton J. C.. 2015. RNA-seq brings new insights to the intra-macrophage transcriptome of Salmonella Typhimurium. PLoS Pathog. 11:e1005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stonehouse E. A., Hulbert R. R., Nye M. B., Skorupski K., Taylor R. K.. 2011. H-NS binding and repression of the ctx promoter in Vibrio cholerae. J. Bacteriol. 193:979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Berg O. G., Roth J. R., Andersson D. I.. 2009. Contribution of gene amplification to evolution of increased antibiotic resistance in Salmonella typhimurium. Genetics 182:1183–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden J., Reynolds L. A., Deng W., Mills A., Scholz R., Imami K., Foster L. J., Duong F., Finlay B. B.. 2016. Salmonella rapidly regulates membrane permeability to survive oxidative stress. mBio 7:e01238–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBogelen R. A., Hutton M. E., Neidhardt F. C.. 1990. Gene-protein database of Escherichia coli K - 12: Edition 3. Electrophoresis 11:1131–1166. [DOI] [PubMed] [Google Scholar]
- Vimberg V., Tats A., Remm M., Tenson T.. 2007. Translation initiation region sequence preferences in Escherichia coli. BMC Mol Biol 8:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt J., Schulz G. E.. 1999. The structure of the outer membrane protein OmpX from Escherichia coli reveals possible mechanisms of virulence. Structure 7:1301–1309. [DOI] [PubMed] [Google Scholar]
- Wagner A. F., Schultz S., Bomke J., Pils T., Lehmann W. D., Knappe J.. 2001. YfiD of Escherichia coli and Y06I of bacteriophage T4 as autonomous glycyl radical cofactors reconstituting the catalytic center of oxygen-fragmented pyruvate formate-lyase. Biochem. Biophys. Res. Commun. 285:456–462. [DOI] [PubMed] [Google Scholar]
- Wang M., Herrmann C. J., Simonovic M., Szklarczyk D., von Mering C.. 2015. Version 4.0 of PaxDb: protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics 15:3163–3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Phillippy A. M., Deng K., Rui X., Li Z., Tortorello M. L., Zhang W.. 2010. Transcriptomic responses of Salmonella enterica serovars Enteritidis and Typhimurium to chlorine-based oxidative stress. Appl. Environ. Microbiol. 76:5013–5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A., Kogl S. A., Jung K.. 2006. Time-dependent proteome alterations under osmotic stress during aerobic and anaerobic growth in Escherichia coli. J. Bacteriol. 188:7165–7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H., Polen T., Heuveling J., Wendisch V. F., Hengge R.. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: S-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187:1591–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Silke J. R., Xia X.. 2017. Elucidating the 16S rRNA 3' boundaries and defining optimal SD/aSD pairing in Escherichia coli and Bacillus subtilis using RNA-Seq data. Sci. Rep. 7:17639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden C. R., Kovac W. K., Dorn L. A., Sandrin T. R.. 2009. Environmental pH affects transcriptional responses to cadmium toxicity in Escherichia coli K-12 (MG1655). FEMS Microbiol. Lett. 293:58–64. [DOI] [PubMed] [Google Scholar]
- Wyborn N. R., Messenger S. L., Henderson R. A., Sawers G., Roberts R. E., Attwood M. M., Green J.. 2002. Expression of the Escherichia coli yfiD gene responds to intracellular pH and reduces the accumulation of acidic metabolic end products. Microbiology 148:1015–1026. [DOI] [PubMed] [Google Scholar]
- Yang T. C., Ma X. C., Liu F., Lin L. R., Liu L. L., Liu G. L., Tong M. L., Fu Z. G., Zhou L.. 2012. Screening of the Salmonella Paratyphi A CMCC 50973 strain outer membrane proteins for the identification of potential vaccine targets. Mol. Med. Rep. 5:78–83. [DOI] [PubMed] [Google Scholar]
- Yim L., Betancor L., Martinez A., Bryant C., Maskell D., Chabalgoity J. A.. 2011. Naturally occurring motility-defective mutants of Salmonella enterica serovar Enteritidis isolated preferentially from nonhuman rather than human sources. Appl. Environ. Microbiol. 77:7740–7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim L., Betancor L., Martinez A., Giossa G., Bryant C., Maskell D., Chabalgoity J. A.. 2010. Differential phenotypic diversity among epidemic-spanning Salmonella enterica serovar Enteritidis isolates from humans or animals. Appl. Environ. Microbiol. 76:6812–6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M., Muneyuki E., Hisabori T.. 2001. ATP synthase — a marvellous rotary engine of the cell. Nat. Rev. Mol. Cell. Biol. 2:669–677. [DOI] [PubMed] [Google Scholar]
- Yu N. Y., Wagner J. R., Laird M. R., Melli G., Rey S., Lo R., Dao P., Sahinalp S. C., Ester M., Foster L. J., Brinkman F. S.. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. M., Frank M. W., Virga K. G., Lee R. E., Rock C. O., Jackowski S.. 2004. Acyl carrier protein is a cellular target for the antibacterial action of the pantothenamide class of pantothenate antimetabolites. J. Biol. Chem. 279:50969–50975. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Zhou J., Wang D., Gao Q., Mu X., Gao S., Liu X.. 2016. Evaluation of ompA and pgtE genes in determining pathogenicity in Salmonella enterica serovar Enteritidis. Vet. J. 218:19–26. [DOI] [PubMed] [Google Scholar]
- Ziveri J., Tros F., Guerrera I. C., Chhuon C., Audry M., Dupuis M., Barel M., Korniotis S., Fillatreau S., Gales L., Cahoreau E., Charbit A.. 2017. The metabolic enzyme fructose-1,6-bisphosphate aldolase acts as a transcriptional regulator in pathogenic Francisella. Nat. Commun. 8:853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.