Abstract
The expanding field of precision gene editing is empowering researchers to directly modify DNA. Gene editing is made possible using synonymous technologies: 1) a DNA targeting platform to molecularly locate user-selected genomic sequences, and 2) an associated biochemical activity that serves as a functional editor. The advent of accessible DNA-targeting molecular systems, like transcription activator like effectors (TALEs) and Clustered Regularly Interspersed Short Palindromic Repeats (CRISPR)-Cas9 gene editing systems, has unlocked the ability to target nearly any DNA sequence with nucleotide-level precision. Progress has also been made in how we can harness endogenous DNA repair machineries to functionally manipulate genetic sequences. The more that is understood about how DNA damage results in deletions, insertions, and modifications the more predictably mutable the genome becomes. These genomic targeting platforms are also useful for locus-specific epigenetic changes and transcriptional enhancement and suppression. This new genome engineering technology builds on a long history of renal science, enabling new animal models of disease as well as novel therapeutic options.
Keywords: Genome Engineering, Gene Editing, DNA Repair, ZFN, TALEN, CRISPR, Cas9, Cpf1, Cas12a, Base Editor, NHEJ, HDR, HR, MMEJ, Renal Disease
Introduction
Engineering DNA is undergoing a far-reaching change with the advent of a new array of genome engineering tools such as CRISPR/Cas9 and TALENs. With the advent of accessible programmable DNA nucleases, genome engineering now includes the growing capability to edit the genome. The initial molecular biologist’s toolbox was largely confined to simple rearrangements of DNA through the use of restriction enzymes and ligases in vitro or in bacteria. These techniques are inherently limited by the intrinsic sequence-specific nature of restriction enzymes and, consequently, by the functional inability to edit higher order organisms. Indeed, the primary options for manipulating eukaryotic genomes were random mutations inducible by chemicals or radiation, directed breeding/evolution through matching of parents with desired alleles, or the use of species-compatible transposon elements or viruses. Despite their limitations, these approaches enabled many advances such as transgenic expression, homologous recombination in embryonic stem (ES) cells to generate knock outs/ins, and led to initial attempts at gene therapy by exogenous gene expression.
The last seven years have, however, been a time of rapid change for the biologists’ toolbox, while also empowering a new way of thinking about DNA editing. This review focuses on the development, mechanism, and accessibility of the different DNA editing platforms as well as their targeted interaction with the genome. We will also cover the current understanding of DNA biochemical functions used to make precise modifications for modern precision gene editing.
Custom programmable gene editors (Fig. 1) started with the advent of the zinc finger nuclease (ZFN)1,2, advanced in precision and usability by the development of the transcription activator-like effector nuclease (TALEN)3,4 and made highly accessible through the development of CRISPR-based systems52. Their shared ability to target unique DNA locations in a targeted genome is an essential core function and continues to be paramount for other editing systems and for others currently in development.
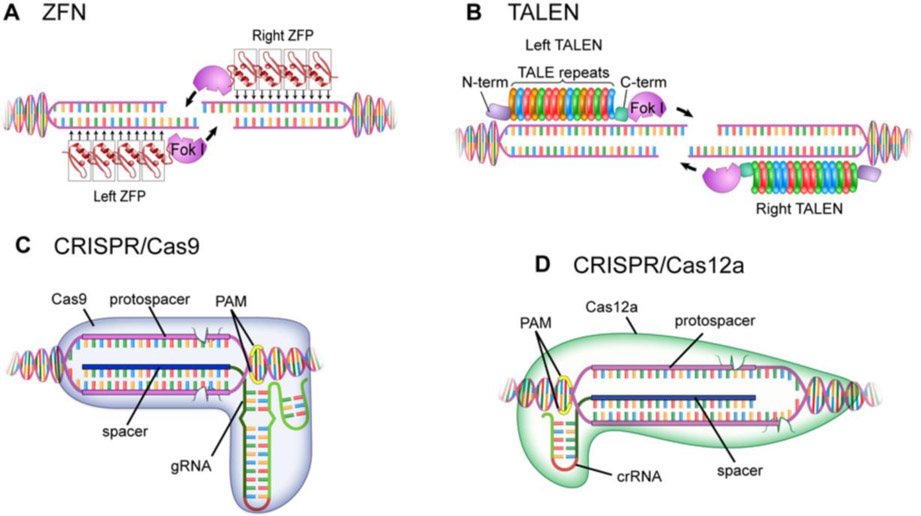
Figure 1. Commonly Used Programmable DNA Platforms:
A diagram showing programmable DNA binding platforms that recognize double-stranded DNA (dsDNA). A. A pair of 4-Finger Zinc Finger proteins binding to each side of the desired double-stranded break (DSB) location in the DNA. Each Zinc Finger (ZF) domain binds three bases of DNA; multiple ZF domains can be stringed together to bind longer stretches of DNA. When bound, the attached FokI nuclease (N) dimers become close enough in proximity to activate and catalyze a double stranded DNA break. B. A pair of Transcription Activator Like Effector domains (TALEs) bound to each side of the preferred DSB position. TALE domains consist of a series of 35 amino acid repeats attached in sequence. Each of these motifs binds to a single specific DNA base and can be strung together to recognize diverse DNA sequences. C. The CRISPR/Cas9 endonuclease system functions through the interaction of a RNA guide with a single protein, Cas9. The RNA guide consists of two domains, a constant poly-hairpin structure that interacts with the Cas9 protein and a programmable guide region that targets DNA through standard Watson-Crick base pairing. Upon binding its target region by interrogating and unwinding (melting) the dsDNA, the Cas9 protein induces a blunt double stranded break. D. Another CRISPR system makes use of different class of guide RNA coupled with a different constant protein, Cas12a. Targeting is again determined by Watson-Crick base pairing between the guide RNA and the DNA, following which Cas12a induces a DSB with its signature overhang.
The versatility of precision gene editing has grown through the addition of various enzymatic activities to these programmable DNA binding platforms. Upon interacting with their defined loci, these functionalized programmable DNA binding platforms activate endogenous DNA repair pathways that serve as the molecular pathways to generate a wide range of DNA sequence edits. Current genomic editing uses at least five different DNA repair pathways. A targeted double-strand break (DSB) from an associated nuclease can induce the nonhomologous end-joining (NHEJ) pathway; classical homologous recombination (HR), the more recently deployed method of single strand template repair (SSTR), or microhomology mediated end joining (MMEJ) to induce DNA sequence changes (Fig. 2). A fifth leverages the endogenous base mismatch repair to edit DNA without rupturing the double strand of DNA (Fig. 2). In turn, each of these mechanisms suggests not only the preferred cellular process used, but also the resultant mutation signatures and their potential uses in genomic sciences.
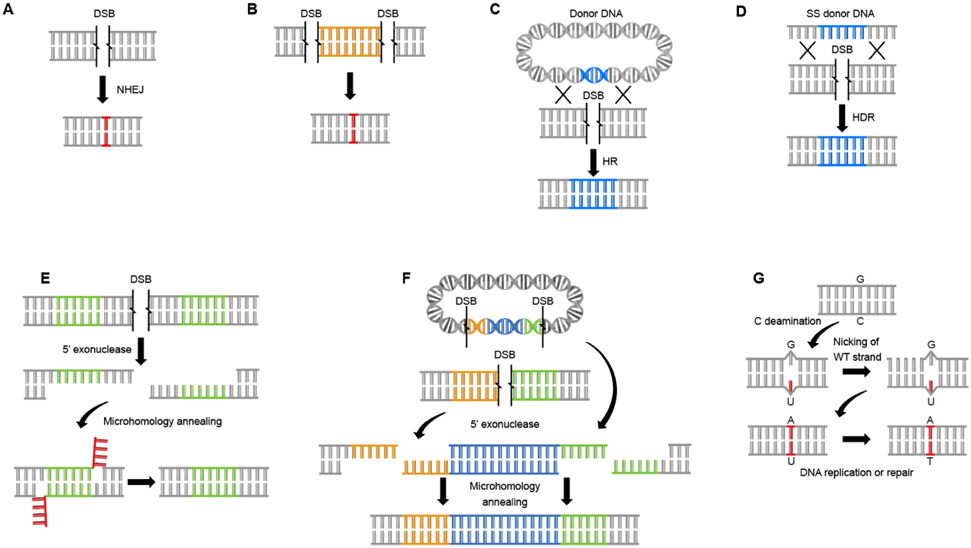
Figure 2. Associated Biochemical Activities Critical For Precision Gene Editing:
Diagram showing the different cellular repair machineries and corresponding biochemical functions critical for gene editing. A. Repair of a DSB by Non-Homologous End Joining (NHEJ) introduces mutation through the creation of small insertions or deletions (indels). B. NHEJ repair can delete long segments of DNA (whole genes) by creating two double stranded breaks and removing the intervening region from the chromosome C. Homologous Recombination (HR) can be used to insert exogenous DNA though the introduction of an exogenous template flanked by large (often >500 base pairs) dsDNA sequences homologous the sequence adjacent to the DSB. D. Oligo-directed HDR can introduce small changes through the introduction of a short single-strand DNA (ssDNA) template flanked with homology matching the regions to either side of the DSB. E. Micro-Homology Mediated End Joining can be used to create small reproducible deletions. This repair pathway functions by annealing small homologous regions on each side of the DSB. Unbound DNA flaps are removed and ends are ligated resulting in the removal of one homology arm and the intervening region. F. MMEJ can also be used to insert exogenous DNA through the introduction of a template with matching microhomology arms. G. Single nucleotide polymorphisms can be introduced without a DSB using a base editor. By targeting a cytosine deaminase to a specific target in the genome it is possible to convert a cytosine to a uracil resulting in a mismatch base. This mismatch is then recognized and repaired, generating a single base change depending on which strand is chosen as repair template, this can be selected by design by repeated nicking of the strand to be modified.
Successful genome engineering depends on the cellular context as well, with editing efficacy being modulated by cell cycle stage, cell type, and other conditions within the cellular environment. In basic science research, DNA editing can be used to study gene function and to create engineered lines of experimental animals. Additionally, this tool represents enormous potential for clinical applications and the generation of accurate disease models in a number of cellular systems. With this new technology, complex genetic disorders can be explored where multiple genetic events and their interactions can be mimicked to understand common renal diseases. Genome engineering allows these diseases to be modeled in diverse cellular and animal models and used to explore both pathology as well as the science underlying health.
Programmable DNA Targeting Platforms (Fig. 1)
The First Precision Edits: ZFNs
The first custom programmable nuclease was developed from the ~30 amino acid Cys2-His2 Zinc-Finger (ZF) domain, the most abundant DNA binding protein motif utilized in eukaryotic transcription factors1,5-7 (Fig 1A). In each ZF domain three amino acid clusters or “fingers” each recognize a single specific DNA base and mediate DNA binding7-9. Therefore, in principle, through manipulation of the amino acids in each finger, one can create a ZF that binds to any of the 64 possible 3 base permutations10. Multiple ZF domains can then be fused together to create a single poly-protein capable of binding long stretches of DNA11-13.
Although ZFs are still in use today, this programmable DNA binding platform has restricted accessibility due to limitations on synthesis and targeting flexibility. For example, the generation of custom ZFs can be a technically arduous task, as synthesized tandem ZF repeats often exhibit unpredictable context-specific interference that mitigate binding and reduce or alter specificity7. Critically, only a subset of the genome has been historically targetable due to these technical limitations for this system14. However, recent progress has improved the ZFN platform15
ZF scientists were also the first to selectively activate DNA repair pathways. Discussed more in detail below, creating an editor from the ZF DNA binding domain required the attachment of a DNA endonuclease since ZFs have no intrinsic editing activity. This was achieved by the fusion of the FokI endonuclease domain to the C-terminal end of a series of tandem ZF domains to create the Zinc-Finger Nuclease (ZFN)1,16. This catalyzes a double-strand break (DSB) between the programmatically targeted ZF binding domains when two ZFNs are bound enabling FokI to dimerize and become catalytically active6. The resulting induced double-stranded break repair (Fig. 2) is then harnessed for precision gene editing17-19. FokI is thus deployed as a nuclease partner for a custom DNA binding protein, as the enzymatic domain can be functionally uncoupled from its inherent, non-programmable binding domain while maintaining activity20,21. Additionally, the FokI nuclease requires the formation of a dimer to be catalytically active. The specificity of the ZF binding platform was thereby enhanced by requiring two ZFNs to bind in close proximity in order to bring together the FokI subunits and enable DNA cutting activity16. The specificity of the ZFN system was further increased by modification of the FokI dimerization interface to create an obligate heterodimer system22-24.
While ZFNs were a critical advancement in the technology of gene editing, the broader deployment of ZFNs has been modest due to the engineering challenges associated with context-dependent assembly constraints25-27. The long-term impact by ZFNs on the field of gene editing cannot be overstated, however. Nearly all of the commonly used approaches today were originally tested using this first generation programmable DNA editing platform, and ZFNs are the first custom nuclease system to be deployed in clinical trials. Oftentimes working in relative obscurity (Fig 4), these innovators developed the core technology many use today for modern gene editing.
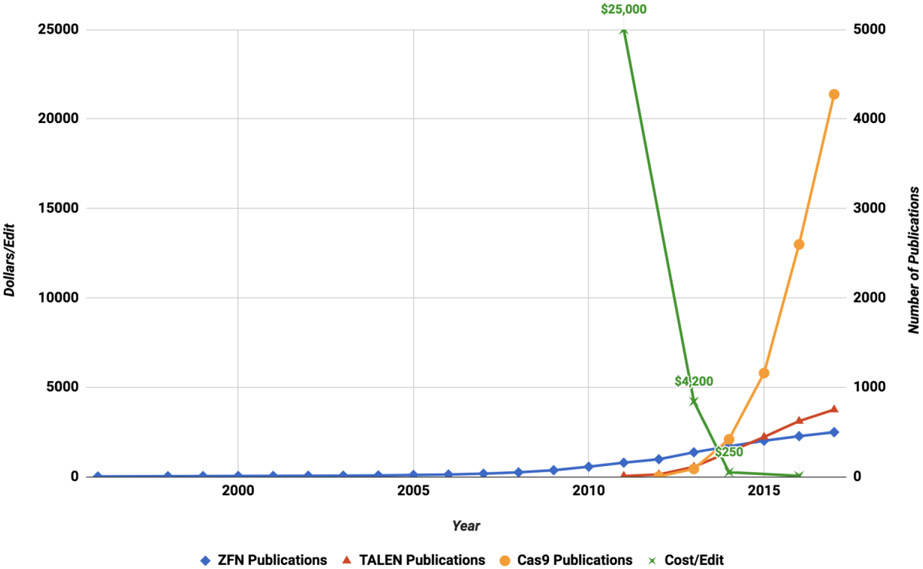
Figure 4. Exponential Reduction in Cost of Gene Editing Tools and Subsequent Rapid Growth of Deployment in Scientific Publications.
Left axis – chart shows representative commercial costs of gene editing tools from ZFNs in 2011 to TALENs in 2013 and CRISPR-Cas9 in 2016.
Right axis -chart shows the number of publications in PubMed using each indicated gene editing platform since 1995. The exponential reduction in cost and the greatly increased access to these new tools is reminiscent of the Moore’s Law of continual reduced cost underlying computer technology.
The Genome Unlocked: TALENs
After nearly two decades of pioneering ZFN work, a new programmable DNA binding platform was developed from DNA binding factors identified in the plant bacterial pathogen Xanthomonas28,29 (Fig 1B). The Transcription Activator-Like Effector (TALE) domains consist of a series of modular 33-35 amino acid repeats, each repeat binding a single specific DNA base28,30. Two hyper-variable residues central to the repeat determine the binding specificity of each repeat28,31. These Repeat Variable Di-residues (RVDs) are flanked on either side by constant amino acid sequences. The 33-35 amino acid repeats can be stitched together to form a long polypeptide capable of binding long stretches of DNA in excess of 20 bases32. TALEs represent a second-generation programmable DNA binding platform.
Based on the prior ZF work, different functional domains were added to the TALE DNA binding system to access the endogenous cellular repair mechanisms (Fig 2B). Nuclease activity is conferred via the fusion of the TALE domain to monomers of the FokI endonuclease creating the TALE Nuclease (TALEN)3,29. A pair of TALEN arms is needed to target the cut site and proximate the FokI monomers3,29. Therefore, a targeted TALEN, made up of two arms and a spacer region between them, can be designed to recognize over 40 bases of DNA29. As a result of both this high base pair recognition count and the high innate specificity of each TALE repeat module, the commonly deployed TALEN systems exhibit high intrinsic binding accuracy with low off-targeting profiles 33-35.
Another important difference between the TALEN and ZFN systems is the TALENs’ lack of any known context-specific assembly constraints4,36. No RVD combinations have yet been identified as complicating either binding ability or specificity, and the targeting constraints of the TALEN system are also very few. The only regular constraint is the common inclusion of a 5’ T recognition motif in front of each TALEN arm for enhanced binding affinity; although this sequence requirement has been eliminated using next generation TALE scaffolds37. In addition, some reports include a differential binding by TALENs in a region of DNA containing a methylated CpG island38,39. TALENs have also proven to be far more easily assembled than ZFNs, with several published techniques for high-throughput assembly of TALEN that use as little as a single tube for synthesis29,39-42. TALEs have the fewest restrictions imposed on the design of the DNA binding system and thus represent the single-most programmable DNA platform to date. The increase in facility of use can be seen in the increase in gene editing publications in the wake of TALEN development (Fig 4). However, the requirement for the synthesis of two custom proteins for each experimental TALEN setup represents a barrier to the use of TALEs in many labs, especially to those with limited experience in genome engineering.
Gene Editing Democratized: CRISPR
Gene editing went mainstream when a naturally occurring ribonucleoprotein (RNP) complex was discovered to function as a new class of DNA recognition domain. In this system, a common protein binds to a short guiding RNA that is used to target the resulting RNP to the specific region in the genome (Fig 1C). Initially identified as an integral component of many bacterial immune systems, the first characterized CRISPR protein (Cas9) was shown to function endogenously to target and degrade invasive phage or plasmid DNA43-47. This defense system functions via the storage of previously encountered viral sequences within the bacterial genome in the form of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs)43,44,48,49. The viral DNA is coded in the spacers while the repeats themselves serve as regulation and processing domains48,50,51. These spacers are then transcribed along with the accompanying repeats and processed into individual CRISPR RNAs (crRNAs) that, together with a constitutive Trans-Activating CRISPR RNA (tracrRNA), bind to a Cas9 endonuclease50,52,53. The crRNA sequence then acts as the guide for the endonuclease, directing it to the complementary foreign DNA sequence, or protospacer. Upon matching its sequence via standard Watson Crick base pairing35,54, the Cas9 induces a DSB50-52,55,56. Additionally, binding specificity is also dependent on the presence of a specific three-nucleotide sequence flanking the 3’ end of the protospacer known as the protospacer adjacent motif (PAM)51,57,58. This sequence is (N)GG (where N is any nucleotide) in the common CRISPR/Cas9 system from Streptococcus pyogenes and is determined by protein-DNA interaction59. This region is important endogenously as it enables the system to avoid cutting the spacers stored in genomic CRISPR DNA, which do not have the appropriate PAM sequence.
The CRISPR-Cas9 system was studied in relative obscurity for several years until this system was shown to cut double strand DNA after DNA binding60-66. One important update that made the Cas9 system highly accessible was the fusion of the crRNAs and tracrRNAs into a single guide RNA (sgRNA). As a result, the only requirement to create a custom DNA interacting complex is to synthesize a single piece of ssRNA co-delivered with Cas9. This simple, two-component system with a single programmable element that can be readily generated by any modern molecular biology laboratory has democratized gene editing.
There are, however, some notable limitations to the targeting specificity of the commonly used CRISPR-Cas9 system67-70. This DNA binding and cutting platform can exhibit notable off-target cutting71. The mechanism underlying DNA/RNA matching with Cas9 and gRNAs often tolerates mismatches in the interaction especially beyond the first 12 bases immediately adjacent to the PAM (known as the seed region), resulting in a measurable proclivity for non-specific cutting67,72-74. Whether this reduced specificity is a significant limitation in a particular gene editing application is highly project-dependent. The search for improvements to CRISPR gene editors is ongoing as exemplified by a 2018 call by the NIH for proposals for improved DNA editing systems and methods to detect off-target effects (https://commonfund.nih.gov/editing/fundingopportunities).
Many potential solutions to the problem of reduced CRISPR-Cas9 specificity have been developed34,57,70,74,75. One option mimics the behavior of the TALEN and ZFN systems, fusing FokI monomers to a pair of catalytically inactive (dead) Cas9 proteins76. The catalytic inactivation of the Cas9 turns this DNA endonuclease into a simple DNA recognition element77. This modification to the system increases specificity by doubling of the binding activity required to create a DSB by the pairing of FokI dimers78. Another specificity enhancing option is to use a pair of partially compromised Cas9 proteins designed to create a pair of trans-strand single-stranded DNA nicks in close proximity to one another79. While this may reduce the risk of potentially toxic off -target cutting69, this solution can attenuate gene editing efficiency79. A third option for increasing specificity is to find or engineer a more specific CRISPR system. Multiple modified Cas9 proteins with enhanced specificity have been reported, with varying levels of success80,81.
The ease of use and lowered cost of engineering with the CRISPR-Cas9 platform has ushered in the rapid acceleration and expansion of gene editing into many laboratories in academia and industry. The technology is now accessible to the life-science community as a whole and, while not without faults, the CRISPR/Cas9 platform is a major step towards the democratization of gene editing and represents the first easily accessible custom DNA endonuclease for gene editing (Fig 4).
CRISPR systems are common to many bacteria, and this rich family of related proteins is being used to identify new DNA editing platforms such as Cas12a (formerly Cpf1)82. The Cas12a family has some features not common to the Cas9 family including the ability to trim its own gRNA and to induce a DSB that has base overhangs (Fig 1D). In contrast to the GG-rich PAM sequence in SpCas9, Cas12a proteins make use of a variety of AT-rich PAM sequences making it easier to target Cas12a in certain areas of the genome. Some Cas12a proteins also appear to have higher innate specificity than many Cas9 proteins. In addition, the generation of the first RNA binding platforms using gRNAs has been reported (C2C2 Cas13)83,84. Other potential programmable RNA binding systems have been characterized reminiscent TALEs consisting of a modular series of protein motifs that each binds a single RNA base85. The ability to target RNA could establish an entirely new branch of genome engineering.
Associated Biochemical Activities Critical For Precision Gene Editing
Binding DNA is only the first step in gene editing. Following the creation of a DNA lesion, various endogenous repair pathways function to create the actual chemical change in the genome (Fig 2). Gene editing thus revolves around the cell’s ability to repair its DNA. The most prominent repair pathway deployed is double-strand break repair, the cellular response to double-strand DNA breaks caused by FokI, Cas9, or Cpf1/Cas12a. Four main categories of DSB repair are used in gene editing and will be discussed below. These mechanisms range in both efficiency and accuracy, and thus possess dynamic mutagenic signatures. The use of different DNA endonuclease platforms can also affect the pathway and the result of DSB repairs86-91. While we are focused on double strand break repair, different genomic insults such as a single-strand breaks have also been explored to diversify and expand the gene editing toolkit92,93
Deletion: NHEJ
Non-Homologous End Joining (NHEJ) is a prolific response to DSBs that functions to maintain genomic integrity92,92. In the process of NHEJ, genomic repair enzymes identify the DSB, following which, either or both DNA strands can be resected or filled in as necessary, usually creating blunt ends. It is then also possible for new bases to be polymerized and incorporated into the DNA sequence. These ends are then ligated to restore continuity to the DNA molecule. The lack of template and the somewhat random resection and polymerization of the DNA ends results in the creation of various length insertions or deletions (indels, Fig 2A). The indels created by this repair can produce variable mutational signatures, so NHEJ is commonly used to create frameshifting mutations in the coding region in order to knockout a protein. Alternatively, two distant cis-DSBs can generated to delete the sequence between them, allowing NHEJ delete whole genes (Fig 2B).
There are many benefits to using NHEJ to make indels or create large deletions. First, NHEJ seems to be active across multiple species so most model systems can be edited. There are also no known context restrictions to NHEJ so edits can be made anywhere in the genome. Though NHEJ is most likely to introduce short indels or large deletions it is also possible to make use it for large insertions by introducing blunt ended DNA template94. These insertions are, however, subject to the same randomness of repair and the ends of the insertion will most often contain indels.
Large Insertions: Homologous Recombination
Operating in tandem, though normally with less frequency than NHEJ, are several distinct yet interconnected mechanisms of repair that rely on the use of a DNA template homologous to the DNA sequence flanking the DSB92,95. Collectively these pathways are referred to as Homology Directed Repair (HDR), with the best-characterized being Homologous Recombination (HR). Following a DSB, HR repair proteins use a template molecule of dsDNA to correct damaged DNA96. The donor contains long stretches of DNA sequences (>500 bps) homologous to either side of the break that is used as a template for repair. The result is a newly synthesized stretch of genomic DNA that is identical to the donor molecule (Fig 2C).
While being useful to the cell in repairing DSBs, HR has been utilized to introduce exogenous DNA to the chromosome97. This is accomplished by designing a donor molecule with long homology arms that flank the integration cargo. By co-delivering this exogenous template with a custom nuclease it is possible to incorporate the desired cargo into the genomic DNA. Unlike NHEJ, however, HR is usually relatively inefficient and often requires antibiotic selection or an equivalent enrichment to identify chromosomes that contain the newly synthesized sequence, limiting its application.
Small Insertion: Oligo-mediated Homology Directed Repair
To address the shortcomings of HR, a variety homologous donors to act as templates for repair have been explored. The simplest of these templates has been single strand oligodeoxynucleotides (ssODNs) with very short homology arms (< 100 bps, Fig 2D) by a repair pathway often called single strand template repair (SSTR). While the precise mechanism of repair used to integrate the ssODN sequence is still unclear, this process harbors both an HDR signature (precision integration of the template) and NHEJ (indel formation) repair outcomes depending on the model system used. Recent in vitro work has implicated the requirement for Rad51 activity98-101. While this method can often lead to higher rates of integration than traditional HR, it is typically deployed for integrating sequences of 50 base pairs or less102.
The wide-ranging mechanisms of HDR combined with the faithful accuracy of template dependent repair empower the creation of almost any desired DNA change. Unfortunately, the lower relative frequency of this event compared to NHEJ often requires exhaustive trials and screens to find the line or cell with the intended change. However, more work is underway to understand what makes a good candidate site for HDR, suggesting that the use of oligo-mediated HDR for gene processing will continue to increase.
Microhomology Mediated End Joining (MMEJ)
Emerging as yet another mechanism of DNA repair to be used for gene editing is a method that shares characteristics with both NHEJ and HDR. Microhomology Mediated End Joining (MMEJ, sometimes called alt-NHEJ or alt-EJ) repairs a DSB by annealing small homologous regions on each side of a DSB, generating a predictable small deletion in between. The precise contextual and cellular conditions that bias a DSB to be repaired via MMEJ rather than classic NHEJ are not fully understood103,104.
During MMEJ and upon the recognition of the DSB, the 5’ strands of both ends of the break are resected leaving overhanging 3’ tails on each molecule105. These overhangs are then aligned through short regions of homology, leaving unmatched “flaps” of DNA at each side of the paired homologous region. The remaining ssDNA regions are used as template for new DNA synthesis while the flaps are cleaved off allowing the nick between the newly synthesized DNA and the homologous region to be ligated resulting in the restoration of continuity within the DNA molecule106. The resection of the DNA flaps created by the homologous matching results in the removal of a short stretch of nucleotides central to the original DSB along with one of the original homology arms. This intrinsic deletion pattern unique to regions predisposed to engage in MMEJ activity makes the creation of specific indels a more predictable and reproducible event (Fig 2E).
The enhanced predictability of this repair pathway also makes it uniquely useful for the precise integration of exogenous DNA88,91,107. The introduction of a double strand template DNA flanked with short microhomology arms (<100 bp) has been shown to result in precise integrations108 (Fig 2F). This integration has the potential to be as accurate as repair with classical homologous recombination and, in some cases, as efficient as NHEJ109. However, the factors that play a role in efficient integration are not currently understood. For example, it is currently unclear how the balance of NHEJ to MMEJ plays a role in the proclivity to integrate a donor molecule using this method. It should here be noted that while the mechanism by which MMEJ proceeds is not yet fully understood, some attempts at characterization have demonstrated that some repair enzymes show preferential activity in different repair pathways.
As MMEJ becomes better characterized and more predictable it may prove incredibly useful for its propensity to produce a small subset of predictable mutations as efficiently as NHEJ, but may also be utilized for precise integration of DNA ranging from point mutations to several kilobases long (Fig 2F). This combination of efficiency and accuracy makes MMEJ an especially potent mechanism that may unlock entirely new options for gene editing.
Direct Editing of Single Nucleotides: Base Editors
Another emerging method for precision gene editing avoids double stranded breaks and instead modifies a single DNA base by activating a series of mismatch repair mechanisms capable of creating single nucleotide substitutions. These base editor systems function via the fusion of a nucleotide-modifying enzyme such as a deaminase to an existent DNA recognition system such as a catalytically inactivated Cas9 protein110. Upon binding their target, the functional enzyme modifies a single DNA base, for example, by deaminating a cytosine to a uracil. Nicking of the complementary strand forces strand excision during repair, resulting in the use of uracil as template, and the net insertion of an adenine. During DNA replication, uracil is read as Thymine resulting it a C to T transition (Fig 2G). To prevent the removal of Uracil before and during replication a UDG (Uracil-DNA Glycosylase) inhibitor is often used. This technique is promising for gene editing as many genetic disorders are caused by missense polymorphisms, and base editing offers a path to repair that does not rely on causing further damage to the DNA via double strand breaks110-118. Recently, a new base editor has been generated through enhanced molecular evolution capable of deaminating adenine119. While improvements to efficiency and specificity are still being developed for base editing, this strategy has the potential to deliver targeted single substitutions much more efficiently than homology-dependent repair integrations.
Locus-Specific Epigenetic Targeting
In addition to using the available DNA binding platforms to modify the actual nucleotide sequence of DNA, it is possible to modulate how the cell interprets DNA through direct chemical modification or through the creation of artificial transcription factors to modify gene expression (Fig 3). These methods confer many potential advantages as they do not require the formation of a DNA lesion such as a DSB or a mismatched base and that these epigenetic modifications are usually reversible. Three primary methods are used to modulate DNA and gene expression: methylation, acetylation, and programmable transcription factors. Each of these can be accomplished by the fusion of the appropriate functional domain to any of the DNA binding systems referred to in this review.
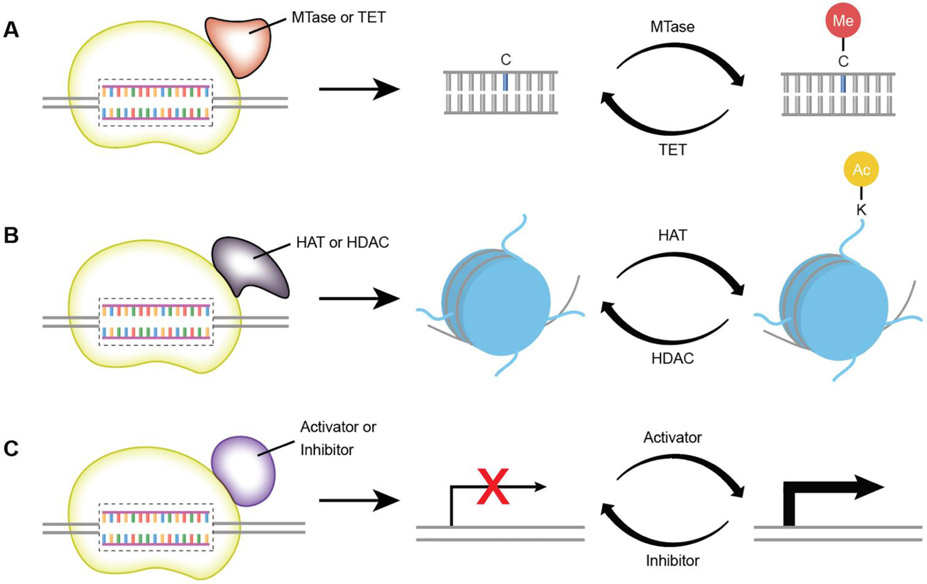
Figure 3. Precision Epigenetic Modulation.
A. Programmable methylation of CpG islands in DNA can be achieved by fusion of any of the DNA binding platforms described in this review to a sequence non-specific methyltranferase (MTase) or Trans-eleven Translocation enzyme (TeT) to methylate or demethylate the DNA, respectively B. Fusion of a DNA binding system to histone acetyltransferases (HATs) or histone deacetylases (HDACs) enables programmable acetylation or deacetylation of specific lysine residues of the histone proteins associated with the target DNA. C. Artificial transcription factors can be created by the fusion of transcription activating or suppressing domains to any DNA binding system. When bound to promoter or enhancer regions, transcription levels of genes can be modulated without any chemical modification to the DNA or associated proteins.
DNA Methylation (methyltransferases and tet proteins)
DNA methylation and the inverse process of demethylation can be used to physically mark genes and distinguish DNA for a range of cellular readouts 120. By fusing DNA binding domains to a sequence non-specific methyltransferase, a methyl group can be added to a cytosine nucleotide that is followed by a guanine nucleotide in the 5’ to 3’ direction, commonly referred to as a CpG site121 (Fig. 3A). Promoter and enhancer regions with high methylation of these sites modulate transcription of the associated genes122. This effect can be reversed by utilizing programmable DNA binding platforms fused to demethylase domains such as the ten eleven translocation (TET) enzymes that oxidize methylated cytosines123. Both of these approaches have been applied with ZFs, TALEs and CRISPR systems to activate and deactivate locus-specific regions within the genome120,124-127.
Histone Acetylation
Acetylation and deacetylation is another common chemical method of modifying expression, although it typically acts using a different mechanism compared to DNA methylation120. Histone acetylatransferases (HATs) and histone deacetylases (HDACs) target lysine residues on the tale chains of histone proteins, leading to altered charge of the lysine128,129. In the case of acetylation, the positive charge of the lysine is neutralized, reducing the histones ability to bind the negatively charged backbone of DNA120. Therefore, acetylation acts indirectly on the DNA expression both by modulating protein-protein interactions and by increasing the accessibility of the targeted regions, which has been shown to increase the steric favorability of promoter and enhancer binding130. Histone deacetylase fused to dCas9 and other programmable DNA binding platforms has been shown to have the opposite effect depending on the cell line used131. The lack of consistent outcomes in targeted deacetylase activity is assumed to be the result of deacetylases having non-specific activity, and altering more histones than expected. Determining the specificity of acetylation and deacetylation targeted epigenetic technology is an active research area.
Artificial Transcription Factors
The concept of using programmable DNA binding platforms as guided and programmable transcription factors returns to the natural origins of both zinc fingers and TALEs. Zinc finger domains can be found in the transcription factors of eukaryotes and even archaea132-134. TALEs have a similar natural history in that they originated in pathogenic bacteria to activate genes within their plant hosts135. Both of these systems can be easily modified to activate or repress genes by the fusion of different transcription regulating domains (Fig 3C). CRISPR systems that have been catalytically inactivated (dCas) can also be used in the creation of artificial transcription factors136. The most commonly used activator domain is from VP16, which is a Herpes simplex 1 viral protein137. This domain is can be used individually but is more often repeated 4 times creating what is referred to as the VP64 activation domain138. In addition to being fused to various DNA recognition elements, KRAB is a commonly used repression domain in synthetic transcription factors and is found on 30% of endogenous zinc finger related proteins132. This domain can be utilized to repress genes by targeting it to the promoter or enhancer region of the gene target139.
History and Current State of Genetic Engineering in Renal Research
Genetic engineering has attracted the attention of Nephrology research for over 20 years (Table 2). Studies as early as the late 1990s showed the potential of gene delivery in vivo in a variety of rodent models of kidney disease including hypertension, cardiovascular disease, glomerulonephritis, renal tubular damage, and renal interstitial fibrosis 140,141 Delivery to the kidney has always been an issue, and in these early studies, with genes mainly provided systemically as naked DNA or in viruses by intramuscular, intravenous, intraportal, or intraperitoneal injection. These non-targeted approaches resulted in widespread expression including the kidney, heart, aorta, lung, liver, muscle, serum, and urine, with relatively short-lasting benefits lasting from 1-6 weeks following injection. An early, more targeted approach was injection of genes packaged in liposomes into the renal pelvis, artery, or parenchyma with expression lasting 3 weeks after injection142. While showing promise, these studies also showed the challenge of obtaining targeted, high-level, long-term expression in the kidney. Furthermore, the relatively short-lasting effects may indicate instability and lack of integration of the transgene. It should here be noted that transgene integration is likely to cause another set of problems due to potential disruption of endogenous genes.
Table 2.
Contributions using new gene editors to renal research.
| Target | Approach | Model | Disorder | Source |
|---|---|---|---|---|
| talpid3 | ZFN | zebrafish | cystogenesis | Ben et al 2011 |
| RAG1 | ZFN | rat | hypertension | Mattson et al 2013 |
| ROMK | ZFN | rat | hypertension | Zhou et al 2013 |
| TGFb1 | ZFN | rat | renal fibrosis | Chen et al 2013 |
| HV1 | ZFN | rat | hypertension | Jin et al 2014 |
| PLEKHA7 | ZFN | rat | hypertension | Endres et al 2014 |
| apol1 | CRISPR/Cas9 | zebrafish | nephropathy | Anderson et al 2015 |
| HSD11B2 | ZFN | rat | hypertension | Mullins et al 2015 |
| PERVs | CRISPR/Cas9 | pig kidney cells | transplant | Yang et al 2015 |
| PKD1 | ZFN | Pig | ADPKD | He et al 2015 |
| PKD1 | CRISPR/Cas9 | kidney organoids | cystogenesis | Freedman et al 2015 |
| PKD2 | CRISPR/Cas9 | kidney organoids | cystogenesis | Freedman et al 2015 |
| PODXL | CRISPR/Cas9 | kidney organoids | glomerular disease | Freedman et al 2015 |
| c21orf29/kurly | CRISPR/Cas9+HR | Xenopus | cilia | Jaffe et al 2016 |
| PDE1A | TALEN | mouse | ADPKD | Ye et al 2016 |
| NOX4 | ZFN | rat | hypertension | Cowley et al 2016 |
| sec61al2 | CRISPR/Cas9 | zebrafish | ADTKD | Bolar et al 2016 |
| Tns2DeltaC | CRISPR/Cas9 | mouse | glomerulonephritis | Marusugi et al 2016 |
| GREB1L | CRISPR/Cas9 | mouse | renal agenesis | Brophy et al 2017 |
| GREB1L | CRISPR/Cas9 | mouse | renal agenesis | De Tomasi et al 2017 |
| greb1l | CRISPR/Cas9 | zebrafish | renal agenesis | Sanna-Cherchi et al 2017 |
| HOXA5 | CRISPR-Cas9-SunTag-DNMT3A | HEK293 | DNA methylation | Huang et al 2017 |
| LAMA5 | TALEN+HR | mouse IMCD cells | ADPKD | Hofherr et al 2017 |
| miR-210-3p | CRISPR/Cas9 | RCC cells | renal cell carcinoma | Yoshino et al 2017 |
| PKD1 | CRISPR/Cas9+HR | mouse IMCD cells | ADPKD | Hofherr et al 2017 |
| PKD1 | TALEN | MDCK cells | ADPKD | Hofherr et al 2017 |
| PKD2 | TALEN | mouse IMCD+MDCK cells | ADPKD | Hofherr et al 2017 |
| PDE1A | TALEN | mouse | ADPKD | Wang et al 2017 |
| wt1 | CRISPR/Cas9 | tilapia | glomerular development | Jiang et al 2017 |
| UMOD | CRISPR/Cas9 | mouse | UAKD | Johnson et al 2017 |
Over time, studies examining the effects of exogenous gene expression have evolved to include modifiers of endogenous gene expression such as DNA enzymes 143, decoy oligonucleotides 144,145, siRNA 146,147, and antisense oligonucleotides (Table 2)148,149. Additionally, multiple delivery methods have been explored including hydrodynamic injection into the tail vein 145,150, renal electroporation 143,151 ultrasound with microbubbles 152-154 and expression of transgenes within mesenchymal stem cells 155. Delivery by injection directly to the kidney has shown benefits in models of diabetic nephropathy, ischemia/reperfusion injury, tubulointerstitial and glomerular fibrosis 144,155-158. These genome engineering systems and cellular delivery knowledge are being deployed anew with gene editing tools as cargo.
Employing Gene Editing for Gene Inactivation to Understand Kidney Disease Pathogenesis
The development of new tools for gene editing, such as ZFN, TALEN, and CRISPR/Cas9, has enabled new approaches including editing endogenous genes in a variety of cellular and in vivo model systems (Table 2). Many studies have applied these technologies to the kidney using renal cell culture 159 and kidney organoids 160. Additionally, gene editing can target multiple models and is not limited to HR in mouse ES cells. Especially valuable is the ability to generate vertebrate models ranging in complexity from zebrafish through pig. Accordingly, kidney diseases are now being modeled using precision gene editing tools in a variety of organisms including zebrafish 161, tilapia 162, Xenopus 163, mouse 164-167, rat 168-170, and pig 171. These studies have primarily involved truncating mutations induced by NHEJ to produce what are effectively gene knockouts that have proven central to characterizing the roles of genes and proteins in a variety of conditions, such as glomerulonephritis, hypertension, polycystic kidney disease, renal agenesis, and renal fibrosis 161,165,168,169,172,173. For example, a TALEN-induced mutation of PDE1A demonstrated its role in polycystic kidney disease in mice 166,167, and three groups used CRISPR/Cas9 to validate identification of a novel renal agenesis gene, GREB1L 165,172,173.
Clearly mouse models have contributed innumerable insights into disease pathophysiology, and the value of obtaining consistent results through these studies cannot be overstated. However, there is also great value to the ability to chose the model that best fits the need, Limited studies to date on non-mouse models make it currently unclear which animal models are closest to human. Indeed this may vary according to the specific kidney disease being studied174. It is clear, however, from the number of failed clinical trials based on mouse studies, that results often poorly translate from mouse to human. For example, studies testing mTOR inhibitors for PKD showed effectiveness in mouse models but not in clinical trials 175-178. Rat and pig models may represent more faithful models than mice, making the advent of gene editing in these animals particularly exciting 179. Use of pigs in particular has the disadvantage of higher cost to raise and maintain lines to achieve genetic homogeneity, which is significant as founders may be mosaics, but the value of this must be considered in comparison to the cost in dollars and human life to that of a failed clinical trial.
Another situation where gene inactivation by CRISPR/Cas9 is potentially valuable for renal health is in generating organs suitable for xenotransplantation. Pigs are a promising source of organs for xenotranplantation provided several obstacles including transmission of porcine endogenous retroviruses (PERVs) can be overcome. Progress has been made on this front using CRISPR/Cas9 inactivation of PERVs in a porcine cell line preventing transmission to human cells in vitro 180. Additionally, PERV-inactivated pigs were recently generated using somatic cell nuclear transfer 181. While several challenges remain such as immunological issues, these studies illustrate the essential contributions of gene editing to this effort 182.
Employing Gene Editing for Engineering Precise Gene Changes Relevant for Kidney Diseases
TALENs and CRISPRs can be employed to make targeted gene edits in cells and model organisms, even enabling the recapitulation of exact disease-causing sequence variations using targeted integration approaches (Fig. 2). An example relevant to the kidney is provided by a study of uromodulin-associated kidney disease (UAKD) 183, where gene inactivation does not recapitulate the human disease phenotype. This study used CRISPR/Cas9 with HR to re-create in mice the p.C147W mutation found in patients. This new mouse model more faithfully reproduced the human phenotype including progressive kidney disease leading to renal failure, provided insight into pathogenic mechanisms of the mutation i.e., the involvement of ER stress, apoptotic signaling, and decreased autophagy, and showed the therapeutic value in mice of blocking TNF alpha to reduce activated caspase-3 and tubule cell death. A few studies have also successfully combined CRISPR with HR for tagging edited genes in Xenopus and renal cell culture 159,163. These tagged proteins allow cellular and subcellular expression and protein trafficking to be better defined by examining the endogenous, albeit modified, protein. This provides an alternative, and potentially more useful approach than standard techniques, such as in situ hybridization and antibody labeling.
The Promise and Challenges of Gene Editing for Targeted Treatments in Nephrology
New technologies for precision genome editing in somatic cells are potentially valuable as therapeutics. Recent studies using CRISPR/Cas9 demonstrate proof of principle as a therapeutic approach in a mouse model of Duchenne muscular dystrophy (DMD). In these studies, CRISPR/Cas9 was used to induce exon skipping to avoid a truncating mutation and partially restore dystrophin expression, building on an approach used in a previous clinical trial using morpholino oligomers 184-187. Possible approaches in Nephrology could be correction of base pair variants in monogenic disease where these types of mutation are common and where a small amount of functional protein may be valuable. In addition, in diseases where gene dosage appears important and a normal allele is present, such as in autosomal dominant polycystic kidney disease (ADPKD), up regulation of the normal copy of the gene may be advantageous.
At present there are questions about the precision of the gene editing and off target effects, which need to be addressed before widespread human treatment is likely. In addition, the extent to which targeted delivery to the kidney is possible is unclear, although, adenovirus, which was used for the DMD studies, has been used previously to deliver genes to the kidney (as well as the liver, aorta and adrenal gland) following injection into the tail vein in rats 188.
The Real Estate Problem for In Vivo Genome Editing in the Kidney.
Despite the promise, there are multiple issues to address to precisely and efficiently perform genome editing in the kidney. These include, effects of pharmacology, vector tropism, and immune responses against vectors. These problems are made worse for renal disease for the simple fact that the kidney is a very effective filter, and it’s effective at preventing vectors delivered in the blood from accessing the kidney.
In addition, the liver and spleen are usually the first organ destinations for most injected particles, leaving little to reach the kidneys. Then, the diameter of the afferent arteriole that feeds the glomerulus is a simple and effective barrier to kidney vector delivery. The arteriole is approximately 10 nm in diameter. In contrast, most viral and non-viral gene delivery vectors are 20 to 200 nm in diameter. If a vector can squeeze through the arteriole, it then confronts the stringent molecular weight cut-off of the glomerulus. Only proteins below 50 kDa are thought to readily pass through this barrier, while most gene therapy vectors are megaDa is size.
For example, adeno-associated virus (AAV) vectors are by far the most popular vectors used for in vivo gene therapy and are only 25 nm in diameter (reviewed in 189). While these small viral vectors can extravasate into other tissues well after intravenous injection to mediate impressive transduction of multiple tissues in mice 146,190, most do a poor job of transducing kidney cells 191. Rare CD31+ endothelial cells and EpCAM+ proximal tubules can be transduced using this approach, but podocytes are not modified. In no cases does one observe saturation of kidney cells after intravenous injection.
While the favorite intravenous route of vector delivery fails for kidney gene therapy, recent developments suggest additional options are forthcoming. For example, delivering non-viral, AAV, and adenovirus vectors by retro-grade injection in the ureter or directly in the capsule can bypass this delivery problem 192.
The DNA Payload Problem for In Vivo Genome Editing in the Kidney.
AAV is popular in part because it is compact. That smallness also has limitations because AAV can also only package 4.5 kilobases (kb) of sequence. This restricted cargo capacity is appropriate for delivering a small gene for renal gene therapy, but it restricts the use of medium to large genes. For example, AAV can easily carry the ~ 3,000 base pair (bp) cDNA for PKD2, but it would take three AAV vectors to carry the ~13,000 bp PKD1 cDNA.
When considering using AAV for renal genome editing in vivo, two ZFNs separated by a 2A peptide and furin cleavage sequence with a DNA repair substrate of 750 nucleotides can be squeezed into one AAV vector 193. A single TALEN expression cassette can easily exceed 3.5 kb, making it impossible to package the two TALENs needed for DNA double strand breaks into one AAV vector 194, let alone both TALENs along with a repair template. CRISPR will require the use of one or two AAVs carrying the sp or saCas9 and gRNA sequences. An additional AAV will likely be needed to carry a repair template restricted to less than 4.7 kb in size. Finally, it is difficult enough to transform many cells with one vector in vivo; delivery approaches needing 2 or 3 vectors in vivo may be difficult unless co-delivery is optimized.
In contrast, adenovirus (Ad) vectors can carry up to 36,000 bp of transgene sequences, reviewed195. Most Ad vectors described in the literature are known as first generation Ads (FG-Ads). FG-Ads can carry ~7 kbp of sequence, since these vectors still retain most adenovirus genes and ORFs. As such, FG-Ads still provoke strong immune responses due to leaky expression of Ad proteins in transduced cells 196. In addition, a similar third generation Ad vector was responsible for the tragic death of Jessie Gelsinger 197making these tools markedly less appealing when considering them for gene therapy.
However, newer Helper Dependent (HD-Ad) vectors have been designed to address these earlier concerns and are appropriate to use for in vivo delivery, particularly if they are confined to the kidney. All adenovirus genes and ORFs are deleted in HD-Ad vectors. This allows sequences as large as 36 kbp to be packaged in the vector. No Ad proteins are produced in HD-Ad vector-transduced cells because all of these viral genes have been removed. Because of this, HD-Ad vectors generate lower Ad T cell responses against transduced cells 198,199. This reduced immunogenicity enables transgene expression over years 200,201. For example, in an ongoing study, baboons were treated once with HD-Ads are still expressing the transgene protein more than 7 years later 195.
The Immune Problem for In Vivo Genome Editing in the Kidney.
Foreign genes usually need to be expressed for gene therapy or genome editing. Treating a genetic disease and repairing the expression of a missing piece of a protein risks targeting and cellular ablation by the immune system. Many of the current proteins used in gene editing are entirely foreign and may stimulate immune responses against the modified cells even if they are expressed only transiently. Finally, since some of these editing proteins are bacterial in origin from human pathogens, this may be a pre-existing immune barrier. For example, humans already have memory antibody and T cell against Cas9 from Staphyloccocus pyogenes (spCas9) and from Staphylococcus aureus (saCas9)202.
Gene Editing is in the Clinic
Despite reservations about human treatments, several CRISPR, TALEN, and Zinc Finger Nuclease trials are ongoing for treatment of such diseases as various forms of cancer (including renal cell carcinoma), beta thalassemia, hemophilia, HIV, mucopolysaccharidosis, and sickle cell disease (clinicaltrials.gov). Several are located in medical centers throughout the US, including some in phase 2. Clearly many hurdles remain, but the implications of recent advances in gene editing for advancing research and medicine in Nephrology holds tremendous promise for advancing medicine.
Table 1.
Major approaches using gene therapy in renal research.
| Substance | DNA packaging |
Delivery | Disorder | Effect Duration |
Source |
|---|---|---|---|---|---|
| Decorin gene | cDNA | IM | glomerulonephritis | 6 days | Isaka et al 1996 |
| Kallikrein gene | naked or adenovirus | IM, IV, IP, intraportal | hypertension | 6 weeks | Chao and Chao 1997 |
| Egr-1 DNA enzyme | NA | ureter+kidney electroporation | UUO tubulointerstitial fibrosis | 7 days | Nakamura et al 2002 |
| Smad7 gene | adenovirus | renal pelvis | UUO tubulointerstitial fibrosis | 14 days | Terada et al 2002 |
| IL-10 gene | recombinant adenoviral vector | renal parenchyma | glomerulosclerosis and renal failure | 20 days | Choi et al 2003 |
| vIL-10 gene | plasmid | tail vein | glomerulonephritis | 7 days | Higuchi et al 2003 |
| Smad7 | plasmid | renal artery and ultrasound with microbubbles | UUO injury inc renal fibrosis | 2 days | Lan et al 2003 |
| AP-1 decoy oligodeoxynucleotides | HVJ-liposome | renal artery | diabetic nephropathy | 15 days | Ahn et al 2004 |
| TGFbeta siRNA | plasmid | renal artery | tubulointerstitial fibrosis | 14 days | Hwang et al 2006 |
| decoy receptor 3 (DCR3) | NA | tail vein | glomerulonephritis | 21 days | Ka et al 2011 |
| ERK2 antisense | adenovirus | perfusion of donor kidneys | renal allograft fibrosis | 24 weeks | Ding et al 2011 |
| HGF gene | mesenchymal stem cells | tail vein | UUO-renal fibrosis | 14 days | Liu et al 2011 |
| Catalase gene | adenovirus | renal artery | I/R injury | ND | Yang et al 2015 |
| CTGF siRNA | plasmid | medulla+kidney electroporation | renal fibrosis | 7 days | Ren et al 2015 |
| AGT antisense oligo | NA | IP | renal fibrosis in PKD2WS25 | 1 week | Ravichandran et al 2015 |
| Intermedin gene | plasmid | renal artery and ultrasound with microbubbles | UUO injury inc renal fibrosis | 7 days | Qiao et al 2015 |
| VEGF gene | mesenchymal stem cells | IV | UUO injury inc renal fibrosis | 14 days | Ozbek et al 2015 |
| siRNA cocktail | NA | perfusion of donor kidneys | I/R injury | NA | Zheng et al 2016 |
Highlights.
Analysis of common genome engineering tools, ZFN, TALEN, and CRISPR systems.
DNA repair processes leveraged to make edits in the genome, NHEJ, HDR, MMEJ SSTR.
The deployment and associated challenges of genome engineering in the study of renal disease.
Acknowledgments
Funding: Supported by the Mayo Foundation and NIH grants GM63904 and P30DK084567 (SCE) and P30DK090728 (CRS, PCH and SCE).
References
- 1.Kim YG, Cha J & Chandrasegaran S Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proceedings of the National Academy of Sciences of the United States of America 93,1156–1160, doi: 10.1073/pnas.93.3.1156 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carroll D Genome Engineering With Zinc-Finger Nucleases. Genetics 188, 773–782, doi: 10.1534/genetics.111.131433 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christian M et al. Targeting DNA Double-Strand Breaks with TAL Effector Nucleases. Genetics 186, 757–U476, doi: 10.1534/genetics.110.120717 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joung JK & Sander JD INNOVATION TALENs: a widely applicable technology for targeted genome editing. Nature Reviews Molecular Cell Biology 14, 49–55, doi: 10.1038/nrm3486 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porteus MH & Baltimore D Chimeric nucleases stimulate gene targeting in human cells. Science 300, 763–763, doi: 10.1126/science.1078395 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Bibikova M, Beumer K, Trautman JK & Carroll D Enhancing gene targeting with designed zinc finger nucleases. Science 300, 764–764, doi: 10.1126/science.1079512 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Wolfe SA, Nekludova L & Pabo CO DNA recognition by Cys(2)His(2) zinc finger proteins. Annual Review of Biophysics and Biomolecular Structure 29, 183–212, doi: 10.1146/annurev.biophys.29.1.183 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Pavletich NP & Pabo CO ZINC FINGER DNA RECOGNITION - CRYSTAL-STRUCTURE OF A ZIF268-DNA COMPLEX AT 2.1-A. Science 252, 809–817, doi: 10.1126/science.2028256 (1991). [DOI] [PubMed] [Google Scholar]
- 9.Desjarlais JR & Berg JM REDESIGNING THE DNA-BINDING SPECIFICITY OF A ZINC FINGER PROTEIN - A DATA BASE-GUIDED APPROACH. Proteins-Structure Function and Genetics 12,101–104, doi: 10.1002/prot.340120202 (1992). [DOI] [PubMed] [Google Scholar]
- 10.Segal DJ, Dreier B, Beerli RR & Barbas CF Toward controlling gene expression at will: Selection and design of zinc finger domains recognizing each of the 5 '-GNN-3 ' DNA target sequences. Proceedings of the National Academy of Sciences of the United States of America 96, 2758–2763, doi: 10.1073/pnas.96.6.2758 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beerli RR & Barbas CF Engineering polydactyl zinc-finger transcription factors. Nature Biotechnology 20,135–141, doi: 10.1038/nbt0202-135 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Kim HJ, Lee HJ, Kim H, Cho SW & Kim J-S Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Research 19,1279–1288, doi: 10.1101/gr.089417.108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhakta MS et al. Highly active zinc-finger nucleases by extended modular assembly. Genome Research 23, 530–538, doi: 10.1101/gr.143693.112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta A et al. An optimized two-finger archive for ZFN-mediated gene targeting. Nature Methods 9, 588–589, doi: 10.1038/nmeth.1994 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laoharawee K et al. Dose-Dependent Prevention of Metabolic and Neurologic Disease in Murine MPS II by ZFN-Mediated In Vivo Genome Editing. Molecular Therapy 26, 1127–1136, doi: 10.1016/j.ymthe.2018.03.002 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bibikova M et al. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Molecular and Cellular Biology 21, 289–297, doi: 10.1128/mcb.21.1.289-297.2001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bibikova M, Golic M, Golic KG & Carroll D Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 161,1169–1175 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Urnov FD et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435, 646–651, doi: 10.1038/nature03556 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Doyon Y et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nature Biotechnology 26, 702–708, doi: 10.1038/nbt1409 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bitinaite J, Wah DA, Aggarwal AK & Schildkraut I FokI dimerization is required for DNA cleavage. Proceedings of the National Academy of Sciences of the United States of America 95,10570–10575, doi: 10.1073/pnas.95.18.10570 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo J, Gaj T & Barbas CF III. Directed Evolution of an Enhanced and Highly Efficient FokI Cleavage Domain for Zinc Finger Nucleases. Journal of Molecular Biology 400, 96–107, doi: 10.1016/j.jmb.2010.04.060 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller JC et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nature Biotechnology 25, 778–785, doi: 10.1038/nbt1319 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Szczepek M et al. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nature Biotechnology 25, 786–793, doi: 10.1038/nbt1317 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Doyon Y et al. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nature Methods 8, 74–U108, doi: 10.1038/nmeth.1539 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Cornu TI et al. DNA-binding specificity is a major determinant of the activity and toxicity of zinc-finger Nucleases. Molecular Therapy 16, 352–358, doi: 10.1038/sj.mt.6300357 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Haendel E-M, Alwin S & Cathomen T Expanding or Restricting the Target Site Repertoire of Zinc-finger Nucleases: The Inter-domain Linker as a Major Determinant of Target Site Selectivity. Molecular Therapy 17, 104–111, doi: 10.1038/mt.2008.233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gabriel R et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nature Biotechnology 29, 816–U872, doi: 10.1038/nbt.1948 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Boch J et al. Breaking the Code of DNA Binding Specificity of TAL-Type III Effectors. Science 326,1509–1512, doi: 10.1126/science.1178811 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Cermak T et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Research 39, doi: 10.1093/nar/gkr218 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng D et al. Structural Basis for Sequence-Specific Recognition of DNA by TAL Effectors. Science 335, 720–723, doi: 10.1126/science.1215670 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moscou MJ & Bogdanove AJ A Simple Cipher Governs DNA Recognition by TAL Effectors. Science 326,1501–1501, doi: 10.1126/science.1178817 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Bogdanove AJ & Voytas DF TAL Effectors: Customizable Proteins for DNA Targeting. Science 333, 1843–1846, doi: 10.1126/science.1204094 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Mussolino C et al. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Research 39, 9283–9293, doi: 10.1093/nar/gkr597 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mali P et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nature Biotechnology 31, 833–838, doi: 10.1038/nbt.2675 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith C et al. Whole-Genome Sequencing Analysis Reveals High Specificity of CRISPR/Cas9 and TALEN-Based Genome Editing in Human iPSCs. Cell Stem Cell 15, 13–14, doi: 10.1016/j.stem.2014.06.011 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hockemeyer D et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nature Biotechnology 29, 731–734, doi: 10.1038/nbt.1927 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamb BM, Mercer AC & Barbas CF Directed evolution of the TALE N-terminal domain for recognition of all 5' bases. Nucleic Acids Research 41, 9779–9785, doi: 10.1093/nar/gkt754 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng D et al. Recognition of methylated DNA by TAL effectors. Cell Research 22, 1502–1504, doi: 10.1038/cr.2012.127 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim Y et al. A library of TAL effector nucleases spanning the human genome. Nature Biotechnology 31, 251–258, doi: 10.1038/nbt.2517 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Zhang F et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nature Biotechnology 29,149–U190, doi: 10.1038/nbt.1775 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reyon D et al. FLASH assembly of TALENs for high-throughput genome editing. Nature Biotechnology 30, 460–465, doi: 10.1038/nbt.2170 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heigwer F et al. E-TALEN: a web tool to design TALENs for genome engineering. Nucleic Acids Research 41, doi: 10.1093/nar/gkt789 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Makarova KS, Grishin NV, Shabalina SA, Wolf YI & Koonin EV A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biology Direct 1, doi: 10.1186/1745-6150-1-7 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barrangou R et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315,1709–1712, doi: 10.1126/science.1138140 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Makarova KS et al. Evolution and classification of the CRISPR-Cas systems. Nature Reviews Microbiology 9, 467–477, doi: 10.1038/nrmicro2577 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sapranauskas R et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Research 39, 9275–9282, doi: 10.1093/nar/gkr606 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barrangou R & Marraffini LA CRISPR-Cas Systems: Prokaryotes Upgrade to Adaptive Immunity. Molecular Cell 54, 234–244, doi: 10.1016/j.molcel.2014.03.011 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bolotin A, Ouinquis B, Sorokin A & Ehrlich SD Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology-Sgm 151, 2551–2561, doi: 10.1099/mic.0.28048-0 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Mojica FJM, Diez-Villasenor C, Garcia-Martinez J & Soria E Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. Journal of Molecular Evolution 60,174–182, doi: 10.1007/s00239-004-0046-3 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Brouns SJJ et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964, doi: 10.1126/science.1159689 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mojica FJM, Diez-Villasenor C, Garcia-Martinez J & Almendros C Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology-Sgm 155, 733–740, doi: 10.1099/mic.0.023960-0 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Gasiunas G, Barrangou R, Horvath P & Siksnys V Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proceedings of the National Academy of Sciences of the United States of America 109, E2579–E2586, doi: 10.1073/pnas.1208507109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jinek M et al. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337, 816–821, doi: 10.1126/science.1225829 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sternberg SH, Redding S, Jinek M, Greene EC & Doudna JA DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67, doi: 10.1038/nature13011 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiedenheft B, Sternberg SH & Doudna JA RNA-guided genetic silencing systems in bacteria and archaea. Nature 482, 331–338, doi: 10.1038/nature10886 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Jinek M et al. Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation. Science 343,1215, doi: 10.1126/science.1247997 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mali P, Esvelt KM & Church GM Cas9 as a versatile tool for engineering biology. Nature Methods 10, 957–963, doi: 10.1038/nmeth.2649 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cencic R et al. Protospacer Adjacent Motif (PAM)-Distal Sequences Engage CRISPR Cas9 DNA Target Cleavage. Plos One 9, doi: 10.1371/journal.pone.0109213 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anders C, Niewoehner O, Duerst A & Jinek M Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513, 569–573, doi: 10.1038/nature13579 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Belhaj K, Chaparro-Garcia A, Kamoun S & Nekrasov V Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9, doi: 10.1186/1746-4811-9-39 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cho SW, Kim S, Kim JM & Kim J-S Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nature Biotechnology 31, 230–232, doi: 10.1038/nbt.2507 (2013). [DOI] [PubMed] [Google Scholar]
- 62.Dickinson DJ, Ward JD, Reiner DJ & Goldstein B Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nature Methods 10,1028–1034, doi: 10.1038/nmeth.2641 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cradick TJ, Fine EJ, Antico CJ & Bao G CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Research 41, 9584–9592, doi: 10.1093/nar/gkt714 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cong L et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339, 819–823, doi: 10.1126/science.1231143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cho SW, Lee J, Carroll D, Kim J-S & Lee J Heritable Gene Knockout in Caenorhabditis elegans by Direct Injection of Cas9-sgRNA Ribonucleoproteins. Genetics 195,1177–1180, doi: 10.1534/genetics.113.155853 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gratz SJ et al. Genome Engineering of Drosophila with the CRISPR RNA-Guided Cas9 Nuclease. Genetics 194,1029–1035, doi: 10.1534/genetics.113.152710 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fu Y et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature Biotechnology 31, 822–826, doi: 10.1038/nbt.2623 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hsu PD et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature Biotechnology 31, 827–832, doi: 10.1038/nbt.2647 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cho SW et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Research 24,132–141, doi: 10.1101/gr.162339.113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsai SQ & Joung JK Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat. Rev. Genet 17, 300–312, doi: 10.1038/nrg.2016.28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peng RX, Lin GG & Li JM Potential pitfalls of CRISPR/Cas9-mediated genome editing. Febs J. 283, 1218–1231, doi: 10.1111/febs.13586 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Duan JZ et al. Genome-wide identification of CRISPR/Cas9 off-targets in human genome. Cell Research 24,1009–1012, doi: 10.1038/cr.2014.87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuscu C, Arslan S, Singh R, Thorpe J & Adli M Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nature Biotechnology 32, 677–683, doi: 10.1038/nbt.2916 (2014). [DOI] [PubMed] [Google Scholar]
- 74.Frock RL et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nature Biotechnology 33, 179–186, doi: 10.1038/nbt.3101 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fu Y, Sander JD, Reyon D, Cascio VM & Joung JK Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nature Biotechnology 32, 279–284, doi: 10.1038/nbt.2808 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guilinger JP, Thompson DB & Liu DR Fusion of catalytically inactive Cas9 to Fokl nuclease improves the specificity of genome modification. Nature Biotechnology 32, 577–582, doi: 10.1038/nbt.2909 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Qi LS et al. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 152,1173–1183, doi: 10.1016/j.cell.2013.02.022 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tsai SQ et al. Dimeric CRISPR RNA-guided Fokl nucleases for highly specific genome editing. Nature Biotechnology 32, 569–576, doi: 10.1038/nbt.2908 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ran FA et al. Double Nicking by RNA-Guided CRISPR Cas9 for Enhanced Genome Editing Specificity. Cell 154,1380–1389, doi: 10.1016/j.cell.2013.08.021 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Slaymaker IM et al. Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88, doi: 10.1126/science.aad5227 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kleinstiver BP et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495, doi: 10.1038/nature16526 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zetsche B et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 163, 759–771, doi: 10.1016/j.cell.2015.09.038 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.East-Seletsky A, O'Connell MR, Burstein D, Knott GJ & Doudna JA RNA Targeting by Functionally Orthogonal Type VI-A CRISPR-Cas Enzymes. Molecular Cell 66, 373–383, doi: 10.1016/j.molcel.2017.04.008 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.East-Seletsky A et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273, doi: 10.1038/nature19802 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yin P et al. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature 504,168–171, doi: 10.1038/nature12651 (2013). [DOI] [PubMed] [Google Scholar]
- 86.Kim Y, Kweon J & Kim J-S TALENs and ZFNs are associated with different mutation signatures. Nature Methods 10,185–185, doi: 10.1038/nmeth.2364 (2013). [DOI] [PubMed] [Google Scholar]
- 87.Xiao A et al. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Research 41, doi: 10.1093/nar/gkt464 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nakade S et al. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nature Communications 5, doi: 10.1038/ncomms6560 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chu VT et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nature Biotechnology 33, 543–U160, doi: 10.1038/nbt.3198 (2015). [DOI] [PubMed] [Google Scholar]
- 90.Maruyama T et al. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nature Biotechnology 33, 538–U260, doi: 10.1038/nbt.3190 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sakuma T, Nakade S, Sakane Y, Suzuki KT & Yamamoto T MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nature Protocols 11, 118–133, doi: 10.1038/nprot.2015.140 (2016). [DOI] [PubMed] [Google Scholar]
- 92.Hoeijmakers JHJ Genome maintenance mechanisms for preventing cancer. Nature 411, 366–374, doi: 10.1038/35077232 (2001). [DOI] [PubMed] [Google Scholar]
- 93.Komor AC, Kim YB, Packer MS, Zuris JA & Liu DR Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424, doi: 10.1038/nature17946 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Auer TO, Duroure K, De Cian A, Concordet J-P & Del Bene F Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Research 24, 142–153, doi: 10.1101/gr.161638.113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Greene EC DNA Sequence Alignment during Homologous Recombination. Journal of Biological Chemistry 291,11572–11580, doi: 10.1074/jbc.R116.724807 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Qi Z et al. DNA Sequence Alignment by Microhomology Sampling during Homologous Recombination. Cell 160, 856–869, doi: 10.1016/j.cell.2015.01.029 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zu Y et al. TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nature Methods 10, 329–331, doi: 10.1038/nmeth.2374 (2013). [DOI] [PubMed] [Google Scholar]
- 98.Sakuma T & Yamamoto T Magic wands of CRISPR-lots of choices for gene knock-in. Cell Biology and Toxicology 33, 501–505, doi: 10.1007/s10565-017-9409-6 (2017). [DOI] [PubMed] [Google Scholar]
- 99.Richardson CD, K. K., Feng SJ, Bray NL, Schaefer AJ, Floor S, Corn J. CRISPR-Cas9 genome editing in human cells works via the Fanconi anemia pathway. bioRxiv. 2017;136028 CRISPR-Cas9 genome editing in human cells works via the Fanconi anemia pathway. bioRxiv (2017). [DOI] [PubMed] [Google Scholar]
- 100.Danner E et al. Control of gene editing by manipulation of DNA repair mechanisms. Mammalian Genome 28, 262–274, doi: 10.1007/s00335-017-9688-5 (2017). [DOI] [PubMed] [Google Scholar]
- 101.Bothmer A et al. Characterization of the interplay between DNA repair and CRISPR/Cas9-induced DNA lesions at an endogenous locus. Nature Communications 8, doi: 10.1038/ncomms13905 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen F et al. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nature Methods 8, 753–U796, doi: 10.1038/nmeth.1653 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kostyrko K & Mermod N Assays for DNA double-strand break repair by microhomology-based end-joining repair mechanisms. Nucleic Acids Research 44, doi: 10.1093/nar/gkv1349 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ahrabi S et al. A role for human homologous recombination factors in suppressing microhomology-mediated end joining. Nucleic Acids Research 44, 5743–5757, doi: 10.1093/nar/gkw326 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McVey M & Lee SE MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends in Genetics 24, 529–538, doi: 10.1016/j.tig.2008.08.007 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lu GQ et al. Ligase I and ligase III mediate the DNA double-strand break ligation in alternative end-joining. Proceedings of the National Academy of Sciences of the United States of America 113,1256–1260, doi: 10.1073/pnas.1521597113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nakamae K et al. Establishment of expanded and streamlined pipeline of PITCh knock-in - a web-based design tool for MMEJ-mediated gene knock-in, PITCh designer, and the variations of PITCh, PITCh-TG and PITCh-KIKO. Bioengineered 8, 302–308, doi: 10.1080/21655979.2017.1313645 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Aida T et al. Gene cassette knock-in in mammalian cells and zygotes by enhanced MMEJ. Bmc Genomics 17, doi: 10.1186/s12864-016-3331-9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yao X et al. Homology-mediated end joining-based targeted integration using CRISPR/Cas9. Cell Research 27, 801–814, doi: 10.1038/cr.2017.76 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yang LH et al. Engineering and optimising deaminase fusions for genome editing (vol 7, 13330, 2016). Nature Communications 8, doi: 10.1038/ncomms16169 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zong Y et al. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nature Biotechnology 35, 438–440, doi: 10.1038/nbt.3811 (2017). [DOI] [PubMed] [Google Scholar]
- 112.Zhang YH et al. Programmable base editing of zebrafish genome using a modified CRISPR-Cas9 system. Nature Communications 8, doi: 10.1038/s41467-017-00175-6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li Z et al. APOBEC signature mutation generates an oncogenic enhancer that drives LMO1 expression in T-ALL. Leukemia 31, 2057–2064, doi: 10.1038/leu.2017.75 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kouno T et al. Crystal structure of APOBEC3A bound to single-stranded DNA reveals structural basis for cytidine deamination and specificity. Nature Communications 8, doi: 10.1038/ncomms15024 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim YB et al. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nature Biotechnology 35, 371–376, doi: 10.1038/nbt.3803 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Brachova P, Alvarez NS, Van Voorhis BJ & Christenson LK Cytidine deaminase Apobec3a induction in fallopian epithelium after exposure to follicular fluid. Gynecologic Oncology, 145 577–583, doi: 10.1016/j.ygyno.2017.02.017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Billon P et al. CRISPR-Mediated Base Editing Enables Efficient Disruption of Eukaryotic Genes through Induction of STOP Codons. Molecular Cell 67,1068–1079, doi: 10.1016/j.molcel.2017.08.008 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yang LH et al. Engineering and optimising deaminase fusions for genome editing. Nature Communications 7, doi: 10.1038/ncomms13330 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gaudelli NM et al. Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage. Nature advance online publication, doi: 10.1038/nature24644 http://www.nature.com/nature/journal/vaap/ncurrent/abs/nature24644.html-supplementary-information (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kungulovski G & Jeltsch A Epigenome Editing: State of the Art, Concepts, and Perspectives. Trends in Genetics 32,101–113, doi: 10.1016/j.tig.2015.12.001 (2016). [DOI] [PubMed] [Google Scholar]
- 121.Smith AE & Ford KG Specific targeting of cytosine methylation to DNA sequences in vivo. Nucleic Acids Research 35, 740–754, doi: 10.1093/nargkl1053 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Thakore PI, Black JB, Hilton IB & Gersbach CA Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nature Methods 13, 127–137, doi: 10.1038/nmeth.3733 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xu XX et al. A CRISPR-based approach for targeted DNA demethylation. Cell Discovery 2, doi: 10.1038/celldisc.2016.9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Carvin CD, Parr RD & Kladde MP Site-selective in vivo targeting of cytosine-5 DNA methylation by zinc-finger proteins. Nucleic Acids Research 31, 6493–6501, doi: 10.1093/nar/gkg853 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Maeder ML et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nature Biotechnology 31, 1137–1142, doi: 10.1038/nbt.2726 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liu YD et al. Zinc Finger Protein 618 Regulates the Function of UHRF2 (Ubiquitin-like with PHD and Ring Finger Domains 2) as a Specific 5-Hydroxymethylcytosine Reader. Journal of Biological Chemistry 291, 13679–13688, doi: 10.1074/jbc.M116.717314 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vojta A et al. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Research 44, 5615–5628, doi: 10.1093/nar/gkw159 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sterner DE & Berger SL Acetylation of histones and transcription-related factors. Microbiology and Molecular Biology Reviews 64, 435–459, doi: 10.1128/mmbr.64.2.435-459.2000(2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Seto E & Yoshida M Erasers of Histone Acetylation: The Histone Deacetylase Enzymes. Cold Spring Harbor Perspectives in Biology 6, doi: 10.1101/cshperspect.a018713 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hilton IB et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nature Biotechnology 33, 510–U225, doi: 10.1038/nbt.3199 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kwon DY, Zhao YT, Lamonica JM & Zhou Z Locus-specific histone deacetylation using a synthetic CRISPR-Cas9-based HDAC. Nature Communications 8, doi: 10.1038/ncomms15315 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Looman C, Abrink M, Mark C & Hellman L KRAB zinc finger proteins: An analysis of the molecular mechanisms governing their increase in numbers and complexity during evolution. Molecular Biology and Evolution 19, 2118–2130, doi: 10.1093/oxfordjournals.molbev.a004037 (2002). [DOI] [PubMed] [Google Scholar]
- 133.Krishna SS, Majumdar I & Grishin NV Structural classification of zinc fingers. Nucleic Acids Research 31, 532–550, doi: 10.1093/nar/gkg161 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Guilliere F et al. Solution Structure of an Archaeal DNA Binding Protein with an Eukaryotic Zinc Finger Fold. Plos One 8, doi: 10.1371/journal.pone.0052908 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Erkes A, Reschke M, Boch J & Grau J Evolution of Transcription Activator-Like Effectors in Xanthomonas oryzae. Genome Biology and Evolution 9,1599–1615, doi: 10.1093/gbe/evx108 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chavez A et al. Highly efficient Cas9-mediated transcriptional programming. Nature Methods 12, 326–U365, doi: 10.1038/nmeth.3312 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shen F, Triezenberg SJ, Hensley P, Porter D & Knutson JR Transcriptional activation domain of the herpesvirus protein VP16 becomes conformationally constrained upon interaction with basal transcription factors. Journal of Biological Chemistry 271, 4827–4837 (1996). [DOI] [PubMed] [Google Scholar]
- 138.Graslund T, Li XL, Magnenat L, Popkov M & Barbas CF Exploring strategies for the design of artificial transcription factors. Journal of Biological Chemistry 280, 3707–3714, doi: 10.1074/jbc.M406809200 (2005). [DOI] [PubMed] [Google Scholar]
- 139.Thakore PI et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nature Methods 12,1143–1149, doi: 10.1038/nmeth.3630 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chao J & Chao L Experimental kallikrein gene therapy in hypertension, cardiovascular and renal diseases. Pharmacol Res 35, 517–522, doi: 10.1006/phrs.1997.0179 (1997). [DOI] [PubMed] [Google Scholar]
- 141.Isaka Y et al. Gene therapy by skeletal muscle expression of decorin prevents fibrotic disease in rat kidney. Nat Med 2, 418–423 (1996). [DOI] [PubMed] [Google Scholar]
- 142.Lai LW, Moeckel GW & Lien YH Kidney-targeted liposome-mediated gene transfer in mice. Gene Ther 4, 426–431, doi: 10.1038/sj.gt.3300406 (1997). [DOI] [PubMed] [Google Scholar]
- 143.Nakamura H et al. Introduction of DNA enzyme for Egr-1 into tubulointerstitial fibroblasts by electroporation reduced interstitial alpha-smooth muscle actin expression and fibrosis in unilateral ureteral obstruction (UUO) rats. Gene Ther 9, 495–502, doi: 10.1038/sj.gt.3301681 (2002). [DOI] [PubMed] [Google Scholar]
- 144.Ahn JD et al. Transcription factor decoy for AP-1 reduces mesangial cell proliferation and extracellular matrix production in vitro and in vivo. Gene Ther 11, 916–923, doi: 10.1038/sj.gt.3302236 (2004). [DOI] [PubMed] [Google Scholar]
- 145.Ka SM et al. Decoy receptor 3 inhibits renal mononuclear leukocyte infiltration and apoptosis and prevents progression of IgA nephropathy in mice. Am J Physiol Renal Physiol 301, F1218–1230, doi: 10.1152/ajprenal.00050.2011 (2011). [DOI] [PubMed] [Google Scholar]
- 146.Hwang M et al. TGF-beta1 siRNA suppresses the tubulointerstitial fibrosis in the kidney of ureteral obstruction. Exp Mol Pathol 81, 48–54, doi: 10.1016/j.yexmp.2005.11.005 (2006). [DOI] [PubMed] [Google Scholar]
- 147.Zheng X et al. Attenuating Ischemia-Reperfusion Injury in Kidney Transplantation by Perfusing Donor Organs With siRNA Cocktail Solution. Transplantation 100, 743–752, doi: 10.1097/TP.0000000000000960 (2016). [DOI] [PubMed] [Google Scholar]
- 148.Ding Z et al. Adenovirus-mediated anti-sense ERK2 gene therapy inhibits tubular epithelial-mesenchymal transition and ameliorates renal allograft fibrosis. Transpl Immunol 25, 34–41, doi: 10.1016/j.trim.2011.04.001 (2011). [DOI] [PubMed] [Google Scholar]
- 149.Ravichandran K, Ozkok A, Wang Q, Mullick AE & Edelstein CL Antisense-mediated angiotensinogen inhibition slows polycystic kidney disease in mice with a targeted mutation in Pkd2. Am J Physiol Renal Physiol 308, F349–357, doi: 10.1152/ajprenal.00478.2014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Higuchi N et al. Hydrodynamics-based delivery of the viral interleukin-10 gene suppresses experimental crescentic glomerulonephritis in Wistar-Kyoto rats. Gene Ther 10, 1297–1310, doi: 10.1038/sj.gt.3301988 (2003). [DOI] [PubMed] [Google Scholar]
- 151.Ren Y et al. CTGF siRNA ameliorates tubular cell apoptosis and tubulointerstitial fibrosis in obstructed mouse kidneys in a Sirt1-independent manner. Drug Des Devel Ther 9, 4155–4171, doi: 10.2147/DDDT.S86748 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Koike H et al. An efficient gene transfer method mediated by ultrasound and microbubbles into the kidney. J Gene Med 7, 108–116, doi: 10.1002/jgm.632 (2005). [DOI] [PubMed] [Google Scholar]
- 153.Lan HY et al. Inhibition of renal fibrosis by gene transfer of inducible Smad7 using ultrasound-microbubble system in rat UUO model. J Am Soc Nephrol 14, 1535–1548 (2003). [DOI] [PubMed] [Google Scholar]
- 154.Qiao X et al. Intermedin is upregulated and attenuates renal fibrosis by inhibition of oxidative stress in rats with unilateral ureteral obstruction. Nephrology (Carlton) 20, 820–831, doi: 10.1111/nep.12520 (2015). [DOI] [PubMed] [Google Scholar]
- 155.Ozbek E et al. Role of Mesenchymal Stem Cells Transfected With Vascular Endothelial Growth Factor in Maintaining Renal Structure and Function in Rats with Unilateral Ureteral Obstruction. Exp Clin Transplant 13, 262–272, doi: 10.6002/ect.2014.0080 (2015). [DOI] [PubMed] [Google Scholar]
- 156.Yang CC, Hsu SP, Chen KH & Chien CT Effect of Adenoviral Catalase Gene Transfer on Renal Ischemia/Reperfusion Injury in Rats. Chin J Physiol 58, 420–430, doi: 10.4077/CJP.2015.BAD324 (2015). [DOI] [PubMed] [Google Scholar]
- 157.Terada Y et al. Gene transfer of Smad7 using electroporation of adenovirus prevents renal fibrosis in post-obstructed kidney. Kidney Int 61, S94–98, doi: 10.1046/j.1523-1755.2002.0610s1094.x (2002). [DOI] [PubMed] [Google Scholar]
- 158.Choi YK et al. Suppression of glomerulosclerosis by adenovirus-mediated IL-10 expression in the kidney. Gene Ther 10, 559–568, doi: 10.1038/sj.gt.3301926 (2003). [DOI] [PubMed] [Google Scholar]
- 159.Hofherr A et al. Efficient genome editing of differentiated renal epithelial cells. Pflugers Arch 469, 303–311, doi: 10.1007/s00424-016-1924-4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Freedman BS et al. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat Commun 6, 8715, doi: 10.1038/ncomms9715 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ben J, Elworthy S, Ng AS, van Eeden F & Ingham PW Targeted mutation of the talpid3 gene in zebrafish reveals its conserved requirement for ciliogenesis and Hedgehog signalling across the vertebrates. Development 138, 4969–4978, doi: 10.1242/dev.070862 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Jiang D et al. CRISPR/Cas9-induced disruption of wt1a and wt1b reveals their different roles in kidney and gonad development in Nile tilapia. Dev Biol 428, 63–73, doi: 10.1016/j.ydbio.2017.05.017 (2017). [DOI] [PubMed] [Google Scholar]
- 163.Jaffe KM et al. c21orf59/kurly Controls Both Cilia Motility and Polarization. Cell Rep 14,1841–1849, doi: 10.1016/j.celrep.2016.01.069 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Marusugi K et al. Functional validation of tensin2 SH2-PTB domain by CRISPR/Cas9-mediated genome editing. J Vet Med Sci 78, 1413–1420, doi: 10.1292/jvms.16-0205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Brophy PD et al. A Gene Implicated in Activation of Retinoic Acid Receptor Targets is a Novel Renal Agenesis Gene in Humans. Genetics, doi: 10.1534/genetics.117.1125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wang X et al. Generation and phenotypic characterization of Pde1a mutant mice. PLoS One 12, e0181087, doi: 10.1371/journal.pone.0181087 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ye H et al. Modulation of Polycystic Kidney Disease Severity by Phosphodiesterase 1 and 3 Subfamilies. J Am Soc Nephrol 27,1312–1320, doi: 10.1681/ASN.2015010057 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chen CC, Geurts AM, Jacob HJ, Fan F & Roman RJ Heterozygous knockout of transforming growth factor-beta1 protects Dahl S rats against high salt-induced renal injury. Physiol Genomics 45,110–118, doi: 10.1152/physiolgenomics.00119.2012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mattson DL et al. Genetic mutation of recombination activating gene 1 in Dahl salt-sensitive rats attenuates hypertension and renal damage. Am J Physiol Regal Integr Comp Physiol 304, R407–414, doi: 10.1152/ajpregu.00304.2012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhou X et al. Heterozygous disruption of renal outer medullary potassium channel in rats is associated with reduced blood pressure. Hypertension 62, 288–294, doi: 10.1161/HYPERTENSIONAHA.111.01051 (2013). [DOI] [PubMed] [Google Scholar]
- 171.He J et al. PKD1 mono-allelic knockout is sufficient to trigger renal cystogenesis in a mini-pig model. Int J Biol Sci 11, 361–369, doi: 10.7150/ijbs.10858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.De Tomasi L et al. Mutations in GREB1L Cause Bilateral Kidney Agenesis in Humans and Mice. Am J Hum Genet 101, 803–814, doi: 10.1016/j.ajhg.2017.09.026 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sanna-Cherchi S et al. Exome-wide Association Study Identifies GREB1L Mutations in Congenital Kidney Malformations. Am J Hum Genet 101, 789–802, doi: 10.1016/j.ajhg.2017.09.018 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hewitson B. a. Animal models of chronic kidney disease: useful but not perfect. Nephrol Dial Transplant 28, 2432 (2013). [DOI] [PubMed] [Google Scholar]
- 175.Walz G et al. Everolimus in Patients with Autosomal Dominant Polycystic Kidney Disease. New England Journal of Medicine 363, 830–840, doi: 10.1056/NEJMoa1003491 (2010). [DOI] [PubMed] [Google Scholar]
- 176.Shillingford JM, Piontek KB, Germino GG & Weimbs T Rapamycin Ameliorates PKD Resulting from Conditional Inactivation of Pkd1. Journal of the American Society of Nephrology 21, 489–497, doi: 10.1681/asn.2009040421 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Serra AL et al. Sirolimus and Kidney Growth in Autosomal Dominant Polycystic Kidney Disease. New England Journal of Medicine 363, 820–829, doi: 10.1056/NEJMoa0907419 (2010). [DOI] [PubMed] [Google Scholar]
- 178.Tao YX, Kim J, Schrier RW & Edelstein CL Rapamycin markedly slows disease progression in a rat model of polycystic kidney disease. Journal of the American Society of Nephrology 16, 46–51, doi: 10.1681/asn.2004080660 (2005). [DOI] [PubMed] [Google Scholar]
- 179.He J et al. PKD1 Mono-Allelic Knockout Is Sufficient to Trigger Renal Cystogenesis in a Mini-Pig Model. International Journal of Biological Sciences 11, 361–369, doi: 10.7150/ijbs.10858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yang LH et al. Genome-wide inactivation of porcine endogenous retroviruses (PERVs). Science 350,1101–1104, doi: 10.1126/science.aad1191 (2015). [DOI] [PubMed] [Google Scholar]
- 181.Niu D et al. Inactivation of porcine endogenous retrovirus in pigs using CRISPR-Cas9. Science 357,1303–1307, doi: 10.1126/science.aan4187 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Petersen B & Niemann H Molecular scissors and their application in genetically modified farm animals. Transgenic Research 24, 381–396, doi: 10.1007/s11248-015-9862-z (2015). [DOI] [PubMed] [Google Scholar]
- 183.Johnson BG et al. Uromodulin p.Cys147Trp mutation drives kidney disease by activating ER stress and apoptosis. J Clin Invest 127, 3954–3969, doi: 10.1172/JCI93817 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Cirak S et al. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study. Lancet 378, 595–605, doi: 10.1016/S0140-6736(11)60756-3 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Long C et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351, 400–403, doi: 10.1126/science.aad5725 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Nelson CE et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351, 403–407, doi: 10.1126/science.aad5143 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Tabebordbar M et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 351, 407–411, doi: 10.1126/science.aad5177 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Chao J & Chao L Experimental kallikrein gene therapy in hypertension, cardiovascular and renal diseases. Pharmacological Research 35, 517–522, doi: 10.1006/phrs.1997.0179 (1997). [DOI] [PubMed] [Google Scholar]
- 189.Samulski RJ & Muzyczka N AAV-Mediated Gene Therapy for Research and Therapeutic Purposes. Annual Review of Virology 1, 427–451 (2014). [DOI] [PubMed] [Google Scholar]
- 190.Zincarelli C, Soltys S, Rengo G & Rabinowitz JE Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Molecular Therapy 16,1073–1080, doi: 10.1038/mt.2008.76 (2008). [DOI] [PubMed] [Google Scholar]
- 191.Hillestad ML, Guenzel AJ, Nath KA & Barry MA A Vector-Host System to Fingerprint Virus Tropism. Human Gene Therapy 23, 1116–1126, doi: 10.1089/hum.2011.116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Chung DC et al. Adeno-Associated Virus-Mediated Gene Transfer to Renal Tubule Cells via a Retrograde Ureteral Approach. Nephron Extra 1, 217–223, doi: 10.1159/000333071 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ellis BL, Hirsch ML, Porter SN, Samulski RJ & Porteus MH Zinc-finger nuclease-mediated gene correction using single AAV vector transduction and enhancement by Food and Drug Administration-approved drugs. Gene Therapy 20, 35–42, doi: 10.1038/gt.2011.211 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Yang J et al. Targeting of macrophage activity by adenovirus-mediated intragraft overexpression of TNFRp55-Ig, IL-12p40, and vIL-10 ameliorates adenovirus-mediated chronic graft injury, whereas stimulation of macrophages by overexpression of IFN-gamma accelerates chronic graft injury in a rat renal allograft model. J Am Soc Nephrol 14, 214–225 (2003). [DOI] [PubMed] [Google Scholar]
- 195.Brunetti-Pierri N & Ng P Gene therapy with helper-dependent adenoviral vectors: lessons from studies in large animal models. Virus Genes 53, 684–691, doi: 10.1007/s11262-017-1471-x (2017). [DOI] [PubMed] [Google Scholar]
- 196.Yang YP, Su Q & Wilson JM Role of viral antigens in destructive cellular immune responses to adenovirus vector-transduced cells in mouse lungs. Journal of Virology 70,7209–7212 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Raper SE et al. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Molecular Genetics and Metabolism 80,148–158, doi: 10.1016/j.ymgme.2003.08.016 (2003). [DOI] [PubMed] [Google Scholar]
- 198.Mitani K, Graham FL, Caskey CT & Kochanek S RESCUE, PROPAGATION, AND PARTIAL-PURIFICATION OF A HELPER VIRUS-DEPENDENT ADENOVIRUS VECTOR. Proceedings of the National Academy of Sciences of the United States of America 92, 3854–3858, doi: 10.1073/pnas.92.9.3854 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Fisher KJ, Choi H, Burda J, Chen SJ & Wilson JM Recombinant adenovirus deleted of all viral genes for gene therapy of cystic fibrosis. Virology 217, 11–22, doi: 10.1006/viro.1996.0088 (1996). [DOI] [PubMed] [Google Scholar]
- 200.Morral N et al. High doses of a helper-dependent adenoviral vector yield supraphysiological levels of alpha(1)-antitrypsin with negligible toxicity. Human Gene Therapy 9, 2709–2716, doi: 10.1089/hum.1998.9.18-2709 (1998). [DOI] [PubMed] [Google Scholar]
- 201.Morral N et al. Administration of helper-dependent adenoviral vectors and sequential delivery of different vector serotype for long-term liver-directed gene transfer in baboons. Proceedings of the National Academy of Sciences of the United States of America 96,12816–12821, doi: 10.1073/pnas.96.22.12816 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Charlesworth CT et al. Identification of Pre-Existing Adaptive Immunity to Cas9 Proteins in Humans. bioRxiv, doi: 10.1101/243345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]