Abstract
Cells are exposed to various endogenous and exogenous insults that induce DNA damage, which, if unrepaired, impairs genome integrity and leads to the development of various diseases, including cancer. Recent evidence has implicated poly(ADP-ribose) polymerase 1 (PARP1) in various DNA repair pathways and in the maintenance of genomic stability. The inhibition of PARP1 is therefore being exploited clinically for the treatment of various cancers, which include DNA repair-deficient ovarian, breast and prostate cancers. Understanding the role of PARP1 in maintaining genome integrity is not only important for the design of novel chemotherapeutic agents, but is also crucial for gaining insights into the mechanisms of chemoresistance in cancer cells. In this Review, we discuss the roles of PARP1 in mediating various aspects of DNA metabolism, such as single-strand break repair, nucleotide excision repair, double-strand break repair and the stabilization of replication forks, and in modulating chromatin structure.
Cells are exposed constantly to various genotoxic stresses that can lead to DNA damage, the repair of which is crucial and requires robust mechanisms for both the detection and repair of the damaged DNA1. One of the earliest events in the DNA damage response (DDR) is the recruitment of poly(ADP-ribose) polymerase 1 (PARP1; also known as ARTD1) to diverse types of DNA lesions. PARP1 is an abundant nuclear protein that post-translationally attaches a negatively charged polymer termed poly(ADP-ribose) (PAR) to itself and to multiple target proteins. This poly(ADP)ribosylation (PARylation) activity contributes to most of the known functions of PARP1 in DNA repair, including repair of single-strand breaks (SSBs) and double-strand breaks (DSBs), in the stabilization of DNA replication forks and in the modification of chromatin structure, the details of which are discussed in this Review.
As targeting the DDR machinery is an attractive strategy for designing novel chemotherapeutics, there has been an intense clinical focus on PARP1 in the past few years. This strategy involves inhibiting the catalytic activity of PARP1 in cancers that are defective in genes that are involved in DNA repair pathways. This approach gives rise to synthetic lethality, a phenomenon in which mutations or perturbations of two genes together result in a loss of cell viability, whereas mutation or perturbation of either gene alone does not result in a loss of viability2. The utilization of targeted PARP inhibition for the treatment of various cancers is likely to give rise to distinct mechanisms of resistance to this inhibition3. Therefore, a comprehensive understanding of the crucial roles of PARP1 in DNA repair is of paramount importance for the development of successful therapeutic regimens for the treatment of different cancers.
Biochemical activities of PARP1
PARP1 was the first member of the PARP family to be identified, which now comprises 18 distinct members4. The main role of PARP1 is to catalyse the polymerization of ADP-ribose units — derived from the ADP donor NAD+ — resulting in the attachment of either linear or branched PAR polymers to itself or other target proteins (FIG. 1). PARP1 is a highly conserved, multifunctional enzyme that is found in eukaryotes. Its structure has been extensively characterized5–9 and it consists of three main domains: an amino-terminal DNA-binding domain (DBD) that consists of zinc finger motifs, a BRCT domain-containing central auto-modification domain and a highly conserved carboxy-terminal catalytic domain10–12. These domains together mediate the response of PARP1 to different kinds of DNA damage. The activity of PARP1 is stimulated markedly in the presence of various activators, including DNA damage11,13 (FIG. 1). Although PARP1 is the dominant member of the PARP family of proteins, some of its functions in DNA repair are shared and overlap with those of PARP2 (REFS 14–16). Indeed, genetic disruption of both PARP1 and PARP2 in mice results in embryonic lethality16, demonstrating the redundancy of these enzymes.
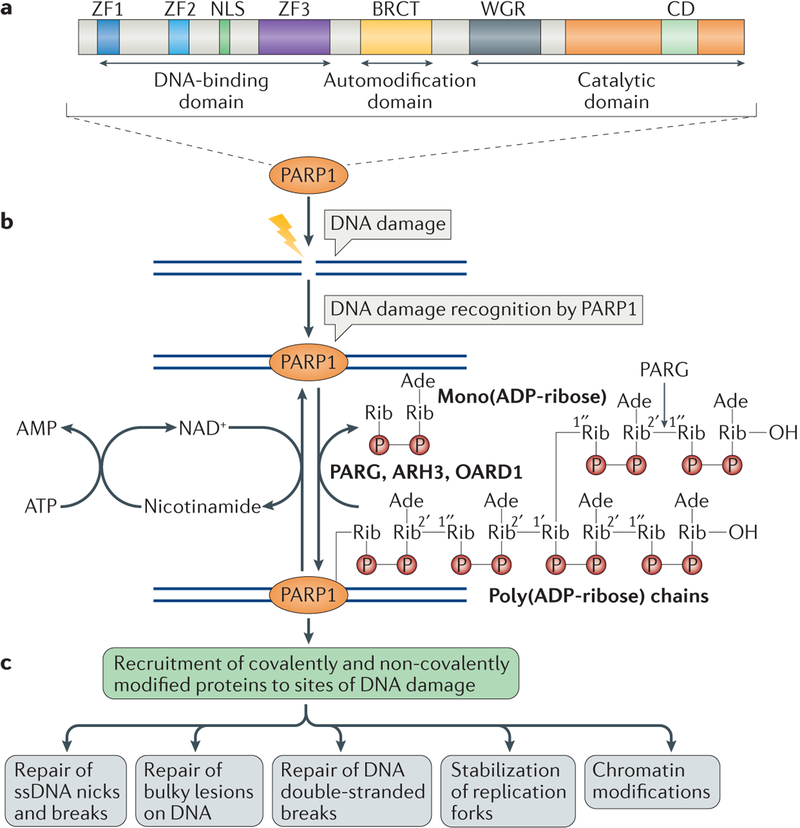
Figure 1 |. The biochemical functions of poly(ADP-ribose) polymerase 1 in DNA damage repair.
a | The domains of poly(ADP-ribose) polymerase 1 (PARP1) are shown. PARP1 has a DNA-binding domain (DBD), which consists of three zinc finger motifs (ZF1–3) and also contains a nuclear localization signal (NLS); a central automodification domain, which contains an interaction motif termed the BRCA1 C terminus (BRCT) domain; and a carboxy-terminal catalytic domain that contains a signature of PARP proteins, the conserved domain (CD). The CD contains the active site and binds to NAD+ and also to the Trp-Gly-Arg (WGR) domain. b | PARP1 detects DNA damage through its DBD, and it is activated by synthesizing poly(ADP-ribose) (PAR) chains mainly on itself but also on some of its target proteins. NAD+ is used as substrate for PAR formation, and is replenished in the cells from nicotinamide, using ATP. PAR chains are rapidly catabolized by PAR glycohydrolase (PARG), ADP-ribosylhydrolase 3 (ARH3) and O-acyl-ADP-ribose deacylase 1 (OARD1). PARG cleaves the 2′,1″-O-glycosidic ribose–ribose bonds of PAR. However, cleavage of the terminal ADP-ribose moiety requires OARD1 and results in the release of mono(ADP-ribose). c | Poly(ADP)ribosylation (PARylation) of PARP1 and other target proteins, both covalently and non-covalently, results in the recruitment of multiple proteins that have roles in different aspects of DNA damage repair. Ade, adenosine; P, phosphate residue; Rib, ribose moiety; ssDNA, single-stranded DNA.
The roles of PARP1 in the DDR have been studied extensively. Induction of various kinds of DNA damage results in rapid recruitment of PARP1 to sites of damage through its DNA-binding ability11,17. This stimulates the catalytic activity of PARP1, which results in the synthesis of PAR chains on itself and on histone and non-histone proteins15,18,19. This enzymatic reaction results in the covalent attachment of ADP-ribose units to Glu, Asp or Lys residues of acceptor proteins by a transesterification reaction. Repeating units of ADP-ribose are attached through 2′,1″-O-glycosidic ribose–ribose bonds to produce long (approximately 200 ADP-ribose units) linear chains of PAR in vitro, which contain branches every 20–50 ADP-ribose units11,20 (FIG. 1). Proteins involved in the DDR and in DNA metabolism can bind to PAR on PARP proteins through non-covalent interactions, which results in their recruitment to sites of DNA damage21. The target proteins that interact non-covalently with PAR generally contain PAR-binding modules, such as PAR-binding consensus motifs (PBMs), PAR-binding zinc finger motifs (PBZs), macrodomain folds, WWE domains and many other modules21,22. The PAR-binding modules in these target proteins frequently overlap with important functional domains, which are activated through binding to PAR and mediate biological functions such as binding to DNA, protein–protein interactions, nuclear localization and other functions18,23–25 (FIG. 1).
The generation of PAR following stresses, such as metabolic, oxidative, oncogenic or genotoxic stresses, is an extremely rapid process; however, PAR is also catabolized rapidly by proteins that promote PAR degradation11,26. The prompt turnover of PAR is crucial for efficient DNA repair27–29. Defects in PAR catabolism result in increased DNA damage and are deleterious to cells30–32 (BOX 1; FIG. 1).
Box 1 |. Catabolism of poly(ADP-ribose).
The best-studied poly(ADP-ribose) (PAR)-degrading enzyme is the endo- and exoglycohydrolase, PAR glycohydrolase (PARG)158,159. In mammalian cells, PARG has three isoforms: a 110 kDa nuclear isoform and two cytoplasmic isoforms of 99 kDa and 102 kDa (REF. 26). PARG contains a catalytic domain and a macrodomain fold, which is its PAR-binding module. PARG cleaves the 2′,1″-O-glycosidic ribose–ribose bonds that covalently link the ADP-ribose monomers of PAR160 (FIG. 1). PARG is crucial for the maintenance of PAR levels in cells and its deregulation results in disruption of multiple cellular processes, such as DNA repair and apoptosis161,162. Knockout mice lacking the nuclear isoform of PARG are highly sensitive to genotoxic stress163, which underscores the importance of PARG in cellular metabolism.
In addition to PARG, other PAR-degrading enzymes include ADP-ribosylhydrolase 3 (ARH3), which is a dinitrogenase reductase-activating glycohydrolase-related protein, and O-acyl-ADP-ribose deacylase 1 (OARD1; also known as TARG1). ARH3 localizes to the nucleus, cytoplasm and the mitochondria. It does not share structural similarity with PARG and it is also able to remove O-acetyl groups from O-acetyl-ADP ribose, which is a product of NAD metabolism164,165. However, its role in the catabolism of PAR in the DNA damage response has not been studied. In contrast to ARH3, which cannot degrade terminal ADP-ribose units, OARD1 is involved in the removal of terminal mono(ADP-ribose) moieties166,167. The catalytic mechanism of OARD1 involves a conserved Lys residue that forms a covalent lysyl-ADP-ribose intermediate, which is then resolved by a closely positioned, catalytic Asp residue. OARD1 seems to have a role in DNA damage signalling, as depletion of the protein results in sensitivity to DNA-damaging agents and reduces the proliferative capacity of cells167.
PARP1 mediates excision repair
Single base modifications or nucleotide damage are the most common type of DNA damage; thousands of new lesions are induced each day by reactive endogenous metabolites and also from spontaneous decay of DNA33. These base or nucleotide modifications are repaired by excision repair mechanisms, which include single-strand break repair (SSBR), base excision repair (BER) and nucleotide excision repair (NER).
The roles of PARP1 in single-strand break repair pathways.
A varied number of DNA lesions can be formed by spontaneous SSBs and by base modifications, which include the oxidation of bases, the generation of abasic sites (also known as apurinic or apyrimidinic sites) and other base modifications (FIG. 2). Single base modifications can change base pairing properties and result in spontaneous mutations that are associated with cancer predisposition33.
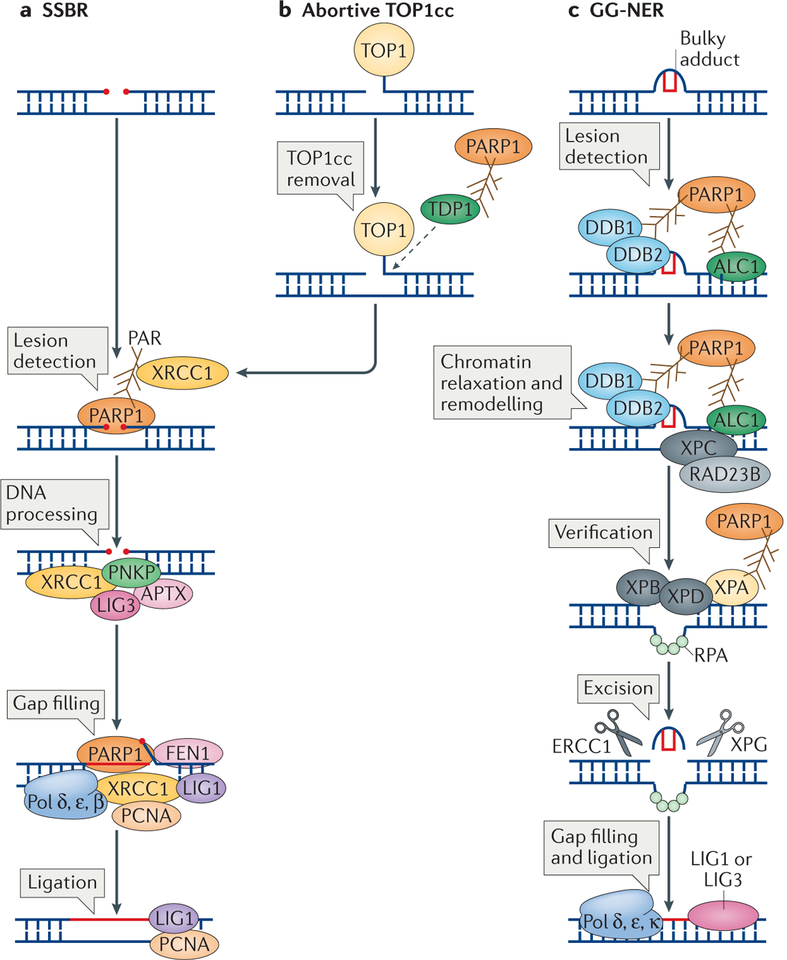
Figure 2 |. The roles of poly(ADP-ribose) polymerase 1 in excision repair.
a | Poly(ADP-ribose) polymerase 1 (PARP1) activity is required in different steps of single-strand break repair (SSBR). It is essential for the detection of the SSBs and is then required for the recruitment of X-ray repair cross-complementing protein 1 (XRCC1), which acts as a scaffold for the recruitment of polynucleotide kinase 3′- phosphatase (PNKP), aprataxin (APTX) and DNA ligase 3 (LIG3) to process the SSB. This is followed by the gap filling step, which is carried out by DNA polymerase δ (Pol δ), Pol ε and Pol β, and also requires PARP1 for stimulating the 5′ flap endonuclease activity of flap endonuclease 1 (FEN1). Finally, the DNA is ligated by LIG1. b | PARP1 is also required for the processing of SSBs at abortive DNA topoisomerase 1 (TOP1) cleavage complexes (TOP1cc). PARP1 recruits and activates tyrosyl-DNA phosphodiesterase 1 (TDP1), which cleaves the TOP1cc from the DNA. The SSB is then repaired by SSBR. c | PARP1 is important for the repair of bulky adducts by global genome nucleotide excision repair (GG-NER). In the initial step, lesion detection, the DNA damage-binding protein 1 (DDB1)–DDB2 complex is recruited to autopoly(ADP) ribosylated (autoPARylated) PARP1. This is followed by the recruitment of amplified in liver cancer protein 1 (ALC1), a step that is also mediated by PARP1. This results in chromatin decondensation, the recruitment of xeroderma pigmentosum group C-complementing protein (XPC) and RAD23B, and lesion verification, which involves recruiting XPB and XPD through the interaction of their binding partner XPA with PARP1. Following lesion verification, the bulky adduct is excised by the concerted action of two nucleases, excision repair cross-complementing group 1 protein (ERCC1) and XPG, and the resulting gap is filled by Pol δ, Pol ε and Pol κ, and ligated by LIG1 or LIG3. PCNA, proliferating cell nuclear antigen; RPA, replication protein A.
SSBs are very rapidly detected and bound by PARP1 (REF. 34). This is followed by the addition of PAR onto PARP1 itself, resulting in its activation, and the addition of PAR on other target proteins. Indeed, using PARP1 inhibition or depletion, PARylation has been shown to increase the rate of SSBR29. The mechanism by which PARP1 activates SSBR could be by promoting the accumulation of SSBR components and/or their stabilization at SSBs. A core factor in SSBR is X-ray repair cross-complementing protein 1 (XRCC1), which acts as a scaffold for SSBR proteins, such as DNA ligase 3 (LIG3), DNA polymerase β and bifunctional polynucleotide kinase 3′-phosphatase (PNKP), thereby stimulating the repair process35–38 (FIG. 2a). The rapid recruitment of XRCC1 to SSBs is dependent on PARP1 or PARP2 (REFS 14,33,35,39,40). A recent study has shown that mutations in XRCC1 in humans and mice lead to defective SSBR and to neuropathological defects owing to futile hyperactivity of PARP1, which arises from unrepaired SSBs41. This hyperactivity of PARP1 results in depletion of cellular NAD+ pools, which leads to cell death. Other possible roles of PARP1 in SSBR may include promotion of the gap-filling step, and an involvement in the final DNA ligation step by mediating the supply of ATP33,42,43 (FIG. 2a).
The repair of another type of single base lesion — single-stranded DNA (ssDNA) nicks — has recently been shown to require the activity of PARP1. ssDNA nicks result from the aborted activity of DNA topoisomerase 1 (TOP1). TOP1 relaxes topological stress in the DNA by transiently nicking one DNA strand, controlling its rotation around the intact strand and, finally, religating the nick. However, the cleavage–religation cycle can be interrupted, resulting in the formation of abortive TOP1–DNA complexes that are known as TOP1 cleavage complexes (TOP1cc). TOP1cc are removed from the DNA by tyrosyl-DNA phosphodiesterase 1 (TDP1), which hydrolyses the phosphodiester bond between the DNA 3′ end and the catalytic TOP1 Tyr moiety44–46. TDP1 is a target of PARP1, and its PARylation not only stabilizes it but also enhances the recruitment of TDP1 to TOP1cc, thereby facilitating their removal (FIG. 2b). The exposed nick can then be a substrate for SSBR, in which the recruitment of XRCC1 is also mediated by the PARP1–TDP1 complex47. PARP1-knockout cells have lower TDP1 activity, strengthening the view that the PARP1–TDP1 complex stimulates the activity of TDP1 (REFS 44,47). This could also explain the epistasis that is observed in the sensitivity of PARP1- or TDP1-deficient cells to TOP1 inhibition, as the TOP1cc that are generated by treatment with TOP1 inhibitors are not repaired owing to the decreased activity of PARP1 (REF. 48).
Does PARP1 function in base excision repair?
BER is one of the major pathways for the repair of oxidative base damage, alkylation damage and abasic sites on the DNA. In BER, damage to bases is detected by apurinic-apyrimidinic (AP) endonucleases that indirectly create SSBs, which are then processed by factors that are shared with SSBR33. Multiple studies have suggested that PARP1 has a role in BER49,50. However, conflicting reports found that PARP1-deficiency or PARP1 inhibition in cells may or may not result in hypersensitivity to agents that are known to cause base lesions50–54. Furthermore, the presence of active PARP1 at sites of damage can reduce the kinetics of the BER process55–57. Together, these reports suggest that PARP1 is not essential for BER, and that the presence of catalytically inactive PARP1 might inhibit the process. This is in contrast to SSBR, in which PARP1 is indeed required for the detection and subsequent repair of SSBs33 (FIG. 2a). Nevertheless, it cannot be ruled out that PARP1 might be required for the downstream processing of the SSBs that arise from the initial steps of BER. In line with this, it has been suggested recently that a subset of DNA lesions, which includes purine base damage that is repaired by the BER pathway, could require PARP1, whereas repair of pyrimidine base damage could be PARP1-independent58.
Chromatin remodelling in global genome nucleotide excision repair.
The NER pathway is responsible for the repair of bulky DNA lesions that arise from multiple sources of mutagenic agents, including ultraviolet (UV) irradiation. Recent studies have shown that PARP1 functions in the initial steps of damage recognition in global genome nucleotide excision repair (GG-NER), which is a dominant subpathway of NER (FIG. 2c). The NER pathway uses multiple proteins for damage recognition, removal of stretches of ssDNA that contain damage, gap filling of the excised region and ligation. UV damage is recognized by xeroderma pigmentosum group C-complementing protein (XPC) and RAD23B, which associate with the DNA damage-binding protein 1 (DDB1)–DDB2 complex59. This complex ubiquitylates core histones, which can lead to nucleosome displacement and can stimulate repair60–62. XPC binds to PAR through its PAR-binding sites, and this is important for XPC recruitment to UV lesions. PARP1 also interacts with DDB2, and its binding to DDB2 at chromatin damaged by UV radiation stimulates its catalytic activity63,64. The stimulation of PARP1 activity results in ubiquitylation-independent chromatin decondensation, which may facilitate repair65 (FIG. 2c). DDB2-stimulated PARylation of histones by PARP1 then mediates the recruitment of the chromatin-remodelling helicase amplified in liver cancer protein 1 (ALC1; also known as CHD1L) through its PAR-binding domains to further stimulate the repair of UV lesions63.
The ensuing UV-lesion recognition by DDB2, which recruits PARP1 to the sites of damage and activates it64,65, results in chromatin remodelling by ALC1, and further recruitment of XPC. Interestingly, recruitment of XPA, which is involved in lesion verification during NER, has also been suggested to be dependent on PAR synthesis66 (FIG. 2d). Whether recruitment of PARP1 to GG-NER sites is mediated only by its interaction with DDB2 or also involves other factors, still needs to be clarified. The role of PARP1 in promoting NER through its interactions with DDB2 and ALC1 appears to be physiologically significant, as PARP inhibition in cells results in sensitivity to UV irradiation63.
DNA double-strand break repair
DSBs are produced following exposure to DNA-damaging agents, such as ionizing irradiation, and endogenously from the collapse of replication forks or during programmed genome rearrangements (which include class-switch recombination (CSR), V(D)J recombination and meiosis)67. DSBs are repaired either through homologous recombination (HR) or through non-homologous end joining (NHEJ), the choice of which is determined by cell cycle phase and chromatin context68,69.
PARP1 as a sensor of DNA double-strand breaks.
PARP1 recognizes DNA breaks, and its activity is involved in the early recruitment of factors to facilitate DSB repair13,70–73. PARP1 deficiency or inhibition results in delayed activation of DDR proteins such as phosphoryl ated histone H2AX, p53 and structural maintenance of chromosomes protein 1 (SMC1), which are targeted to DSBs by the apical DDR kinase ataxia telangiectasia mutated (ATM)74 (FIG. 3). ATM contains PAR-binding domains, and its interactions with PAR stimulate its activity in vitro74,75. However, mice deficient in both PARP1 and ATM display embryonic lethality, suggesting that, despite their interaction, they have redundant roles in the DDR76. PARP1 is also involved in the rapid recruitment of the DNA damage sensors meiotic recombination 11 (MRE11) and Nijmegen breakage syndrome protein 1 (NBS1; also known as nibrin) to sites of DSBs77. MRE11 has a putative PAR-binding domain, suggesting that its recruitment could be dependent on the activity of PARP1 (REF. 77) (FIG. 3). However, there might be other redundant pathways for the activation of the DDR, as PARP1-deficiency only delays the recruitment of certain proteins, but does not completely abolish it74.
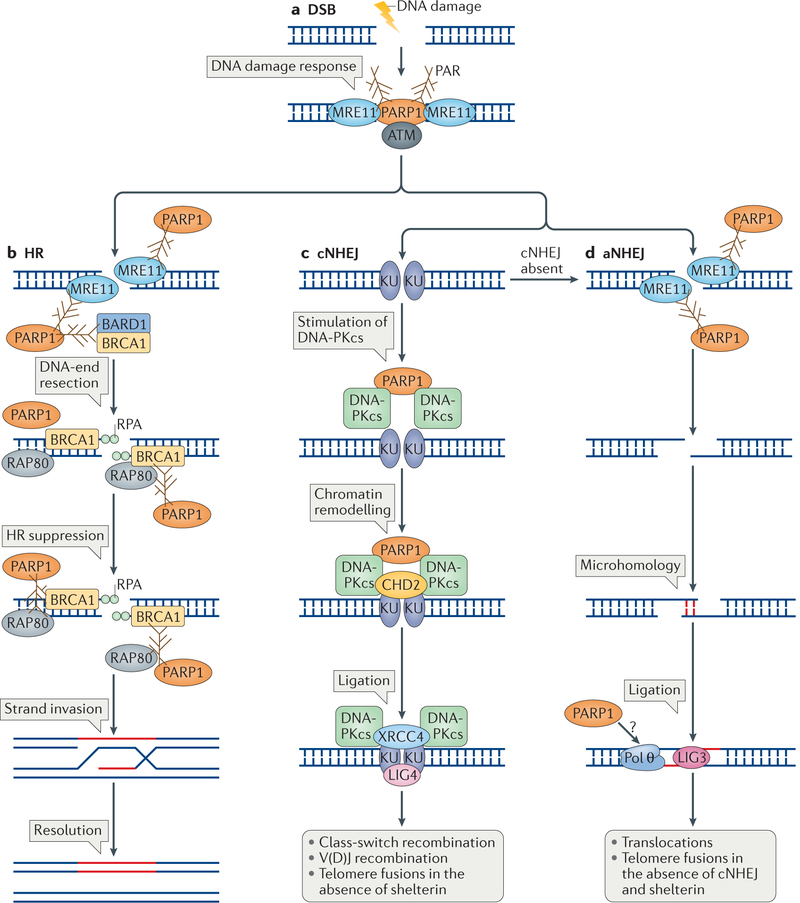
Figure 3 |. The roles of poly(ADP-ribose) polymerase 1 in detection and repair of DNA double-strand breaks.
a | Poly(ADP-ribose) polymerase 1 (PARP1) is required for the robust detection of DNA double-strand breaks (DSBs) and for the initial DNA damage response through its interaction with meiotic recombination 11 (MRE11) and the apical checkpoint kinase ataxia telangiectasia mutated (ATM). b | PARP1 has a role in DNA end resection during the homologous recombination (HR) process through the recruitment of MRE11 to DSBs, which is followed by binding of the single strand by replication protein A (RPA). This reaction also requires breast cancer type 1 susceptibility protein (BRCA1), and PARP1 may stimulate it by indirectly recruiting BRCA1 through its interaction with BRCA1-associated RING domain protein 1 (BARD1). PARP1 is also required at later steps to suppress HR, probably by limiting the extent of DNA end resection. Resection is limited by the stabilization of poly(ADP)ribosylated (PARylated) BRCA1, which is then bound by receptor-associated protein 80 (RAP80). The interaction with RAP80 stabilizes BRCA1 and suppresses HR, which results in limited strand invasion and in the subsequent repair of the DSB. c | PARP1 is also involved in DSB repair by stimulating classical non-homologous end joining (cNHEJ). When DSBs are channelled for repair by cNHEJ, they are bound by KU70–KU80 dimers, which activate DNA-dependent protein kinase catalytic subunit (DNA-PKcs). PARP1 interacts with DNA-PKcs and probably stimulates its activity without the requirement of KU proteins. PARP1 is also involved in chromatin remodelling during cNHEJ by facilitating the recruitment of chromodomain helicase DNA-binding protein 2 (CHD2), which then facilitates the recruitment of X-ray repair cross-complementing protein 4 (XRCC4) and DNA ligase 4 (LIG4) for DNA ligation. d | PARP1 has a role in alternative NHEJ (aNHEJ), which is active in the absence of cNHEJ. aNHEJ requires processing of the DNA ends by MRE11, which is recruited by PARP1. The resected ends are then joined though sequence microhomology and the gap is filled by DNA polymerase θ (Pol θ) and ligated by LIG3. Whether PARP1 is required for the recruitment of Pol θ is unknown (question mark).
The role of PARP1 in homologous recombination.
Early recruitment of the nuclease MRE11 by PARP1 activity could contribute to DNA-end processing at DSBs and could determine DNA repair-pathway choice by channelling the repair of DSBs towards HR. In support of this idea, HR following treatment with DSB-inducing genotoxic agents was reduced in PARP1-deficient chicken DT40 cells78. Similarly to the MRE11–RAD50–NBS1 (MRN) complex, PARP1 has been implicated in the control and recruitment of important HR proteins. Most important among them is breast cancer type 1 susceptibility protein (BRCA1), which controls not only the initial steps of DSB resection but has also been implicated in the loading of RAD51 onto DNA, which is essential for strand exchange during HR79–81. PARP1 is important for the early and rapid recruitment of BRCA1 to DSBs82 (FIG. 3a). However, there are also PARylation-independent mechanisms through which BRCA1 can be recruited to DSBs, such as DNA damage-mediated ubiquitylation83. It remains to be tested whether the PARP1- and ubiquitin-dependent mechanisms are interdependent.
PARP inhibition or PARP1 loss results in a hyper-recombinogenic phenotype, which is evident from the increase in sister chromatid exchanges (SCEs) and the increased level of RAD51 foci formation at sites of DNA damage51,54,84–87. These data suggest that PARP1 limits HR, and indeed it was shown that PARylation of BRCA1 by PARP1 is recognized by receptor-associated protein 80 (RAP80), which stabilizes the BRCA1–RAP80 complex and limits HR88 (FIG. 3b). Loss of PARylation of BRCA1 results in elevated recombination, suggesting that the requirement for PARP1 is to control and fine-tune the HR process in cells88. An alternative explanation for the increased level of SCEs that is observed in PARP1-deficient cells could be the loss of SSBR in these cells, which leads to increased levels of one-ended DSBs and thus a higher frequency of HR. In support of this hypothesis, deficiency of either XRCC1 or TDP1 also results in increased levels of SCEs89,90. This is of clinical interest owing to the synthetic lethality that results from the loss of BRCA1 or BRCA2 in combination with inhibition of PARP activity79,91–93. PARP inhibitors are therefore widely used in preclinical and clinical settings for the treatment of individuals with mutations in BRCA1 or BRCA2 (REFS 92–95). The proposed mechanism for the observed genotoxicity of PARP inhibition in BRCA1- or BRCA2-deficient cells is that PARP inhibition results in an increase in SSBs owing to the role of PARP1 in SSBR. The SSBs are processed into DSBs during DNA replication and are particularly cytotoxic owing to the reduced capacity for DSB repair in BRCA1- or BRCA2-deficient cells92,93. However, alternative but non-mutually exclusive models have been proposed to explain this synthetic lethality, as discussed below.
PARP1 function in non-homologous end joining.
NHEJ is active throughout the cell cycle but is the preferred mechanism for DSB repair in G1, when a DNA template that could be used for HR is absent. In vitro studies have shown that PARP1 can PARylate DNA-dependent protein kinase catalytic subunit (DNA-PKcs), which is an important NHEJ factor. PARylation stimulates the kinase activity of DNA-PKcs, without the requirement for the KU70–KU80 complex96 (FIG. 3c). Furthermore, PARP1 and DNA-PKcs form a complex in vivo, suggesting that they act in the same pathway97; however, cells with a combined deficiency of PARP1 and DNA-PKcs have increased genomic instability and hypersensitivity to ionizing radiation compared with single knockouts, suggesting that the two proteins may act in parallel to repair DSBs98. A recent study showed that PARP1 can influence the classical NHEJ (cNHEJ) pathway by recruiting the chromatin remodeller chromodomain helicase DNA-binding protein 2 (CHD2) to sites of DSBs through the recruitment of XRCC4 (REF. 99) (FIG. 3c). Thus, PARP1 activity could increase the efficiency of cNHEJ.
Alternative NHEJ (aNHEJ) utilizes sequence micro-homologies between two DNA ends to facilitate ligation independently of cNHEJ factors such as LIG4 and the KU proteins, and instead requires processing of the DSB by the MRN complex100. aNHEJ is inherently mutagenic, as it generates insertions or deletions at sites of repair101. For example, aNHEJ can facilitate CSR in the absence of cNHEJ, but this results in deletions and leads to translocations102. PARP1 may promote aNHEJ by competing with the KU complex for access to DNA ends. In the absence of the KU complex, PARP1 binding at DSB sites mediates repair by the aNHEJ pathway103–106. The reduction in the level of translocations following PARP1 inhibition in cells treated with DNA-damaging agents is further evidence that PARP1 mediates aNHEJ107. Similarly, in the absence of shelterin complex proteins, cNHEJ facilitates telomere end-joining and fusions108,109. However, in the absence of cNHEJ, these fusions are still observed and were shown to depend on PARP1-mediated aNHEJ110 (FIG. 3d). PARP1 is also involved in the recruitment of DNA polymerase θ (Pol θ), which mediates aNHEJ through its terminal transferase activity111–113.
Mechanistically, PARP1 may aid in the recruitment of the MRN complex to sites of DSBs. In the absence of KU70 or KU80, the MRN complex has access to the DNA ends and may initiate end processing, giving rise to DNA substrates that channel the repair towards aNHEJ77,103 (FIG. 3d). This could be clinically relevant, as BRCA1-deficient tumours are heavily dependent on Pol θ-mediated, error-prone aNHEJ repair for their survival111. Therefore, it is possible that the synthetic lethal interaction between PARP1 inhibition and BRCA1 deficiency could also be due to the loss of aNHEJ following PARP inhibition in BRCA1-deficient cells, which lack the ability to repair DSBs by HR.
PARP1 at DNA replication forks
Early biochemical studies have shown increased activity of PARP1 upon initiation of DNA synthesis and also at newly replicated chromatin, which suggest that PARP1 is associated with the DNA replication process114,115. In vitro studies have also implicated PARP1 in binding to DNA replication fork-like structures116. Indeed, PARP1 interacts with and stimulates the activity of multiple proteins that are involved in DNA replication117–119. PARP1 is also activated in response to replicative stress, as observed by increased PARylation in cells following treatment with hydroxyurea116. Recent high-throughput analysis of protein dynamics at replication forks has identified PARP1 at normal forks and at stalled forks. PARP1 protein levels were not observed to increase upon fork stalling but the levels of PAR glycohydrolase (PARG), which catalyses the degradation of PAR, are decreased at stalled forks120. Therefore, the increased PARylation observed upon fork stalling could be a result of PARG dissociating from the stalled forks.
Replication fork reversal.
PARP1 activity also regulates the rate of DNA replication fork progression in replication stress. PARP inhibition or its loss result in the abrogation of fork slowdown following treatment with various agents that poison the DNA replication process121–123. This phenomenon has been linked to the role of PARP1 in modulating replication fork reversal, which is a general mechanism of fork stabilization in conditions of replication stress121,124–126. Replication fork reversal is a mechanism in which a three-way junction at a replication fork is converted to a four-way junction by the annealing of the two newly replicated strands into a regressed arm at the forks125 (FIG. 4a). Fork reversal was long proposed to be a mechanism for lesion bypass and DNA damage tolerance in replication stress in bac teria127. Recent studies in human cells have uncovered that fork reversal is an evolutionarily conserved mechanism that stabilizes replication forks that are exposed to multiple sources of DNA replication stress125. Inhibition of PARP1 activity in conditions of mild TOP1 inhibition prevents the formation of reversed forks, giving rise to increased genomic instability121. Subsequently, ATP-dependent DNA helicase Q1 (RECQ1) was identified as a target of PARP1 activity and shown to be involved in the restart of reversed replication forks128. RECQ1 binds to PARylated PARP1, and this interaction inhibits the heli-case activity of RECQ1, thereby preventing the untimely restart of stalled replication forks and allowing repair of lesions ahead of the fork. Inhibition of PARP1 results in the activation of RECQ1-mediated fork restart in conditions of replication stress128 (FIG. 4a). This mechanism can explain the abrogation of fork slowdown observed upon PARP inhibition during replicative stress, as stalled forks could be restarted in an untimely manner owing to hyperactivity of RECQ1. This untimely restart could then lead to replication fork ‘run-off’ into unrepaired lesions ahead of the fork, resulting in fork collapse and subsequent DSB formation121,128.
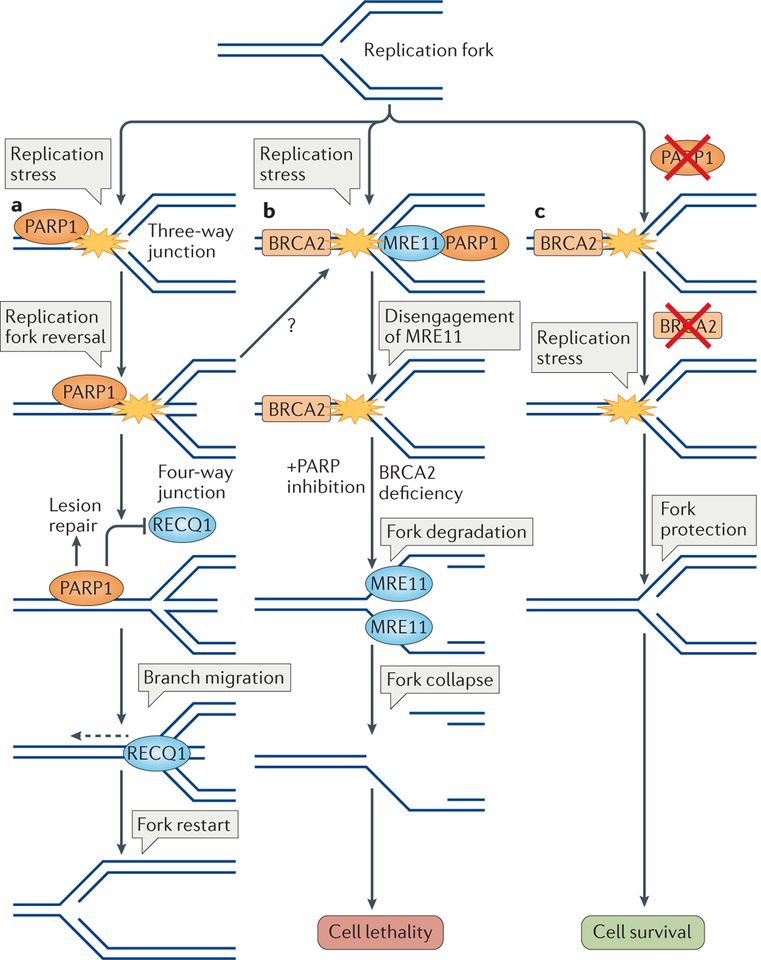
Figure 4 |. Poly(ADP-ribose) polymerase 1 helps maintain the stability of replication forks.
a | Replication fork reversal and restart. Various types of replication stress-inducing agents cause replication fork reversal to stabilize stalled replication forks. Poly(ADP-ribose) polymerase 1 (PARP1) is recruited to stalled replication forks for lesion detection and repair, in coordination with the replication machinery (not shown). PARP1 stabilizes reversed forks by inhibiting the activity of ATP-dependent DNA helicase Q1 (RECQ1), which can otherwise restart the fork. Following the repair of the lesion (not shown), RECQ1 can be activated, which allows branch migration of the reversed fork (dashed arrow), thereby mediating replication restart. b | Synthetic lethality. Stalled replications forks are detected by PARP1, which recruits the nuclease meiotic recombination 11 (MRE11) to mediate fork restart through limited processing of DNA at the forks. Disengagement of MRE11 from replication forks is required for their protection, which might be mediated by breast cancer type 2 susceptibility protein (BRCA2). In the absence of BRCA2, MRE11 has unlimited access to stalled forks. In combination with PARP inhibition, which limits PARP1 control of MRE11, this results in excessive DNA degradation, which might result in fork collapse and DSB formation. As cells lacking BRCA2 are deficient in DSB repair by homologous recombination (HR), PARP1 inhibition-mediated fork collapse and DSB formation results in cellular lethality. c | Synthetic viability. Loss of PARP1 activity results in defective MRE11 recruitment. If BRCA2 activity is lost after the loss of PARP1, MRE11 accessibility is diminished at stalled replication forks and they are therefore protected from degradation. Such cells are defective in DSB repair by HR, but they are viable owing to replication fork protection.
PARP1-mediated fork reversal and stabilization is adopted by cells following the introduction of a variety of replication stress-inducing agents123, and it is becoming increasingly clear that PARP1-mediated fork reversal is important for the maintenance of genome stability, not only upon treatment of cells with replication poisons but also in the face of endogenous replication stress, such as during replication of repeat regions and in conditions of oncogenic stress. In line with this idea, depletion of PARG results in elevated levels of PAR in regions in which there is a high frequency of fork reversal, probably by limiting RECQ1-mediated restart of reversed forks31. Furthermore, PARP1-dependent replication slowdown and reversal was found to occur frequently in proliferating mouse embryonic stem cells129. Taken together, these results signify that PARP1 is a crucial mediator of the stabilization of replication forks through mediating fork reversal, both during unperturbed DNA replication and in the face of exogenous replicative stress.
Replication fork processing.
PARP1 has also been implicated in the restart of stalled forks after release from replication blocks. PARP1-dependent replication restart is proposed to be mediated by the recruitment of MRE11 to stalled forks to allow for limited resection of the forks to initiate restart116. Indeed, PARP1 and PAR have been shown to interact with and recruit MRE11 to stalled forks77,116,130. However, PARP1 has also been reported to prevent excessive processing of DNA at stalled replication forks by MRE11 (REF. 131). Excessive fork degradation by MRE11 is observed in the context of BRCA deficiency, in which replication forks become highly unstable upon fork stalling131–133. In addition, BRCA2-deficient cells display increased degradation of stalled forks following PARP inhibition, suggesting that PARP1 could limit the amount of fork degradation in BRCA2-deficient cells. This additive effect of PARP1 inhibition on the amount of fork degradation could in part explain the synthetic lethality seen in BRCA2-deficient tumours that are subjected to PARP inhibition (FIG. 4b). Whether this additional degradation of forks upon PARP inhibition affects replication restart still remains to be tested, as cells with BRCA2 deficiency do not display fork restart defects132.
PARP inhibition can also trap PARP1 on the DNA, resulting in the formation of a DNA–protein complex (DPC)134. DPCs can potentially block the replication machinery, which then requires BRCA1 or BRCA2 for its stabilization132,133. Furthermore, because PARP1 has also been implicated in the protection and restart of stalled forks131,135, PARP inhibition would be expected to result in fork collapse owing to the combined effects of forming DPCs (replication barriers) and destabilizing forks by preventing their restart. These DSBs would not be repaired in the absence of BRCA proteins, thus causing synthetic lethality.
Interestingly, BRCA2-deficient cells have increased PARP1 and MRE11 activity131,136,137. The high activity of MRE11 is correlated with the high levels of PAR that have been observed in BRCA2-deficient cells and has been implicated in the increased genomic instability and loss of viability observed in these cells130,132,137. Increased levels of PAR in PARG-depleted cells has also been linked to replicative stress and growth defects, which suggests that a tight control over cellular PAR levels is essential for the maintenance of genomic stability31,138. Endogenous fork stalling in BRCA2-deficient cells could result in increased PARP1 activity and MRE11 recruitment at sites of stalled forks. Although in normal conditions MRE11 would disengage from forks, a deficiency in BRCA2 could result in the retention of MRE11 at stalled forks, resulting in increased fork degradation and genomic instability (FIG. 4b). One implication of such a model is that a reduction of MRE11 levels at stalled forks could result in a rescue of genomic instability and in synthetic viability — a genetic interaction in which a cell that is non-viable owing to the loss of a gene becomes viable when the original loss is combined with the loss of another gene. Recent studies have shown that either depletion or inhib ition of PARP1 can result in a rescue of the viability of BRCA2-deficient mouse embryonic stem (mES) cells and can result in genomic stability in differentiated cells130.
The timing of PARP1 depletion or inhibition appears to be important in this context, as the loss of PARP1 must take place before the loss of BRCA2. Viable mES cells in which PARP1 is depleted before deletion of BRCA2 are deficient in HR-mediated DSB repair and have proliferation defects. However, replication forks in these HR-deficient cells do not undergo degradation upon replication stress. Therefore, protection of replication forks by limiting PARP1-mediated MRE11 access to forks would stabilize replication forks and result in lower levels of genomic instability and in synthetic viability and chemoresistance in BRCA-deficient cells130,137 (FIG. 4c). However, the rescue of viability upon BRCA2 deficiency is limited to mES cells, and a loss of PARP1 in BRCA2-deficient embryos results in embryonic lethality. This is in contrast to loss of TP53BP1, which rescues HR and viability in BRCA1-deficient mice79. A loss of other chromatin-associated proteins, such as PAX transactivation activation domain-interacting protein (PTIP; also known as PAXIP1) and CHD4, has also been implicated in replication fork protection-mediated chemoresistance in BRCA-deficient cells137,139. However, an understanding of the link between these factors and PARP1 requires further study. Although both replication fork protection and HR contribute to genome integrity and chemo-resistance, HR appears to be the dominant pathway that promotes fitness.
PARP1 modulates chromatin structure
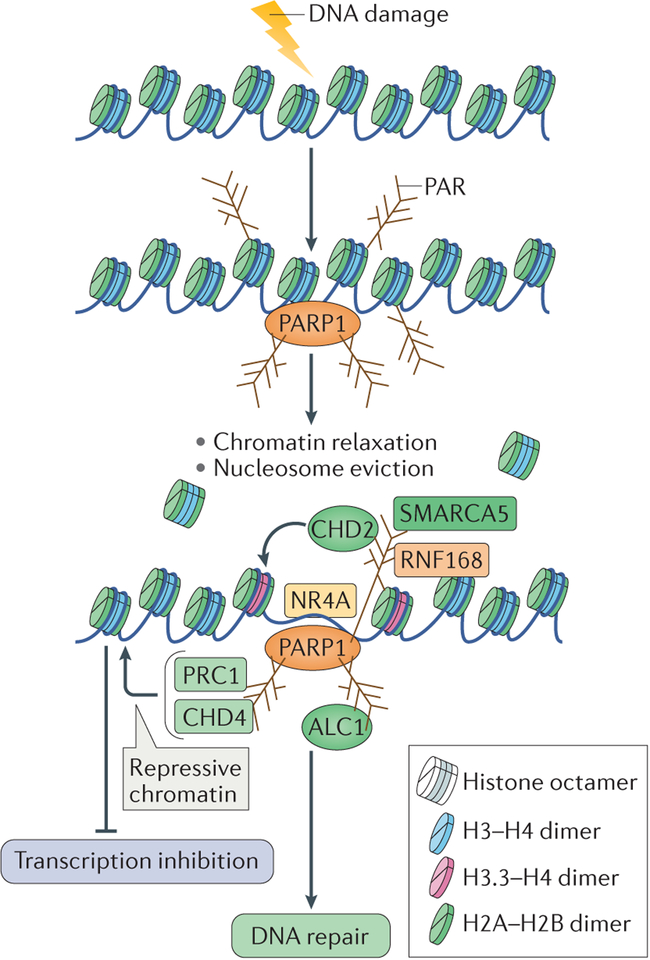
The response to DNA damage and its repair in cells necessitates a coordination of multiple cellular processes, such as transcription and replication, which is dependent on the chromatin environment. Chromatin structure is highly reorganized in response to DNA damage to facilitate a favourable environment for the accurate repair of damage140. In addition to its role in controlling transcription12,141–143 (BOX 2), PARP1 has been implicated in the modulation of chromatin structure that facilitates repair in response to various kinds of DNA damage. Biochemical studies have shown that PARP1 can facilitate nucleosome disassembly by PARylating histones, which results in chromatin relaxation144,145. This in turn supports the recruitment of multiple chromatin remodellers, thereby making chromatin accessible for DNA repair (FIG. 5). The relaxation of chromatin could explain the global impact of PARP1 in mediating the different DNA repair processes that are described above.
Box 2 |. The role of poly(ADP-ribose) polymerase 1 in transcription.
In addition to its role in DNA repair and replication, poly(ADP-ribose) polymerase 1 (PARP1) also has varied roles in controlling the functions of various components of the transcription machinery. PARP1 localizes to the promoters of actively transcribed genes and limits the binding of histone H1 to DNA, to promote an ‘open’ chromatin environment that is favourable for transcription168. The transcription regulation function of PARP1 may not always require its enzymatic activity and can include binding to enhancers, modulation of chromatin structure or direct regulation of transcription factors169. PARP1 can regulate the activity of transcription factors by recruiting transcription co-regulators; for example, it recruits the Lys acetyltransferase p300 to nuclear factor-κB to stimulate its transcriptional activity169. Poly(ADP) ribosylation (PARylation) of transcription regulators can also have a role in gene regulation. For example, PARP1 has been shown to PARylate subunits of the negative elongation factor (NELF), NELFA and NELFE. PARylation of NELF proteins promotes transcription elongation by RNA polymerase II, by releasing it from its paused state170. However, PARP1 binding at regulatory regions of genes can also correlate with transcription repression12,171, which may potentially occur in concert with the role of PARP1 in mediating DNA repair at actively transcribing regions to prevent interference of the transcription machinery with the DNA repair process.
Figure 5 |. Chromatin changes induced by poly(ADP-ribose) polymerase 1 — integrating DNA repair.
DNA damage in the context of chromatin results in poly(ADP-ribose) polymerase 1 (PARP1) activation and autopoly(ADP)ribosylation (autoPARylation). PARP1 also PARylates histone tails, which results in chromatin relaxation and in nucleosome eviction from the DNA. This also allows the recruitment of several chromatin remodellers through their binding to PAR, which further relax chromatin to facilitate DNA repair. These chromatin remodellers include amplified in liver cancer protein 1 (ALC1), SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 (SMARCA5), chromodomain helicase DNA-binding protein 2 (CHD2), which also incorporates the histone variant H3.3 into nucleosomes, and the transcription factor NR4A (a member of the nuclear orphan receptors). Conversely, repressive chromatin modifiers, such as CHD4 and Polycomb repressive complex 1 (PRC1), are also recruited to sites of DNA damage by binding to PAR. They might be recruited to inhibit transcription at flanking regions to facilitate DNA repair. RNF168, RING finger protein 168.
One of the best-studied chromatin remodellers that is recruited by PARP1 activity to sites of DNA damage is ALC1, a member of the sucrose non-fermenting 2 (SNF2) family of chromatin remodellers, which is overexpressed in hepatocellular carcinomas146,147. The recruitment of ALC1 to sites of DNA damage by its binding to PAR results in nucleosome repositioning and increased accessibility of DNA repair factors to sites of damage148,149. Similarly to the loss of PARP1, loss of ALC1 leads to sensitivity to DNA-damaging agents and to delayed repair, thereby substantiating a role for PARP1-mediated chromatin remodelling in the repair of DNA damage63,148 (FIG. 5).
The catalytic subunit of imitation switch (ISWI) chromatin-remodelling complexes, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 (SMARCA5), is also recruited to DSBs in a PARP1-dependent manner150 (FIG. 5). In vivo studies showed that SMARCA5 interacts with the PARylated E3 ubiquitin ligase RING finger protein 168 (RNF168), and thus is involved in the NHEJ repair pathway. The physiological significance of this interaction is highlighted by the fact that depletion of SMARCA5 in cells results in their sensitivity to ionizing radiation150. CHD2 is another chromatin remodeller that is recruited to DSBs in a PARP1-dependent manner; this triggers the deposition of the histone variant H3.3 at these regions and results in chromatin relaxation, which promotes efficient repair by NHEJ99 (FIG. 5).
In addition to chromatin remodellers, the transcription factor NR4A (a member of the nuclear orphan receptors) also translocates to sites of DSBs following PARP1-mediated chromatin relaxation (FIG. 5). This results in its phosphorylation by DNA-PKcs and facilitates DNA repair151. The DNA repair activity of NR4A is thought to be independent of its role in mediating transcription.
Finally, faithful repair of DSBs also requires the inhibition of transcription at sites of DSB repair152,153. Transcription repression complexes are recruited to sites of DNA damage by PAR — prominent among them are the nucleosome remodelling and deacetylase (NuRD) complex proteins CHD4 and metastasis-associated protein 1 (MTA1)154,155, and members of Polycomb repressive complex 1 (PRC1)154. The localization of the components of the NuRD and PRC1 complexes to sites of DNA damage has been linked to repression of transcription at these regions, as shown by the absence of RNA polymer-ase II from these sites. Therefore, the local chromatin environment at sites of DNA damage could be shaped by PARP1 through two complementary mechanisms: first, chromatin relaxation through nucleosome eviction and the recruitment of chromatin remodellers; and second, repression of transcriptional activity by the recruitment of repressive chromatin modifiers, to prevent interference with the DNA repair machinery by the transcription machinery (BOX 2; FIG. 5).
Future perspectives
PARP1 regulates many cellular functions, which include multiple pathways of DNA repair. Thus, it is not surprising that PARP1 has been used as a pharmacological target for the treatment of multiple types of cancers that have DNA repair defects, by exploiting the phenomenon of synthetic lethality. However, the intricate mechanisms by which PARP1 regulates the different DNA repair processes, and the interplay between the repair process and the transcription programme, require further study. This has been difficult, in part because DNA-damaging agents that are used to study the role of PARP1 can have pleiotropic effects. For example, ionizing irradiation, which is a standard laboratory tool to induce DSBs in cells, also induces other lesions, such as SSBs and base damage, all of which may require PARP1 for their efficient repair. Furthermore, some lesions might be converted to others; for example, SSBs may be converted to DSBs during replication. Therefore, there is a need to develop targeted tools, such as CRISPR–Cas9 technology: the use of different versions of the Cas9 protein could be exploited to generate a specific type of DNA damage — single-stranded nicks, DSBs or DPCs — at targeted genomic loci. The development of such tools will then allow the dissection of the roles of PARP1 in the repair of specific lesions. Furthermore, a major focus of future studies of PARP1 should be to understand the relationship between distinct but interrelated processes such as DNA replication and repair, which are coordinated to bring about the accurate repair of damaged DNA.
Pharmacological inhibitors that target PARP1 (and other PARPs) are now either being used in the clinic or are in the final phases of clinical trials for the treatment of breast (https://www.breastcancertrials.org) and ovarian cancers that contain mutations in BRCA2, taking advantage of the synthetic lethality caused by a deficiency in both PARP1 and BRCA2. Recent phase II clinical trials, which evaluated the efficacy of PARP inhibitors for the treatment of metastatic prostate cancer that contains defects in DNA repair genes, have also shown promising results156. These data indicate that PARP inhibitors could be used to treat different types of cancer that display defects in DNA repair genes. Therefore, it has been suggested that PARP inhibitor treatment should be used as a preventive agent in high-risk populations157. However, recent studies have uncovered synthetic viability between PARP1 loss and BRCA1 and/or BRCA2 loss, which occurs if PARP1 is depleted before the loss of BRCA2 (REF. 130). This raises concern about the use of PARP inhibitors as a preventive agent in high-risk populations, as early inhibition of PARP may facilitate the survival of cells that contain hetero zygous BRCA2 mutations, which can then undergo loss of heterozygosity.
In conclusion, additional studies of the role of PARP1 in modulating DNA repair processes will provide valuable insights into the basic biological mechanisms that are governed by PARP1 and could also result in the future development of novel cancer therapies.
DNA damage response.
(DDR). The collection of cellular pathways that detect, signal and repair DNA damage.
BRCT domain.
(BRCA1 C terminus domain). An evolutionarily conserved protein domain that has DNA repair functions; it contains phosphoprotein-binding sites.
Abasic sites.
DNA sites that lack either a purine or a pyrimidine base owing to endogenous and/or exogenous DNA damage.
Epistasis.
A genetic interaction in which a mutation in one gene masks the effects of a mutation in another gene.
Oxidative base damage.
Damage to DNA bases caused by oxidation, which mostly modifies guanine to produce 8-hydroxyguanine.
Alkylation damage.
DNA damage mediated by transfer of a single methyl group to a DNA base (mostly to N or O atoms of guanine), which results in the formation of a methyl adduct on the base.
Lesion verification.
Verification of a chemical modification on the DNA by the transcription and repair factor transcription factor IIH (TFIIH) during nucleotide excision repair.
Class-switch recombination.
(CSR). A process in B cells that involves switching the type of antibody that is produced by changing the constant region of the antibody heavy chain.
V(D)J recombination.
A DNA recombination process that occurs during B cell or T cell activation, in which the variable domain exons of antigen receptors are assembled from sub-exonic segments called V, D and J to ultimately generate an immunoglobulin gene or T cell receptor, respectively.
One-ended DSBs.
(One-ended DNA double-strand breaks). DSBs formed during collision of ongoing replications forks with a lesion on one strand of the template DNA.
Shelterin complex.
A complex of six proteins that binds to TTAGGG repeats at telomeres and protects them from recognition as DNA double-strand breaks.
Terminal transferase.
An enzyme that catalyses the addition of nucleotides to 3′ DNA overhangs at double-stranded DNA.
Acknowledgements
The authors are especially grateful to A. Tubbs, S. John and G. Poirier for comments on the manuscript and also for discussions. This work was supported by the Intramural Research Program of the US National Institutes of Health (NIH), the US National Cancer Institute and the Center for Cancer Research. A.N. was also supported by the US Department of Defense (BCRP DOD Idea Expansion Award BC133858 and BCRP Breakthrough Award BC151331), the Ellison Foundation Award for Aging Research and Alex’s Lemonade Stand Foundation Reach Award. A.R.C. has been supported by a Human Frontier Science Program Long-term Fellowship (LT000393/2013).
Footnotes
Competing interests statement
The authors declare no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tubbs A & Nussenzweig A Endogenous DNA damage as a source of genomic instability in cancer. Cell 168, 644–656 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lord CJ, Tutt AN & Ashworth A Synthetic lethality and cancer therapy: lessons learned from the development of PARP inhibitors. Annu. Rev. Med 66, 455–470 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Montoni A, Robu M, Pouliot E & Shah GM Resistance to PARP-inhibitors in cancer therapy. Front. Pharmacol 4, 18 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ame JC, Spenlehauer C & de Murcia G The PARP superfamily. Bioessays 26, 882–893 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Buki KG & Kun E Polypeptide domains of ADP-ribosyltransferase obtained by digestion with plasmin. Biochemistry 27, 5990–5995 (1988). [DOI] [PubMed] [Google Scholar]
- 6.Froelich CJ et al. Granzyme B/perforin-mediated apoptosis of Jurkat cells results in cleavage of poly(ADP-ribose) polymerase to the 89-kDa apoptotic fragment and less abundant 64-kDa fragment. Biochem. Biophys. Res. Commun 227, 658–665 (1996). [DOI] [PubMed] [Google Scholar]
- 7.Kameshita I, Matsuda Z, Taniguchi T & Shizuta Y Poly (ADP-ribose) synthetase. Separation and identification of three proteolytic fragments as the substrate-binding domain, the DNA-binding domain, and the automodification domain. J. Biol. Chem 259, 4770–4776 (1984). [PubMed] [Google Scholar]
- 8.Langelier MF, Planck JL, Roy S & Pascal JM Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science 336, 728–732 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article reports the crystal structure of PARP1 bound to a DNA DSB and proposes a mechanism for the DNA-dependent activation of PARP1.
- 9.Nishikimi M, Ogasawara K, Kameshita I, Taniguchi T & Shizuta Y Poly(ADP-ribose) synthetase. The DNA binding domain and the automodification domain. J. Biol. Chem 257, 6102–6105 (1982). [PubMed] [Google Scholar]
- 10.Bork P et al. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 11, 68–76 (1997). [PubMed] [Google Scholar]
- 11.D’Amours D, Desnoyers S, D’Silva I & Poirier GG Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J 342, 249–268 (1999). [PMC free article] [PubMed] [Google Scholar]
- 12.Kraus WL & Lis JT PARP goes transcription. Cell 113, 677–683 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Kim MY, Zhang T & Kraus WL Poly(ADP-ribosyl) ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 19, 1951–1967 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Hanzlikova H, Gittens W, Krejcikova K, Zeng Z & Caldecott KW Overlapping roles for PARP1 and PARP2 in the recruitment of endogenous XRCC1 and PNKP into oxidized chromatin. Nucleic Acids Res. 45, 2546–2557 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isabelle M et al. Investigation of PARP-1, PARP-2, and PARG interactomes by affinity-purification mass spectrometry. Proteome Sci. 8, 22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menissier de Murcia J et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 22, 2255–2263 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huambachano O, Herrera F, Rancourt A & Satoh MS Double-stranded DNA binding domain of poly(ADP-ribose) polymerase-1 and molecular insight into the regulation of its activity. J. Biol. Chem 286, 7149–7160 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gagne JP et al. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 36, 6959–6976 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jungmichel S et al. Proteome-wide identification of poly(ADP-ribosyl)ation targets in different genotoxic stress responses. Mol. Cell 52, 272–285 (2013). [DOI] [PubMed] [Google Scholar]; This article reports the high-throughput identification of targets of PARylation in response to different genotoxic stresses.
- 20.Hassa PO & Hottiger MO The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front. Biosci 13, 3046–3082 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Krietsch J et al. Reprogramming cellular events by poly(ADP-ribose)-binding proteins. Mol. Aspects Med 34, 1066–1087 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teloni F & Altmeyer M Readers of poly(ADP-ribose): designed to be fit for purpose. Nucleic Acids Res. 44, 993–1006 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Althaus FR et al. Poly ADP-ribosylation: a DNA break signal mechanism. Mol. Cell. Biochem 193, 5–11 (1999). [PubMed] [Google Scholar]
- 24.Malanga M, Pleschke JM, Kleczkowska HE & Althaus FR Poly(ADP-ribose) binds to specific domains of p53 and alters its DNA binding functions. J. Biol. Chem 273, 11839–11843 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Pleschke JM, Kleczkowska HE, Strohm M & Althaus FR Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem 275, 40974–40980 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Meyer-Ficca ML, Meyer RG, Coyle DL, Jacobson EL & Jacobson MK Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Exp. Cell Res 297, 521–532 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Erdelyi K et al. Dual role of poly(ADP-ribose) glycohydrolase in the regulation of cell death in oxidatively stressed A549 cells. FASEB J. 23, 3553–3563 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng X & Koh DW Inhibition of poly(ADP-ribose) polymerase-1 or poly(ADPribose) glycohydrolase individually, but not in combination, leads to improved chemotherapeutic efficacy in HeLa cells. Int. J. Oncol 42, 749–756 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fisher AEO, Hochegger H, Takeda S & Caldecott KW Poly(ADP-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol. Cell. Biol 27, 5597–5605 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koh DW et al. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc. Natl Acad. Sci. USA 101, 17699–17704 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article reports the essential role of PARG in degradation of PAR.
- 31.Ray Chaudhuri A, Ahuja AK, Herrador R & Lopes M Poly(ADP-ribosyl) glycohydrolase prevents the accumulation of unusual replication structures during unperturbed S phase. Mol. Cell. Biol 35, 856–865 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Y, Feng X & Koh DW Enhanced DNA accessibility and increased DNA damage induced by the absence of poly(ADP-ribose) hydrolysis. Biochemistry 49, 7360–7366 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Caldecott KW Single-strand break repair and genetic disease. Nat. Rev. Genet 9, 619–631 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Satoh MS & Lindahl T Role of poly(Adp-ribose) formation in DNA-repair. Nature 356, 356–358 (1992). [DOI] [PubMed] [Google Scholar]
- 35.Caldecott KW, McKeown CK, Tucker JD, Ljungquist S & Thompson LH An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol. Cell. Biol 14, 68–76 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loizou JI et al. The protein kinase CK2 facilitates repair of chromosomal DNA single-strand breaks. Cell 117, 17–28 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Marintchev A et al. Domain specific interaction in the XRCC1-DNA polymerase beta complex. Nucleic Acids Res. 28, 2049–2059 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitehouse CJ et al. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell 104, 107–117 (2001). [DOI] [PubMed] [Google Scholar]
- 39.El-Khamisy SF, Masutani M, Suzuki H & Caldecott KW A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 31, 5526–5533 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article shows the requirement for PARP1 in the recruitment of XRCC1, which is an essential factor in the repair of SSBs.
- 40.Schreiber V et al. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem 277, 23028–23036 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Hoch NC et al. XRCC1 mutation is associated with PARP1 hyperactivation and cerebellar ataxia. Nature 541, 87–91 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oei SL & Ziegler M ATP for the DNA ligation step in base excision repair is generated from poly(ADP-ribose). J. Biol. Chem 275, 23234–23239 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Petermann E, Ziegler M & Oei SL ATP-dependent selection between single nucleotide and long patch base excision repair. DNA Repair 2, 1101–1114 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Pommier Y Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer 6, 789–802 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Pouliot JJ, Yao KC, Robertson CA & Nash HA Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science 286, 552–555 (1999). [DOI] [PubMed] [Google Scholar]
- 46.Yang SW et al. A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc. Natl Acad. Sci. USA 93, 11534–11539 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Das BB et al. PARP1-TDP1 coupling for the repair of topoisomerase I-induced DNA damage. Nucleic Acids Res. 42, 4435–4449 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article elucidates the interaction of PARP1 and TDP1 and its important role in the repair of TOP1-abortive complexes.
- 48.Patel AG et al. Enhanced killing of cancer cells by poly(ADP-ribose) polymerase inhibitors and topoisomerase I inhibitors reflects poisoning of both enzymes. J. Biol. Chem 287, 4198–4210 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dantzer F et al. Base excision repair is impaired in mammalian cells lacking poly(ADP-ribose) polymerase-1. Biochemistry 39, 7559–7569 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Dantzer F et al. Involvement of poly(ADP-ribose) polymerase in base excision repair. Biochimie 81, 69–75 (1999). [DOI] [PubMed] [Google Scholar]
- 51.de Murcia JM et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc. Natl Acad. Sci. USA 94, 7303–7307 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pachkowski BF et al. Cells deficient in PARP-1 show an accelerated accumulation of DNA single strand breaks, but not AP sites, over the PARP-1-proficient cells exposed to MMS. Mutat. Res 671, 93–99 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vodenicharov MD, Sallmann FR, Satoh MS & Poirier GG Base excision repair is efficient in cells lacking poly(ADP-ribose) polymerase 1. Nucleic Acids Res. 28, 3887–3896 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang ZQ et al. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 11, 2347–2358 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Allinson SL, Dianova II & Dianov GL Poly(ADP-ribose) polymerase in base excision repair: always engaged, but not essential for DNA damage processing. Acta Biochim. Pol 50, 169–179 (2003). [PubMed] [Google Scholar]
- 56.Orta ML et al. The PARP inhibitor Olaparib disrupts base excision repair of 5-aza-2′-deoxycytidine lesions. Nucleic Acids Res. 42, 9108–9120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strom CE et al. Poly (ADP-ribose) polymerase (PARP) is not involved in base excision repair but PARP inhibition traps a single-strand intermediate. Nucleic Acids Res. 39, 3166–3175 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reynolds P, Cooper S, Lomax M & O’Neill P Disruption of PARP1 function inhibits base excision repair of a sub-set of DNA lesions. Nucleic Acids Res. 43, 4028–4038 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marteijn JA, Lans H, Vermeulen W & Hoeijmakers JHJ Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol 15, 465–481 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Guerrero-Santoro J et al. The cullin 4B-based UV-damaged DNA-binding protein ligase binds to UV-damaged chromatin and ubiquitinates histone H2A. Cancer Res. 68, 5014–5022 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Kapetanaki MG et al. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc. Natl Acad. Sci. USA 103, 2588–2593 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang H et al. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol. Cell 22, 383–394 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Pines A et al. PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1. J. Cell Biol. 199, 235–249 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]; This report shows the role of PARP1 in mediating NER through the recruitment of DDB2 and the chromatin modifier ALC1.
- 64.Robu M et al. Role of poly(ADP-ribose) polymerase-1 in the removal of UV-induced DNA lesions by nucleotide excision repair. Proc. Natl Acad. Sci. USA 110, 1658–1663 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luijsterburg MS et al. DDB2 promotes chromatin decondensation at UV-induced DNA damage. J. Cell Biol. 197, 267–281 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.King BS, Cooper KL, Liu KJ & Hudson LG Poly(ADP-ribose) contributes to an association between poly(ADP-ribose) polymerase-1 and xeroderma pigmentosum complementation group A in nucleotide excision repair. J. Biol. Chem 287, 39824–39833 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mehta A & Haber JE Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol 6, a016428 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chapman JR, Taylor MRG & Boulton SJ Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 47, 497–510 (2012). [DOI] [PubMed] [Google Scholar]
- 69.Price BD & D’Andrea AD Chromatin remodeling at DNA double-strand breaks. Cell 152, 1344–1354 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ali AA et al. The zinc-finger domains of PARP1 cooperate to recognize DNA strand breaks. Nat. Struct. Mol. Biol 19, 685–692 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Langelier MF & Pascal JM PARP-1 mechanism for coupling DNA damage detection to poly(ADP-ribose) synthesis. Curr. Opin. Struct. Biol 23, 134–143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Polo SE & Jackson SP Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 25, 409–433 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sukhanova MV et al. Single molecule detection of PARP1 and PARP2 interaction with DNA strand breaks and their poly(ADP-ribosyl)ation using high-resolution AFM imaging. Nucleic Acids Res. 44, e60 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haince JF et al. Ataxia telangiectasia mutated (ATM) signaling network is modulated by a novel poly(ADP-ribose)-dependent pathway in the early response to DNA-damaging agents. J. Biol. Chem 282, 16441–16453 (2007). [DOI] [PubMed] [Google Scholar]
- 75.Aguilar-Quesada R et al. Interaction between ATM and PARP-1 in response to DNA damage and sensitization of ATM deficient cells through PARP inhibition. BMC Mol. Biol 8, 29 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Menisser-de Murcia J, Mark M, Wendling O, Wynshaw-Boris A & de Murcia G Early embryonic lethality in PARP-1 Atm double-mutant mice suggests a functional synergy in cell proliferation during development. Mol. Cell. Biol 21, 1828–1832 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Haince JF et al. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem 283, 1197–1208 (2008). [DOI] [PubMed] [Google Scholar]
- 78.Hochegger H et al. Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. EMBO J. 25, 1305–1314 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bunting SF et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cruz-Garcia A, Lopez-Saavedra A & Huertas P BRCA1 accelerates CtIP-mediated DNA-end resection. Cell Rep. 9, 451–459 (2014). [DOI] [PubMed] [Google Scholar]
- 81.Scully R et al. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 88, 265–275 (1997). [DOI] [PubMed] [Google Scholar]
- 82.Li M & Yu X Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell 23, 693–704 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schwertman P, Bekker-Jensen S & Mailand N Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat. Rev. Mol. Cell Biol 17, 379–394 (2016). [DOI] [PubMed] [Google Scholar]
- 84.Morgan WF & Cleaver JE 3-Aminobenzamide synergistically increases sister-chromatid exchanges in cells exposed to methyl methanesulfonate but not to ultraviolet light. Mutat. Res 104, 361–366 (1982). [DOI] [PubMed] [Google Scholar]
- 85.Oikawa A, Tohda H, Kanai M, Miwa M & Sugimura T Inhibitors of poly(adenosine diphosphate ribose) polymerase induce sister chromatid exchanges. Biochem. Biophys. Res. Commun 97, 1311–1316 (1980). [DOI] [PubMed] [Google Scholar]
- 86.Schultz N, Lopez E, Saleh-Gohari N & Helleday T Poly(ADP-ribose) polymerase (PARP-1) has a controlling role in homologous recombination. Nucleic Acids Res. 31, 4959–4964 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang YG, Cortes U, Patnaik S, Jasin M & Wang ZQ Ablation of PARP-1 does not interfere with the repair of DNA double-strand breaks, but compromises the reactivation of stalled replication forks. Oncogene 23, 3872–3882 (2004). [DOI] [PubMed] [Google Scholar]
- 88.Hu Y et al. PARP1-driven poly-ADP-ribosylation regulates BRCA1 function in homologous recombination-mediated DNA repair. Cancer Discov. 4, 1430–1447 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.El-Khamisy SF et al. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature 434, 108–113 (2005). [DOI] [PubMed] [Google Scholar]
- 90.Fan J et al. XRCC1 down-regulation in human cells leads to DNA-damaging agent hypersensitivity, elevated sister chromatid exchange, and reduced survival of BRCA2 mutant cells. Environ. Mol. Mutagen 48, 491–500 (2007). [DOI] [PubMed] [Google Scholar]
- 91.Bouwman P et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol 17, 688–695 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bryant HE et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 (2005). [DOI] [PubMed] [Google Scholar]
- 93.Farmer H et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005). [DOI] [PubMed] [Google Scholar]; References 92 and 93 are the first reports of the synthetic lethality of the combined loss of PARP1 and BRCA2.
- 94.Evers B, Helleday T & Jonkers J Targeting homologous recombination repair defects in cancer. Trends Pharmacol. Sci 31, 372–380 (2010). [DOI] [PubMed] [Google Scholar]
- 95.Rottenberg S et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc. Natl Acad. Sci. USA 105, 17079–17084 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ruscetti T et al. Stimulation of the DNA-dependent protein kinase by poly(ADP-ribose) polymerase. J. Biol. Chem 273, 14461–14467 (1998). [DOI] [PubMed] [Google Scholar]
- 97.Spagnolo L, Barbeau J, Curtin NJ, Morris EP & Pearl LH Visualization of a DNA-PK/PARP1 complex. Nucleic Acids Res. 40, 4168–4177 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rybanska I et al. PARP1 and DNA-PKcs synergize to suppress p53 mutation and telomere fusions during T-lineage lymphomagenesis. Oncogene 32, 1761–1771 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Luijsterburg MS et al. PARP1 links CHD2-mediated chromatin expansion and H3.3 deposition to DNA repair by non-homologous end-joining. Mol. Cell 61, 547–562 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Truong LN et al. Microhomology-mediated end joining and homologous recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc. Natl Acad. Sci. USA 110, 7720–7725 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Deriano L & Roth DB Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu. Rev. Genet 47, 433–455 (2013). [DOI] [PubMed] [Google Scholar]
- 102.Yan CT et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature 449, 478–482 (2007). [DOI] [PubMed] [Google Scholar]
- 103.Cheng Q et al. Ku counteracts mobilization of PARP1 and MRN in chromatin damaged with DNA double-strand breaks. Nucleic Acids Res. 39, 9605–9619 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fattah F et al. Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells. PLoS Genet. 6, e1000855 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mansour WY, Rhein T & Dahm-Daphi J The alternative end-joining pathway for repair of DNA double-strand breaks requires PARP1 but is not dependent upon microhomologies. Nucleic Acids Res. 38, 6065–6077 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang M et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 34, 6170–6182 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wray J et al. PARP1 is required for chromosomal translocations. Blood 121, 4359–4365 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Celli GB & de Lange T DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 7, 712–718 (2005). [DOI] [PubMed] [Google Scholar]
- 109.Celli GB, Denchi EL & de Lange T Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat. Cell Biol. 8, 885–890 (2006). [DOI] [PubMed] [Google Scholar]
- 110.Sfeir A & de Lange T Removal of shelterin reveals the telomere end-protection problem. Science 336, 593–597 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ceccaldi R et al. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature 518, 258–262 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kent T, Mateos-Gomez PA, Sfeir A & Pomerantz RT Polymerase theta is a robust terminal transferase that oscillates between three different mechanisms during end-joining. eLife 5, e13740 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mateos-Gomez PA et al. Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature 518, 254–257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; References 111 and 113 discuss the requirement for Pol θ in aNHEJ.
- 114.Anachkova B, Russev G & Poirier GG DNA replication and poly(ADP-ribosyl)ation of chromatin. Cytobios 58, 19–28 (1989). [PubMed] [Google Scholar]
- 115.Lehmann AR, Kirk-Bell S, Shall S & Whish WJ The relationship between cell growth, macromolecular synthesis and poly ADP-ribose polymerase in lymphoid cells. Exp. Cell Res. 83, 63–72 (1974). [DOI] [PubMed] [Google Scholar]
- 116.Bryant HE et al. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 28, 2601–2615 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dantzer F, Nasheuer HP, Vonesch JL, de Murcia G & Menissier-de Murcia J Functional association of poly(ADP-ribose) polymerase with DNA polymerase alpha-primase complex: a link between DNA strand break detection and DNA replication. Nucleic Acids Res. 26, 1891–1898 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Simbulan-Rosenthal CM et al. Regulation of the expression or recruitment of components of the DNA synthesome by poly(ADP-ribose) polymerase. Biochemistry 37, 9363–9370 (1998). [DOI] [PubMed] [Google Scholar]
- 119.Smirnova M & Klein HL Role of the error-free damage bypass postreplication repair pathway in the maintenance of genomic stability. Mutat. Res 532, 117–135 (2003). [DOI] [PubMed] [Google Scholar]
- 120.Dungrawala H et al. The replication checkpoint prevents two types of fork collapse without regulating replisome stability. Mol. Cell 59, 998–1010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ray Chaudhuri A et al. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat. Struct. Mol. Biol 19, 417–423 (2012). [DOI] [PubMed] [Google Scholar]; This report demonstrates the essential role of PARP1 in replication fork reversal.
- 122.Sugimura K, Takebayashi S, Taguchi H, Takeda S & Okumura K PARP-1 ensures regulation of replication fork progression by homologous recombination on damaged DNA. J. Cell Biol 183, 1203–1212 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zellweger R et al. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J. Cell Biol 208, 563–579 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Follonier C, Oehler J, Herrador R & Lopes M Friedreich’s ataxia-associated GAA repeats induce replication-fork reversal and unusual molecular junctions. Nat. Struct. Mol. Biol 20, 486–494 (2013). [DOI] [PubMed] [Google Scholar]
- 125.Neelsen KJ & Lopes M Replication fork reversal in eukaryotes: from dead end to dynamic response. Nat. Rev. Mol. Cell Biol 16, 207–220 (2015). [DOI] [PubMed] [Google Scholar]
- 126.Neelsen KJ, Zanini IM, Herrador R & Lopes M Oncogenes induce genotoxic stress by mitotic processing of unusual replication intermediates. J. Cell Biol 200, 699–708 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Atkinson J & McGlynn P Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res. 37, 3475–3492 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Berti M et al. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat. Struct. Mol. Biol 20, 347–354 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ahuja AK et al. A short G1 phase imposes constitutive replication stress and fork remodelling in mouse embryonic stem cells. Nat. Commun 7, 10660 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ding X et al. Synthetic viability by BRCA2 and PARP1/ARTD1 deficiencies. Nat. Commun 7, 12425 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; This report shows that synthetic viability results from deficiency of both PARP1 and BRCA2.
- 131.Ying S, Hamdy FC & Helleday T Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer Res. 72, 2814–2821 (2012). [DOI] [PubMed] [Google Scholar]
- 132.Schlacher K et al. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145, 529–542 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schlacher K, Wu H & Jasin M A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 22, 106–116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Murai J et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 72, 5588–5599 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Petermann E, Orta ML, Issaeva N, Schultz N & Helleday T Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell 37, 492–502 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gottipati P et al. Poly(ADP-ribose) polymerase is hyperactivated in homologous recombination-defective cells. Cancer Res. 70, 5389–5398 (2010). [DOI] [PubMed] [Google Scholar]
- 137.Ray Chaudhuri A et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 535, 382–387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; This report demonstrates that a loss of PARP1 that precedes BRCA1 loss results in relative genome stability owing to replication fork protection.
- 138.Illuzzi G et al. PARG is dispensable for recovery from transient replicative stress but required to prevent detrimental accumulation of poly(ADP-ribose) upon prolonged replicative stress. Nucleic Acids Res. 42, 7776–7792 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Guillemette S et al. Resistance to therapy in BRCA2 mutant cells due to loss of the nucleosome remodeling factor CHD4. Genes Dev. 29, 489–494 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Escargueil AE, Soares DG, Salvador M, Larsen AK & Henriques JA What histone code for DNA repair? Mutat. Res 658, 259–270 (2008). [DOI] [PubMed] [Google Scholar]
- 141.Gibson BA & Kraus WL New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol 13, 411–424 (2012). [DOI] [PubMed] [Google Scholar]
- 142.Kraus WL & Hottiger MO PARP-1 and gene regulation: progress and puzzles. Mol. Aspects Med 34, 1109–1123 (2013). [DOI] [PubMed] [Google Scholar]
- 143.Schiewer MJ & Knudsen KE Transcriptional roles of PARP1 in cancer. Mol. Cancer Res 12, 1069–1080 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Messner S et al. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 38, 6350–6362 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Poirier GG, de Murcia G, Jongstra-Bilen J, Niedergang C & Mandel P Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc. Natl Acad. Sci. USA 79, 3423–3427 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chen M et al. Transgenic CHD1L expression in mouse induces spontaneous tumors. PLoS ONE 4, e6727 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ma NF et al. Isolation and characterization of a novel oncogene, amplified in liver cancer 1, within a commonly amplified region at 1q21 in hepatocellular carcinoma. Hepatology 47, 503–510 (2008). [DOI] [PubMed] [Google Scholar]
- 148.Ahel D et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 325, 1240–1243 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]; This report shows that PARP1-dependent chromatin remodelling by ALC1 is essential for DNA repair.
- 149.Gottschalk AJ et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc. Natl Acad. Sci. USA 106, 13770–13774 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Smeenk G et al. Poly(ADP-ribosyl)ation links the chromatin remodeler SMARCA5/SNF2H to RNF168-dependent DNA damage signaling. J. Cell Sci 126, 889–903 (2013). [DOI] [PubMed] [Google Scholar]
- 151.Malewicz M et al. Essential role for DNA-PK-mediated phosphorylation of NR4A nuclear orphan receptors in DNA double-strand break repair. Genes Dev. 25, 2031–2040 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kruhlak M et al. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature 447, 730–734 (2007). [DOI] [PubMed] [Google Scholar]
- 153.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM & Greenberg RA ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141, 970–981 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chou DM et al. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl Acad. Sci. USA 107, 18475–18480 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Polo SE, Kaidi A, Baskcomb L, Galanty Y & Jackson SP Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 29, 3130–3139 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mateo J et al. DNA-repair defects and olaparib in metastatic prostate cancer. N. Engl. J. Med 373, 1697–1708 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Vinayak S & Ford JM PARP inhibitors for the treatment and prevention of breast cancer. Curr. Breast Cancer Rep. 2, 190–197 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gagne JP, Hendzel MJ, Droit A & Poirier GG The expanding role of poly(ADP-ribose) metabolism: current challenges and new perspectives. Curr. Opin. Cell Biol. 18, 145–151 (2006). [DOI] [PubMed] [Google Scholar]
- 159.Min W & Wang ZQ Poly (ADP-ribose) glycohydrolase (PARG) and its therapeutic potential. Front. Biosci. (Landmark Ed.) 14, 1619–1626 (2009). [DOI] [PubMed] [Google Scholar]
- 160.Barkauskaite E, Jankevicius G & Ahel I Structures and mechanisms of enzymes employed in the synthesis and degradation of PARP-dependent protein ADP-ribosylation. Mol. Cell 58, 935–946 (2015). [DOI] [PubMed] [Google Scholar]
- 161.Andrabi SA et al. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc. Natl Acad. Sci. USA 103, 18308–18313 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mortusewicz O, Fouquerel E, Ame JC, Leonhardt H & Schreiber V PARG is recruited to DNA damage sites through poly(ADP-ribose)- and PCNA-dependent mechanisms. Nucleic Acids Res. 39, 5045–5056 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Cortes U et al. Depletion of the 110-kilodalton isoform of poly(ADP-ribose) glycohydrolase increases sensitivity to genotoxic and endotoxic stress in mice. Mol. Cell. Biol 24, 7163–7178 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Mueller-Dieckmann C et al. The structure of human ADP-ribosylhydrolase 3 (ARH3) provides insights into the reversibility of protein ADP-ribosylation. Proc. Natl Acad. Sci. USA 103, 15026–15031 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Oka S, Kato J & Moss J Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J. Biol. Chem 281, 705–713 (2006). [DOI] [PubMed] [Google Scholar]
- 166.Rosenthal F et al. Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat. Struct. Mol. Biol 20, 502–507 (2013). [DOI] [PubMed] [Google Scholar]
- 167.Sharifi R et al. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 32, 1225–1237 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Krishnakumar R et al. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science 319, 819–821 (2008). [DOI] [PubMed] [Google Scholar]
- 169.Hassa PO, Buerki C, Lombardi C, Imhof R & Hottiger MO Transcriptional coactivation of nuclear factor-kappaB-dependent gene expression by p300 is regulated by poly(ADP)-ribose polymerase-1. J. Biol. Chem 278, 45145–45153 (2003). [DOI] [PubMed] [Google Scholar]
- 170.Gibson BA et al. Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science 353, 45–50 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Soldatenkov VA et al. Transcriptional repression by binding of poly(ADP-ribose) polymerase to promoter sequences. J. Biol. Chem 277, 665–670 (2002). [DOI] [PubMed] [Google Scholar]