SUMMARY
Replication origins, fragile sites, and rDNA have been implicated as sources of chromosomal instability. However, the defining genomic features of replication origins and fragile sites are among the least understood elements of eukaryote genomes. Here, we map sites of replication initiation and breakage in primary cells at high resolution. We find that replication initiates between transcribed genes within nucleosome-depleted structures established by long asymmetrical poly(dA:dT) tracts flanking the initiation site. Paradoxically, long (>20 bp) (dA:dT) tracts are also preferential sites of polar replication fork stalling and collapse within early-replicating fragile sites (ERFSs) and late-replicating common fragile sites (CFSs) and at the rDNA replication fork barrier. Poly(dA:dT) sequences are fragile because long single-strand poly(dA) stretches at the replication fork are unprotected by the replication protein A (RPA). We propose that the evolutionary expansion of poly(dA:dT) tracts in eukaryotic genomes promotes replication initiation, but at the cost of chromosome fragility.
Graphical Abstract
In Brief
Mammalian replication origins are fragile sites defined by poly-dA/dT stretches that are nucleosome free and devoid of the single-strand DNA-protecting protein RPA.
INTRODUCTION
In eukaryotic cells, DNA replication initiates from multiple origins distributed throughout the genome. Replication origins are marked by the assembly of the replicative helicase (MCM2–7), which unwinds the parental DNA duplex to establish bidirectional replication. Discrete origins of replication (ORI) were identified in yeast Saccharomyces cerevisiae (S. cerevisiae) as 150-bp DNA elements comprising an 11-bp T-rich consensus sequence recognized by the origin recognition complex (ORC) and flanked by an A-rich sequence that augments origin activity (Eaton et al., 2010). ORI in Schizosaccaromyces pombe do not have consensus sequences but exhibit an asymmetric distribution of dA and dT mononucleotide tracts (Leonard and Méchali, 2013; Mojardín et al., 2013) that constitutes a strong nucleosome excluding signal (Struhl and Segal, 2013).
Despite intensive investigation, the identification of replication origins in mammalian cells is complicated by the fact that large genomes are replicated by thousands of forks initiating at degenerate, redundant, and inefficient origins scattered throughout the genome (Prioleau and MacAlpine, 2016). Accordingly, human origins mapped at high resolution have not yielded predictive sequences motifs. Origin mapping approaches that measure short nascent strands (SNS-seq) have identified narrow and localized initiation sites with preferential enrichment at CpG islands and G-quadruplexes (Besnard et al., 2012; Cayrou et al., 2011). In contrast, genome-wide directional sequencing of Okazaki fragments (OK-seq) revealed broad zones of initiation, which did not overlap with origins mapped by SNS-seq (Petryk et al., 2016). While there is no unified view of mammalian origins, the establishment of nucleosome-free regions is speculated to be a general property of replication initiation in all eukaryotes (Prioleau and MacAlpine, 2016).
Depletion of dNTP pools (e.g., by hydroxyurea [HU] or deficiency in Ataxia telangiectasia mutated-related kinase [ATR]) can induce fork stalling, arrest, or chromosomal breakage (Glover et al., 2017; Técher et al., 2017). Replication stress frequently leads to the accumulation of single-stranded DNA (ssDNA) generated through uncoupling of the helicase and DNA polymerase activities of the replisome (Byun et al., 2005). It has been proposed that replication fork (RF) stability is dependent on the protection of ssDNA by replication protein A (RPA), whereby insufficient RPA loading onto ssDNA triggers DNA breakage and replication catastrophe (Toledo et al., 2013).
RFs can also stall when they encounter impediments, including non-histone proteins bound to DNA, damaged bases, or DNA sequences that fold into non-canonical structures (e.g., hairpins, triplexes, quadruplexes) (Mirkin and Mirkin, 2007). RFs pause during normal replication at so-called “replication fork barriers” (RFBs), where specific proteins impede the RF by binding tightly to DNA. For example, polar barriers at the 30 end of the rDNA transcription unit are necessary to ensure that replication and transcription are co-directional, thereby preventing head-on collisions (Tsang and Carr, 2008). Nevertheless, RFBs are often associated with increased frequency of recombination and instability, leading to the idea that some fraction of stalled forks may collapse and recombine at specific sites (Tsang and Carr, 2008).
While many natural impediments to DNA replication occur randomly, a considerable number of recurrent chromosomal rearrangements arise from breakage within fragile hotspot regions, which have thus far been mapped at low resolution. Early-replicating fragile sites (ERFSs) and common fragile sites (CFSs) are genomic regions spanning tens to hundreds of kilobases that manifest as breaks in metaphase chromosome spreads upon replication stress (Barlow et al., 2013; Glover et al., 2017; Técher et al., 2017). Although impaired replication has emerged as a universal contributor to chromosome fragility, the mechanisms that account for breakage at ERFSs and CFSs remain unclear.
Here, we map replication associated break sites genome wide at nucleotide resolution. We find that large homopolymeric (dA/dT) tracts are preferential sites of polar replication fork stalling and collapse within ERFSs, CFSs, and rDNA. We propose a unifying mechanism of instability at replication stress-induced fragile sites and natural RFBs.
RESULTS
Replication Origins Are Prone to Fork Collapse in Early S Phase
ERFSs are defined as regions bound by DNA repair proteins and the ssDNA binding protein RPA upon treatment of cells with HU (Barlow et al., 2013). Upon release from the HU arrest, DNA breaks were detected at ERFS hotspots in the subsequent metaphase (Barlow et al., 2013). To map recurrent sites of replication associated DSBs genome wide, we isolated mouse splenic B cells (>97% of which reside in G0/G1) and activated them with lipopolysaccharide (LPS)/interleukin-4 (IL-4)/RP105 so that cells would synchronously enter the cell cycle (Figure S1A). Activated B cells begin to enter S phase at 16 hr, and by 28 hr, more than 69% of cells were in cycle (Figure S1A). The presence of HU does not affect B cell activation, as measured by nascent RNA (nsRNA) synthesis (Figure S1B, left panel), but it does stall replication forks soon after origins fire (Figures S1A and S1C). We activated splenic B cells in the presence of 10 mM HU for 28 hr, then captured and sequenced DNA ends using END-seq, a method that maps DSBs at nucleotide resolution (Canela et al., 2016).
Recurrent DSBs were detected in S phase (Figures 1Aiii and S1D) in independent experiments (Figures S1E and S1F), which did not require long HU exposure, as 6- or 28-hr treatment produced similar results (Figures S1E and S1G). Overall, 43,585 DSBs peaks were detected, 24% of which localized within 69% of ERFSs, overlapping significantly (Figure S1H). We conclude that ERFSs represent zones of recurrent DSBs during S phase, prior to mitosis.
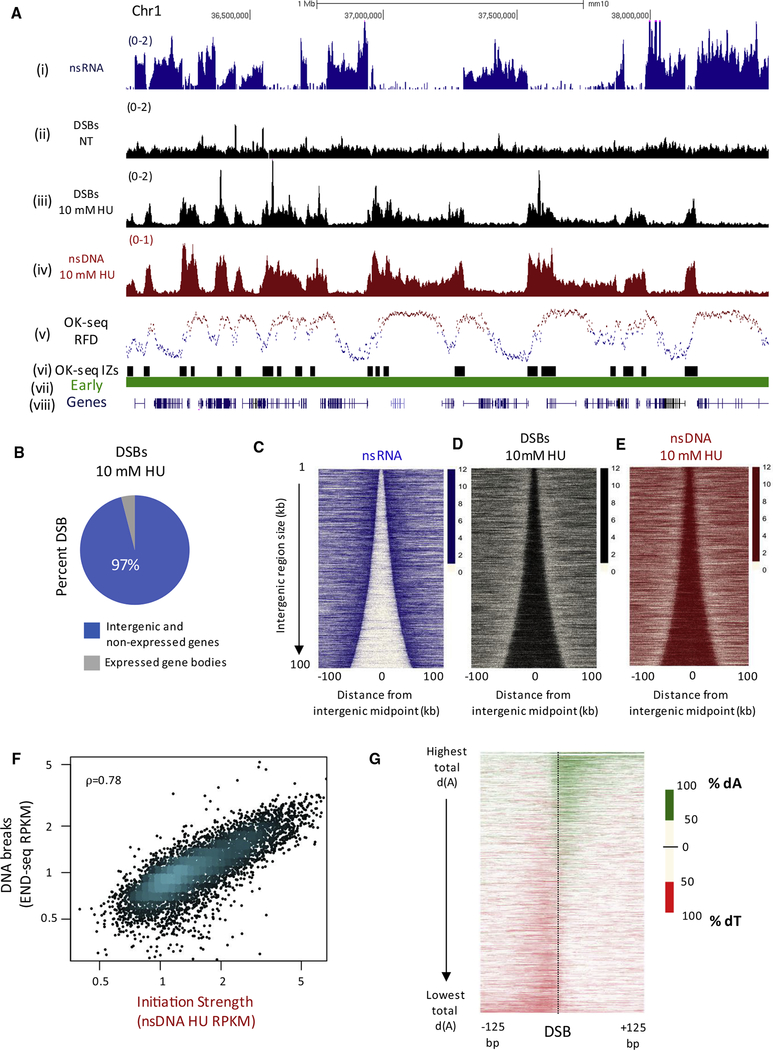
Figure 1. Replication Initiation Zones Are Prone to Fork Collapse.
(A) Genome browser shots illustrating DSBs and replication initiation zones for activated B cells. (Top to bottom) Normalized read density (reads per million, RPM) for (i) nascent RNA (nsRNA), (ii and iii) DSBs from END-seq in activated B cells in non-treated (NT) and treated with 10 mM HU for 28 hr, (iv) nascent DNA from HU-EdU-seq (nsDNA) in activated B cells treated with 10 mM HU for 28 hr, (v) OK-seq replication fork direction (RFD), (vi) replication initiation zones from 48-hr activated B cells using OK-seq, (vii) replication timing determined using TimEX ratios from resting and activated B cells, and (viii) gene annotations from UCSC.
(B) Genomic distribution of 43,585 DNA DSB sites in activated B cells treated with 10 mM HU for 28 hr.
(C) Heatmaps showing the antagonistic relationship between transcription (nsRNA, blue), replication initiation, and HU-induced DSBs in activated B cells. For all heatmaps, gene pairs are ordered by the size of the intergenic region and centered on the midpoint of the intergenic region between the two active genes.
(D) Heatmap of 10 mM HU-induced DSBs (black) in activated B cells.
(E) Heatmap of replication initiation (nsDNA, red) in activated B cells.
(F) Scatterplot demonstrating correlation between replication initiation strength (nsDNA levels) and DNA DSBs in activated B cells treated with 10 mM HU. ρ = 0.78, Spearman’s rank correlation.
(G) Heatmap of asymmetric dA and dT distribution in a 20-bp sliding window around 43,585 DSB sites found in activated B cells treated with 10 mM HU.
Transcribed Regions and DSBs Exist in a Mutually Exclusive Genomic Space
DSB peaks were distributed across 6,189 broad zones, with sizes ranging from 0.2–450 kb and a median size of 14 kb (Figures 1A and S1I). To determine DNA replication timing for these regions, we performed whole-genome sequencing in resting and proliferating B cells (Figure S1J) (Koren et al., 2014). We found that the majority (98%) of DSB-containing zones clustered within early replication domains (Figure S1K). Early-replicating regions are gene dense, suggesting the breakage might be due to transcription-replication conflicts. However, almost 100% of HU-induced DSBs were found between expressed genes (Figures 1Ai, 1Aiii, and 1B). Genome-wide representation of nascent transcription and DSBs as a heatmap showed that clustering of DSBs within intergenic zones was conserved across different cell types including activated B cells (Figures 1C and 1D) and T cells (Figure S2A). The same trend held for the human HCT116 cell line synchronized using CDK4/6i and released into 10 mM HU (Figures S2B and S2C). To understand whether levels of transcription generally correlated with levels of DSBs over large (1 Mb) genomic regions, we compared nsRNA and DSB levels in primary B cells in the presence of HU. A significant correlation was observed between overall levels of nsRNA and DSBs (r = 0.84), indicating that transcription modulates DNA breakage over large domains (Figure S2D).
DSBs could silence gene transcription, thereby explaining their mutual exclusivity (Kruhlak et al., 2007; Shanbhag et al., 2010). Alternatively, transcription could specify the intergenic localization of replication origins (Gros et al., 2015; Macheret and Halazonetis, 2018; Petryk et al., 2016), in which case DSBs would represent sites of fork collapse sites near replication origins. To map replication initiation zones without HU-associated breakage, we performed directional sequencing of Okazaki fragments (OK-seq) (Petryk et al., 2016) in asynchronously growing primary B cells. Initiation zones are identified as regions of transition from high Watson-strand OK-seq signal (blue) to regions of high Crick-strand OK-seq signal (red) (Figures 1Av– 1Avi). Overall, 54% of the DSB-associated zones determined by END-seq overlapped with 8,897 initiation zones identified by OK-seq (Figure S2E). Similar results were obtained for human cells, where 50% of the DSB-associated zones mapped using END-seq in HCT116 (5,274) overlapped with 9,836 initiation zones mapped by OK-seq in asynchronous HeLa cells (Figure S2F). Given that DSBs largely overlap with initiation zones, we favor the idea that ERFSs represent zones of RF collapse after origin firing.
To confirm that DNA breakage is caused by DNA replication initiation, we labeled all newly synthesized DNA with EdU in the presence of 10 mM HU. After EdU labeling, we isolated nascent DNA (nsDNA) and performed high-throughput sequencing (HU-EdU-seq) (Macheret and Halazonetis, 2018). The position and relative intensities of EdU peaks were highly reproducible among three different experiments (Figure S2G). Merging nearby EdU peaks within 20 kb of each other revealed that replication initiation in primary B cells was distributed across 5,422 broad zones, with sizes ranging from 0.2–450 kb and a median size of 24 kb (Figures 1Aiv and S2H). 77% of initation zones mapped by EdU-seq overlapped with 72% of DSB zones mapped by END-seq (Figure S2I). Genome-wide representation of HU- EdU-seq and END-seq as heatmaps indicated that replication initiation and DSBs overlapped (Figures 1D and 1E), and their integrated intensities within a zone were correlated (Figure 1F).
To determine whether DSBs were dependent on initiation, we blocked replication origin firing by inhibiting cyclin-dependent kinases CDK2 and CDC7 (CDK2/CDC7i) at 22 hr after B cell activation and 30 min prior to HU treatment (Figures S2J–S2L). DSB formation decreased significantly within the initiation zones (Figures S2K and S2L). These data indicate that, upon exposure to high-dose HU, a subset of forks collapse close to the sites of initiation.
To study how differences in transcription influence DSB sites at a local scale, we mapped DSBs, nsDNA, and nsRNA in primary T cells. Like in B cells, the presence of HU did not affect T cell activation as measured by nsRNA (Figure S1B, right panel). Despite highly similar transcription programs (Figures S3A), for genes that were expressed in one cell type, but not in the other, DSBs and nsDNA were specifically excluded from the active gene (Figure S3B). Notably, however, even low levels of transcription (>10-fold lower in one cell type) were sufficient to exclude DSBs and replication initiation from gene bodies (Figure S3C). These results are consistent with the idea that RNA polymerase can redistribute MCM2–7 helicase into intergenic regions prior to its activation in S phase (Gros et al., 2015).
Because DSBs are mapped at nucleotide resolution (Canela et al., 2016), we wanted to determine whether there were preferential sites of fork collapse within the broad initiation zones. Analysis of the 43,585 DSB peaks within initiation zones revealed that they were bordered by either homopolymeric runs of dA on the 3’side or dT on the 5’ side (Figure 1G). As discussed below, this suggests the possibility that under conditions of stress, replication forks preferentially collapse into DSBs when the fork reaches poly(dA:dT) sequences in the vicinity of individual replication origins.
Progressing Forks Break at Poly(dA:dT) within CFSs
To determine whether, similar to replication fork collapse near origins, traveling forks are subject to DNA breakage, we allowed forks to advance beyond their initiation sites. To do so, we lowered the concentration of HU by 20-fold, to 0.5 mM, during B cell activation, then performed HU-EdU- seq to map fork progression and END-seq to map DSBs. In contrast to 10 mM HU, EdU incorporation was readily detected at 0.5 mM HU by flow cytometry (Figure S4A), and nsDNA was detected beyond initiation zones and within the flanking genes (Figures 2Aii and 2Aiii). Strikingly, DNA breaks were constrained to precise locations that did not mirror the 0.5 mM HU nsDNA profile (Figures 2Aiii and 2Av), distinct from 10 mM HU where they overlapped (Figures 2Aii and 2Aiv). 82% of DSB sites remained within intergenic initiation zones, while 18% were located within transcribed genes flanking the initiation sites (Figure 2B), indicating that forks travel further before they collapse at preferred sites.
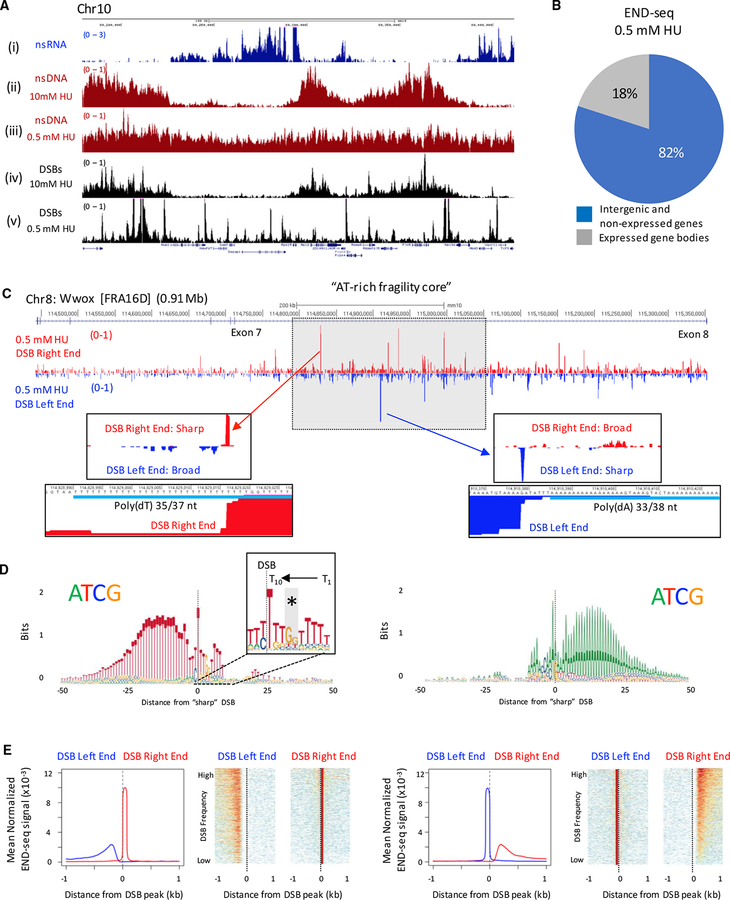
Figure 2. Asymmetric Collapse of Progressing Forks at Poly(dA:dT) within Common Fragile Sites.
(A) Genome browser shots showing (i) nsRNA, (ii and iii) nsDNA, and (iv and v) DSBs in activated B cells treated with 10 mM or 0.5 mM HU.
(B) Genomic distribution of 76,382 DNA DSB sites in activated B cells treated with 0.5 mM HU.
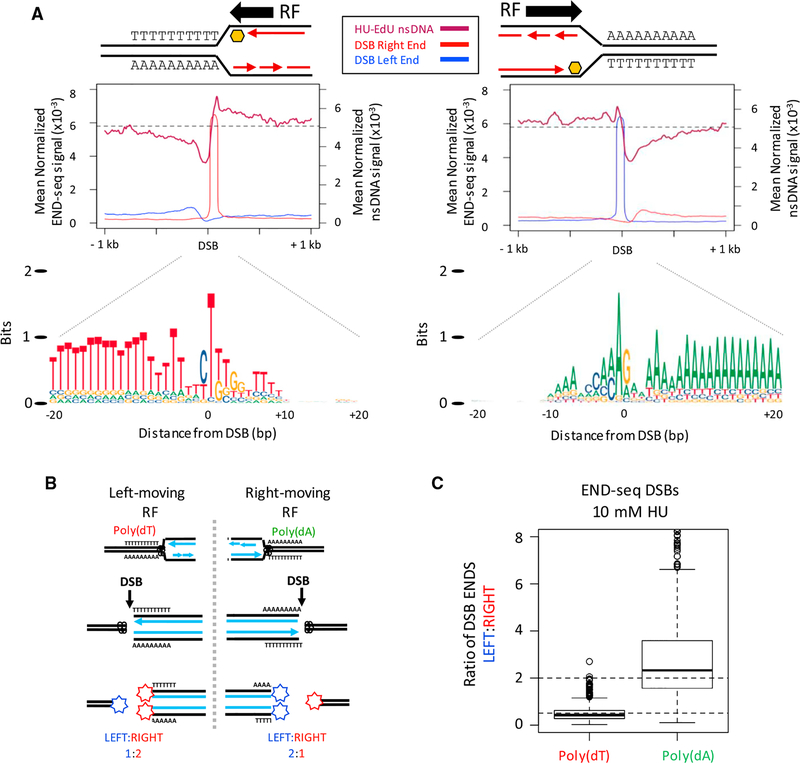
(C) Genome browser shots showing DSBs in the Wwox “AT-rich fragility core” in activated B cells treated with 0.5 mM HU. END-seq reads were separated by strand: bottom strand sequences represent a DSB left end (blue), and top strand sequences represent a DSB right end (red). DSB peaks with a sharp right end (middle left panel) or sharp left end (middle right panel) are displayed along with the poly(dA:dT) sequences associated with the sharp DSB end (bottom panels).
(D) Composite motif at the sharp DSB end for the top-1,000 DSBs associated with poly(dT) (left panel) and poly(dA) (right panel) in activated B cells treated with 0.5 mM HU. The sharp DSB end is indicated with a vertical line at position 0, preferentially positioned 10 nt from the beginning of the dA/dT tract. Inset box shows conserved 10 nt dT tract at the sharp DSB end and GG interruption (*).
(E) Composite plots and heatmaps depicting asymmetry between DSB ends for the top-1,000 DSB sites associated with poly(dT) (left panels) and poly(dA) (right panels) in activated B cells treated with 0.5 mM HU.
To confirm that 0.5 mM HU DSB sites are associated with progressing forks rather than the firing of dormant origins, we inhibited replication initiation with CDK2/CDC7 inhibitors 22 hr after B cell stimulation, a time at which 35% of cells had already entered S phase (Figure S2J). In cells pre-incubated for 30 min with CDK2/CDC7i followed by 0.5 mM HU, DSBs within initiation zones largely disappeared, whereas DSBs outside of zones remained largely intact (Figures S4B and S4C). This suggests that the DSBs within expressed genes result from collapsed forks that have traveled outside of the initiation zones and not due to firing of dormant origins at these sites.
Since a fraction of breaks at low-dose HU occurred within gene bodies, we wondered whether CFSs were affected (Arlt et al., 2011). In human lymphocytes, the late-replicating CFS FRA16D is one of the loci most sensitive to replication stress (Glover et al., 2017; Técher et al., 2017). In the presence of 0.5 mM HU, we detected several break sites clustered within the 0.91-Mb Wwox gene (Figure 2C), which spans FRA16D. An apparent DSB hotspot region was found within intron 7 of Wwox (Figure 2C), which corresponds to the “AT-rich fragility core” described in human cells (Madireddy et al., 2016; Zlotorynski et al., 2003).
Asymmetric DNA Break Structures at Poly(dA:dT)
For each DSB within the AT-rich fragility core of Wwox, sequence reads were present from both strands, indicating that two-ended DSBs were generated in response to 0.5 mM HU (Figure 2C). However, the two ends were not symmetric, consisting of one sharp peak and one broader peak, separated by a gap (Figure 2C). For a two-ended DSB, END-seq reads originating from the left and right end of the break map to opposite strands (Canela et al., 2016). The sequencing is performed such that reads to the left side of the DSB map to the bottom strand (Figure 2C; DSB left end). Conversely, reads to the right side of the DSB map to the top strand (Figure 2C, DSB right end).
To determine whether specific sequences were associated with the asymmetric CFS breakage, we examined the nucleotide composition at Wwox breaks orientated by DNA strands. When the sharp peak was present on the top strand (DSB right end), homopolymeric dT tracts were at the break site; conversely, when the sharp peak was present on the bottom strand (DSB left end), homopolymeric dA tracts were present (Figure 2C). A similar asymmetric DSB structure was found within the Fhit gene (FRA3B) (Figure S4D). Additionally, we predicted the fragility of five genomic regions in activated B cells based on active transcription, late replication timing, and large gene size (>500 kb), similar to Wwox and Fhit (Glover et al., 2017) (Figure S4E). The breakage frequency at poly(dA:dT) sequences at these predicted CFSs in B cells was significantly higher than similarly sized, late-replicating regions that were not expressed (Figure S4F). Thus, under conditions of low-dose HU, replication forks that travel into CFSs break preferentially at poly (dA:dT) sequences, similar to forks that break near replication origins with high-dose HU (ERFSs).
Analysis of 76,382 total break sites detected with 0.5 mM HU showed that 37,514 had at least 50% dA (dT) content within 20 bp to right (left) of the DSB (Figure S5A), a frequency significantly higher than expected by random (p < 10−10, Fisher’s Exact test). Analysis of the strongest DSBs (top 2,000) showed an even higher fraction (83%) of sites having poly(dA:dT) near the break site (Figure S5A). Moreover, genome-wide, the DNA end structures were similar to those observed at CFSs: for the top-1,000 DSBs associated with poly(dT), the DSB Right end was sharp (Figures 2D and 2E, left panels); for the top-1,000 DSBs associated with poly(dA), the DSB Left end was sharp (Figures 2D and 2E, right panels). Heatmaps showed that the width of the broad DSB peak, ranging from 200–500 bp, correlated with the intensity of the sharp peak (r = 0.46) (Figure 2E). Aggregate analysis of DSBs at the same poly(dA) sites yielded a similar asymmetric pattern with 10 mM HU (Figure S5B). Thus, under conditions of replication stress induced by lowor high-dose HU, replication fork collapse is characterized by two-endedness and asymmetry between the two ends at poly(dA:dT) tracts.
We hypothesized that naturally occurring polymorphisms in poly(dA:dT) tracts between distinct mouse strains (C57BL6/NJ vs. CAST/EiJ) would provide a physiological context to examine the impact of poly(dA:dT) mutations on DSBs induced by HU in activated B cells. Thus, we activated B cells from C57BL6/NJ and CAST/EiJ mice and examined poly(dA:dT) sites with conserved flanking sequences that differed only in the length of the poly(dA:dT) tract between these two strains. We found that conserved poly(dA:dT) sites lead to DSBs in both strains after HU treatment (Figure S5C, left panel), but poly(dA:dT) sites unique to one strain lead to DSBs only in that strain harboring the poly(dA:dT) tract (Figure S5C, middle and right panels).
We considered the possibility that HU might preferentially deplete dATP or dTTP, leading to fork collapse at these sites specifically. Notably, in actively replicating B cells without HU treatment, the same poly(dA:dT) sites were found to be broken, and the asymmetry between sharp and broad peaks was preserved at these spontaneous break sites (Figure 3A). Spontaneous DSBs occurred equally within replication initiation zones and within gene bodies (Figure 3B). These same sites were not broken in resting B cells, indicating that they are replication dependent (Figure 3B). We also observed preferential breakage at poly(dA:dT) upon treatment of cells with a specific inhibitor of the ATR kinase (Figures 3C and 3D), which is necessary for maintaining chromosome stability at ERFSs and CFSs (Barlow et al., 2013; Casper et al., 2002). Aggregate analysis of these sites also showed an asymmetrical break pattern similar to that in CFSs (Figure 3D). We conclude that homopolymeric stretches of dA or dT are susceptible to spontaneous-and replication-stress-induced DSBs.
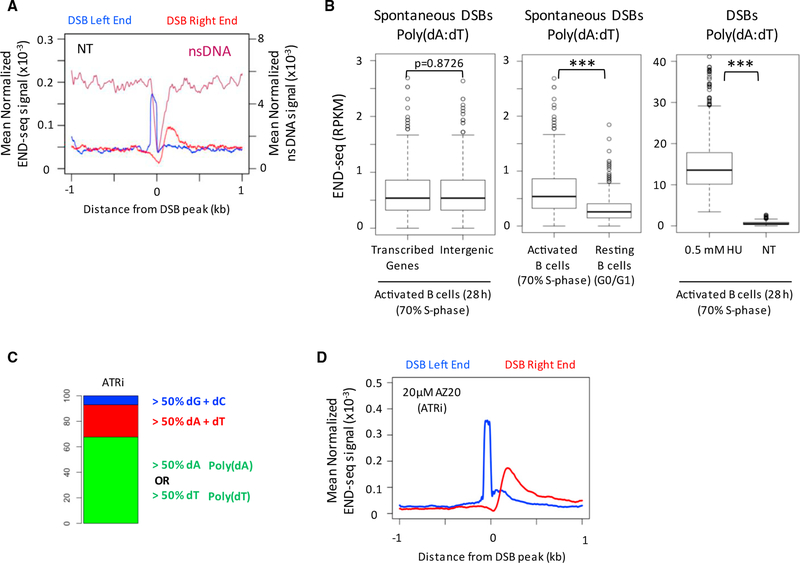
Figure 3. Spontaneous Replication Fork Breakage at Poly(dA:dT).
(A) Composite plot showing DSBs and nsDNA depletion at poly(dA) tracts in non-HU-treated (NT) 28-hr activated B cells.
(B) Boxplots showing END-seq reads at the 2,000 poly(dA:dT) sites most frequently broken with 0.5 mM HU in activated B cells. (Right) Bar plot comparing the frequency of spontaneous DSBs to DSBs induced by 0.5 mM HU. *** p < 10−12; Wilcoxon rank-sum test.
(C) Nucleotide composition ± 20 bp from the “sharp” DSB end for the top-1,000 DSBs in B cells activated for 28 hr with ATR inhibitor.
(D) Composite plot showing DSBs at poly(dA) tracts in B cells activated for 28 hr with ATR inhibitor.
CFS instability can lead to copy number variations (CNV) in normal and tumor cells (Glover et al., 2017). Interestingly, breakpoint junction sequences associated with de novo CNV in mouse embryonic stem cells (ESCs) treated with the replication inhibitor aphidicolin are enriched for poly(dA:dT) (Figure S5D) (Arlt et al., 2012). Moreover, MCM2-deficient lymphomas are characterized by focal deletions in gene-rich early-replicating regions (similar to ERFSs), with breakpoints enriched for poly(dA:dT) tracts (Rusiniak et al., 2012). Taken together with our analysis of S phase breaks in primary cells, these data suggest that poly(dA:dT) sequences can contribute to the fragility of both origin-rich ERFSs and origin-poor CFSs.
Poly(dA:dT) Tracts are Polar RFBs upon Replication Stress
Previous in vitro studies have shown that human DNA polymerase strongly pauses when the template DNA contains poly(dT), but not on a poly(dA) template (Hile and Eckert, 2008). Furthermore, this stalling occurs preferentially at position T9 or T10 of the poly(dT) template (Hile and Eckert, 2008). Motif analysis of poly(dT)-associated breaks indicated that the “sharp” end of the DSB occurred at position T10 (Figure 2D, insert). As such, we propose the “sharp” DSB end represents the precise site of DNA polymerase stalling at poly(dT) prior to fork breakage (See Figure 6A, model). Profiles of nsDNA in B cells support a unidirectional stalling model at poly(dA:dT), as nsDNA precipitously drops at dA10 at right-moving forks and dT10 at left-moving forks with, and even without, HU (Figures 3A and 4A). Combining these in vitro and in vivo data, we propose a model in which replication forks encountering a leading strand template of poly(dT) will stall around T10, producing DSBs at poly(dA:dT) sequences (see Figure 6A, model).
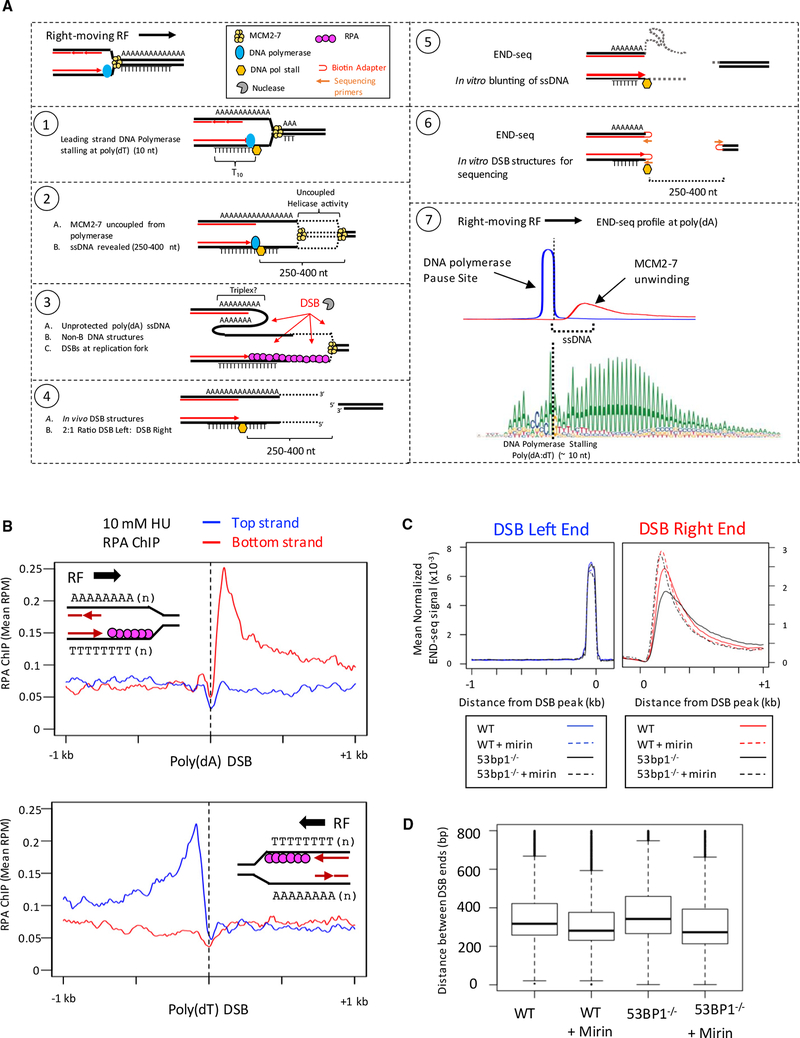
Figure 6. Model for Replication Fork Breakage at Poly(dA:dT) Tracts.
(A) Model for replication fork breakage at poly(dA:dT) and detection of replication fork DSBs by END-seq. END-seq reveals the site of DNA polymerase stalling on one end and the location where MCM2–7 helicase stops ahead of the polymerase on the other end.
(B) ChIP-seq separated by strands at poly(dA) and poly(dT) sequences. (Top panel) Composite plot for RPA ChIP at poly(dA) sites broken in activated B cells treated with 10 mM HU. (Bottom panel) Composite plot for RPA ChIP at poly(dT) sites broken in activated B cells treated with 10 mM HU.
(C) Composite plots showing the distribution of DSB ends at poly(dA) tracts in activated B cells treated with 0.5 mM HU.
(D) Boxplots showing the distance between left and right DSB ends in activated B cells treated with 0.5 mM HU. Distances are divided into quartiles (box), with the line indicating the median. Median distances: wild-type (WT) = 328 bp; WT + mirin = 288 bp; 53BP−/− = 357 bp; 53BP1−/−+ mirin = 287 bp.
Figure 4. Unidirectional Replication Fork Stalling and Breakage Are Associated with poly(dA:dT).
(A) (Top panels) Composite plots for nsDNA and strand-separated END-seq reads (DSB left/right end) at the top-1,000 poly(dA) or poly(dT) DSB sites in activated B cells treated with 10 mM HU. Replication fork (RF) direction is inferred from the nsDNA signal surrounding the DSB. (Bottom panel) Composite motif ± 20 bp of the sharp DSB end for the regions analyzed in top panels, illustrating a unidirectional RF stall at poly(dA:dT) containing poly(dT) on the leading strand.
(B) Proposed ratios of DSB ends at a poly(dA:dT) polar replication fork barrier.
(C) Boxplots showing the ratio of DSB ends (left:right) for the top-1,000 DSB sites with poly(dA) or poly(dT) near the sharp DSB end in activated B cells treated with 10 mM HU.
We reasoned that such unidirectional replication fork stalling and breakage would produce a skewed ratio of DSB ends from parental and daughter DNA molecules. A single DSB end derived from the unreplicated parental DNA would contribute one-half of the END-seq signal relative to the two DSB ends derived from the replicated daughters (Figure 4B). For example, when a right-moving fork collapses at a poly(dA) sequence, the DNA to the right end of the DSB has yet to be copied, resulting in a DSB ratio of 2:1 (LEFT:RIGHT); conversely, a left moving fork that collapses at poly(dT) sequences produces a DSB ratio of 1:2 (LEFT:RIGHT) (Figure 4B). Indeed, these ratios were observed (Figure 4C), supporting a model of a polar replication fork barrier at poly(dA:dT) upon replication stress.
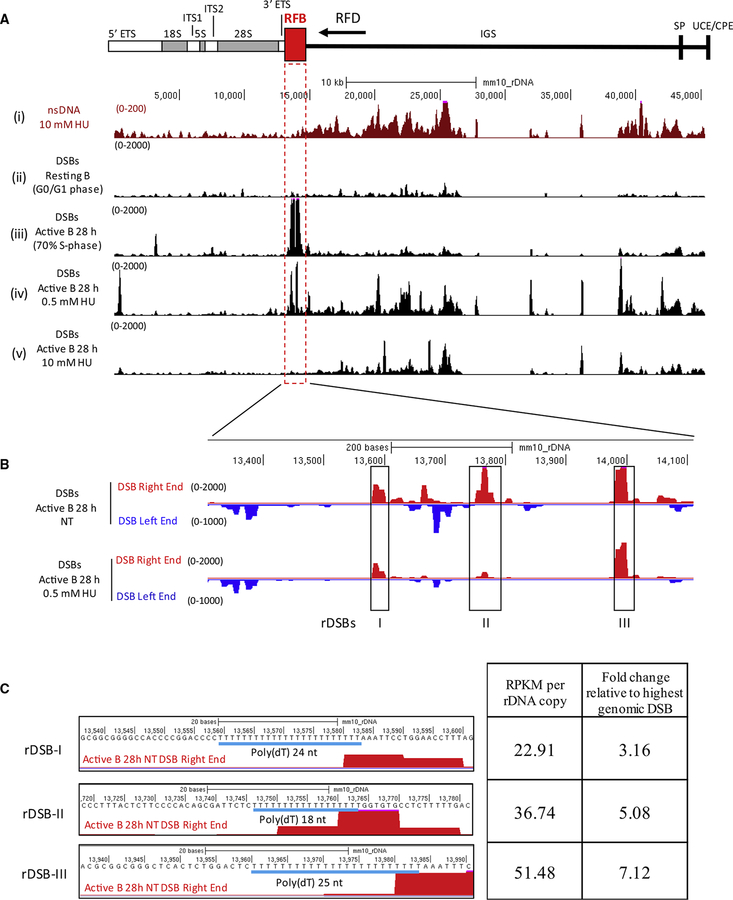
RFB in rDNA Has an Asymmetrical DSB Break Pattern
We wondered whether the natural polar RFB within rDNA, which blocks fork progression in one direction (Gerber et al., 1997), would harbor spontaneous DSBs. During rDNA replication, initiation sites are contained within the intergenic spacer (IGS), and replication forks arrest at the 3’ end of the transcription unit (Figure 5A), presumably to avoid transcription-replication conflicts.
Figure 5. Spontaneous Asymmetric DSBs at the rDNA Replication Fork Barrier.
(A) (Top) Diagram of the rDNA locus. Replication fork barrier (RFB) and predicted replication fork direction (arrow, RFD) are indicated. (Bottom) UCSC genome browser shots for (i) replication initiation (nsDNA), DSBs in (ii) resting B cells, (iii) activated B cells (28 hr), (iv) activated B cells treated with 0.5 mM HU, and (v) activated B cells treated with 10 mM HU.
(B) Genome browser shots of the three spontaneous DSBs at the rDNA RFB, separated by DSB ends.
(C) Left panel: poly(dA:dT) sequences at the spontaneous rDSBs. Right panel: DSB frequency as determined by RPKM, relative to the highest spontaneous genomic DSB (RPKM = 7.2) in activated B cells.
To interrogate replication and DSBs at the repetitive rDNA loci, we utilized a previously constructed build of the mouse genome that combined all copies of rDNA into a single chromosomal locus to which we aligned our HU-EdU-seq and END- seq data (Zentner et al., 2014). In resting B cells, we observed low levels of DSBs (Figure 5ii); in contrast, actively replicating B cells showed intense DSB peaks precisely at the RFB (Figure 5Aiii). Interestingly, when cells were treated with 0.5 and 10 mM HU, DSB peaks occurred at lower frequency than non-treated cells, suggesting that fewer forks reached the 3’ end of the rDNA transcription unit upon HU treatment (Figures 5Aiv and 5Av). A zoomed-in view of DSB sites revealed that the three DSB sites present within the RFB that were retained upon 0.5 mM HU overlapped precisely with the same sites in cycling non-treated cells (Figure 5B). Based on our whole genome sequencing of non-replicating B cells we estimate that rDNA is present at approximately 101 copies per haploid B cell genome (Gibbons et al., 2014). Normalizing by rDNA copy number, we found that the intensity of DSB peaks at the RFB was 3- to 7-fold higher than the highest spontaneous DSB peak within the rest of the genome (Figure 5C). Moreover, all three rDNA peaks exhibited sharp right-ended breaks and poly(dT) tracts characteristic of left moving forks (Figure 5C) consistent with our data from elsewhere in the genome (Figure 4A). Together, these data suggest poly(dA:dT) tracts at the polar RFB within rDNA are particularly fragile, even in the absence of replication stress. We speculate that these well-positioned DSB sites may be the source of recombination at rDNA, which is known to be particularly vulnerable to copy number changes (Tsang and Carr, 2008).
Unprotected ssDNA at the Replication Fork at Poly(dA:dT) Tracts
Our data are consistent with a model in which a DSB is made ahead of the stalled polymerase, likely within the ssDNA region where the MCM2–7 helicase has unwound the template DNA (Figure 6A). MCM helicase unwinding exposes unprotected ssDNA stretches spanning the polymerase pause site on the leading strand (~T10) up to the point where the helicase stops (Figure 6A, steps 1 and 2). One possibility is that the unwinding of poly(dA:dT) tracts into long ssDNA upon HU treatment makes them prone to fold into intramolecular triplexes (Fox, 1990), particularly if one of the ssDNA tracts remains unprotected.
In vitro binding experiments have shown that RPA association with long ssDNA stretches of poly(dA) is approximately 50-fold less efficient than at poly(dT) (Kim et al., 1992). To examine whether there is asymmetric RPA association with poly(dA:dT) tracts in vivo, we performed RPA ChIP-seq in B cells treated with HU. We found that RPA was preferentially bound between the sharp and broad DNA end peaks where we predicted uncoupling between the polymerase and helicase (Figure 6B). Moreover, RPA was bound only to the strand containing the poly(dT) tracts, while the strand containing poly(dA) was unbound by RPA (Figure 6B). Such unprotected ssDNA devoid of RPA is predicted to lead to replication catastrophe and DSBs (Figure 6A, step 3), which could involve a broad spectrum of nucleases (Toledo et al., 2013).
DNA End Structure Reveals MCM Helicase/DNA Polymerase Uncoupling
Our model predicts that fork breakage could result in long stretches of ssDNA in vivo. A critical aspect of END-seq is the in vitro blunting of ssDNA overhangs (Canela et al., 2016) (Figure 6A, steps 4–6), revealing the precise nucleotide where the polymerase pauses on one end (Figure 6A, step 6, left end), and also the location where the helicase stops ahead of the polymerase (Figure 6A, step 5, right end). This model would explain why poly(dA:dT) sequences reside between the sharp and broad peaks (Figure 6A, step 7).
DSBs also could be subject to resection by MRE11 to generate long 30 ssDNA overhangs, which could mask the true amount of ssDNA associated with polymerase/MCM uncoupling. To limit DSB resection after DSB formation, we exposed cells to 0.5 mM HU in combination with the MRE11 nuclease in-hibitor mirin, which blocks ssDNA resection at DSBs (Shibata et al., 2014). Whereas the sharp peak (site of polymerase stalling) was unaffected by MRE11 inhibition, the position of the opposite DNA end, where we propose the MCM helicase stops, moved slightly (~40 bp) closer (Figures 6C and 6D). Interestingly,53BP1 / cells, which are prone to DSB resection (Bunting et al., 2010), exhibited a (~50 bp) longer gap between the two DSB ends that was dependent on MRE11 activity (Figures 6C and 6D). This suggests that the collapsed fork is minimally processed by MRE11, and the 250–400-nt gap that we observed between the two ends in the presence of mirin (Figure 6D) reveals the extent of uncoupled MCM2–7 helicase activity ahead of the stalled DNA polymerase (Figure 6A, step 7).
Poly(dA:dT) Tracts Are Associated with Efficient Replication Initiation Sites
Long poly(dA:dT) tracts >10 bp are strongly overrepresented within non-coding DNA in various eukaryotic genomes (Dechering et al., 1998). In yeast, poly(dA:dT) tracts are associated with a consensus motif at replication origins; thus, we considered the possibility these sequences might also positively influence replication origin firing in mammalian cells.
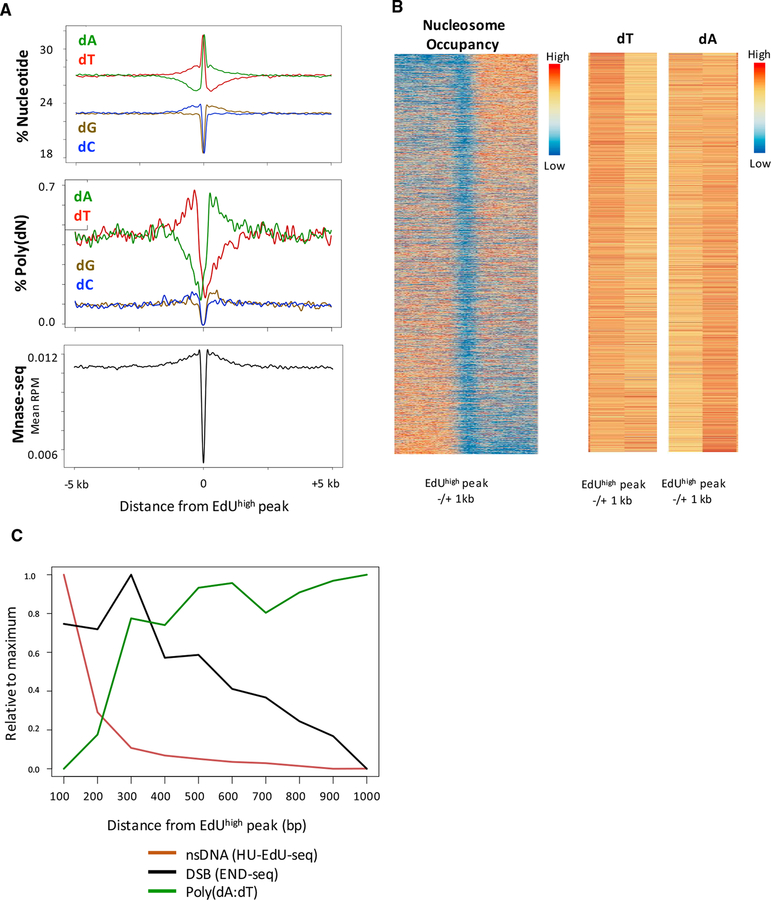
The linear increase in replication fork direction (RFD) measured by OK-seq suggests an equal probability of replication initiation at any location within a given zone (Petryk et al., 2016). In contrast, HU-EdU-seq revealed regions of different EdU intensity (EdUhigh and EdUlow) within the zones themselves (Figure S6A). We identified 21,527 EdUhigh peaks (>2.2-fold higher intensity than the zone on average) (Figure S6B) within the 5422 initiation zones, which we interpret are the most efficient sites of replication initiation. Characterization of these initiation peaks revealed that they have higher AT-content than regions with little or no initiation (Figure S6A). Aggregating EdUhigh peaks showed that they exhibit a sharp increase in AT-content compared to a 10-kb neighboring region and random regions of the same size (Figure S6C). Interestingly, the EdUhigh peaks had an asymmetric nucleotide distribution flanking the peak, with higher dA and poly(dA) content on the 30 side and higher dT and poly(dT) content on the 5’ side (Figure 7A). A similar feature was observed at EdUhigh peaks in HCT116 cells (Figure S6D). The asymmetric poly(dA:dT) tracts, defined as containing at least 15 dA’s or dT’s in a 20-bp window on the top strand (Figures 7A and S6D), are preferentially positioned 0.1–1 kb from the center of the EdUhigh peaks.
Figure 7. Poly(dA:dT) Tracts Are Associated with Replication Initiation.
(A) Composite plots for nucleotide frequency, polynucleotide frequency, and nucleosome occupancy around 21,527 EdUhigh peaks in activated B cells. Poly(dN) is defined as a 20-bp window containing at least 75% dN. MNase-seq data are obtained from 72-hr activated B cells (Kieffer-Kwon et al., 2017).
(B) (Left) Heatmap showing nucleosome occupancy (MNase-seq) centered on 21,527 EdUhigh peaks, sorted by nucleosome asymmetry around the EdUhigh peak. (Right) Heatmaps for total dT and dA frequency within 1 kb to the left or right of the EdUhigh peak. Higher frequency of dT (to the left of the EdUhigh peak) and dA (to the right of the EdUhigh peak) correlates with nucleosome depletion.
(C) Histogram showing distributions of DNA DSBs, poly(dA:dT) tracts, and nsDNA relative to the summit of 21,527 EdUhigh peaks, for activated B cells treated with 10 mM HU. DNA DSB frequencies are coincident with increases in poly(dA:dT) tracts 100–1,000 bp downstream of EdUhigh peaks. Poly(dN) is defined as any 20-bp sequence with at least 75% dN.
To control for the possibility that the observed poly(dA:dT) enrichment might result from EdU incorporation opposite dA residues, we substituted EdU with the cytosine analog EdC, and similarly mapped nsDNA in primary B cells (HU-EdC-seq). We found that 82% of HU-EdU-seq initiation zones overlapped with those from HU-EdC-seq (Figure S6E). Moreover, nsDNA levels from HU-EdC-seq strongly correlated with nsDNA from HU-EdU-seq (Figure S6F) and exhibited similar AT enrichment flanking the EdChigh peaks (Figure S6G).
In yeast, replication origins are nucleosome-free regions flanked by precisely positioned nucleosomes (Eaton et al., 2010). Accordingly, we examined MNase-seq profiles near EdU-high initiation sites in mouse and human cells (Guzman and D’Orso, 2017; Kieffer-Kwon et al., 2017). We observed a strong nucleosome-depleted region centered precisely on the EdUhigh initiation sites in mouse B cells (Figures 7A and 7B) and the human HCT116 cell line (Figure S6D). The finding that the most efficient initiation sites are nucleosome depleted and comprise AT-rich sequences with an asymmetric distribution of poly(dA:dT) suggests that the genetic and structural elements controlling replication origin selection are non-random and conserved.
As shown above, DSB peaks within initiation zones were bordered by homopolymeric runs of dA at the 30 end or dT at the 5’ end (Figure 1G), which may reflect the relationship between replication origins and fork collapse at flanking poly(dA:dT) tracts. Thus, we analyzed the distribution of HU DSBs and poly(dA:dT) tracts surrounding EdUhigh peaks. HU-induced DSBs were most frequent at the nearest poly(dA:dT) tracts, located several hundred base-pairs away from EdUhigh peaks, with lower DSB frequency at subsequent poly(dA:dT) tracts (Figure 7C). Thus, efficient replication origins are structured in a way that makes them prone to fork stalling nearby. Under conditions of replication stress, this could expose long unprotected ssDNA that can form secondary structures. Even without replication stress, it is possible that replication origin firing is naturally followed by “pausing,” prior to bona fide elongation, to coordinate replication across the genome in early S phase.
DISCUSSION
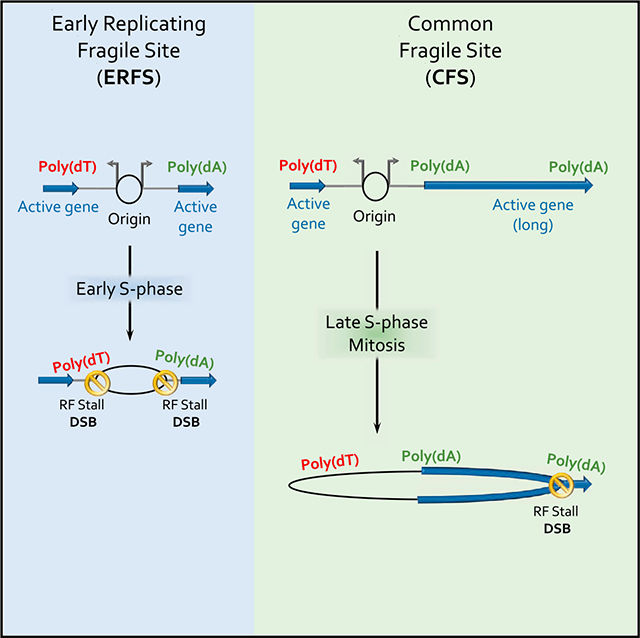
The majority of cytogenetic lesions resulting from impediments in replication fork progression occur at recurrent non-random fragile sites (Glover et al., 2017; Mirkin and Mirkin, 2007; Técher et al., 2017). The mechanisms responsible for instability at these specific regions remain unclear. The diametrically opposite properties of CFSs (late replicating, AT-rich, within large isolated genes, replication origin poor) and ERFSs (early replicating, GC- rich, within gene clusters, replication origin rich) suggested that they represent distinct classes of fragility (Barlow et al., 2013; Glover et al., 2017). However, our high-resolution approaches that independently map sites of DNA synthesis and DSBs indicate that poly(dA:dT) sequence are a causal factor for stalling and breakage at both CFSs and ERFSs in response to HU. Upon unwinding at the replication fork, poly(dA:dT) tracts may be prone to form non-B DNA structures (Fox, 1990). Based on our finding that poly(dA) sequences remain unprotected by RPA under conditions of stress, we presume that the long dA-rich strand could fold back on itself during polymerization provoking triplex formation (Figure 6A, step 3). Such a configuration could, in turn, prevent further DNA synthesis, resulting in selftermination and fork collapse (Samadashwily et al., 1993).
Mechanism of Breakage at CFSs and ERFSs
While ERFSs represent sites of RF collapse in S phase (Figure 1), CFSs are thought to arise in mitosis as a result of underreplication (Naim et al., 2013; Ying et al., 2013). Because of their proximity to strong origins surrounded by poly(dA:dT) tracts, ERFSs represent sites at which most replication forks stall and collapse upon S phase entry. Similarly, we find that when the replication fork encounters the CFS fragility core prior to mitosis, these regions are unstable and may serve as hubs of fork collapse during S phase (Figures 2C and S4D). In addition to S phase breaks, DSBs at CFSs may persist into mitosis because the probability that they complete replication is intrinsically low, and further decreases upon replication stress and fork stalling at poly(dA:dT). While transcription is a key determinant of cell-type-specific breakage at CFSs and ERFSs (Tubbs and Nussenzweig, 2017), we propose that the main impact of transcription is the displacement of origins from gene bodies (Macheret and Halazonetis, 2018; Petryk et al., 2016). While replication initiation and concomitant ERFS breakage is shaped by transcription in such a way that avoids replication/transcription conflicts, replication fork progression within CFSs may be exacerbated by transcription. For example, long poly(dA:dT) tracts are not only replication pause sites, but also a major contributor to R-loop formation with gene bodies (Wahba et al., 2016), which promote DSBs (Hamperl et al., 2017).
Signature of Polymerase Kappa at Fragile Poly(dA:dT) DNA Structures
It has been suggested that specialized translesion polymerases, such as polymerase eta and polymerase kappa, are needed to complete replication at CFSs (Barnes et al., 2017). Notably, DNA polymerase kappa has a unique signature of interrupted mutations and polar pausing at mononucleotide dT repeats (Hile and Eckert, 2008). For example, 71% of polymerase kappa errors observed at T11 runs are dG insertions, and most of these occur between positions T6 and T8 (Hile and Eckert, 2008). Interestingly, we found that 57% of the top-2,000 HU-induced DSBs show recurrent interruptions of the poly(dA:dT) tract by a CC:GG dinucleotide sequence (Figures 2D, insert, and S7A). Motif analysis of poly(dA:dT) broken in 0.5mM HU revealed that these have a stronger CC:GG signature within 10 bp of the DSB compared to all poly(dA:dT) in the genome (Figure S7B). The CC:GG signature frequently appeared between T6 and T8 (Figures 2D, 4A, and S7B), indicative of a highly mutable hotspot within mononucleotide dT repeats. Altogether, these data suggest that throughout evolution, polymerase kappa has been employed during replication of poly(dA:dT) repeats, which we show are preferential sites of polar RF stalling and collapse.
Conserved Genomic Features of Origins
Poly(dA:dT) repeats are the most abundant simple repetitive sequence motif in the human genome (Dechering et al., 1998). The preferred maintenance and expansion of long poly(dA:dT) tracts that contribute to chromosomal fragility therefore presents an evolutionary enigma. While origins of replication in S. pombe lack a specific consensus sequence analogous to S. cerevisiae, S. pombe origins are located in intergenic regions, and poly(dA:dT) tracts are the strongest predictors of origin function (Mojardín et al., 2013). Using two independent approaches (HU-EdU-seq and END-seq) with high precision and sensitivity to detect strong initiation events, we find recurrent peaks of nsDNA synthesis and DSBs within zones of initiation in mouse and human cells, suggesting non-uniformity of origin efficiency.
The strongest initiation events are characterized by AT-enrichment, nucleosome depletion, and compositional asymmetry of poly(dA:dT) motifs flanking these sites. Therefore, we propose that AT richness and poly(dA:dT) motifs are significant determinants of origin usage in mammalian cells.
The association of AT-rich sequences at origins is reminiscent of previously described DNA Unwinding Elements (DUEs), characterized by sequences with dA/dT skew which facilitate replication origin unwinding (Kowalski and Eddy, 1989). These DUEs have been described at bacteria, yeast, and mammalian replication origins (Kowalski and Eddy, 1989; Liu et al., 2003; Umek and Kowalski, 1990), indicating structural conservation throughout evolution. A similar dA/dT asymmetry is seen at origins in this study (Figure 7A, top panel) that are distinct from the flanking poly(dA:dT) tracts (Figure 7A, middle panel) that are rigid and nucleosome-free (Aymami et al., 1989). We reason that the central AT-rich sequence at the origin and flanking poly(dA:dT) tracts may synergize to form a nucleosome-free region that is easily unwound to promote origin firing. Upon replication stress, the same sequences may expose long stretches of naked DNA, which could induce fork stalling and collapse.
The trans-activating proteins that initiate replication are highly conserved. In addition to other DNA-related processes facilitated by nucleosome positioning, replication origin specification by long poly(dA:dT) tracts may contribute to the exponential expansion of these simple repeats during evolution, but at the cost of increasing chromosome fragility.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, André Nussenzweig (andre_nussenzweig@nih.gov).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
B cells were obtained from 6–12 week old wild-type C57BL/6 and 53BP1−/−mice and wild-type CAST/EiJ. Both male and female mice were indiscriminately used for these studies, as both male and female have similar DSB profiles in response to replication stress. Mice were housed and handled according to regulatory standards set by the NCI Animal Care and Use Committee with unrestricted access to regular mouse chow and water.
Cells culture and cell lines
Mature resting B cells were isolated from wild-type and 53BP1 −/−mouse spleens with anti-CD43 MicroBeads (Miltenyi Biotech). Bcells were activated with LPS (25 mg/ml; Sigma), IL-4 (5 ng/ml; Sigma) and RP105 (0.5 mg/ml; BD). Cells were cultured in RPMI media with 10% FBS as described (Callen et al., 2013) until cells were collected. For 28 hour HU treatments, 10 mM or 0.5 mM HU was added immediately upon activation. For 6 hour HU treatments, 10 mM HU was added after 22 hours and cultured for an additional 6 hours. For ATR inhibitor experiments, ATRi (AZ20, 10 μM) was added after 20 hours and cultured for an additional 8 hours. For CDK2i/CDC7i experiments, 10 μM Roscovitine, 10 μM PHA-767491, and HU (10 mM or 0.5 mM) were added after 22 hours and cultured for an additional 6 hours. For experiments with Mirin, 50 μM Mirin was added after 21 hours, and 0.5 mM HU was added 1 hour later (at 22 hours), and cells were cultured for an additional 6 hours. Mature T cells were isolated from wild-type C57BL6/J mouse lymph nodes and activated for 28 hr with plate-bound anti-TCR beta and anti-CD28 antibodies in RPMI media with 10% FBS. For 28 hour HU treatments, 2 mM HU was added immediately upon activation. HCT116 cells (gift from Masato Kanemaki) were cultured in McCoys 5A Medium (GIBCO) + 10% FBS. Cells were synchronized using CDK4/6 inhibitor (Palbociclib, 2 μM) for 20 hours. Arrested HCT116 cells were released from G1 arrest by washing out 3X with fresh medium and cultured for 8 hours in the presence or absence of 10 mM HU. In all experiments, 15 million cells were harvested for END-seq or HU-EdU-seq.
METHOD DETAILS
Flow Cytometry
To measure DNA synthesis, B cell cultures were stimulated for 28 hours, pulsed with 10 mM of EdU (5-ethynyl-2’ -deoxyuridine) for 15 min at 37°C and stained using the Click-IT EdU Alexa Fluor 488/647 Flow Cytometry Assay Kit according to the manufacturer’s specifications (ThermoFisher). DNA content was measured with DAPI or Propidium Iodide. Samples were acquired on a FACSCantoll (BD biosciences) or Accuri C6 (BD biosciences). To measure H3S10p, cells were fixed overnight with 70% ethanol, permeabilized with 0.25% Triton X-100 for 10 minutes, washed in PBS, incubated for 3 hours with H3S10p antibody (1:200, Millipore), washed, stained with anti-rabbit Alexa Fluor 488 antibody (1:2000, Thermofisher). Propidium iodide + RNAase A was added before FACS analysis on BD Accuri C6 (BD Biosciences). Data was analyzed using FlowJo, gating on live cells by FSC,SSC and single cells by FSC-H,FSC-A.
Molecular Combing of DNA Fibers
Dynamic molecular combing was performed as described (Fu et al., 2015). Cells were labeled with with 20 μmol/l IdU for 20 min, then washed and allowed to grow in the presence or absence of HU as indicated in the figure legend. CldU (50 μmol/l) was added to the growth medium for 20 minutes or 6 hours prior to harvesting. DNA fibers were combed onto silanized microscope slides, incorporation of IdU and CldU was detected on the stretched fibers and the ratios of CldU and IdU labels were calculated as in Ray Chaudhuri et al. (2016). Coverslips with combed DNA were incubated at 60°C for 2 h and denatured in 0.5 N NaOH for 20 min. Coverslips with DNA were incubated with primary antibodies (mouse IgG1 anti-BrdU, Becton Dickinson, cat.347580, 1:25, for IdU; rat anti-BrdU, Accurate chemical, cat. OBT0030, 1:50 for CldU; mouse IgG2a anti-ssDNA, Chemicon, MAB3034, 1:100) for 1 h at room temperature. Slides were washed and stained with secondary antibodies (Alexa Fluor 594, 488, 647, 1:100, Thermofisher) for 45 minutes at room temperature. Slides were scanned using BD pathway 855 controlled by AttoVision. Fluorescent signals were measured using ImageJ. Statistical sigificance was calculated using the non-parametric Mann-Whitney test.
Replication Timing
Replication timing profiles were determined using TimEX as described (Bartholdy et al., PMID: 25987481) with minor modifications. The S/G1 TimEX ratio, proportional to the replication time during S phase, was measured in mature splenic B cells from wild-type C57BL/6NCr mice. The G1 population was obtained from mature resting B cells (100% G0/G1 arrested), which were isolated using anti-CD43 MicroBeads (Miltenyi Biotech) for negative selection. To obtain the S-phase population, the resting B cells were activated with LPS (25 mg/ml; Sigma), IL-4 (5 ng/ml; Sigma) and RP105 (0.5 mg/ml; BD) for 48 hours (~70% S-phase). 5 million cells were collected for both populations, and whole genome sequencing was performed using standard lllumina library prep and sequenced on HiSeq 2000.
Nascent DNA sequencing (HU-EdU/EdC-seq)
DNA labeling and Fixation
Cells were incubated with 20 uM EdU/EdC for 28 hours (B cells, T Cells) or 8 hours (HCT116 cells). Cells were pelleted and fixed in 90% methanol for 15 minutes on ice. Cells were washed and permeablized with 0.2% Triton X-100 in PBS, on ice, for 10 minutes. Cells were then washed 1X in PBS.
Biotin-labeling of EdU using Click-IT
For Click-IT reaction, cell pellets were resuspended in PBS, 10 μM Biotin Azide (ThermoFisher Cat# B10184), 200 mM CuSO4 (Sigma), and 10 mM sodium ascorbate (Sigma) for 2 hours, at room temperature, in the dark.
DNA sonication
To recover DNA, cell pellets were washed 2x with PBS, lysed with 50 mM Tris pH 8.0, 1% SDS before phenol chloroform extraction of DNA. Purification of DNA was performed using UltraPure Phenol:Chloroform:Isoamyl Alcohol (25:24:1, v/v) (Invitrogen), according to manufacturer’s instructions. DNA was resuspended in 13 μL TE buffer, then sheared to 150–200 bp fragments using Covaris S220 sonicator at 10% duty cycle, peak incident power 175, 200 cycles per burst, 240 s.
DNA purification
Biotin-EdU labeled DNA fragments were purified using MyOne Streptavidin C1 Beads (ThermoFisher #650–01). 35 μL of Dynabeads were washed twice with 1 mL Binding and Wash Buffer (1xBWB) (10 mM Tris-HCl pH8.0, 1 mM EDTA, 1 M NaCl, 0.1% Tween20). Beads were recovered using a DynaMag-2 magnetic separator (12321D, Invitrogen). Supernatants were discarded. Washed beads were resuspended in 130 μL 2xBWB (10 mM Tris-HCl pH8.0, 2 mM EDTA, 2 M NaCl) combined with the 130 μL of sonicated DNA and incubated at 24°C for 30 min in a ThermoMixer C at 400 rpm.
End-Repair, A-tailing, and Library Amplification
Following the 30 min of mixing (above), the supernatant was removed and the bead bound biotinylated DNA was washed 3 times with 1 mL 1xBWB, twice with 1 mL EB buffer, once with 1 mL T4 ligase reaction buffer (NEB) and then resuspended in 50 μL of end-repair reaction mix (0.4 mM of dNTPs, 2.7 U of T4 DNA polymerase (NEB), 9 U of T4 Polynucleotide Kinase (NEB) and 1 U of Klenow fragment (NEB)). The end-repair reaction was incubated at 24°C for 30 min in a ThermoMixer C at 400 rpm (tubes were vortexed every 10 min). The supernatant was removed using a magnetic separator and beads were then washed once with 1 mL 1xBWB, twice with 1 mL EB buffer, once with 1 mL NEBNext dA-Tailing reaction buffer (NEB) and then resuspended in 50 μL ofA-tailing reaction with NEBNext dA-Tailing reaction buffer (NEB) and 20 U of Klenow fragment exo- (NEB). The A-tailing reaction was incubated at 37°C for 30 min in a ThermoMixer C at 400 rpm (tubes were vortexed every 10 min). The supernatant was removed using a magnetic separator and washed once with 1 mL NEBuffer 2 and then resuspended in 115 mL of Ligation reaction with Quick Ligase buffer (NEB), 6,000 U of Quick Ligase (NEB) and 5 nM annealed adaptor (Truseq truncated adaptor) and incubated at 25°C for 30 min in a ThermoMixer C at 400 rpm. Ligation reaction was stopped by adding 50 mM of EDTA, then DNA was purified with 1.8X volume AMPure XP beads and eluted in 15 μL of EB. PCR amplification was performed in 50 μL reaction with 10 mM primers 5’-CAAGCAGAAGACGGCATACGAGATxrefGTGACTGGAGTTCAGACGTGTGCTCTTCCGATC*T- 30 and 5’-AATGATACGGCGACCACCGAGATCTACACTCTTTCC CTACACGACGCTCTTCCGATC*T- 30, and 2X Kapa HiFi HotStart Ready mix (Kapa Biosciences). * represents a phosphothioratebond and NNNNNN a Truseq index sequence. PCR program: 98°C, 45 s; 15 cycles [98°C, 15 s; 63°C, 30 s; 72°C, 30 s]; 72°C, 5 min. PCR reactions were cleaned with AMPure XP beads, and 200–500 bp fragments were isolated after running on 2% agarose gel. Libraries were purified using QIA-quick Gel Extraction Kit (QIAGEN). Library concentration was determined with KAPA Library Quantification Kit for lllumina Platforms (Kapa Biosystems). Sequencing was performed on using Illumina NextSeq 500 or 550 (75bp single end reads).
Strand-specific Nascent RNA Sequencing (nsRNA-seq)
Nascent RNA sequencing was performed as previously described (Canela et al., 2017). B cell cultures were stimulated for 28 hours ± 10 mM HU, and 5 million cells were labeled with 0.5 mM 5-ethynyl uridine (EU) for the final 30 min. Total RNA extraction was performed using TRIzol (Invitrogen) and 2 μg was rRNA depleted using the NEBNext rRNA Depletion kit (human/mouse/rat) (New England Biosciences). rRNA-depleted RNA was used for biotinylation via the Click-IT reaction (Click-iT Nascent RNA Capture Kit, ThermoFisher C10365) using manufacturer’s specification. First-strand cDNA synthesis of the captured nascent RNA was done using SuperScript VILO cDNA synthesis kit (Invitrogen), followed by purification with 1.8X volume of AMPure XP beads (1.8X) and elution in 20 μL. Second-strand synthesis was performed using 0.6 mM dATP, 0.6 mM dCTP, 0.6 mM dGTP, and 1.2 mM dUTP in the presence of 2 Units of RNase H (Invitrogen) and 20 Units of E. coli DNA polymerase I (Invitrogen) in a total volume of 30 μL for 2.5 hours at 16°C. cDNA was purified using 1.8X volume AMPure XP beads and eluted in 20 μL of EB. Sequencing libraries were then prepared. End-repair was performed in 50 μL of T4 ligase reaction buffer (1X), dNTPs (0.4 nM), T4 DNA polymerase (NEB, 3 Units), T4 Polynucleotide Kinase (NEB, 9 Units) and Klenow fragment (NEB, 1 Unit) at 24°C for 30 min in a ThermoMixer at 400 rpm. End-repair reaction was cleaned using 1.8X volume AMPure XP beads and eluted in 15 mL of EB. A-tailing was performed using NEBNext dA-Tailing reaction buffer (NEB, 1X) with Klenow fragment exo- (NEB, 7.5 U) at 37°C for 30 min. A-tailing reaction was mixed with Quick Ligase Buffer (NEB), Quick ligase (NEB, 3000 Units) and 5 nM of annealed adaptor (Illumina truncated adaptor) in a volume of 75 μL and incubated at 25°C for 20 min. Ligation reaction was stopped by adding 50 mM of EDTA, then DNA was purified with 1.8X volume AMPure XP beads and eluted in 15 μL of EB. 0.5 Units of Uracil-DNA glycosylase (ThermoFisher) was added for 15 min at 37°C. PCR amplification was performed in 50 μL reaction with 10 mM primers 5’-CAAGCAGAAGACGGCATACGAGATxrefGTGACTGGAGTTCAGACGTGTGCTCTTCCGATC*T- 30 and 5’-AATGATACGGCGACCACCGAGATCTACACTCTTTCC CTACACGACGCTCTTCCGATC*T- 30, and 2X Kapa HiFi HotStart Ready mix (Kapa Biosciences). * represents a phosphothioratebond and NNNNNN a Truseq index sequence. PCR program: 98°C, 45 s; 15 cycles [98°C, 15 s; 63°C, 30 s; 72°C, 30 s]; 72°C, 5 min. PCR reactions were cleaned with AMPure XP beads, and 200–500 bp fragments were isolated after running on 2% agarose gel. Libraries were purified using QIA-quick Gel Extraction Kit (QIAGEN). Library concentration was determined with KAPA Library Quantification Kit for Illumina Platforms (Kapa Biosystems). Sequencing was performed on using Illumina NextSeq 500 or 550 (75bp single end reads).
OK-seq
OK-seq was performed in asynchronous mouse B cells activated for 48 hours, as previously described in a detailed protocol Here, the exponentially growing B cells were pulsed with 20 mM EdU (5-ethynyl-2’-deoxyuridine, Jenabioscience) for 2 minutes to label newly synthesized DNA. DNA was extracted according to a standard proteinase K/phenol chloroform protocol. The extracted DNA was dissolved in TE buffer (10mM Tris-HCl, PH 8.0, 1mM EDTA) for at least 48 hours, heat denatured, and size fractionated by sucrose gradient centrifugation. To isolate Okazaki fragments, fractions of ≤ 200 nt length were collected. EdU-labeled DNA was covalently linked to biotin azide (Invitrogen, #B10184) via Click-iT reaction (10 mM Tris-HCl pH 8.0, 2 mM CuSO4, 1 mM biotin-azide, 10 mM sodium ascorbate) for 45 min at room temperature. RNA was hydrolyzed using NaOH. After RNA hydrolysis, the biotinylated DNA was captured on Dynabeads (ThermoFisher) and was ligated to adaptors A1 and A2. Ligation mixes were amplified with KAPA HIFI DNA polymerase (12 cycles) using the oligonucleotides “PE PCR Primer 1.0 Meyer” and “TruSeq PCR Primer,” listed in the reagents section. PCR products were separated from beads and gel-purified to select for fragments 150–300 bp. Resulting library were quantified and sequenced on Illumina NextSeq 500. Adaptors have been removed using Cutadapt-1.15 and reads > 10nt have been mapped on reference genome (GRCm38/mm10) with Bwa 0.6.2-r126. Three biological replication samples were sequenced, and data was pooled. The OK-seq procedure has been described in detail (Petryk et al., 2016).
END-seq
For B cells and T cells, 15 million cells in single cell suspension were embedded in a single agarose plug. For HCT116 cells, 7.5 million cells in single suspension were embedded in an agarose plug (1% agarose final), and DNA from two plugs were combined after DNA shearing, prior to DNA purification for library prep. Embedded cells were lysed and digested using Proteinase K (50°C, 1 hour then 37°C for 7 hours). Plugs were rinsed in TE buffer and treated with RNase A at 37°C, 1 hour. DNA ends were then blunted. DNA was retained in agarose plugs to prevent shearing throughout the ssDNA blunting reactions. The first blunting reaction was performed using ExoVII (NEB, M0379S) for 1hr, 37C. Agarose plugs were washed 2X in NEB Buffer 4 (1X), followed by the second blunting reaction using ExoT (NEB, M0265S) for 1 hour, 24C. After blunting, two washes were performed in NEBNext dA-Tailing Reaction Buffer (NEB, B6059S), followed by A-tailing to attach dA to the free 30-OH (Klenow 30− > 5’ exo-, NEB, M0212S). After A-tailing, ligation of “END-seq hairpin adaptor 1,” listed in reagents section, using NEB Quick Ligation Kit (NEB, M2200S).
DNA sonication
Agarose plugs were then melted and dissolved, and DNA was sonicated using to a median shear length of 170bp using Covaris S220 sonicator for 4 min at 10% duty cycle, peak incident power 175, 200 cycles per burst, 4°C. DNA was ethanol-precipitated and dissolved in 70 μL TE buffer. 35 μL of Dynabeads were washed twice with 1 mL Binding and Wash Buffer (1xBWB) (10 mM Tris-HCl pH8.0, 1 mM EDTA, 1 M NaCl, 0.1% Tween20). Beads were recovered using a DynaMag-2 magnetic separator (12321D, Invitrogen). Supernatants were discarded. Washed beads were resuspended in 130 μL 2xBWB (10 mM Tris-HCl pH8.0, 2 mM EDTA, 2 M NaCl) combined with the 130 μL of sonicated DNA and incubated at 24°C for 30 min in a ThermoMixer C at 400 rpm.
End-Repair, A-tailing, and Library Amplification
Following the 30 min of mixing (above), the supernatant was removed and the bead bound biotinylated DNA was washed 3 times with 1 mL 1xBWB, twice with 1 mL EB buffer, once with 1 mL T4 ligase reaction buffer (NEB) and then resuspended in 50 μL of end-repair reaction mix (0.4 mM of dNTPs, 2.7 U of T4 DNA polymerase (NEB), 9 U of T4 Polynucleotide Kinase (NEB) and 1 U of Klenow fragment (NEB)). The end-repair reaction was incubated at 24°C for 30 min in a ThermoMixer C at 400 rpm (tubes were vortexed every 10 min). The supernatant was removed using a magnetic separator and beads were then washed once with 1 mL 1xBWB, twice with 1 mL EB buffer, once with 1 mL NEBNext dA-Tailing reaction buffer (NEB) and then resuspended in 50 μL ofA-tailing reaction with NEBNext dA-Tailing reaction buffer (NEB) and 20 U of Klenow fragment exo- (NEB). The A-tailing reaction was incubated at 37°C for 30 min in a ThermoMixer C at 400 rpm (tubes were vortexed every 10 min). The supernatant was removed using a magnetic separator and washed once with 1 mL NEBuffer 2 and then resuspended in 115 mL of Ligation reaction with Quick Ligase buffer (NEB), 6,000 U of Quick Ligase (NEB) and ligated to “END-seq hairpin adaptor 2” and incubated at 25°C for 30 min in a ThermoMixer C at 400 rpm. Ligation reaction was stopped by adding 50 mM of EDTA, then beads were washed 3X BWB and 3X EB, and eluted in 8 μL of EB. Hairpin adaptors were digested using USER enzyme (NEB, M5505S) at 37C, 30 minutes. PCR amplification was performed in 50 μL reaction with 10 mM primers 5’-CAAGCAGAAGACGGCATACGA-GATxrefGTGACTGGAGTTCAGACGTGTGCTCTTCCGATC*T- 30 and 5’-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATC*T- 30, and 2X Kapa HiFi HotStart Ready mix (Kapa Biosciences). * represents a phosphothioratebond and NNNNNN a Truseq index sequence. PCR program: 98°C, 45 s; 15 cycles [98°C, 15 s; 63°C, 30 s; 72°C, 30 s]; 72°C, 5 min. PCR reactions were cleaned with AMPure XP beads, and 200–500 bp fragments were isolated after running on 2% agarose gel. Libraries were purified using QIA-quick Gel Extraction Kit (QIAGEN). Library concentration was determined with KAPA Library Quantification Kit for Illumina Platforms (Kapa Biosystems). Sequencing was performed on using Illumina NextSeq 500 or 550 (75bp single end reads).
The second end was ligated to “END-seq hairpin adaptor 2” using NEB Quick Ligase. Hairpins were digested using USER (NEB), and the resulting DNA fragments were PCR amplified using “TruSeq barcoded primer” and “TruSeq multiplex primer,” listed in reagents. PCR fragments were isolated by size selection from agarose gel, selecting 200–500 bp fragments followed by DNA purification using QIAquick Gel Extraction Kit. Libraries were quantified and sequenced using Illumina NextSeq 500 or 550. A detailed END-seq rationale and protocol can be found in Canela et al. (2016, 2017).
RPA ChIP-seq
Splenic B cells were isolated and activated for 28 hours in the presence of 10 mM HU. Cells were fixed adding 37% formaldehyde (F1635, Sigma) to a final concentration of 1% and incubating them at 37°C for 10 min. Fixation was quenched by addition of 1M glycine (Sigma) in PBS at a final concentration of 125 mM. Twenty million fixed cells were washed twice with cold PBS and pellets were snap frozen in dry ice and stored at —80°C. Fixed cell pellets of 20 million cells were thawed on ice and resuspended in 1 mL of cold RIPA buffer (10 mM TrisHCl pH 7.5, 1 mM EDTA, 0.1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100, 1 3 Complete Mini EDTA free proteinase inhibitor (Roche)). Sonication was performed using the Covaris S220 sonicator at duty cycle 20%, peak incident power 175, cycle/burst 200 for 30 min. Chromatin were clarified by centrifugation at 21,000 g at 4°C for 10 min and precleared with 80 mL prewashed Dynabeads protein A (ThermoFisher) for 30 min at 4°C. 40 μL prewashed Dynabeads protein A were incubated with 10 mg of Anti-RPA32/RPA2 antibody (Abcam, ab10359) in 100 mL of PBS for 20 min at room temperature in continuous mixing, washed twice in PBS for 5 min and added to 1 mL of chromatin followed by overnight incubation at 4°C on a rotator. Beads were then collected in a magnetic separator (DynaMag-2 Invitrogen), washed twice with cold RIPA buffer, twice with RIPA buffer containing 0.3M NaCl, twice with LiCl buffer (0.25 M LiCl, 0.5% Igepal-630, 0.5% sodium deoxycholate), once with TE (10 mM Tris pH 8.0, 1mM EDTA) plus 0.2% Triton X-100, and once with TE. Crosslinking was reversed by incubating the beads at 65°C for 4 hr in the presence of 0.3% SDS and 1mg/ml of Proteinase K (Ambion). DNA was purified by standard phenol-chloroform method and eluted in 20 μl. 100ng ChIP DNA was used to prepare Illumina sequencing libraries. End-repair was performed in 50 μL volume containing 1X T4 ligase reaction buffer, 0.4 mM of dNTPs, 3 U of T4 DNA polymerase (NEB), 10 U of T4 Polynucleotide Kinase (NEB) and 1 U of Klenow fragment (NEB) at 20°C for 30 min in a ThermoMixer. End-repair reaction was cleaned using MinElute PCR cleanup (QIAGEN) and eluted in 12 μL of EB that was used for A-tailing reaction in 50 μL volume consisting of 1X NEBuffer 2 (NEB), 5 U of Klenow fragment exo-(NEB) and 0.2mM dATP at 37°C for 30 min. The reaction was cleaned using MinElute PCR cleanup (QIAGEN) and eluted in 22 μL of EB. The DNA is subsequently incubated at 95°C for 3 min to enrich for ssDNA. The reaction were cooled at room temperature and mixed with Quick Ligase buffer 2X (NEB), 3,000 U of Quick ligase and 5 nM of annealed adaptor (Illumina truncated adaptor) in a volume of 70 μL and incubated at 20°C for 20 min. Adaptor was prepared by annealing the following HPLC oligos: 5’-Phos/GATCG GAAGAGCACACGTCT- 30and 5’-ACACTCTTTCCCTACACGACGCTCTTCCGATC*T- 30 (*phosphorothioate bond). Ligation was stopped by adding 50mM of EDTA and cleaned with MinElute PCR cleanup (QIAGEN) and eluted in 17 μL of EB that was used for PCR amplification in a 50 μL reaction with 10 μM primers 5’-CAAGCAGAAGACGGCATACGAGATNNNNNNGTGACTGGAGTTCAGACGTGTGCTCTTCCGATC*T- 30 and 5’-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGC TCTTCCGATC*T- 30, and 2X Kapa HiFi HotStart Ready mix (Kapa Biosciences), * represents a phosphothiorate bond and NNNNNN a Truseq index sequence. The temperature settings during the PCR amplification were 45 s at 98°C followed by 15 cycles of 15 s at 98°C, 30 s at 63°C, 30 s at 72°C and a final 5 min extension at 72° C. PCR reactions were cleaned with Agencourt AMPure XP beads (Beckman Coulter), run on a 2% agarose gel and a smear of 200–500bp was cut and gel purified using QIAquick Gel Extraction Kit (QIAGEN). Library concentration was determined with KAPA Library Quantification Kit for Illumina Platforms (Kapa Biosystems). Sequencing was performed on the Illumina Nextseq500 (75bp single end reads).
QUANTIFICATION AND STATISTICAL ANALYSIS
Genome Alignment
Tags were aligned to the mouse (mm10) or human (hg19) genome using Bowtie (version 1.1.2) (Langmead et al., 2009) with the options -l 25 -n 3–best–all–strata -M 1–tryhard –t (END-seq, nascent RNA) and with the options -l 50 -n 2–best–all–strata -m 1 (EdU-seq). Alignment of OK-seq tags was done as follows: adaptor sequences were removed by Cutadapt (version 1.15). Reads > 10 nt were aligned to the mouse reference genome (mm10) using the BWA (version 0.6.2-r126) software with default parameters. Repliseq tags were aligned using Bowtie2 (version 2.3.4.1) (Langmead and Salzberg, 2012) with parameters -N 0 -k 1.The alignment output sam files were converted and sorted into bam files using samtools (Li et al., 2009).
Alignment of END-seq data to CAST/EiJ (Mus musculus castaneus) mouse genome
Whole genome fasta of CAST/EiJ mice was obtained from https://useast.ensembl.org/info/data/ftp/index.html. Using this whole genome fasta we built a new index for alignment to CAST/EiJ genome using bowtie-build command. END-seq data from CAST/ EiJ mice was aligned to the CAST/EiJ genome using the same parameters explained previously. For aligning C57BL6/NJ samples to this CAST/EiJ genome, we clipped the first 10 bases of every read. These clipped reads were aligned using the same parameters explained previously. Data visualization was performed using UCSC genome browser track hubs from 16 different mice strain including the CAST/EiJ genome.
Peak Calling
Peaks were called for END-seq data using MACS 1.4.3 (Zhang et al., 2008) using the parameters -p 1e-5–nolambda–nomodel–keep- dup = all (keep all redundant reads). For EdU-seq peak-calling, the default parameters of MACS were used. The corresponding nontreated samples were used as control, and peaks enriched over control by a certain threshold are retained for further downstream analysis.
Defining EdUhigh peaks
B cells: The results of peak-calling was filtered to retain peaks enriched over control sample by at least 18-fold. Peaks within blacklisted regions were discarded. HCT116: The results of peak-calling were filtered to retain peaks enriched over random background by at least 4 fold. No control sample was used in peak-calling. Peaks within blacklisted regions were discarded.
Defining Replication Initiation Zones by HU-EdU-seq
The EdU peaks within 20kb of each other were merged together using bedtools merge –d function. The resulting merged peaks were defined as the HU-EdU-seq initiation zones in the respective cell types.
Defining Replication Initiation Zones by OK-seq
A four-state HMM was used to detect within the RFD profiles the ascending, descending and flat segments representing regions of predominant initiation (‘Up’ state), predominant termination (‘Down’ state) and constant RFD (‘Flat1’ and ‘Flat2’ states). The RFD values were computed within 15 kb sliding windows (stepped by 1 kb across the autosomes). The HMM used the ΔRFD values between adjacent windows (that is, ΔRFDn = (RFD(n+1)-RFD(n))/2 for window n). Windows with < 30 reads on one strand were masked. The ΔRFD values were divided into five quantiles and used the transition and emission probabilities from Petryk et al. to build the initial HMM model. Inferred the parameters of the HMM model and perform HMM prediction by the HMM package of R (http://www.r-project.org/). Identification of replication initiation zones by OK-seq procedure has been described in rigorous detail (Petryk et al., 2016).
Defining END-seq DSB sites
The results of peak-calling were filtered to retain peaks enriched over control sample by at least 5 fold. The DSB site is defined as edge of sharp peak flanking the gap between sharp and broad ends of the DSBs.
Defining HU-DSB zones
The END-seq DSB sites within 20 kb of each other were merged together using bedtools merge –d function. The resulting merged peaks were defined as the HU-DSB zones in the mouse samples. For HCT116 cells, only zones overlapping with at least two END- seq break-sites were retained.
Motif Finding
Motifs were plotted centering on the DSB site. The nucleotide sequence in a window of interest around the DSB site was obtained using bedtools getfasta function, and ggseqlogo in R was used to plot the resulting motif.
To show GG insertion as a signature of polymerase kappa genome-wide, we used the nucleotide sequence around all the DSB sites to make a position weight matrix (PWM). This PWM was used in fimo (part of MEME package) to identify sites with cutoff value (p value < 10−12), resulting in 174,419 sites genome-wide. The resulting motif is shown in Figure S7B. From the 174,419 sites genomewide, sites with END-seq RPKM > 2 (21,801 sites) were selected and the corresponding motif was generated to show enrichment of GG insertions near the DSB site.
Visualization of Sequencing Data
To make genome tracks, we first used bedtools genomecov (Quinlan and Hall, 2010) to convert the aligned bed files to bedgraph, and then bedGraphToBigWig to make a bigwig file. Values were normalized to show RPM. Visualization of genomic profiles was done by the UCSC browser (Kent et al., 2002). Heatmaps were produced using the R package ‘pheatmap’.
Composite plots for sequencing reads around sites of interest (genes, initiation sites, and DSBs) were performed as follows: A window was defined around the sites of interest genome-wide. This window was further divided into smaller windows using bedtools makewindows function. The number of feature overlapping each smaller window was calculated using bedtools coverage –counts function. The aggregate signal was smoothed using smooth.spline function in R.
Statistical Analysis
To test for significance of overlaps between OK-seq versus HU-EdU-seq, HU-EdU-seq versus HU-END-seq, ERFS hotspots versus HU-END-seq, and enrichment of the poly (dA/dT) tracts at the DSB site we used Fisher’s Exact Test over a random genomic background. To test for significant changes in a) DSB within initiation zones and gene bodies upon CDK2i/CDC7i treatment, and b) DSBs in WT versus MCM2Δ/Δ B cells within initiation zones, we used the Wilcoxon Rank Sum Test. The p value is reported for each set of comparison. Pearson (r) correlation was used to report when comparing similar types of experiments (nsRNA versus nsRNA, EdU-seq versus EdC-seq, EdU-seq versus EdC-seq). Spearman (ρ) correlation was used to report correlations when comparing different types of experiments (END-seq versus EdU-seq, EdU-seq versus nsRNA).
Replication Timing by TimEX
TimEX ratio (ratio between number of reads observed in the S and G1 phase of the cell cycle) was calculated in 100 kb windows genome-wide using bedtools coverage function. This ratio was converted to a log scale and windows with the log ratio greater (less) than 0 was designated as early (late) replicating windows. The early and late replicating windows are merged using bedtools merge function to yield early and late replicating regions genome-wide.
Alignment of END-seq and HU-EdU-seq data to ribosomal DNA
As rDNA is not included in mm10 reference genome, we built a custom mm10 genome which contained a single copy mouse rDNA sequence (GenBank accession number BK000964). Because of repetitive sequences within rDNA, a weighting method based on unique reads was used to the multiple mapped reads. First, END-seq reads were mapped to the custom genome by Bowtie (version 1.1.2) using the “-a -m 50 -n 3 -l 50.” Functions “view” and “sort” of samtools (version 1.6) were used to convert and sort the mapping output to sorted bam file. Then, multiple mapped reads were weighted by the counts of unique mapped reads within 10bp around an alignment. The bigWig file for visualization was built for each 10bp bin using RPKM value.
Estimation of ribosomal DNA dosage
We used the read depth of 45S ribosomal DNA coding region (18S, 5.8S and 28S) relative to read depth of single copy sequences to estimate the ribosomal DNA dosage by using a whole genome sequencing data from resting B cells. The background read depth (BRD) which represent the depth of single copy DNA sequences acts as a normalized factor to estimate the copy number of the 45S ribosomal DNA. First, several filtering steps to build a reference single copy exon set as a representation for BRD: 1) Homologous exons were identified by BLAST (E-value < 1×10–3) and then removed. 2) Exons from chromosome X and chromosome Y were removed. 3) Only the largest exon of each gene and longer than 400bp were retained. Second, reads were mapped to ribosomal DNA added genome by Bowtie2 (version 2.3.4) and per-base BRD was calculated by “depth” function of samtools (version 1.6). As fewer reads were mapped to the beginning and end of references, the first and last 152 bp depth values are excluded. In addition, depth value in the upper 5% were excluded. Next, samtools “depth” function was used to get the average read depth of ribosomal DNA coding region as well. At last, ribosomal DNA dosage was calculated like this: ribosomal DNA dosage = (average read depth of ribosomal DNA coding region)/(average BRD).
DATA AND SOFTWARE AVAILABILITY
All sequencing data for this study was deposited at NCBI GEO under the accession number GEO: GSE116321.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-RPA32/RPA2 antibody (ab10359) | Abcam | Cat# ab10359; RRID: AB_297095 |
| RP105 (Purified Rat Anti-Mouse CD180) | BD Biosciences | Cat# 552128; RRID: AB_394343 |
| Purified Hamster Anti-Mouse TCR beta chain | BD Biosciences | Cat# H57–597; RRID: AB_394679 |
| Purified Hamster Anti-Mouse CD28 | BD Biosciences | Cat# 553294; RRID: AB_394763 |
| Anti-phospho-Histone H3 (Ser10) Antibody | Millipore-Sigma | Cat# 06–570; RRID: AB_310177 |
| Goat anti-Rabbit IgG (H+L) Alexa Fluor 488 | ThermoFisher | Cat# A-11008; RRID: AB_143165 |
| Mouse IgG1 anti-BrdU | BD Biosciences | Cat# 347580; RRID: AB_400326 |
| Rat anti-BrdU | Accurate chemical | Cat# OBT0030; RRID: AB_2313756 |
| Mouse IgG2a anti-ssDNA | Chemicon | Cat# MAB3034; RRID: AB_94645 |
| Alexa Fluor 594 donkey anti-rat | ThermoFisher | Cat# A21209; RRID: AB_2535795 |
| Alexa Fluor 488 goat anti mouse IgG1 | ThermoFisher | Cat# A21121; RRID: AB_2535764 |
| Alexa Fluor 647 goat anti mouse IgG2a | ThermoFisher | Cat# A21241; RRID: AB_2535810 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| HU | Millipore-Sigma | Cat# H8627 |
| TRIzol Reagent | Thermo Fisher | Cat# 15596026 |
| EdU (5-ethynyl-2′-deoxyuridine) | Thermo Fisher | Cat# A10044 |
| 5-Ethynyl-2′-deoxycytidine, (EdC) | Millipore-Sigma | Cat# T511307 |
| AZ20 (ATR inhibitor) | Selleckchem | Cat# S7050 |
| Palbociclib (PD-0332991) HCl | Selleckchem | Cat# S1116 |
| Roscovitine (Seliciclib,CYC202) | Selleckchem | Cat# S1153 |
| PHA-767491 | Selleckchem | Cat# S2742 |
| Biotin Azide (PEG4 carboxamide-6-Azidohexanyl Biotin) | Invitrogen | Cat# B10184 |
| Exonuclease T (ExoT) | NEB | Cat# M0265S |
| Exonuclease VII (ExoVII) | NEB | Cat# M0379S |
| Klenow Fragment (3′→5′ exo-) | NEB | Cat# M0212S |
| Quick Ligation Kit | NEB | Cat# M2200S |
| USER enzyme | NEB | Cat# M5505S |
| KAPA HiFi HotStart ReadyMix (2X) | KAPA Biosystems | Cat# KK2600 |
| Lipopolysaccharide (LPS) | Millipore-Sigma | Cat# L-2630 |
| IL-4 | Millipore-Sigma | Cat# I1020 |
| UltraPure Phenol:Chloroform:Isoamyl Alcohol (25:24:1, v/v) | Invitrogen | Cat# 15593031 |
| Critical Commercial Assays | ||
| Click-IT Nascent RNA Capture Kit | Invitrogen | Cat# C10365 |
| Click-iT EdU Alexa Fluor 647 Flow Cytometry Assay Kit | Invitrogen | Cat# C10424 |
| Anti-CD43 (Ly-48) MicroBeads mouse | Miltenyi Biotech | Cat# 130–049-80 |
| NEBNext rRNA Depletion kit | NEB | Cat# E6310S |
| KAPA Library Quantification Kit | Kapa Biosystems | Cat# KK4824 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GEO: GSE116321 |
| HCT116 MNase-seq | Guzman and D’Orso, 2017 | GEO: GSE89871 |
| B cell MNase-seq | Kieffer-Kwon et al., 2017 | GEO: GSE82144 |
| OK-seq: HeLa | Petryk et al., 2016 | SRA: SRP065949 |
| ERFS hotspots | Barlow et al., 2013 | GEO: GSE43504 |
| Experimental Models: Cell Lines | ||
| HCT116 | Gift from Masato Kanemaki | |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6NCr | Charles River | Strain code# 027 |
| Mouse: CAST/EiJ | Jackson Laboratory | Stock No: 000928 |
| Oligonucleotides | ||
| TruSeq barcoded primer, 5′-Phos- CAAGCAGAAGACGGCA TACGAGATNNNNNNGTGACTGG AGTTCAGACGTGTGCT CTTCCGATC*T-5′ |
Canela et al., 2017 | N/A |
| TruSeq mulitplex primer, 5′-Phos- AATGATACGG CGACC ACCGAGATCTACACTCTTTCCCTA CACGACGCTCTTC CGATC |
Canela et al., 2017 | N/A |
| END-seq hairpin adaptor 1, 5′-Phos-GATCGGAAGAGC GTCGTGTAGGGAAAGAGTGUU[Biotin-dT]U[Biotin-dT] UUACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′ |
Canela et al., 2017 | N/A |
| END-seq hairpin adaptor 2, | Canela et al., 2017 | |
| OK-seq Adaptor 1 - Watson, 5′-ACA CTC TTT CCC TAC ACG ACG CTC TTC C-3′ |
Petryk et al. 2016 | |
| OK-seq Adaptor 1- Crick, 5′- NNN NNN G GAA GAG CGT CGT GTA GGG AAA GAG TG-3′ |
Petryk et al. 2016 | |
| OK-seq Adaptor 2- Watson, 5′ AGA TCG GAA GAG CAC ACG TCT GAA CTC CAG TCA [ddC]-3′ |
Petryk et al. 2016 | |
| OK-seq Adaptor 2- Crick, 5′-TGA CTG GAG TTC AGA CGT GTG CTC TTC CGA TCT NNN NNN [DDC]-3′ |
Petryk et al. 2016 | |
| PE PCR Primer 1.0 Meyer - Forward, 5′-ATGATACGGC GACCACCGAGATCTACACTCTTTCCCTACACGACGC TCTTCC-3′ |
Petryk et al. 2016 | |
| TruSeq PCR Primer - Reverse, 5′-CAAGCAGAAGACGG CATACGAGAT-INDEX-GTGACTGGAGTTCAGACGTGT GCTCTTCCGATCT-3′ |
Petryk et al. 2016 | |
| Software and Algorithms | ||
| Bowtie 1.1.2 | Langmead et al., 2009 | https://sourceforge.net/projects/bowtie-bio/files/bowtie/1.1.2/ |
| Bowtie 2.3.4.1 | Langmead and Salzberg, 2012 | https://sourceforge.net/projects/bowtie-bio/files/bowtie2/2.3.4.1/ |
| MACS 1.4.3 | Zhang et al., 2008 | https://pypi.org/project/MACS/1.4.3/ |
| UCSC Database | Karolchick et al., 2004 | https://genome.ucsc.edu |
| UCSC Genome Browser | Kent et al., 2002 | https://genome.ucsc.edu |
| Bedtools | Quinlan and Hall, 2010 | https://github.com/arq5x/bedtools2 |
| Samtools | Li et al., 2009 | https://github.com/samtools/samtools |
| R | https://www.r-project.org/ | |
| FlowJo (10.1) | FlowJo | https://www.flowjo.com/solutions/flowjo/ |
Highlights.
Genome-wide map of DNA breaks due to replication stress in mammalian cells
Poly(dA:dT) tracts are natural polar replication barriers and fragile sites
Common mechanism for DNA breakage at early- and late- replicating fragile sites
AT richness and poly(dA:dT) motifs are determinants of origin usage in mammals
ACKNOWLEDGMENTS
We thank Kristin Eckert, Sergei Mirkin, Thomas Kunkel, Thomas Glover, and Steve West for stimulating discussions and sequencing support from David Goldstein and the CCR Genomics Core. This work utilized the computational resources of the NIH HPC Biowulf cluster. The A.N. laboratory is supported by the Intramural Research Program of the NIH; A.N. was supported by an Ellison Medical Foundation Senior Scholar in Aging Award (AG-SS-2633–11), the Department of Defense Idea Expansion (W81XWH-15–2-006) and Breakthrough (W81XWH-16–1-0599) Awards, Alex Lemonade Stand Foundation Award, and an NIH Intramural FLEX Award. A.T. was supported by a fellowship from the American Cancer Society (PF-16–037-01-DMC). Work in the O.H. lab was supported by the Ligue Nationale Contre le Cancer, the Association pour la Recherche sur le Cancer, the Fondation pour la Recherche Médicale (FRM DEI201512344404), and the Canceropole Ile-de France and the INCa (2016–1-PL-BIO-13-CNRS DR B-1). We dedicate this manuscript to the late Ruth Nussenzweig.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and can be found with this article online at https://doi.org/10.1016/j.cell.2018.07.011.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Arlt MF, Ozdemir AC, Birkeland SR, Wilson TE, and Glover TW (2011). Hydroxyurea induces de novo copy number variants in human cells. Proc. Natl. Acad. Sci. USA 108, 17360–17365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt MF, Rajendran S, Birkeland SR, Wilson TE, and Glover TW (2012). De novo CNV formation in mouse embryonic stem cells occurs in the absence of Xrcc4-dependent nonhomologous end joining. PLoS Genet. 8, e1002981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymami J, Coll M, Frederick CA, Wang AH, and Rich A (1989). The propeller DNA conformation of poly(dA).poly(dT). Nucleic Acids Res. 17, 3229–3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow JH, Faryabi RB, Callén E, Wong N, Malhowski A, Chen HT, Gutierrez-Cruz G, Sun HW, McKinnon P, Wright G, et al. (2013). Identification of early replicating fragile sites that contribute to genome instability. Cell 152, 620–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes RP, Hile SE, Lee MY, and Eckert KA (2017). DNA polymerases eta and kappa exchange with the polymerase delta holoenzyme to complete common fragile site synthesis. DNA Repair (Amst.) 57, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard E, Babled A, Lapasset L, Milhavet O, Parrinello H, Dantec C, Marin JM, and Lemaitre JM (2012). Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat. Struct. Mol. Biol. 19, 837–844. [DOI] [PubMed] [Google Scholar]
- Bunting SF, Callén E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. (2010). 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, and Cimprich KA (2005). Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 19, 1040–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen E, Di Virgilio M, Kruhlak MJ, Nieto-Soler M, Wong N, Chen HT, Faryabi RB, Polato F, Santos M, Starnes LM, et al. (2013). 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell 153, 1266–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela A, Sridharan S, Sciascia N, Tubbs A, Meltzer P, Sleckman BP, and Nussenzweig A (2016). DNA breaks and end resection measured genome-wide by end sequencing. Mol. Cell 63, 898–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela A, Maman Y, Jung S, Wong N, Callen E, Day A, Kieffer-Kwon KR, Pekowska A, Zhang H, Rao SSP, et al. (2017). Genome organization drives chromosome fragility. Cell 170, 507–521.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper AM, Nghiem P, Arlt MF, and Glover TW (2002). ATR regulates fragile site stability. Cell 111, 779–789. [DOI] [PubMed] [Google Scholar]
- Cayrou C, Coulombe P, Vigneron A, Stanojcic S, Ganier O, Peiffer I, Rivals E, Puy A, Laurent-Chabalier S, Desprat R, and Méchali M (2011). Genome-scale analysis of metazoan replication origins reveals their organization in specific but flexible sites defined by conserved features. Genome Res. 21, 1438–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechering KJ, Cuelenaere K, Konings RN, and Leunissen JA (1998). Distinct frequency-distributions of homopolymeric DNA tracts in different genomes. Nucleic Acids Res. 26, 4056–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton ML, Galani K, Kang S, Bell SP, and MacAlpine DM (2010). Conserved nucleosome positioning defines replication origins. Genes Dev. 24, 748–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox KR (1990). Long (dA)n.(dT)n tracts can form intramolecular triplexes under superhelical stress. Nucleic Acids Res. 18, 5387–5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Martin MM, Regairaz M, Huang L, You Y, Lin CM, Ryan M, Kim R, Shimura T, Pommier Y, and Aladjem MI (2015). The DNA repair endonuclease Mus81 facilitates fast DNA replication in the absence of exogenous damage. Nat. Commun. 6, 6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber JK, Gögel E, Berger C, Wallisch M, Müller F, Grummt I, and Grummt F (1997). Termination of mammalian rDNA replication: polar arrest of replication fork movement by transcription termination factor TTF-I. Cell 90, 559–567. [DOI] [PubMed] [Google Scholar]
- Gibbons JG, Branco AT, Yu S, and Lemos B (2014). Ribosomal DNA copy number is coupled with gene expression variation and mitochondrial abundance in humans. Nat. Commun. 5, 4850. [DOI] [PubMed] [Google Scholar]
- Glover TW, Wilson TE, and Arlt MF (2017). Fragile sites in cancer: more than meets the eye. Nat. Rev. Cancer 17, 489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros J, Kumar C, Lynch G, Yadav T, Whitehouse I, and Remus D (2015). Post-licensing specification of eukaryotic replication origins by facilitated Mcm2–7 sliding along DNA. Mol. Cell 60, 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman C, and D’Orso I (2017). CIPHER: a flexible and extensive workflow platform for integrative next-generation sequencing data analysis and genomic regulatory element prediction. BMC Bioinformatics 18, 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamperl S, Bocek MJ, Saldivar JC, Swigut T, and Cimprich KA (2017). Transcription-replication conflict orientation modulates R-loop levels and activates distinct DNA damage responses. Cell 170, 774–786.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hile SE, and Eckert KA (2008). DNA polymerase kappa produces interrupted mutations and displays polar pausing within mononucleotide microsatellite sequences. Nucleic Acids Res. 36, 688–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchick D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, and Kent WJ (2004). The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32, D493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, and Haussler D (2002). The human genome browser at UCSC. Genome Res. 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer-Kwon KR, Nimura K, Rao SSP, Xu J, Jung S, Pekowska A, Dose M, Stevens E, Mathe E, Dong P, et al. (2017). Myc regulates chromatin decompaction and nuclear architecture during B cell activation. Mol. Cell 67, 566–578.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Snyder RO, and Wold MS (1992). Binding properties of replication protein A from human and yeast cells. Mol. Cell. Biol. 12, 3050–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren A, Handsaker RE, Kamitaki N, KarliĆ R, Ghosh S, Polak P, Eggan K, and McCarroll SA (2014). Genetic variation in human DNA replication timing. Cell 159, 1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski D, and Eddy MJ (1989). The DNA unwinding element: a novel, cisacting component that facilitates opening of the Escherichia coli replication origin. EMBO J. 8, 4335–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak M, Crouch EE, Orlov M, Montaño C, Gorski SA, Nussenzweig A, Misteli T, Phair RD, and Casellas R (2007). The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature 447, 7 30–734. [DOI] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard AC, and Méchali M (2013). DNA replication origins. Cold Spring Harb. Perspect. Biol. 5, a010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Malott M, and Leffak M (2003). Multiple functional elements comprise a Mammalian chromosomal replicator. Mol. Cell. Biol. 23, 1832–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macheret M, and Halazonetis TD (2018). Intragenic origins due to short G1 phases underlie oncogene-induced DNA replication stress. Nature 555, 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madireddy A, Kosiyatrakul ST, Boisvert RA, Herrera-Moyano E, GarcíaRubio ML, Gerhardt J, Vuono EA, Owen N, Yan Z, Olson S, et al. (2016). FANCD2 facilitates replication through common fragile sites. Mol. Cell 64, 388–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin EV, and Mirkin SM (2007). Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. 71, 13–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojardín L, Vázquez E, and Antequera F (2013). Specification of DNA replication origins and genomic base composition in fission yeasts. J. Mol. Biol. 425, 4706–4713. [DOI] [PubMed] [Google Scholar]
- Naim V, Wilhelm T, Debatisse M, and Rosselli F (2013). ERCC1 and MUS81-EME1 promote sister chromatid separation by processing late replication intermediates at common fragile sites during mitosis. Nat. Cell Biol. 15, 1008–1015. [DOI] [PubMed] [Google Scholar]
- Petryk N, Kahli M, d’Aubenton-Carafa Y, Jaszczyszyn Y, Shen Y, Silvain M, Thermes C, Chen CL, and Hyrien O (2016). Replication landscape of the human genome. Nat. Commun. 7, 10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prioleau MN, and MacAlpine DM (2016). DNA replication origins-where do we begin? Genes Dev. 30, 1683–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri A, Callen E, Ding X, Gogola E, Duarte AA, Lee JE, Wong N, Lafarga V, Calvo JA, Panzarino NJ, et al. (2016). Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 535, 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusiniak ME, Kunnev D, Freeland A, Cady GK, and Pruitt SC (2012). Mcm2 deficiency results in short deletions allowing high resolution identification of genes contributing to lymphoblastic lymphoma. Oncogene 31, 4034–4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samadashwily GM, Dayn A, and Mirkin SM (1993). Suicidal nucleotide sequences for DNA polymerization. EMBO J. 12, 4975–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, and Greenberg RA (2010). ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141, 970–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata A, Moiani D, Arvai AS, Perry J, Harding SM, Genois MM, Maity R, van Rossum-Fikkert S, Kertokalio A, Romoli F, et al. (2014). DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol. Cell 53, 7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K, and Segal E (2013). Determinants of nucleosome positioning. Nat. Struct. Mol. Biol. 20, 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Técher H, Koundrioukoff S, Nicolas A, and Debatisse M (2017). The impact of replication stress on replication dynamics and DNA damage in vertebrate cells. Nat. Rev. Genet. 18, 535–550. [DOI] [PubMed] [Google Scholar]
- Toledo LI, Altmeyer M, Rask MB, Lukas C, Larsen DH, Povlsen LK, Bekker-Jensen S, Mailand N, Bartek J, and Lukas J (2013). ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 155, 1088–1103. [DOI] [PubMed] [Google Scholar]
- Tsang E, and Carr AM (2008). Replication fork arrest, recombination and the maintenance of ribosomal DNA stability. DNA Repair (Amst.) 7, 1613–1623. [DOI] [PubMed] [Google Scholar]
- Tubbs A, and Nussenzweig A (2017). Endogenous DNA damage as a source of genomic instability in cancer. Cell 168, 644–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umek RM, and Kowalski D (1990). Thermal energy suppresses mutational defects in DNA unwinding at a yeast replication origin. Proc. Natl. Acad. Sci. USA 87, 2486–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L, Costantino L, Tan FJ, Zimmer A, and Koshland D (2016). S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev. 30, 1327–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying S, Minocherhomji S, Chan KL, Palmai-Pallag T, Chu WK, Wass T, Mankouri HW, Liu Y, and Hickson ID (2013). MUS81 promotes common fragile site expression. Nat. Cell Biol. 15, 1001–1007. [DOI] [PubMed] [Google Scholar]
- Zentner GE, Balow SA, and Scacheri PC (2014). Genomic characterization of the mouse ribosomal DNA locus. G3 (Bethesda) 4, 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Modelbased analysis of ChIP-seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotorynski E, Rahat A, Skaug J, Ben-Porat N, Ozeri E, Hershberg R, Levi A, Scherer SW, Margalit H, and Kerem B (2003). Molecular basis for expression of common and rare fragile sites. Mol. Cell. Biol. 23, 7143–7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data for this study was deposited at NCBI GEO under the accession number GEO: GSE116321.