Abstract
Background/aims
Studies have shown that miR-146a-5p was differentially expressed in diverse cancers, but the associations between miR-146a-5p expression and prognosis across multiple types of cancer as well its potential targets and downstream pathways have not been comprehensively analyzed. In this study, we performed the first meta-analysis of the prognostic value of miR-146a-5p expression in diverse malignancies and explored prospective targets of miR-146a-5p and related signaling pathways.
Methods
A thorough search for articles related to miR-146a-5p was performed, and RNA-seq data from The Cancer Genome Atlas (TCGA) and microarray data from gene expression omnibus profiles were used to collect information about the prognostic value of miR-146a-5p. A comprehensive meta-analysis was conducted. Twelve platforms in miRWalk 2.0 were applied to predict targets of miR-146a-5p. TCGA RNA-seq data were used to validate the inverse relationships between miR-146a-5p and its likely targets. Subsequently, gene ontology and pathway analyses were conducted using Funrich version 3.1.3. Potential protein–protein interaction (PPI) networks were constructed. Potential target genes of miR-146a-5p in lung cancer were validated by RT-qPCR.
Results
We included 10 articles in the meta-analysis. In a pooled analysis, the high miR-146a-5p expression group showed a better overall survival in solid cancers, particularly in reproductive system cancers and digestive system cancers. A total of 120 predicted target genes were included in a bioinformatics analysis. Five pathways involving phospholipase C (PLC) and aquaporins (AQPs) were the most significantly enriched Kyoto Encyclopedia of Genes and Genomes pathways. Moreover, the PPI network displayed the related signaling pathways and interactions among proteins. AQP1 and FYN were validated by RT-qPCR to be potential targets of miR-146a-5p in lung cancer.
Conclusion
There is a close link between high miR-146a-5p expression and better overall survival in 21 types of solid cancer, especially in reproductive system and digestive system cancers. Furthermore, miR-146a-5p could inhibit diverse malignancies by modulating pathways linked to PLC or AQPs. In summary, miR-146a-5p is a potential prognostic biomarker and therapeutic target for various cancers.
Keywords: miR-146a-5p, Cancer, Prognosis, Meta-analysis, Molecular mechanism
Background
Cancer is a serious threat to human health, with almost 1,735,350 newly diagnosed cancer cases and nearly 609,640 cancer-related deaths in the United States in 2018 [1, 2]. Gene sequencing technologies are frequently utilized to explore correlations between genomic changes and morbidity from various cancers [2–4]. Multiple factors regulate tumorigenesis and tumor development by altering DNA replication, transcription, and translation. The discovery of these factors, including microRNAs (miRNAs) and long non-coding RNAs, was considered a breakthrough for the early diagnosis and prevention of cancers. The identification of new molecular biomarkers and studies of their underlying mechanisms are valuable for the development of more effective treatment strategies.
miRNAs are single-stranded non-coding RNAs (18–22 nucleotides) with vital roles in the regulation of biological processes, including transcription, translation, cell cycle, and organismal development [5]. These molecules are linked to post-transcriptional regulation by interacting with corresponding messenger RNAs (mRNAs). Deregulated expression by miRNAs could increase the risk of metabolic diseases, such as diabetes and obesity, by disrupting signaling pathways [6]. Moreover, numerous studies have reported associations between altered miRNAs and cancers in different systems, suggesting that miRNAs are potential biomarkers for early diagnosis, treatment, and prognosis in cancers [7–14]. Furthermore, miRNAs are promising therapeutic candidates for metastatic cancer [5].
MiR-146a-5p, a member of the microRNA-146 family, has a crucial role in a series of cancer-related processes, including tumorigenesis, tumor progression, and outcomes. The expression of miR-146a-5p is increased in tissue samples from breast cancer and thyroid carcinoma [15, 16]. By contrast, levels of miR-146a-5p are decreased in gastric cancer, lung cancer, and prostate cancer [17–20]. Decreased miR-146a-5p levels are associated with biological activities in the latter cancers (gastric, lung, and prostate cancers), such as the growth, invasion, and migration of cancer cells [21]. It has been demonstrated that miR-146a-5p is a powerful suppressor in breast cancer, lung cancer, and prostate cancer [18–20]. Increasing studies have provided insight into the mechanism underlying the effects of abnormal miR-146a-5p on various cancers. A recent study has shown that down-regulated miR-146a-5p in gastric cancer is associated with poor prognosis and WASF2 might be a target gene of miR-146a-5p [17]. However, more detailed analyses of the correlation between differentially expressed miR-146a-5p and prognosis for other solid tumors are needed. Therefore, we performed a comprehensive and thorough analysis of its prognostic significance by utilizing integrated data extracted from the literature, RNA-seq data from The Cancer Genome Atlas (TCGA) datasets, and the SurvMicro website. Additionally, to examine the mechanism underlying the effects of aberrant miR-146a-5p in solid cancers, a pathway analysis and protein interaction network analysis were conducted.
Materials and methods
Literature search strategy
A systematic searched for literature related to the prognostic value of miR-146a-5p in cancer was performed using the PubMed, EBSCO, CNKI, VIP, and WanFang databases. The most recent search was performed on July 16, 2017. The search terms for English-language databases included “miR146”, “miRNA146”, “microRNA146”, “microRNA146a”, “miR146a”, “miRNA146a”, “microRNA-146a-5p”, “miRNA-146a-5p”, and “miR-146a-5p” as well as “cancer”, “carcinoma”, “adenocarcinoma”, “sarcoma”, “tumor”, “neoplas*”, and “malignan*”, using “OR” to connect terms. Finally, “AND” was used to link the two classes of terms. For searches against Chinese databases, similar terms were input. Two authors performed the search independently to ensure the accuracy.
Eligibility criteria
Only studies that satisfied the following criteria were included in the meta-analysis: (1) samples were obtained from human tissues or blood; (2) clearly described analysis of miR-146a-5p; (3) explored the prognostic value of miR-146a-5p expression levels in cancers; (4) provided sufficient information to extract hazard ratios (HRs) and 95% confidence intervals (CIs). The exclusion criteria were as follows: (1) unrelated to humans; (2) neither Chinese nor English, reviews, conference abstracts, case reports; (3) unable to extract HR and 95% CIs; (4) not satisfying the inclusion criteria.
Data extraction
Data extraction was performed by two reviewers independently. The following information was extracted: the name of the first author, country, publication year, tumor type, number of cases, tumor stage, lymph node metastasis, time of follow-up, sample type, miR-146a level, cut-off values, HR and corresponding 95% CI. Comprehensive discussions were conducted to resolve any disagreements.
Prognostic data for 21 human cancers downloaded from RNA-seq data
Expression levels of miR-146a-5p and corresponding prognostic data were obtained for 21 types of solid cancers from RNA-seq data. Expression values of less than 1 were removed. The median expression values were calculated using SPSS 22.0. Then, each cohort was separated into an experimental group (high expression level) and control group (low expression level). GraphPad Prism 7.0 was utilized to draw survival curves for 21 solid cancers. Cox regression was employed to calculate HRs.
Statistical analysis
Stata12.0 and SPSS 22.0 were used to conduct statistical analyses. HRs and 95% CIs were extracted from the literature, RNA-seq data, and microarray profiles to obtain pooled results. HRs were directly collected if the they were reported in studies. HRs and 95% CIs were calculated based on the methods described by Tierney when the information provided was insufficient. Otherwise, Engauge Digitizer version 4.1 was used for studies that did not report concrete data. This software could determine HRs and 95% CIs based on Kaplan–Meier survival curves. The prognostic data downloaded from TCGA and SurvMicro were analyzed using SPSS 22.0 to estimate HRs and 95% CI. An HR greater than 1 indicated that patients with high miR-146a-5p expression were more likely to have a poor prognosis.
An integrated meta-analysis of relevant literature studies, RNA-seq data, and microarray profiles as well as separate meta-analysis for literature studies and RNA-seq data were performed. Heterogeneity was evaluated by the I2 statistic. Subgroup analysis stratified by human systems was used to identify the source of heterogeneity. Depending on the heterogeneity results, two distinct models were adopted, a random-effects model or fixed-effects model. A random-effects model was applied when there was obvious statistical heterogeneity; a fixed-effects model was applied in other cases. A significant difference was observed when the two-sided p-value was less than 0.05.
Target prediction and validation for miR-146a-5p
Target genes for miR-146a-5p were predicted using MirWalk2.0. Twelve algorithms were used, including miRWalk, Microt4, miRanda, mirBridge, miRDB, miRMap, miRNAMap, Pictar2, PITA, RNA22, RNAhybrid, and TargetScan [22, 23]. According to the miRWalk2.0 algorithm, genes would be recognized as potential targets if there was a seed sequence that binds to miRNAs. The sequence of the genes could be complementary to that of miRNAs in the 3′-untranslated region, 5′-untranslated region, promoter, or coding sequence [22, 23]. To improve the accuracy of the prediction analysis, genes that overlapped in more than seven databases were chosen. Another criterion was applied to select the potential targets based on expression levels. A list of differentially expressed genes (DE genes) for 21 types of solid cancer was obtained from the TCGA and GTEx databases using GEPIA (|Log2FC| > 2, q-value < 0.05, limma methods) [24]. Because miR-146a-5p targets might show differential expression in diverse tumors, the intersection of predicted genes and DE genes in solid cancers contained the most likely potential targets of miR-146a-5p.
To further validate the relationships between miR-146a-5p and the potential targets, the correlations between their expression levels were assessed using R with miRNA sequence data [log2 (RPM + 1), miRNA mature strand expression] and RNA sequence data [log2 (TPM + 0.001), gene expression RNAseq] for the TCGA Pan-Cancer cohort with 7965 tumor samples and 639 non-tumor adjacent-tumor samples. To obtain reliable results, these data were first processed as follows: Relative expression = Expression in the cancer tissue group—mean expression in the non-cancer tissue group. Ultimately, the genes were selected as candidate targets when their expression levels were inversely associated with levels of miR-146a-5p (correlation coefficient < 0, p < 0.05), and these genes were used to explore the mechanism underlying the effects of miR-146a-5p in diverse cancers.
Bioinformatics analysis
The molecular mechanism underlying the effect of aberrant miR-146a-5p levels in solid cancers was evaluated using Funrich version 3.1.3, with the assistance of gene ontology (GO) and pathway analyses. By observing the p-value for each term, significant pathways were identified and the molecular functions, biological processes, and cellular components involved in the effects of miR-146a-5p were explored. The search tool for the retrieval of interacting genes/proteins (STRING) database (http://string-db.org/) was used for the construction of the protein–protein interaction (PPI) network, which displayed potential signaling pathways and connections among proteins in a dimensional way. The potential binding sites of miR-146a-5p and candidate targets were determined using miRWalk2.0. Additionally, scatter diagrams were constructed to visualize correlations between the expression levels of miR-146a-5p and candidate targets in diverse malignant tumors, utilizing miRNA and RNA sequence data.
Validation of potential targets of miR-146a-5p through RT-qPCR
Previously, we found that miR-146a-5p showed a decreased expression in lung cancer tissues compared to normal lung tissues [25]. Then we collected 55 lung cancer tissues (32 lung adenocarcinoma and 23 lung squamous cell carcinoma) and their matched non-tumor lung tissues to further test the expression of miR-146a-5p potential targets (AQP1 and FYN). The mean age of the patients was 56.9 years old. The Ethical Committees of First Affiliated Hospital, Guangxi Medical University, China have approved the study protocol. And all of the patients have signed the informed consent. According to the methods described in our previous studies [26–32], we isolated total RNA and performed relative quantification analysis. RT–qPCR was executed using the 7900HT PCR system of USA (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA). The internal control in the analysis was GAPDH, with the forward primer (5′-TGCACCACCAACTGCTTA-3′) and the reverse primer (5′-GGATGCAGGGATGATGTTC-3′). The thermocycling steps included hot start at 95 °C for 10 min, 95 °C for 10 s, 60 °C for 5 s and annealing at 72 °C for 5 s, totally 40 PCR cycles. And the 2−∆Cq method was utilized to calculate expression of candidate miR-146a-5p targets.
Results
Characteristics of the studies included in the meta-analysis
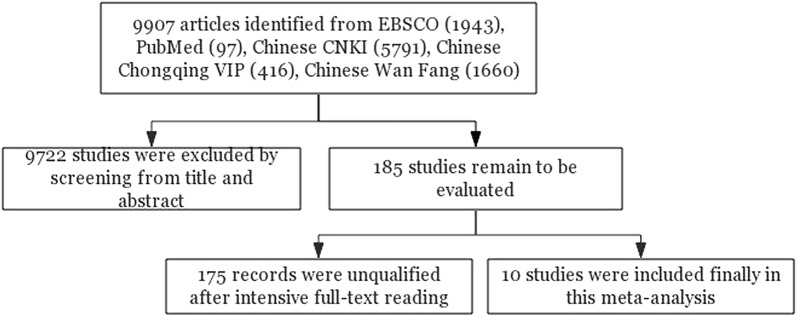
A total of 9907 references were collected in the initial search, of which 9722 were removed based on titles and abstracts. After full-text screening, we included ten studies published between 2010 and 2017 in the meta-analysis, with 783 patients in total (Fig. 1) [25, 33–41]. Tumor tissues were mostly utilized to detect miR-146a-5p expression, and bone marrow samples were employed in one of the included studies. The necessary information about the ten studies is outlined in Table 1.
Fig. 1.
Flow diagram of the process for searching for articles
Table 1.
Main characteristics of the articles included in the meta-analysis
| Study | Year | Country | Tumour type | N | Detected sample | Stage I/II/III/IV | Lymph node −/+ | Follow-up months | miR-146a assay | Cut-off | HR (95% CI) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Hess | 2017 | Germany | HNSCC | 149 | Tissue | 0/0/0/149 | NR | Median 61 | qRT-PCR | Median | 0.450 (0.230–0.830) |
| LUO | 2017 | China | GC | 93 | Tissue | 0/0/42/51 | 17/76 | NR | qRT-PCR | 2.0 | 0.129 (0.028–0.601) |
| Zavala | 2016 | Chile | TNBC | 39 | Tissue | NR | 20/19 | NR | qRT-PCR | Median | 0.130 (0.026–0.650) |
| Li | 2014 | China | NSCLC | 43 | Tissue | NR | NR | NR | MISH & qRT-PCR | Median | 2.930 (1.440–5.960) |
| Chen | 2013 | China | NSCLC | 101 | Tissue |
(I/II:28) (III/IV:73) |
35/66 | NR | qRT-PCR | Mean | 0.420 (0.180–0.950) |
| Zhong | 2012 | China | DLBCL | 90 | Tissue | 9/31/24/26 | NR | Mean 26.3 | qRT-PCR | Optimal | 1.119 (0.977–1.282) |
| Hou | 2012 | China | GC | 43 | Tissue | 8/11/18/6 | 14/29 | NR | qRT-PCR | Median | 0.280 (0.100–0.750) |
| Paik | 2011 | Korea | NK/T | 50 | Tissue | 26/11/0/13 | 43/7 | NR | qRT-PCR | Optimal | 0.076 (0.019–0.300) |
| Kogo | 2011 | Japan | GC | 90 | Tissue | 27/18/23/22 | 30/60 | Mean 36 | qRT-PCR | Median | 0.650 (0.440–0.940) |
| Wang | 2010 | China | ALL | 32 | Bone marrow | NR | NR | NR | qRT-PCR | Median | 1.690 (1.040–2.760) |
| Wang | 2010 | China | AML | 53 | Bone marrow | NR | NR | NR | qRT-PCR | Median | 1.560 (1.120–2.170) |
HNSCC: head and neck squamous cell carcinoma; GC: gastric cancer; TNBC: triple negative breast cancer; NSCLC: non-small cell lung cancer; DLBCL: diffuse large B-cell lymphoma; NK/T: NK/T cell lymphoma; ALL: acute lymphoblastic leukemia; AML: acute myeloid leukemia; NR: not reported; median: middle value; mean: average value; qRT-PCR: quantitative real-time polymerase chain reaction; MISH: miRNA in situ hybridization; optimal: cut-off point associated with a minimum p-value; HR: hazard ratio
Survival curves
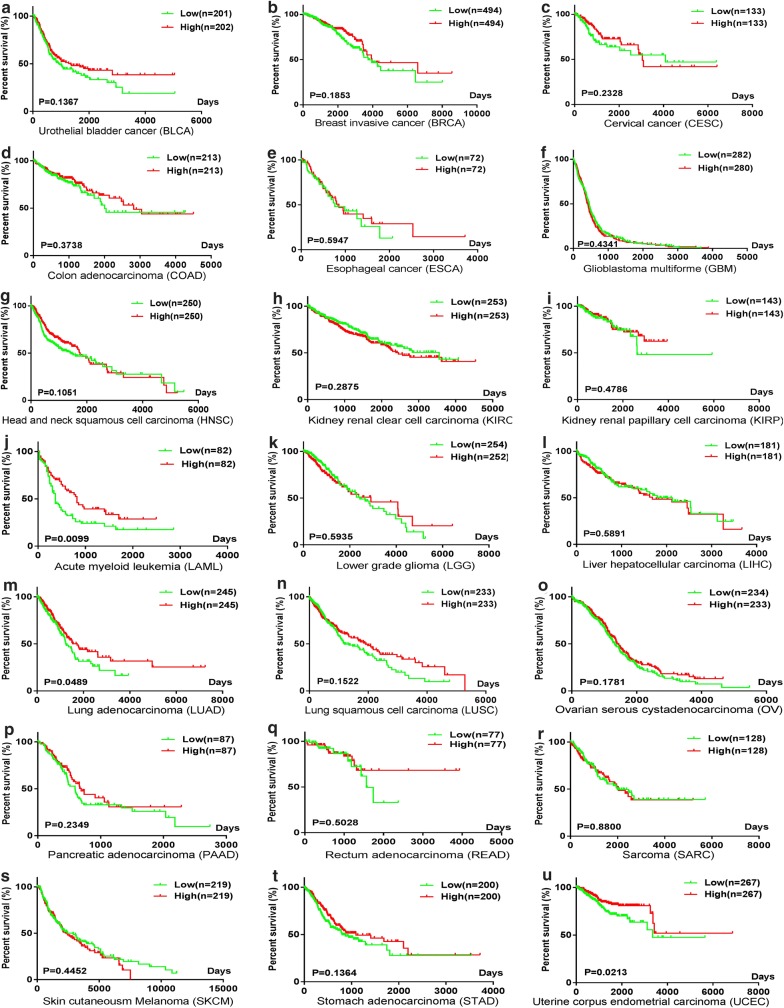
The expression and survival data for miR-146a-5p in 21 types of solid cancers were extracted from RNA-seq data, including data for 8519 patients. HRs were estimated using SPSS 22.0 and the results are summarized in Table 2. Kaplan–Meier survival curves were utilized to compare survival between low and high miR-146a-5p expression groups using GraphPad Prism 7.0 (Fig. 2).
Table 2.
Characteristics of eligible studies from RNA-seq data and microarray profiles
| Study | Cancer type | Group | Number | HR |
|---|---|---|---|---|
| TCGA BLCA | Urothelial bladder cancer | Urinary system | 404 | 0.800 (0.595–1.074) |
| TCGA BRCA | Breast invasive cancer | Endocrine system | 988 | 0.793 (0.562–1.119) |
| TCGA CESC | Cervical cancer | Reproductive system | 266 | 0.736 (0.444–1.220) |
| TCGA COAD | Colon adenocarcinoma | Digestive system | 426 | 0.834 (0.559–1.245) |
| TCGA ESCA | Esophageal cancer | Digestive system | 144 | 0.869 (0.517–1.460) |
| TCGA GBM | Glioblastoma multiforme | Nervous system | 562 | 1.075 (0.896–1.290) |
| TCGA HNSC | Head and neck squamous cell carcinoma | Other | 500 | 0.800 (0.611–1.048) |
| TCGA KIRC | Kidney renal clear cell carcinoma | Urinary system | 506 | 1.177 (0.871–1.592) |
| TCGA KIRP | Kidney renal papillary cell carcinoma | Urinary system | 286 | 0.805 (0.442–1.468) |
| TCGA LAML | Acute myeloid leukemia | Other | 164 | 0.596 (0.400–0.890) |
| TCGA LGG | Lower grade glioma | Nervous system | 506 | 1.101 (0.773–1.569) |
| TCGA LIHC | Liver hepatocellular carcinoma | Digestive system | 262 | 1.102 (0.775–1.567) |
| TCGA LUAD | Lung adenocarcinoma | Respiratory system | 262 | 0.743 (0.552–1.000) |
| TCGA LUSC | Lung squamous cell carcinoma | Respiratory system | 466 | 0.815 (0.615–1.079) |
| TCGA OV | Ovarian serous cystadenocarcinoma | Reproductive system | 470 | 0.855 (0.680–1.075) |
| TCGA PAAD | Pancreatic adenocarcinoma | Digestive system | 174 | 0.776 (0.509–1.181) |
| TCGA READ | Rectum adenocarcinoma | Digestive system | 154 | 0.765 (0.348–1.680) |
| TCGA SARC | Sarcoma | Other | 258 | 1.031 (0.691–1.539) |
| TCGA SKCM | Skin Cutaneous Melanoma | Other | 438 | 1.113 (0.845–1.467) |
| TCGA STAD | Stomach adenocarcinoma | Digestive system | 400 | 0.790 (0.579–1.078) |
| TCGA UCEC | Uterine corpus endometrial carcinoma | Reproductive system | 534 | 0.608 (0.396–0.932) |
| GSE10694 | Liver hepatocellular carcinoma | Digestive system | 156 | 0.900 (0.620–1.310) |
| GSE13937 | Lung cancer | Respiratory system | 152 | 1.360 (0.830–2.220) |
| GSE16025 | Lung cancer | Respiratory system | 61 | 0.620 (0.310–1.220) |
| GSE21849 | Acute myeloid leukemia | Other | 37 | 0.340 (0.070–1.750) |
| GSE27290 | Ovarian serous cystadenocarcinoma | Reproductive system | 62 | 0.870 (0.460–1.640) |
| GSE27705 | Lung cancer | Respiratory system | 20 | 0.250 (0.070–0.960) |
| GSE31384 | Liver hepatocellular carcinoma | Digestive system | 166 | 1.410 (0.890–2.250) |
| GSE36682 | Nasopharyngeal carcinoma | Respiratory system | 62 | 1.030 (0.450–2.380) |
| GSE37405-GPL13703 | Breast invasive cancer | Endocrine system | 60 | 1.560 (0.740–3.330) |
| GSE37405-GPL14149 | Breast invasive cancer | Endocrine system | 40 | 0.800 (0.300–2.130) |
Fig. 2.
Kaplan–Meier survival curves for miR-146a-5p in diverse cancers using RNA-seq data
Pooled meta-analysis results
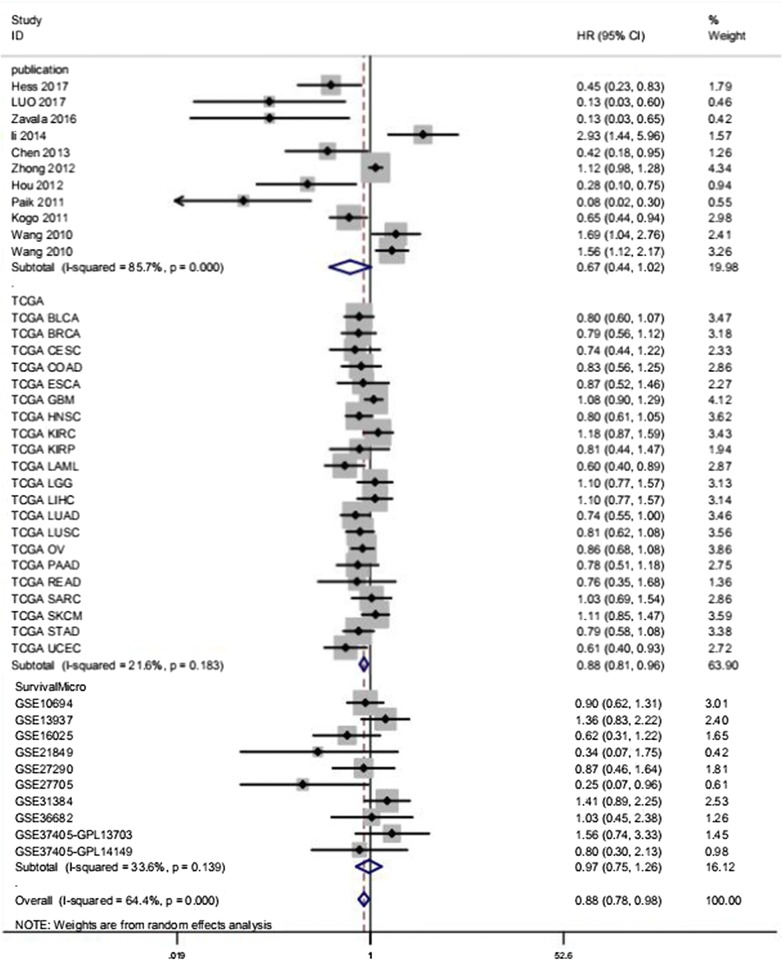
A comprehensive meta-analysis of relevant studies, RNA-seq data, and microarray expression profiles was conducted to analyze the prognostic value of miR-146a-5p in solid cancers. The results obtained using the total data (HR 0.875, 95% CI 0.784–0.976, I2 = 64.40%) suggested that the high miR-146a-5p group had a better overall survival than that of the low expression group (Fig. 3). Since significant heterogeneity was observed, we used the random-effects model. We obtained similar pooled results in subgroup analyses stratified by human systems (Table 3). Moreover, we found that increased miR-146a-5p levels were closely related to a better overall survival in reproductive system cancers (HR 0.791, 95% CI 0.661–0.947, I2 = 0.00%) and digestive system cancers (HR 0.844, 95% CI 0.738–0.965, I2 = 49.00%) (Table 3). Additionally, an association between high expression miR-146a-5p and a better prognosis was observed in gastric cancer (HR 0.535, 95% CI 0.327–0.877, I2 = 64.40%, random-effects model) (Table 3, Fig. 4).
Fig. 3.
Subgroup analysis of total records to determine the prognostic value of miR-146a-5p in solid cancers. Generally, miR-146a-5p was expressed at low levels in diverse solid cancers and was related to a worse prognosis for patients with cancer (HR 0.875, 95% CI 0.784–0.976). Due to obvious heterogeneity (I2 = 64.4%), a random-effects model was used, as indicated in the note shown at the bottom
Table 3.
Summary of meta-analysis results for overall survival
| Analysis | No. of studies | HR (95% CI) | Heterogeneity | ||
|---|---|---|---|---|---|
| Fix | Random | I2 (%) | p | ||
| Overall pooled result | 42 | 0.940 (0.888–0.995) | 0.875 (0.784–0.976) | 64.40 | 0.000 |
| Data resource | |||||
| The included studies | 11 | 1.050 (0.941–1.172) | 0.667 (0.438–1.016) | 85.70 | 0.000 |
| TCGA data | 21 | 0.89 (0.830–0.956) | 0.880 (0.810–0.956) | 21.60 | 0.183 |
| SurvMicro GSE | 10 | 1.005 (0.825–1.224) | 0.971 (0.747–1.261) | 33.60 | 0.139 |
| Human system | |||||
| Urinary system | 3 | 0.947 (0.776–1.156) | 0.938 (0.708–1.244) | 43.40 | 0.171 |
| Endocrine System | 4 | 0.828 (0.618–1.111) | 0.774 (0.406–1.474) | 61.90 | 0.049 |
| Reproductive system | 4 | 0.791 (0.661–0.947) | 0.791 (0.661–0.947) | 0.00 | 0.560 |
| Digestive system | 11 | 0.844 (0.738–0.965) | 0.821 (0.672–1.003) | 49.00 | 0.033 |
| Nervous system | 2 | 1.08 (0.919–1.270) | 1.080 (0.919–1.270) | 0.00 | 0.906 |
| Respiratory system | 8 | 0.855 (0.723–1.011) | 0.872 (0.611–1.244) | 69.80 | 0.002 |
| Other | 10 | 1.036 (0.942–1.140) | 0.895 (0.688–1.164) | 79.60 | 0.000 |
| Tumour type | |||||
| HNSC | 2 | 0.734 (0.572–0.941) | 0.648 (0.376–1.116) | 61.90 | 0.105 |
| GC | 4 | 0.668 (0.530–0.841) | 0.535 (0.327–0.877) | 64.40 | 0.038 |
| BRCA | 4 | 0.828 (0.618–1.111) | 0.774 (0.406–1.474) | 61.90 | 0.049 |
| NSCLC | 7 | 0.848 (0.715–1.006) | 0.853 (0.578–1.260) | 73.90 | 0.001 |
| AML | 2 | 1.055 (0.818–1.362) | 0.971 (0.378–2.492) | 92.40 | 0.000 |
| Overall pooled result | 0.816 (0.737–0.904) | 0.762 (0.614–0.945) | 71.30 | 0.000 | |
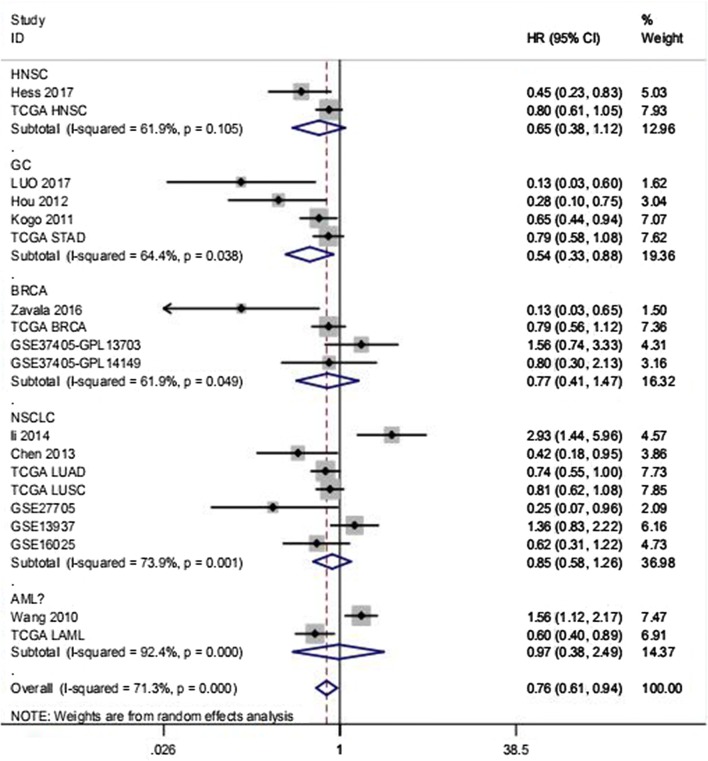
Fig. 4.
Subgroup analysis by tumor type to show the prognostic value of miR-146a-5p in diverse tumors. In the subgroup analysis, decreased miR-146a-5p expression in gastric cancer (GC) was associated with shorter overall survival (HR 0.535, 95% CI 0.327–0.877). The random-effects model was applied as noted above (I2 = 64.40%)
The pooled results for the 10 articles identified in the literature search showed no statistical significance, with obvious heterogeneity (HR 0.667, 95% CI 0.438–1.016, I2 = 85.70%) (Table 3). Thus, we used the random-effects model. However, a subgroup analysis by human system indicated that high expression of miR-146a-5p was associated with better prognosis in digestive system cancers (HR 0.355, 95% CI 0.143–0.881, I2 = 66.1%, random-effects model).
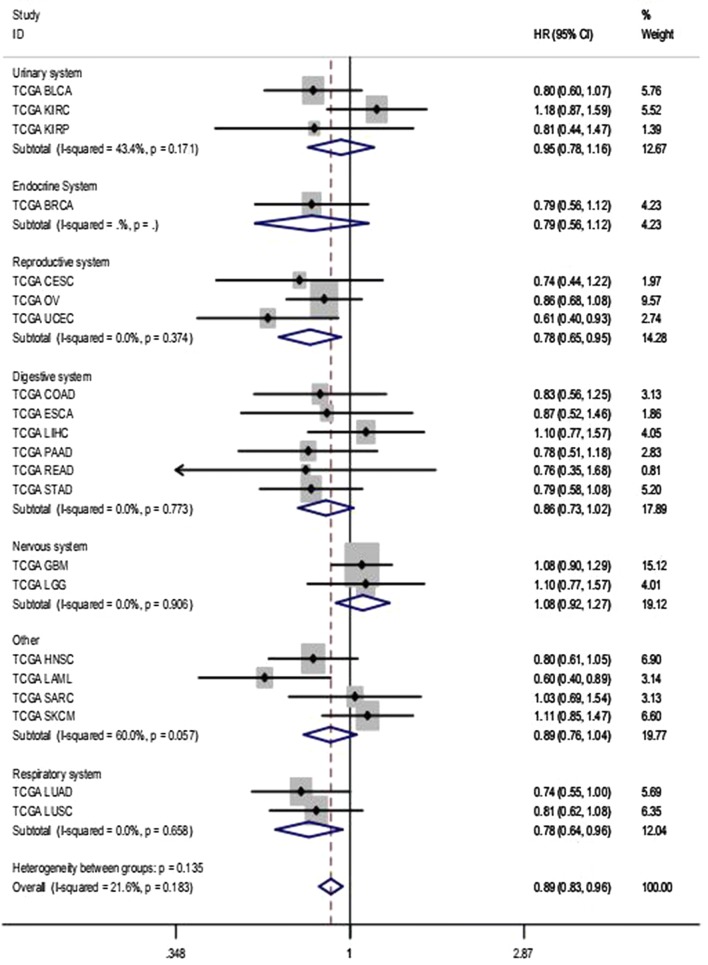
The aggregated results for RNA-seq data indicated that high miR-146a-5p expression was associated with a better prognosis in solid cancers without considerable heterogeneity (HR 0.880, 95% CI 0.810–0.956, I2 = 21.60%) (Table 3, Fig. 5). The association between high miR-146a-5p expression and improved survival was found in reproductive system cancers (HR 0.784, 95% CI 0.650–0.946, I2 = 0.00%) and respiratory system cancers (HR 0.780, 95% CI 0.636–0.957, I2 = 0.00%) (Fig. 5). There was no obvious link between miR-146a-5p and prognosis in other cancer types or systems.
Fig. 5.
Subgroup analysis of TCGA data by human system. In combination with Table 3, lower miR-146a-5p expression was associated with poorer prognosis for patients with reproductive system cancers (HR 0.791, 95% CI 0.661–0.947, I2 = 0.00%) and digestive system cancers (HR 0.844, 95% CI 0.738–0.965, I2 = 49.00%). The final heterogeneity could be considered slight (I2 = 21.6%); Thus, a fixed-effect model was used
Target genes of miR-146a-5p
A total of 14,280 genes were identified as candidate targets using 12 prediction platforms, and only those targets identified by more than seven platforms were included in subsequent analyses. A total of 926 target genes were obtained, of which 429 were abnormally expressed in solid cancers (Fig. 6). Finally, expression levels of 120 genes were confirmed to be inversely correlated with the level of miR-146a-5p and thus these genes were identified as potential targets for subsequent analyses (Fig. 7).
Fig. 6.
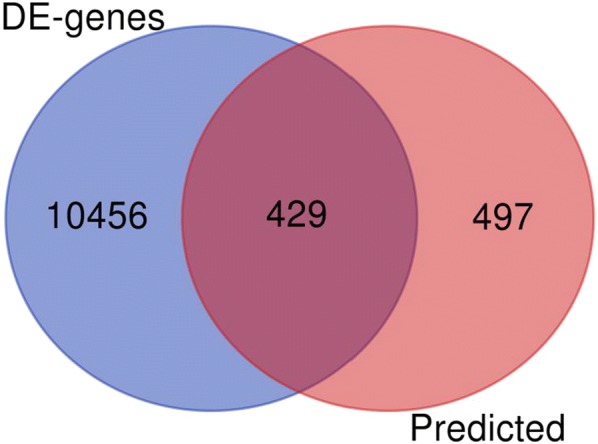
The 429 potential targets of miR-146a-5p. The genes were selected as potential targets based on sequence complementation and their expression levels in solid cancers
Fig. 7.
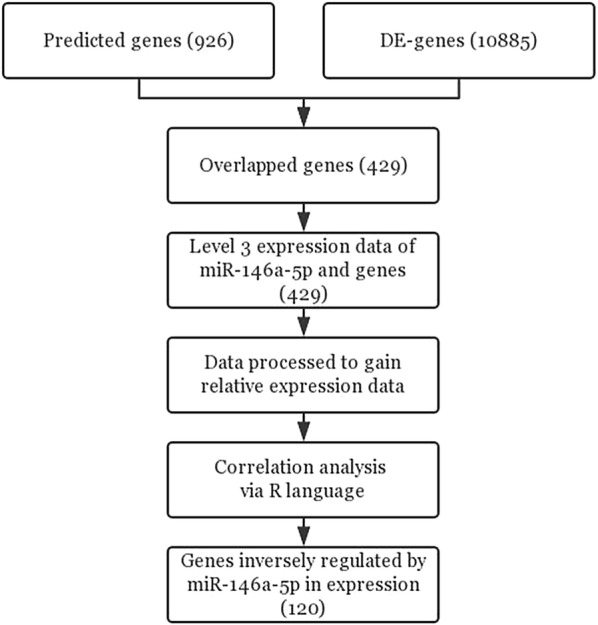
Flow chart of miR-146a-5p target prediction and validation. By utilizing twelve prediction platforms, DE genes from the GEPIA website and miRNA and RNA sequencing data for the TCGA Pan-Cancer cohort were used to identify 120 likely targets of miR-146a-5p
Bioinformatics analysis
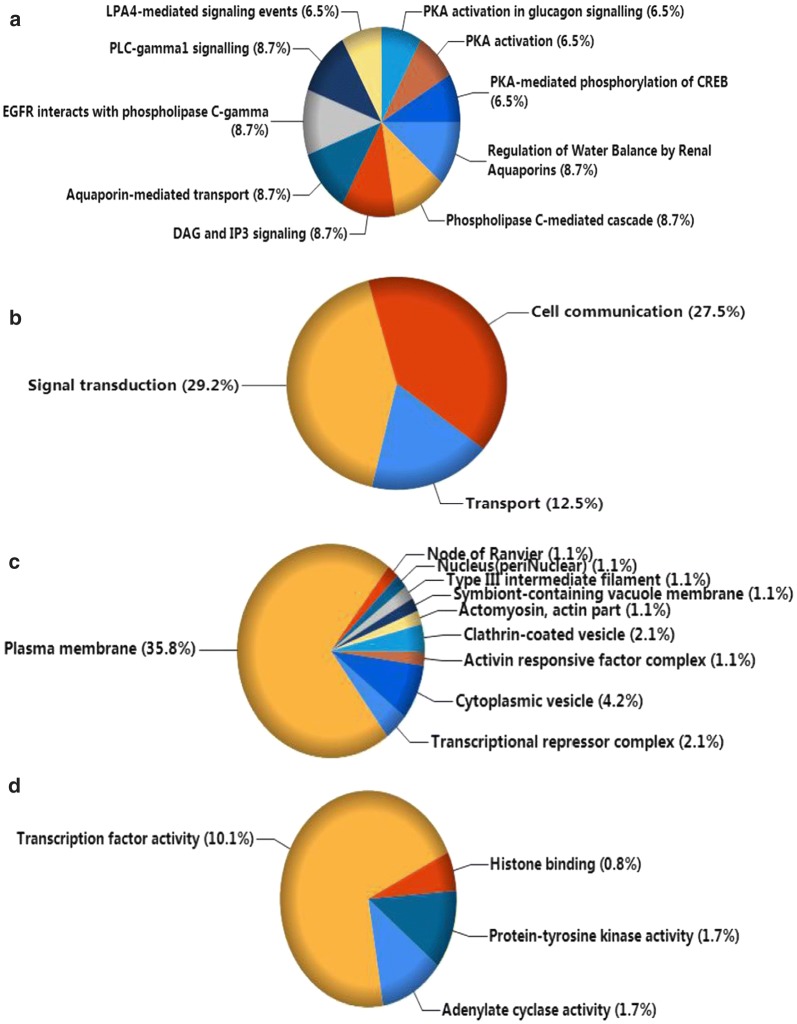
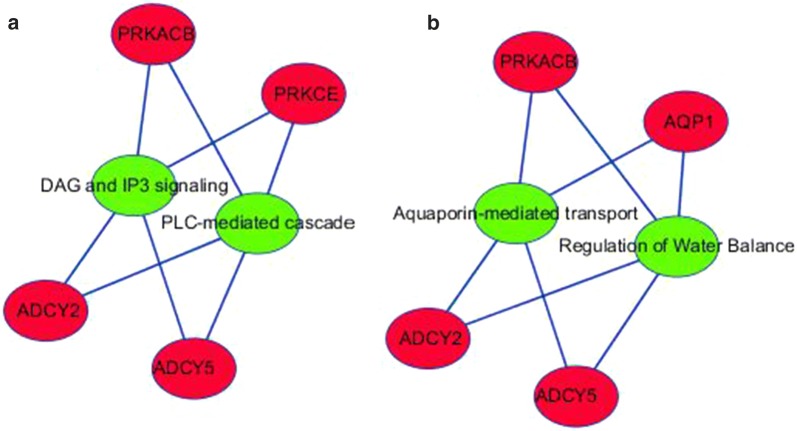
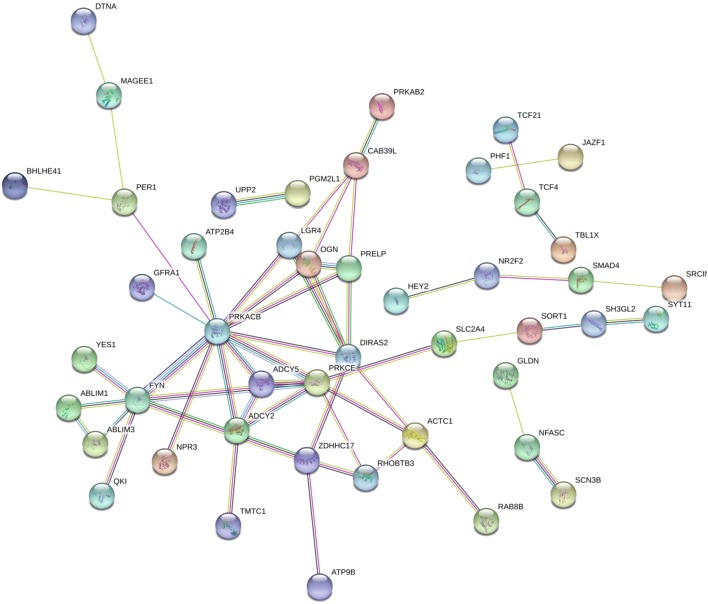
Funrich version 3.1.3 was utilized to perform GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses using the 120 potential targets. In total, 126 biological functions were obtained in the GO analysis, of which 25 were significant (p < 0.05). Several processes might function as essential activities regulated by miR-146a-5p in various malignancies, including Transport, Signal transduction, and Cell communication in the biological processes category; Transcriptional repressor complex and Plasma membrane in the cellular components category; and adenylate cyclase activity, transcription factor activity, histone binding, and protein-tyrosine kinase activity in the molecular functions category (Fig. 8). A total of 291 critical pathways were identified by KEGG, and pathways involving phospholipase C (PLC) and aquaporins (AQPs) were the most statistically significant (p < 0.001), as shown in Table 4 and Fig. 8. In particular, PRKACB, PRKCE, ADCY5, ADCY2, and AQP1 were identified as five key genes in the above pathways (Fig. 9). A protein network was constructed using STRING with 120 predicted targets, as shown in Fig. 10. The genes that frequently interacted with other genes were recognized as hub genes of miR-146a-5p, including PRKACB, FYN, and PRKCE. Overall, PRKACB, PRKCE, ADCY5, ADCY2, AQP1, and FYN were recognized as the most likely targets of miR-146a-5p. The predicted binding sites for the six genes and miR-146a-5p are displayed in Table 5. There were significant correlations between levels of the six likely targets and miR-146a-5p in solid cancers, as shown in Fig. 11.
Fig. 8.
Visualization of the most highly enriched pathways and GO processes for miR-146a-5p targets. As described in Table 4, 120 potential targets were evaluated. miR-146a-5p was potentially involved in the regulation of following activities in various cancers: a KEGG pathways: PLC-mediated cascade, DAG and IP3 signaling, AQP-mediated transport, EGFR interactions with PLC-γ, and PLC-γ1 signaling; b biological processes: transport, signal transduction, cell communication; c cellular components: plasma membrane; d molecular function: transcription factor activity and protein-tyrosine kinase activity
Table 4.
Most highly enriched pathways and processes for miR-146a-5p sorted by p-values
| Terms | Count | Percentage (%) | p value |
|---|---|---|---|
| KEGG pathways | |||
| Regulation of water balance by renal aquaporins | 4 | 8.696 | < 0.001 |
| Phospholipase C-mediated cascade | 4 | 8.696 | < 0.001 |
| DAG and IP3 signaling | 4 | 8.696 | < 0.001 |
| Aquaporin-mediated transport | 4 | 8.696 | < 0.001 |
| EGFR interacts with phospholipase C-gamma | 4 | 8.696 | < 0.001 |
| PLC-gamma1 signalling | 4 | 8.696 | < 0.001 |
| LPA4-mediated signaling events | 3 | 6.522 | < 0.001 |
| PKA activation in glucagon signalling | 3 | 6.522 | < 0.001 |
| PKA activation | 3 | 6.522 | < 0.001 |
| PKA-mediated phosphorylation of CREB | 3 | 6.522 | < 0.001 |
| Biological processes (BP) of GO | |||
| Transport | 15 | 12.500 | 0.014 |
| Signal transduction | 35 | 29.167 | 0.033 |
| Cell communication | 33 | 27.500 | 0.040 |
| Cellular components (CC) of GO | |||
| Transcriptional repressor complex | 2 | 2.105 | 0.005 |
| Plasma membrane | 34 | 35.789 | 0.006 |
| Actomyosin, actin part | 1 | 1.053 | 0.007 |
| Type III intermediate filament | 1 | 1.053 | 0.007 |
| Node of Ranvier | 1 | 1.053 | 0.007 |
| Symbiont-containing vacuole membrane | 1 | 1.053 | 0.007 |
| Nucleus (periNuclear) | 1 | 1.053 | 0.007 |
| Clathrin-coated vesicle | 2 | 2.105 | 0.007 |
| Activin responsive factor complex | 1 | 1.053 | 0.019 |
| Cytoplasmic vesicle | 4 | 4.211 | 0.021 |
| Molecular function (MF) of GO | |||
| Adenylate cyclase activity | 2 | 1.681 | 0.002 |
| Transcription factor activity | 12 | 10.084 | 0.009 |
| Histone binding | 1 | 0.840 | 0.020 |
| Protein-tyrosine kinase activity | 2 | 1.681 | 0.026 |
The predicted genes that overlapped in at least 7 online platforms were validated by RNA-seq data, and only 120 targets were confirmed to be inversely correlated with miR-146a-5p in expression. Enrichment analysis was conducted, adopting the 120 potential targets to identify statistically significant pathways and GO processes (p < 0.05). Most of the above terms have been reported to play essential roles in the regulation of tumorigenesis and other biological activities in cancers
Fig. 9.
Connections among potential targets of miR-146a-5p in crucial pathways. a Pathways of PLC (green nodes); b pathways of AQPs (green nodes). PRKACB, PRKCE, ADCY5, ADCY2, and AQP1 are five potentially vital genes (red nodes) in the above pathways
Fig. 10.
PPI network of miR-146a-5p potential target genes drawn using STRING. The most frequent genes were identified as hub genes, including PRKACB, FYN, and PRKCE
Table 5.
Predicted complementary sequence of the candidate targets and miR-146a-5p
| Terms | The predicted binding sites |
|---|---|
| miR-146a-5p | 3′ UUGGGUACCUUAAGUCAAGAGU |
| Position 1955-1961 of PRKCE 3′ UTR | 5′…CUAAUGAUGACAUUCAGUUCUCU… |
| Position 2501-2508 of PRKCE 3′ UTR | 5′…GCCUCUGGCUGGUGAAGUUCUCA… |
| Position 821-827 of FYN 3′ UTR | 5′…AACUUACUGCGAUUUGUUCUCAA… |
| Position 1131-1137 of FYN 3′ UTR | 5′…UUCUAUAUGUCCAGGAGUUCUCC… |
| Position 959-965 of AQP1 3′ UTR | 5′…UCUUGCUCAUUCUUCAGUUCUCU… |
| Position 2566-2572 of PRKACB 3′ UTR | 5′…UAUGAUCUCAGCCUCAGUUCUCU… |
| Position 2040-2047 of ADCY2 3′ UTR | 5′…CUGGGUGGUCCACCAAGUUCUCA… |
| Position 666-672 of ADCY5 3′ UTR | 5′…AAAUAAAACAAAACAAGUUCUCC… |
| Position 1994-2000 of ADCY5 3′ UTR | 5′…UUUCUUCCAGCUGUUGUUCUCAA… |
We identified potential binding sites of the likely targets and miR-146a-5p using TargetScan 6.1, one of the platforms available in miRWalk2.0
Fig. 11.
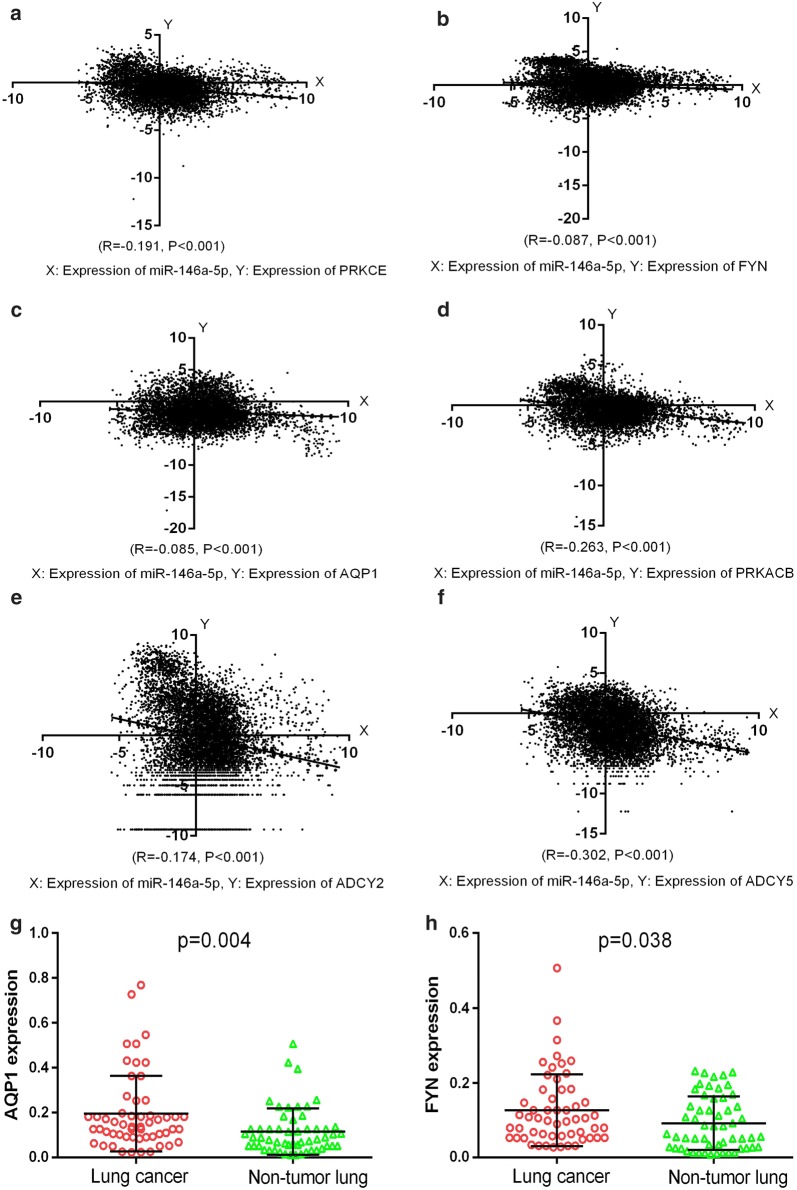
Correlations between expression levels of candidate targets and miR-146a-5p in pan-cancer. Expression data for miR-146a-5p and the six selected targets were obtained from the TCGA Pan-Cancer cohort, as above described. Expression levels of all candidate targets were inversely associated with miR-146a-5p expression. GraphPad Prism 6.0 was employed to generate scatter plots. a PRKCE; b FYN; c AQP1; d PRKACB; e ADCY2; f ADCY5. g AQP1 expression in lung cancer. h FYN expression in lung cancer
AQP1 and FYN were validated by RT-qPCR to be potential targets of miR-146-5p
It has been previously proven by us that miR-146a-5p was down-regulated in lung cancer tissues [25]. Currently, we found that two likely miR-146a-5p targets, AQP1 and FYN, showed higher levels in tissues obtained from lung cancer, compared to noncancerous lung tissues (Fig. 11). The primers of AQP1 were GGACACCTCCTGGCTATTGACTAC (the forward primer) and GTTGCTGAAGTTGTGTGTGATCAC (the reverse primer); the primers of FYN were CTCAGCACTACCCCAGCTTC (the forward primer) and ATCTCCTTCCGAGCTGTTCA (the reverse primer). Hence, the expression of AQP1 and FYN tend to be inversely modulated by miR-146a-5p in lung cancer.
Discussion
Using an integrated meta-analysis, we found that lower miR-146a-5p expression was correlated with worse outcomes in solid cancers, especially in reproductive system cancers and digestive system cancers. In particular, we observed that low expression of miR-146a-5p in gastric cancer is significantly associated with poor prognosis. Additionally, five critical pathways [PLC-mediated cascade, diacylglycerol (DAG) and inositol 1,4,5-triphosphate (IP3) signaling, AQP-mediated transport, epidermal growth factor receptor (EGFR) interaction with PLC-γ, and PLC-γ1 signaling] and several GO processes were identified as potential mechanisms underlying the effects of miR-146a-5p in cancers. Moreover, we identified six potential targets of miR-146a-5p, including PRKCE, FYN, AQP1, PRKACB, ADCY2, and ADCY5. Specifically, we have verified the expression of AQP1 and FYN in lung cancer. The high expression of AQP1 and FYN in lung cancer tissues increased the possibility of them as true targets of miR-146a-5p.
Cancer is one of the most common causes of death in humans, and rapid invasiveness and metastasis lead to an unfavorable prognosis. More than half of recent deaths in the United States are caused by lung cancer, breast cancer, prostate cancer, colorectal cancer, or pancreatic cancer in both males and females, and this can be attributed to the inability to detect tumor growth, limitations of modern imaging technologies, and insufficient bio-markers [2]. Extensive research has indicated that quite a few molecules (long non-coding RNAs, miRNAs, circRNAs, and genes) participate in the modulation of biological activities in cancers, providing a basis for improving detection and treatment [42, 43]. Therefore, we explored the clinical significance and underlying mechanisms of miR-146a-5p in diverse cancers, a miRNA with an indispensable role in the regulation of tumor progression. Our pooled results suggested that miR-146a-5p functions as a protective factor or tumor suppressor in solid cancers.
Notably, significant pooled results were found for reproductive system cancers and digestive system cancers, indicating that miR-146a-5p has prognostic value in these cancers. Several studies have demonstrated that miR-146a-5p could function as an inhibitory factor in reproductive system cancers. Sun et al. found that miR-146a-5p levels are decreased in prostate cancer, leading to tumor progression and poor prognosis; miR-146a-5p might be useful for the treatment for prostate cancer as a suppressor [20]. Additionally, miR-146a-5p has been reported to function as a powerful inhibitor in cervical cancer [44]. Similar results have been obtained for epithelial ovarian cancer [45, 46]. In this work, we found that elevated miR-146a-5p expression was indeed correlated with a better prognosis for patients with reproductive system cancers. We speculated that miR-146a-5p might be a protective factor in reproductive system cancers and is a novel therapeutic target for the improved management of tumor development. Future research is needed to evaluate this hypothesis.
Additionally, a series of studies have shown that miR-146a-5p has an indispensable role in retarding tumor progression and prolonging overall survival for patients with cancers of the digestive system. In gastric cancer, growing evidence has revealed the anti-tumor function of miR-146a-5p; it functions as a protective factor to repress neoplasm metastasis and tissue infiltration and to improve overall survival [47, 48]. Recent studies have revealed that miR-146a-5p expression is lower in pancreatic cancer tissues than in non-tumor tissues based on analyses of miR-146a-5p levels in human tissue samples, cell lines, and mouse models by real-time PCR [49, 50]. Our previous studies have confirmed that miR-146a-5p could clearly inhibit the deterioration of hepatocellular carcinoma [51, 52]. A similar protective role of miR-146a-5p has been suggested in other digestive system cancers, including colorectal cancer [53] and esophageal squamous cell carcinoma [54–56]. The results of our study were consistent with the notion that higher miR-146a-5p expression is linked to a favorable prognosis in the above cancers, especially gastric cancer. The results of our study could provide support for further research on the clinical application of miR-146a-5p in digestive system cancers.
Increased miR-146a-5p expression is beneficial for repressing the development of other cancers, especially non-small cell lung cancer (NSCLC). A recent meta-analysis concluded that the outcome of NSCLC was better in the experimental group with high levels of miR-146a-5p than in a group with low levels of expression, and indicated the potential prognostic role of miR-146a-5p [57]. Our previous work also showed that the loss of miR-146a-5p might lead to the deterioration of NSCLC with a shorter progression-free survival [25]. In head and neck squamous cell carcinoma (HNSCC) and breast cancer, miR-146a-5p was identified as a tumor suppressor [34, 58, 59]. However, we only observed a trend in which the over-expression of miR-146a-5p was related to longer survival times in NSCLC, breast cancer, and HNSCC. These associations should be explored in further studies.
We next explored the functions of predicted targets, validated predicted targets, analyzed KEGG pathways and GO enrichment, and constructed a PPI network. We found that miR-146a-5p might play a prominent role in modulating a series of biological pathways closely associated with PLC, a key enzyme on cytomembranes, including the PLC-mediated cascade, DAG and IP3 signaling, EGFR interactions with PLC-γ, and PLC-γ1 signaling. DAG and IP3 are important for the generation of phosphatidylinositol 4,5-bisphosphate and the regulation of PLC, which are linked to the release of calcium and activation of protein kinase C (PKC), two vital second messengers in signal transduction [60, 61]. Prior studies have noted the importance of PLC in modulating proliferation, invasion, and metastasis, suggesting its role in carcinogenesis and tumor progression in various malignancies [62, 63]. It has been reported that abnormal expression levels or mutations in PLC-γ (a PLC member) are related to the occurrence of breast cancer, gastric cancer, and oral squamous cell carcinoma [64–67]. We found potential interactions between EGFR and PLC-γ activities, which have a role in breast cancer, whereby EGFR/human epidermal growth factor receptor 2/PLC-γ1 signaling results in tumor cell invasion and migration [25, 68, 69]. Additionally, we found that miR-146a-5p clearly participates in the regulation of signal transduction on plasma membranes and protein-tyrosine kinase activity. We hypothesized that the specific binding sites of miR-146a-5p are located on the plasma membrane, and miR-146a-5p has a protective role, potentially by repressing the invasion and migration of tumor cells via the regulation of the above processes in solid cancers.
We found that miR-146a-5p could modulate two pathways involving AQP (regulation of water balance by renal AQPs and AQP-mediated transport). AQPs are a group of specific proteins related to the transport of water and glycerol across cell membranes; they act as promoters in cell proliferation and cell motility [70, 71]. AQPs play an indispensable role in maintaining water homeostasis in the kidney and are closely related to the regulation of urine osmolality [72]. It has been suggested that the abnormal regulation of renal AQPs would lead to diseases correlated with water balance disorders in the kidney, such as diabetes insipidus and hyponatremia [73]. Moreover, as mentioned in literature reviews, AQP expression is closely associated with tumor angiogenesis and dissemination in the majority of human malignancies [74, 75]. The application of AQP inhibitors might be beneficial to improve prognosis in diverse cancers [76]. Overall, these findings suggest that miR-146a-5p could be used to treat water balance disorders in the kidney and diverse cancers via the regulation of AQP-associated pathways.
PRKCE and AQP1 were identified as two likely targets of miR-146a-5p involved in the pathways of PLC and AQPs, respectively. We also found that FYN is a potential hub gene in the miR-146a-5p PPI network. PRKCE, also known as PKC ε, is a novel member of the PKC isozyme family involved in the regulation of complex cellular processes in diverse cancers, as a paramount bridge between protein networks [77]. PKC ε is widely considered an oncogene, with increased expression in diverse malignant tumors, including lung cancer [78, 79], breast cancer [80], prostate cancer [81], clear cell renal carcinoma [81, 82], and HNSCC [83]. Associations have been detected between up-regulated PKC ε levels and metastatic outcomes in general cancers [84, 85]. Zhang et al. elucidated the negative modulation by miR-146a-5p f PKC ε expression, as a tumor suppressor in papillary thyroid carcinoma; PKC ε was identified as a direct target of miR-146a-5p based on a dual luciferase assay [86]. Inverse correlations between miRNAs and PKC ε were also revealed in lung cancer and HNSCC [79, 83]. Moreover, PKC ε inhibitors have been prospectively applied as novel therapies for patients with cancers [80]. In our study, PRKCE was identified as a potential target for miR-146a-5p; higher miR-146a-5p expression might decrease PRKCE levels to repress cellular activities in solid cancers, but further studies are needed to confirm this.
FYN, a member of Src family tyrosine kinases, is a proto-oncogene responsible for regulating the expression of protein-tyrosine kinases on membranes [87]. FYN is closely related to cancer development, tumor progression, and even dissemination in diverse cancers. Early in 2010, Yoshihito and colleagues revealed that FYN is highly expressed in prostate cancer and possibly leads to a more advanced tumor stage [88]. Elias et al. confirmed the intensive effects of FYN in promoting breast cancer development, suggesting that the over-expression of FYN is related to worse outcomes [89]. Researchers have also revealed a prospective strategy to better manage breast cancer by targeting FYN. Additionally, several studies have demonstrated that FYN functions as a promoter in diverse cancers and is associated with a poor prognosis [90–92]. In this study, we discovered higher expression levels of FYN in lung cancer than in normal tissues by conducting RT-qPCR for clinical tissues. Consequently, we speculated that miR-146a-5p might suppress the malignant transformation of cancers by controlling protein-tyrosine kinases activity, targeting FYN.
AQP1, a member of the AQP family, could modulate water transport across the plasma membrane and thereby is related to water balance in the kidney [73, 93]. Previous studies have revealed the positive regulation of AQP1 in tumor progression and demonstrated that AQP1 suppressors could repress biological activities of various tumors, including the growth of tumor cells, cell motility, and angiogenesis [94, 95]. Positive correlations between AQP1 expression and tumor development have been detected in cancers of the reproductive system (ovarian cancer [96] and prostate cancer [97]), digestive system (cancers of the stomach [98], colon [99], and esophagus [100]), and other systems (astrocytoma [101], and cancers of the lung [102], breast [103, 104], bladder [105], and pleura [106]). In our study, the high expression of AQP1 in lung cancer was proved by RT-qPCR, contrasting with low miR-146a-5p levels in lung cancer tissues in previous work [25], which strengthened the reliability of our prediction results and implied that AQP1 might be a target of miR-146a-5p in diverse cancers, especially lung cancer. Thus, it is possible that miR-146a-5p is an effective target for the management of tumor progression via AQP1, as a novel anti-cancer therapy.
As for PRKACB, ADCY2, and ADCY5. PRKACB encodes the catalytic subunit β of protein kinase A, a type of protein mainly depending on cyclic AMP (cAMP) [107]. Current studies have found the likely tumorigenic roles of PRKACB in diverse malignant tumors, including gastrointestinal cancer (gastric, colon and pancreatic tumors) and others (breast, ovary, leukemia and brain tumors) [107–109]. Moreover, PRKACB might be a promising target in cancer treatment by increasing drugs responsiveness to tumors [108, 109]. Adenylate cyclase 2 (ADCY2), a gene related to the production of cAMP, is greatly implied in acceleration of phosphor-acidification and metabolic processes of glycogen [110, 111]. Duerr et al. found that ADCY2 expression was obviously elevated in pancreatic neuroendocrine malignant tumors, possibly linked with tumor invasiveness [112]. Similar to ADCY2, ADCY5 is a catalyzer in the formation of cAMP. Takashi et al. and Chen et al. discovered that abnormal ADCY5 expression was correlated to tumor aggressiveness and DNA Methylation of ADCY5 might lead to unsatisfied outcomes for patients with lung cancer [113, 114]. We assumed that the participation of these genes in human cancers may correlate with miR-146a-5p. More biological investigation should be conducted in future to validate the correlation of three potential miR-146a-5p targets (PRKACB, ADCY2, and ADCY5) and miR-146a-5p in cancers.
However, there are a few limitations that should also be pointed out. Because hematological tumors may have different biological characteristics from solid tumors, the clinical significance and mechanism of miR-146a-5p in hematological tumors and solid tumors may be different. Because of the small number of blood tumors included in this study, the above problems could not be analyzed in detail. More samples need to be added to clarify the problem in future work. Moreover, the biological roles of miR-146a-5p in cancers and the targeting regulatory relationship between miR-146a-5p and target genes needed to be validated through further experiments in future studies. In the present study, we only chose AQP1 and FYN for qRT-PCR validations. RT-qPCR experiments for all potential targets in various cancer types should be performed in future studies to achieve a comprehensive verification.
Conclusion
Based on a comprehensive meta-analysis and bioinformatics analysis, we concluded that miR-146a-5p might serve as an inhibitory factor in general cancers (reproductive system cancers and digestive system cancers, especially gastric cancer). We speculated that miR-146a-5p might inhibit the progression of solid cancers via pathways involving PLC (candidate targets: PRKCE and FYN) or AQPs (candidate target: AQP1) by inversely regulating target genes. Moreover, miR-146a-5p could be used to treat certain diseases correlated with water balance disorders in the kidney by regulating kidney AQP pathways. In addition, miR-146a-5p could repress certain biological activities in tumor cells by the modulation of cell communication on plasma membranes. Overall, miR-146a-5p could be utilized as a prognostic biomarker, with implications for the prediction and treatment of diverse cancers.
Acknowledgements
Not applicable.
Abbreviations
- TCGA
The Cancer Genome Atlas
- GEO
gene expression omnibus
- HRs
extract hazard ratios
- 95% CIs
95% confidence intervals
- DE genes
differentially expressed genes
- GO
gene ontology
- STRING
search tool for the retrieval of interacting genes/proteins
- PPI
protein–protein interaction
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- PLC
pathways involving phospholipase C
- AQPs
aquaporins
- DAG
diacylglycerol
- EGFR
epidermal growth factor receptor
- PKC
protein kinase C
- HNSCC
head and neck squamous cell carcinoma
- GC
gastric cancer
- TNBC
triple negative breast cancer
- NSCLC
non-small cell lung cancer
- DLBCL
diffuse large B cell lymphoma
- NK/T
NK/T cell lymphoma
- ALL
acute lymphoblastic leukemia
- AML
acute myeloid leukemia
- qRT-PCR
quantitative real-time polymerase chain reaction
- MISH
miRNA in situ hybridization
- BLCA
urothelial bladder cancer
- BRCA
breast invasive cancer
- CESC
cervical cancer
- COAD
colon adenocarcinoma
- ESCA
esophageal cancer
- GBM
glioblastoma multiforme
- HNSC
head and neck squamous cell carcinoma
- KIRC
kidney renal clear cell carcinoma
- KIRP
kidney renal papillary cell carcinoma
- LAML
acute myeloid leukemia
- LGG
lower grade glioma
- LIHC
liver hepatocellular carcinoma
- LUAD
lung adenocarcinoma
- LUSC
lung squamous cell carcinoma
- OV
ovarian serous cystadenocarcinoma
- PAAD
pancreatic adenocarcinoma
- READ
rectum adenocarcinoma
- SARC
sarcoma
- SKCM
skin cutaneous mMelanoma
- STAD
stomach adenocarcinoma
- UCEC
uterine corpus endometrial carcinoma
Authors’ contributions
M-wL and LG analyzed and interpreted the patient data, and were two major contributor in writing the manuscript. Y-wD and PL participated in literature search and data collection. GC and D-zL participated in the design of the study and funds collection. All authors read and approved the final manuscript.
Funding
This study was supported by funds from the National Natural Science Foundation of China (NSFC81560469, NSFC81360327, NSFC81760420), Natural Science Foundation of Guangxi, China (2017GXNSFAA198016, 2016GXNSFAA380255), and Guangxi Medical University Training Program for Distinguished Young Scholars and Medical Excellence Award Funded by the Creative Research Development Grant from the First Affiliated Hospital of Guangxi Medical University.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The Ethical Committees of First Affiliated Hospital, Guangxi Medical University, China have approved the study protocol. And all of the patients have signed the informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Mei-wei Li and Li Gao contributed equally as first authors.
Contributor Information
Mei-wei Li, Email: 2783638224@qq.com.
Li Gao, Email: 2339352006@qq.com.
Yi-wu Dang, Email: dangyiwu@126.com.
Ping Li, Email: liping@gxmu.edu.cn.
Zu-yun Li, Email: li_zuyun_gxmu@163.com.
Gang Chen, Email: chengang@gxmu.edu.cn.
Dian-zhong Luo, Email: 13878802796@163.com.
References
- 1.Mandelker D, Zhang L, Kemel Y, Stadler ZK, Joseph V, Zehir A, Pradhan N, Arnold A, Walsh MF, Li Y, et al. Mutation detection in patients with advanced cancer by universal sequencing of cancer-related genes in tumor and normal dna vs guideline-based germline testing. JAMA. 2017;318(9):825–835. doi: 10.1001/jama.2017.11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics. Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Felsenstein M, Hruban RH, Wood LD. New developments in the molecular mechanisms of pancreatic tumorigenesis. Adv Anatom Pathol. 2018;25(2):131–142. doi: 10.1097/PAP.0000000000000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mack SC, Northcott PA. Genomic analysis of childhood brain tumors: methods for genome-wide discovery and precision medicine become mainstream. J Clin Oncol. 2017;35(21):2346–2354. doi: 10.1200/JCO.2017.72.9921. [DOI] [PubMed] [Google Scholar]
- 5.Jafri MA, Al-Qahtani MH, Shay JW. Role of miRNAs in human cancer metastasis: implications for therapeutic intervention. Semin Cancer Biol. 2017;44:117–131. doi: 10.1016/j.semcancer.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Del Carmen Martinez-Jimenez V, Mendez-Mancilla A, Patricia Portales-Perez D. miRNAs in nutrition, obesity, and cancer: the biology of miRNAs in metabolic disorders and its relationship with cancer development. Mol Nutr Food Res. 2018;62(1):1600994. doi: 10.1002/mnfr.201600994. [DOI] [PubMed] [Google Scholar]
- 7.Sun JJ, Chen GY, Xie ZT. MicroRNA-361-5p inhibits cancer cell growth by targeting CXCR7 in hepatocellular carcinoma. Cell Physiol Biochem. 2016;38(2):777–785. doi: 10.1159/000443033. [DOI] [PubMed] [Google Scholar]
- 8.Shi W, Bruce J, Lee M, Yue S, Rowe M, Pintilie M, Kogo R, Bissey PA, Fyles A, Yip KW, et al. MiR-449a promotes breast cancer progression by targeting CRIP2. Oncotarget. 2016;7(14):18906–18918. doi: 10.18632/oncotarget.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Z, Liu K, Wang Y, Xu Z, Meng J, Gu S. Upregulation of microRNA-96 and its oncogenic functions by targeting CDKN1A in bladder cancer. Cancer Cell Int. 2015;15:107. doi: 10.1186/s12935-015-0235-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizuno K, Seki N, Mataki H, Matsushita R, Kamikawaji K, Kumamoto T, Takagi K, Goto Y, Nishikawa R, Kato M, et al. Tumor-suppressive microRNA-29 family inhibits cancer cell migration and invasion directly targeting LOXL2 in lung squamous cell carcinoma. Int J Oncol. 2016;48(2):450–460. doi: 10.3892/ijo.2015.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qu J, Li M, An J, Zhao B, Zhong W, Gu Q, Cao L, Yang H, Hu C. MicroRNA-33b inhibits lung adenocarcinoma cell growth, invasion, and epithelial-mesenchymal transition by suppressing Wnt/beta-catenin/ZEB1 signaling. Int J Oncol. 2015;47(6):2141–2152. doi: 10.3892/ijo.2015.3187. [DOI] [PubMed] [Google Scholar]
- 12.D’Angelo E, Zanon C, Sensi F, Digito M, Rugge M, Fassan M, Scarpa M, Pucciarelli S, Nitti D, Agostini M. miR-194 as predictive biomarker of responsiveness to neoadjuvant chemoradiotherapy in patients with locally advanced rectal adenocarcinoma. J Clin Pathol. 2018;71(4):344–350. doi: 10.1136/jclinpath-2017-204690. [DOI] [PubMed] [Google Scholar]
- 13.D’Angelo E, Fassan M, Maretto I, Pucciarelli S, Zanon C, Digito M, Rugge M, Nitti D, Agostini M. Serum miR-125b is a non-invasive predictive biomarker of the pre-operative chemoradiotherapy responsiveness in patients with rectal adenocarcinoma. Oncotarget. 2016;7(19):28647–28657. doi: 10.18632/oncotarget.8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perilli L, Vicentini C, Agostini M, Pizzini S, Pizzi M, D’Angelo E, Bortoluzzi S, Mandruzzato S, Mammano E, Rugge M, et al. Circulating miR-182 is a biomarker of colorectal adenocarcinoma progression. Oncotarget. 2014;5(16):6611–6619. doi: 10.18632/oncotarget.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Labbaye C, Testa U. The emerging role of MIR-146A in the control of hematopoiesis, immune function and cancer. J Hematol Oncol. 2012;5:13. doi: 10.1186/1756-8722-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Czajka AA, Wojcicka A, Kubiak A, Kotlarek M, Bakula-Zalewska E, Koperski L, Wiechno W, Jazdzewski K. Family of microRNA-146 regulates RARbeta in papillary thyroid carcinoma. PLoS ONE. 2016;11(3):e0151968. doi: 10.1371/journal.pone.0151968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao Q, Tu C, Lu D, Zou Y, Liu H, Zhang S. Clinicopathological significance of the microRNA-146a/WASP-family verprolin-homologous protein-2 axis in gastric cancer. Cancer Sci. 2017;108(7):1285–1292. doi: 10.1111/cas.13254. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Hurst DR, Edmonds MD, Scott GK, Benz CC, Vaidya KS, Welch DR. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Can Res. 2009;69(4):1279–1283. doi: 10.1158/0008-5472.CAN-08-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li YL, Wang J, Zhang CY, Shen YQ, Wang HM, Ding L, Gu YC, Lou JT, Zhao XT, Ma ZL, et al. MiR-146a-5p inhibits cell proliferation and cell cycle progression in NSCLC cell lines by targeting CCND1 and CCND2. Oncotarget. 2016;7(37):59287–59298. doi: 10.18632/oncotarget.11040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Q, Zhao X, Liu X, Wang Y, Huang J, Jiang B, Chen Q, Yu J. miR-146a functions as a tumor suppressor in prostate cancer by targeting Rac1. Prostate. 2014;74(16):1613–1621. doi: 10.1002/pros.22878. [DOI] [PubMed] [Google Scholar]
- 21.Bleau AM, Redrado M, Nistal-Villan E, Villalba M, Exposito F, Redin E, de Aberasturi AL, Larzabal L, Freire J, Gomez-Roman J, et al. miR-146a targets c-met and abolishes colorectal cancer liver metastasis. Cancer Lett. 2018;414:257–267. doi: 10.1016/j.canlet.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 22.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44(5):839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12(8):697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 24.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen G, Umelo IA, Lv S, Teugels E, Fostier K, Kronenberger P, Dewaele A, Sadones J, Geers C, De Greve J. miR-146a inhibits cell growth, cell migration and induces apoptosis in non-small cell lung cancer cells. PLoS ONE. 2013;8(3):e60317. doi: 10.1371/journal.pone.0060317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gan BL, He RQ, Zhang Y, Wei DM, Hu XH, Chen G. Downregulation of HOXA3 in lung adenocarcinoma and its relevant molecular mechanism analysed by RT-qPCR, TCGA and in silico analysis. Int J Oncol. 2018;53(4):1557–1579. doi: 10.3892/ijo.2018.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang WT, He RQ, Li XJ, Ma J, Peng ZG, Zhong JC, Hu XH, Chen G. miR146a5p targets TCSF and influences cell growth and apoptosis to repress NSCLC progression. Oncol Rep. 2019;41(4):2226–2240. doi: 10.3892/or.2019.7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo YN, Luo B, Chen WJ, Chen X, Peng ZG, Wei KL, Chen G. Comprehensive clinical implications of homeobox A10 in 3,199 cases of non-small cell lung cancer tissue samples combining qRT-PCR, RNA sequencing and microarray data. Am J Transl Res. 2019;11(1):45–66. [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X, Deng Y, He RQ, Li XJ, Ma J, Chen G, Hu XH. Upregulation of HOXA11 during the progression of lung adenocarcinoma detected via multiple approaches. Int J Mol Med. 2018;42(5):2650–2664. doi: 10.3892/ijmm.2018.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang R, Zhang TT, Zhai GQ, Guo XY, Qin Y, Gan TQ, Zhang Y, Chen G, Mo WJ, Feng ZB. Evaluation of the HOXA11 level in patients with lung squamous cancer and insights into potential molecular pathways via bioinformatics analysis. World J Surg Oncol. 2018;16(1):109. doi: 10.1186/s12957-018-1375-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng Y, He R, Zhang R, Gan B, Zhang Y, Chen G, Hu X. The expression of HOXA13 in lung adenocarcinoma and its clinical significance: a study based on The Cancer Genome Atlas, Oncomine and reverse transcription-quantitative polymerase chain reaction. Oncol Lett. 2018;15(6):8556–8572. doi: 10.3892/ol.2018.8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Li XJ, He RQ, Wang X, Zhang TT, Qin Y, Zhang R, Deng Y, Wang HL, Luo DZ, et al. Upregulation of HOXA1 promotes tumorigenesis and development of nonsmall cell lung cancer: a comprehensive investigation based on reverse transcription-quantitative polymerase chain reaction and bioinformatics analysis. Int J Oncol. 2018;53(1):73–86. doi: 10.3892/ijo.2018.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo Z, Li X, Zhao Z, Yang X, Xiao S, Zhou Y. MicroRNA-146a affects the chemotherapeutic sensitivity and prognosis of advanced gastric cancer through the regulation of LIN52. Oncol Lett. 2017;13(3):1386–1392. doi: 10.3892/ol.2016.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zavala V, Perez-Moreno E, Tapia T, Camus M, Carvallo P. miR-146a and miR-638 in BRCA1-deficient triple negative breast cancer tumors, as potential biomarkers for improved overall survival. Cancer Biomark. 2016;16(1):99–107. doi: 10.3233/CBM-150545. [DOI] [PubMed] [Google Scholar]
- 35.Hess AK, Muer A, Mairinger FD, Weichert W, Stenzinger A, Hummel M, Budach V, Tinhofer I. MiR-200b and miR-155 as predictive biomarkers for the efficacy of chemoradiation in locally advanced head and neck squamous cell carcinoma. Eur J Cancer. 2017;77:3–12. doi: 10.1016/j.ejca.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 36.Hou Z, Xie L, Yu L, Qian X, Liu B. MicroRNA-146a is down-regulated in gastric cancer and regulates cell proliferation and apoptosis. Medical oncology (Northwood, London, England) 2012;29(2):886–892. doi: 10.1007/s12032-011-9862-7. [DOI] [PubMed] [Google Scholar]
- 37.Kogo R, Mimori K, Tanaka F, Komune S, Mori M. Clinical significance of miR-146a in gastric cancer cases. Clin Cancer Res. 2011;17(13):4277–4284. doi: 10.1158/1078-0432.CCR-10-2866. [DOI] [PubMed] [Google Scholar]
- 38.Li J, Yang H, Li Y, Liu Y, Chen S, Qi C, Zhang Q, Lan T, He X, Guan XY, et al. microRNA-146 up-regulation predicts the prognosis of non-small cell lung cancer by miRNA in situ hybridization. Exp Mol Pathol. 2014;96(2):195–199. doi: 10.1016/j.yexmp.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 39.Paik JH, Jang JY, Jeon YK, Kim WY, Kim TM, Heo DS, Kim CW. MicroRNA-146a downregulates NFkappaB activity via targeting TRAF6 and functions as a tumor suppressor having strong prognostic implications in NK/T cell lymphoma. Clin Cancer Res. 2011;17(14):4761–4771. doi: 10.1158/1078-0432.CCR-11-0494. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Li Z, He C, Wang D, Yuan X, Chen J, Jin J. MicroRNAs expression signatures are associated with lineage and survival in acute leukemias. Blood Cells Mol Dis. 2010;44(3):191–197. doi: 10.1016/j.bcmd.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhong H, Xu L, Zhong JH, Xiao F, Liu Q, Huang HH, Chen FY. Clinical and prognostic significance of miR-155 and miR-146a expression levels in formalin-fixed/paraffin-embedded tissue of patients with diffuse large B-cell lymphoma. Exp Ther Med. 2012;3(5):763–770. doi: 10.3892/etm.2012.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease. J Clin Investig. 2017;127(3):761–771. doi: 10.1172/JCI84424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eichmuller SB, Osen W, Mandelboim O, Seliger B. Immune modulatory microRNAs involved in tumor attack and tumor immune escape. J Natl Cancer Inst. 2017 doi: 10.1093/jnci/djx034. [DOI] [PubMed] [Google Scholar]
- 44.Peta E, Sinigaglia A, Masi G, Di Camillo B, Grassi A, Trevisan M, Messa L, Loregian A, Manfrin E, Brunelli M, et al. HPV16 E6 and E7 upregulate the histone lysine demethylase KDM2B through the c-MYC/miR-146a-5p axys. Oncogene. 2018;37(12):1654–1668. doi: 10.1038/s41388-017-0083-1. [DOI] [PubMed] [Google Scholar]
- 45.Cui Y, She K, Tian D, Zhang P, Xin X. miR-146a inhibits proliferation and enhances chemosensitivity in epithelial ovarian cancer via reduction of SOD2. Oncol Res. 2016;23(6):275–282. doi: 10.3727/096504016X14562725373798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilczynski M, Zytko E, Szymanska B, Dzieniecka M, Nowak M, Danielska J, Stachowiak G, Wilczynski JR. Expression of miR-146a in patients with ovarian cancer and its clinical significance. Oncol Lett. 2017;14(3):3207–3214. doi: 10.3892/ol.2017.6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shomali N, Mansoori B, Mohammadi A, Shirafkan N, Ghasabi M, Baradaran B. MiR-146a functions as a small silent player in gastric cancer. Biomed Pharmacother. 2017;96:238–245. doi: 10.1016/j.biopha.2017.09.138. [DOI] [PubMed] [Google Scholar]
- 48.Li H, Xie S, Liu M, Chen Z, Liu X, Wang L, Li D, Zhou Y. The clinical significance of downregulation of mir-124-3p, mir-146a-5p, mir-155-5p and mir-335-5p in gastric cancer tumorigenesis. Int J Oncol. 2014;45(1):197–208. doi: 10.3892/ijo.2014.2415. [DOI] [PubMed] [Google Scholar]
- 49.Papaconstantinou IG, Manta A, Gazouli M, Lyberopoulou A, Lykoudis PM, Polymeneas G, Voros D. Expression of microRNAs in patients with pancreatic cancer and its prognostic significance. Pancreas. 2013;42(1):67–71. doi: 10.1097/MPA.0b013e3182592ba7. [DOI] [PubMed] [Google Scholar]
- 50.Ali S, Ahmad A, Aboukameel A, Ahmed A, Bao B, Banerjee S, Philip PA, Sarkar FH. Deregulation of miR-146a expression in a mouse model of pancreatic cancer affecting EGFR signaling. Cancer Lett. 2014;351(1):134–142. doi: 10.1016/j.canlet.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rong M, He R, Dang Y, Chen G. Expression and clinicopathological significance of miR-146a in hepatocellular carcinoma tissues. Upsala J Med Sci. 2014;119(1):19–24. doi: 10.3109/03009734.2013.856970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang S, He R, Rong M, Dang Y, Chen G. Synergistic effect of MiR-146a mimic and cetuximab on hepatocellular carcinoma cells. Biomed Res Int. 2014;2014:384121. doi: 10.1155/2014/384121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu D, Yao Q, Zhan C, Le-Meng Z, Liu H, Cai Y, Tu C, Li X, Zou Y, Zhang S. MicroRNA-146a promote cell migration and invasion in human colorectal cancer via carboxypeptidase M/src-FAK pathway. Oncotarget. 2017;8(14):22674–22684. doi: 10.18632/oncotarget.15158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu H, Ren G, Zhu L, Liu X, He X. The upregulation of miRNA-146a inhibited biological behaviors of ESCC through inhibition of IRS2. Tumour Biol. 2016;37(4):4641–4647. doi: 10.1007/s13277-015-4274-5. [DOI] [PubMed] [Google Scholar]
- 55.Wang C, Guan S, Liu F, Chen X, Han L, Wang D, Nesa EU, Wang X, Bao C, Wang N, et al. Prognostic and diagnostic potential of miR-146a in oesophageal squamous cell carcinoma. Br J Cancer. 2016;114(3):290–297. doi: 10.1038/bjc.2015.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang C, Zhang W, Zhang L, Chen X, Liu F, Zhang J, Guan S, Sun Y, Chen P, Wang D, et al. miR-146a-5p mediates epithelial-mesenchymal transition of oesophageal squamous cell carcinoma via targeting Notch2. Br J Cancer. 2018;118(6):e12. doi: 10.1038/bjc.2017.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhan B, Lu D, Luo P, Wang B. Prognostic value of expression of MicroRNAs in non-small cell lung cancer: a systematic review and meta-analysis. Clin Lab. 2016;62(11):2203–2211. doi: 10.7754/Clin.Lab.2016.160426. [DOI] [PubMed] [Google Scholar]
- 58.Lerner C, Wemmert S, Bochen F, Kulas P, Linxweiler M, Hasenfus A, Heinzelmann J, Leidinger P, Backes C, Meese E, et al. Characterization of miR-146a and miR-155 in blood, tissue and cell lines of head and neck squamous cell carcinoma patients and their impact on cell proliferation and migration. J Cancer Res Clin Oncol. 2016;142(4):757–766. doi: 10.1007/s00432-015-2087-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kumaraswamy E, Wendt KL, Augustine LA, Stecklein SR, Sibala EC, Li D, Gunewardena S, Jensen RA. BRCA1 regulation of epidermal growth factor receptor (EGFR) expression in human breast cancer cells involves microRNA-146a and is critical for its tumor suppressor function. Oncogene. 2015;34(33):4333–4346. doi: 10.1038/onc.2014.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fukami K, Inanobe S, Kanemaru K, Nakamura Y. Phospholipase C is a key enzyme regulating intracellular calcium and modulating the phosphoinositide balance. Prog Lipid Res. 2010;49(4):429–437. doi: 10.1016/j.plipres.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 61.Kadamur G, Ross EM. Mammalian phospholipase C. Annu Rev Physiol. 2013;75:127–154. doi: 10.1146/annurev-physiol-030212-183750. [DOI] [PubMed] [Google Scholar]
- 62.Raimondi C, Falasca M. Phosphoinositides signalling in cancer: focus on PI3K and PLC. Adv Biol Regul. 2012;52(1):166–182. doi: 10.1016/j.advenzreg.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 63.Lattanzio R, Piantelli M, Falasca M. Role of phospholipase C in cell invasion and metastasis. Adv Biol Regul. 2013;53(3):309–318. doi: 10.1016/j.jbior.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 64.Cai S, Sun PH, Resaul J, Shi L, Jiang A, Satherley LK, Davies EL, Ruge F, Douglas-Jones A, Jiang WG, et al. Expression of phospholipase C isozymes in human breast cancer and their clinical significance. Oncol Rep. 2017;37(3):1707–1715. doi: 10.3892/or.2017.5394. [DOI] [PubMed] [Google Scholar]
- 65.Ertao Z, Jianhui C, Chuangqi C, Changjiang Q, Sile C, Yulong H, Hui W, Shirong C. Autocrine Sonic hedgehog signaling promotes gastric cancer proliferation through induction of phospholipase Cgamma1 and the ERK1/2 pathway. J Exp Clin Cancer Res. 2016;35:63. doi: 10.1186/s13046-016-0336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ma LW, Zhou ZT, He QB, Jiang WW. Phospholipase C-gamma1 expression correlated with cancer progression of potentially malignant oral lesions. J Oral Pathol Med. 2013;42(1):47–52. doi: 10.1111/j.1600-0714.2012.01179.x. [DOI] [PubMed] [Google Scholar]
- 67.Zhu D, Tan Y, Yang X, Qiao J, Yu C, Wang L, Li J, Zhang Z, Zhong L. Phospholipase C gamma 1 is a potential prognostic biomarker for patients with locally advanced and resectable oral squamous cell carcinoma. Int J Oral Maxillofac Surg. 2014;43(12):1418–1426. doi: 10.1016/j.ijom.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 68.Paris L, Cecchetti S, Spadaro F, Abalsamo L, Lugini L, Pisanu ME, Iorio E, Natali PG, Ramoni C, Podo F. Inhibition of phosphatidylcholine-specific phospholipase C downregulates HER2 overexpression on plasma membrane of breast cancer cells. BCR. 2010;12(3):R27. doi: 10.1186/bcr2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Balz LM, Bartkowiak K, Andreas A, Pantel K, Niggemann B, Zanker KS, Brandt BH, Dittmar T. The interplay of HER2/HER3/PI3 K and EGFR/HER2/PLC-gamma1 signalling in breast cancer cell migration and dissemination. J Pathol. 2012;227(2):234–244. doi: 10.1002/path.3991. [DOI] [PubMed] [Google Scholar]
- 70.Verkman AS. Aquaporins in clinical medicine. Annu Rev Med. 2012;63:303–316. doi: 10.1146/annurev-med-043010-193843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beitz E, Golldack A, Rothert M, von Bulow J. Challenges and achievements in the therapeutic modulation of aquaporin functionality. Pharmacol Ther. 2015;155:22–35. doi: 10.1016/j.pharmthera.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 72.Noda Y, Sohara E, Ohta E, Sasaki S. Aquaporins in kidney pathophysiology. Nat Rev Nephrol. 2010;6(3):168–178. doi: 10.1038/nrneph.2009.231. [DOI] [PubMed] [Google Scholar]
- 73.Kortenoeven ML, Fenton RA. Renal aquaporins and water balance disorders. Biochem Biophys Acta. 2014;1840(5):1533–1549. doi: 10.1016/j.bbagen.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 74.Wang J, Feng L, Zhu Z, Zheng M, Wang D, Chen Z, Sun H. Aquaporins as diagnostic and therapeutic targets in cancer: how far we are? J Transl Med. 2015;13:96. doi: 10.1186/s12967-015-0439-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang L, Zhang Y, Wu X, Yu G. Aquaporins: new targets for cancer therapy. Technol Cancer Res Treat. 2016;15(6):821–828. doi: 10.1177/1533034615607693. [DOI] [PubMed] [Google Scholar]
- 76.Ribatti D, Ranieri G, Annese T, Nico B. Aquaporins in cancer. Biochem Biophys Acta. 2014;1840(5):1550–1553. doi: 10.1016/j.bbagen.2013.09.025. [DOI] [PubMed] [Google Scholar]
- 77.Garg R, Benedetti LG, Abera MB, Wang H, Abba M, Kazanietz MG. Protein kinase C and cancer: what we know and what we do not. Oncogene. 2014;33(45):5225–5237. doi: 10.1038/onc.2013.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Caino MC, Lopez-Haber C, Kissil JL, Kazanietz MG. Non-small cell lung carcinoma cell motility, rac activation and metastatic dissemination are mediated by protein kinase C epsilon. PLoS ONE. 2012;7(2):e31714. doi: 10.1371/journal.pone.0031714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang N, Su Y, Xu L. Targeting PKCepsilon by miR-143 regulates cell apoptosis in lung cancer. FEBS Lett. 2013;587(22):3661–3667. doi: 10.1016/j.febslet.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 80.Toton E, Ignatowicz E, Skrzeczkowska K, Rybczynska M. Protein kinase Cepsilon as a cancer marker and target for anticancer therapy. PR. 2011;63(1):19–29. doi: 10.1016/s1734-1140(11)70395-4. [DOI] [PubMed] [Google Scholar]
- 81.Basu A. PKCepsilon paves the way for prostate cancer. Cell Cycle. 2011;10(3):378. doi: 10.4161/cc.10.3.14739. [DOI] [PubMed] [Google Scholar]
- 82.Huang B, Cao K, Li X, Guo S, Mao X, Wang Z, Zhuang J, Pan J, Mo C, Chen J, et al. The expression and role of protein kinase C (PKC) epsilon in clear cell renal cell carcinoma. J Exp Clin Cancer Res. 2011;30:88. doi: 10.1186/1756-9966-30-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Datta J, Smith A, Lang JC, Islam M, Dutt D, Teknos TN, Pan Q. microRNA-107 functions as a candidate tumor-suppressor gene in head and neck squamous cell carcinoma by downregulation of protein kinase Cvarepsilon. Oncogene. 2012;31(36):4045–4053. doi: 10.1038/onc.2011.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gorin MA, Pan Q. Protein kinase C epsilon: an oncogene and emerging tumor biomarker. Molecular cancer. 2009;8:9. doi: 10.1186/1476-4598-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brownlow N, Pike T, Crossland V, Claus J, Parker P. Regulation of the cytokinesis cleavage furrow by PKCepsilon. Biochem Soc Trans. 2014;42(6):1534–1537. doi: 10.1042/BST20140240. [DOI] [PubMed] [Google Scholar]
- 86.Zhang X, Li D, Li M, Ye M, Ding L, Cai H, Fu D, Lv Z. MicroRNA-146a targets PRKCE to modulate papillary thyroid tumor development. Int J Cancer. 2014;134(2):257–267. doi: 10.1002/ijc.28141. [DOI] [PubMed] [Google Scholar]
- 87.Schenone S, Brullo C, Musumeci F, Biava M, Falchi F, Botta M. Fyn kinase in brain diseases and cancer: the search for inhibitors. Curr Med Chem. 2011;18(19):2921–2942. doi: 10.2174/092986711796150531. [DOI] [PubMed] [Google Scholar]
- 88.Saito YD, Jensen AR, Salgia R, Posadas EM. Fyn: a novel molecular target in cancer. Cancer. 2010;116(7):1629–1637. doi: 10.1002/cncr.24879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Elias D, Ditzel HJ. Fyn is an important molecule in cancer pathogenesis and drug resistance. Pharmacol Res. 2015;100:250–254. doi: 10.1016/j.phrs.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 90.Lewin B, Siu A, Baker C, Dang D, Schnitt R, Eisapooran P, Ramos DM. Expression of Fyn kinase modulates EMT in oral cancer cells. Anticancer Res. 2010;30(7):2591–2596. [PubMed] [Google Scholar]
- 91.Chen JS, Hung WS, Chan HH, Tsai SJ, Sun HS. In silico identification of oncogenic potential of fyn-related kinase in hepatocellular carcinoma. Bioinformatics. 2013;29(4):420–427. doi: 10.1093/bioinformatics/bts715. [DOI] [PubMed] [Google Scholar]
- 92.Alexanian A, Miller B, Chesnik M, Mirza S, Sorokin A. Post-translational regulation of COX2 activity by FYN in prostate cancer cells. Oncotarget. 2014;5(12):4232–4243. doi: 10.18632/oncotarget.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Verkman AS, Anderson MO, Papadopoulos MC. Aquaporins: important but elusive drug targets. Nat Rev Drug Discov. 2014;13(4):259–277. doi: 10.1038/nrd4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Papadopoulos MC, Saadoun S, Verkman AS. Aquaporins and cell migration. Pflugers Arch. 2008;456(4):693–700. doi: 10.1007/s00424-007-0357-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tomita Y, Dorward H, Yool AJ, Smith E, Townsend AR, Price TJ, Hardingham JE. Role of Aquaporin 1 signalling in cancer development and progression. Int J Mol Sci. 2017;18(2):299. doi: 10.3390/ijms18020299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang Y, Fan Y, Zheng C, Zhang X. Knockdown of AQP1 inhibits growth and invasion of human ovarian cancer cells. Mol Med Rep. 2017;16(4):5499–5504. doi: 10.3892/mmr.2017.7282. [DOI] [PubMed] [Google Scholar]
- 97.Park JY, Yoon G. Overexpression of Aquaporin-1 is a prognostic factor for biochemical recurrence in prostate adenocarcinoma. POR. 2017;23(1):189–196. doi: 10.1007/s12253-016-0145-7. [DOI] [PubMed] [Google Scholar]
- 98.Sun WJ, Hu DH, Wu H, Xiao H, Lu MD, Guo WJ, Huang H, Yu YJ, Hu TY, Zheng ZQ. Expression of AQP1 was associated with apoptosis and survival of patients in gastric adenocarcinoma. Dig Surg. 2016;33(3):190–196. doi: 10.1159/000443843. [DOI] [PubMed] [Google Scholar]
- 99.Kang BW, Kim JG, Lee SJ, Chae YS, Jeong JY, Yoon GS, Park SY, Kim HJ, Park JS, Choi GS, et al. Expression of aquaporin-1, aquaporin-3, and aquaporin-5 correlates with nodal metastasis in colon cancer. Oncology. 2015;88(6):369–376. doi: 10.1159/000369073. [DOI] [PubMed] [Google Scholar]
- 100.Yamazato Y, Shiozaki A, Ichikawa D, Kosuga T, Shoda K, Arita T, Konishi H, Komatsu S, Kubota T, Fujiwara H, et al. Aquaporin 1 suppresses apoptosis and affects prognosis in esophageal squamous cell carcinoma. Oncotarget. 2018;9(52):29957–29974. doi: 10.18632/oncotarget.25722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.El Hindy N, Bankfalvi A, Herring A, Adamzik M, Lambertz N, Zhu Y, Siffert W, Sure U, Sandalcioglu IE. Correlation of aquaporin-1 water channel protein expression with tumor angiogenesis in human astrocytoma. Anticancer Res. 2013;33(2):609–613. [PubMed] [Google Scholar]
- 102.Wei X, Dong J. Aquaporin 1 promotes the proliferation and migration of lung cancer cell in vitro. Oncol Rep. 2015;34(3):1440–1448. doi: 10.3892/or.2015.4107. [DOI] [PubMed] [Google Scholar]
- 103.Esteva-Font C, Jin BJ, Verkman AS. Aquaporin-1 gene deletion reduces breast tumor growth and lung metastasis in tumor-producing MMTV-PyVT mice. FASEB J. 2014;28(3):1446–1453. doi: 10.1096/fj.13-245621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Qin F, Zhang H, Shao Y, Liu X, Yang L, Huang Y, Fu L, Gu F, Ma Y. Expression of aquaporin1, a water channel protein, in cytoplasm is negatively correlated with prognosis of breast cancer patients. Oncotarget. 2016;7(7):8143–8154. doi: 10.18632/oncotarget.6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu J, Zhang WY, Ding DG. Expression of aquaporin 1 in bladder uroepithelial cell carcinoma and its relevance to recurrence. APJCP. 2015;16(9):3973–3976. doi: 10.7314/apjcp.2015.16.9.3973. [DOI] [PubMed] [Google Scholar]
- 106.Jagirdar RM, Apostolidou E, Molyvdas PA, Gourgoulianis KI, Hatzoglou C, Zarogiannis SG. Influence of AQP1 on cell adhesion, migration, and tumor sphere formation in malignant pleural mesothelioma is substratum- and histological-type dependent. Am J Physiol Lung Cell Mol Physiol. 2016;310(6):L489–L495. doi: 10.1152/ajplung.00410.2015. [DOI] [PubMed] [Google Scholar]
- 107.Ihara T, Hosokawa Y, Kumazawa K, Ishikawa K, Fujimoto J, Yamamoto M, Muramkami T, Goshima N, Ito E, Watanabe S, et al. An in vivo screening system to identify tumorigenic genes. Oncogene. 2017;36(14):2023–2029. doi: 10.1038/onc.2016.351. [DOI] [PubMed] [Google Scholar]
- 108.Furuta K, Arao T, Sakai K, Kimura H, Nagai T, Tamura D, Aomatsu K, Kudo K, Kaneda H, Fujita Y, et al. Integrated analysis of whole genome exon array and array-comparative genomic hybridization in gastric and colorectal cancer cells. Cancer Sci. 2012;103(2):221–227. doi: 10.1111/j.1349-7006.2011.02132.x. [DOI] [PubMed] [Google Scholar]
- 109.Makondi PT, Chu CM, Wei PL, Chang YJ. Prediction of novel target genes and pathways involved in irinotecan-resistant colorectal cancer. PloS ONE. 2017;12(7):e0180616. doi: 10.1371/journal.pone.0180616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li YX, Jin HG, Yan CG, Ren CY, Jiang CJ, Jin CD, Seo KS, Jin X. Molecular cloning, sequence identification, and gene expression analysis of bovine ADCY2 gene. Mol Biol Rep. 2014;41(6):3561–3568. doi: 10.1007/s11033-014-3167-9. [DOI] [PubMed] [Google Scholar]
- 111.Shen JX, Cooper DM. AKAP79, PKC, PKA and PDE4 participate in a Gq-linked muscarinic receptor and adenylate cyclase 2 cAMP signalling complex. Biochem J. 2013;455(1):47–56. doi: 10.1042/BJ20130359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Duerr EM, Mizukami Y, Ng A, Xavier RJ, Kikuchi H, Deshpande V, Warshaw AL, Glickman J, Kulke MH, Chung DC. Defining molecular classifications and targets in gastroenteropancreatic neuroendocrine tumors through DNA microarray analysis. Endocrine Relat Cancer. 2008;15(1):243–256. doi: 10.1677/ERC-07-0194. [DOI] [PubMed] [Google Scholar]
- 113.Sato T, Arai E, Kohno T, Tsuta K, Watanabe S, Soejima K, Betsuyaku T, Kanai Y. DNA methylation profiles at precancerous stages associated with recurrence of lung adenocarcinoma. PloS ONE. 2013;8(3):e59444. doi: 10.1371/journal.pone.0059444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen H, Cai W, Chu ESH, Tang J, Wong CC, Wong SH, Sun W, Liang Q, Fang J, Sun Z, et al. Hepatic cyclooxygenase-2 overexpression induced spontaneous hepatocellular carcinoma formation in mice. Oncogene. 2017;36(31):4415–4426. doi: 10.1038/onc.2017.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.