Abstract
The poly(ADP-ribose) polymerases (PARP) play important roles in repairing damaged DNA during intrinsic cell death. We recently linked PARP-1 to death receptor (DR)-activated extrinsic apoptosis, the present studies sought to elucidate the function of cytoplasmic PARP-1 in pancreatic cancer tumorigenesis and therapy. Using human normal and pancreatic cancer tissues, we analyzed the prevalence of cytoplasmic PARP-1 expression. In normal human pancreatic tissues, PARP-1 expression was present in the nucleus; however, cytoplasmic PARP-1 expression was identified in pancreatic cancers. Therefore, cytoplasmic PARP-1 mutants were generated by site-direct mutagenesis, to determine a causative effect of cytoplasmic PARP-1 on pancreatic cancer tumorigenesis and sensitivity to therapy with TRA-8, a humanized DR5 antibody. PARP-1 cytoplasmic mutants rendered TRA-8 sensitive pancreatic cancer cells, BxPc-3 and MiaPaCa-2, more resistant to TRA-8-induced apoptosis; whereas wild-type PARP-1, localizing mainly in the nucleus, had no effects. Additionally, cytoplasmic PARP-1, but not wild-type PARP-1, increased resistance of BxPc-3 cells to TRA-8 therapy in a mouse xenograft model in vivo. Inhibition of PARP enzymatic activity attenuated cytoplasmic PARP-1-mediated TRA-8 resistance. Furthermore, increased cytoplasmic PARP-1, but not wild-type PARP-1, was recruited into the TRA-8-activated death-inducing signaling complex and associated with increased and sustained activation of Src-mediated survival signals. In contrast, PARP-1 knockdown inhibited Src activation. Taken together, we have identified a novel function and mechanism underlying cytoplasmic PARP-1, distinct from nuclear PARP-1, in regulating DR5-activated apoptosis. Our studies support an innovative application of available PARP inhibitors or new cytoplasmic PARP-1 antagonists to enhance TRAIL therapy for TRAIL-resistant pancreatic cancers.
Keywords: cytoplasmic PARP-1, pancreatic cancer, death receptor, apoptosis, survival
Introduction
Pancreatic cancer is the fourth leading cause of cancer deaths in the USA.1 The American Cancer Society estimates 53,670 new cases and 43,090 deaths from pancreatic cancer in 2017 in the USA. It is the most lethal type of digestive cancer. The 5-year survival rate has only improved from 2% to 6% in the past 30 years.2 The only potentially curative therapy for pancreatic cancer is surgical resection. Unfortunately, only 20% of patients are resectable at the time of diagnosis. Even among those patients who undergo resection for pancreatic cancer and have tumor-free margins, the 5-year survival rate after re-section is 10% to 25%.3 Gemcitabine or 5-fluorouracil chemo-therapy coupled with radiotherapy may improve the quality of life of patients, but their survival benefit is very limited.4,5 Resistance to chemotherapy and radiotherapy remains a major obstacle to obtaining a better prognosis for patients with pancreatic cancer. Thus, better understanding of the molecular mechanisms underlying pancreatic cancer pathogenesis and their resistance to therapy will lead to developing novel strategies and therapeutic targets to prevent, diagnose and treat this highly fatal tumor.
Dysregulation of apoptosis of tumor cells plays an important role in the pathogenesis of many cancers and their resistance to therapies. At the cellular level, there are two main apoptotic pathways, the extrinsic and intrinsic pathways.6 The intrinsic pathway, also called the mitochondrial pathway, mediates apoptosis initiated by internal signals, such as DNA damage,7 induced by stress signals, including chemotherapy agents and radiation. Poly(adenosine diphosphate [ADP]-ribose) polymerase (PARP) is a family of enzymes that play important roles in DNA damage repair.8 Inhibition of PARPs results in accumulation of irreparable damaged DNA, which triggers intrinsic apoptosis in DNA repair-deficient cancers.9 In a meta-analysis performed on a large public retrospective gene expression data set from breast cancers, PARP-1 mRNA expression correlated with high grade, medullary histological type, tumor size, worse metastasis-free survival and poor overall patient survival.10 Accordingly, targeting PARP’s DNA-repairing function by PARP inhibitors had been tested in cancer therapy, either as a single agent in tumor cells with defects in DNA repair or combining with DNA damaging agents. Single-agent activity with PARP inhibitors has been shown to be effective in BRCA-deficient breast and ovarian cancer and BRCA2-associated pancreatic cancer.8,11 Combination therapy of PARP-1 inhibitor with the DNA-damaging drug, oxaliplatin, has enhanced pancreatic cancer therapy in a preclinical model.12
Our recent studies have provided the first evidence linking PARP-1 to the extrinsic apoptosis pathway,13 which appears to be independent of the known PARP-1 function of DNA repair in the nucleus. The extrinsic apoptosis pathway is initiated by activation of death receptors (DRs) present on the cell surface, including the Fas receptor, tumor necrosis factor (TNF) receptor, and death receptor 4 and 5 that are activated by the TNF-related apoptosis inducing ligand (TRAIL).14 Activating the DR4 or 5-mediated extrinsic apoptotic signaling pathway by TRAIL has been demonstrated in a wide variety of tumor cells in vitro and in vivo, and has been consistently selective for tumor cells over normal cells.15,16 Recombinant TRAIL or anti-human DR4/5 monoclonal antibodies have been tested in stage I-III clinical trials for their anti-tumor efficacy. These include trails with conatumumab (AMG655)17 and tigatuzumab (CS-1008, TRA-8, monoclonal antibodies for DR5)18 for pancreatic cancer (Clinicaltrials. gov). Although TRAIL agonist therapies have shown low toxicity in patients19 and many types of tumors are sensitive to TRAIL-induced apoptosis, substantial numbers of cancers are resistant to TRAIL, especially some highly malignant tumors such as pancreatic cancer.20 Pancreatic cancer cells show a variable susceptibility to cell death induced by TRAIL, despite expressing intact receptors and components of the apoptotic signaling pathway.20,21 Our finding of a potential new role of PARP-1 in TRAIL-induced apoptosis prompted us to design the present studies to determine the function and elucidate the underlying mechanisms of cytoplasmic PARP-1 in regulating pancreatic cancer tumorigenesis and their sensitivity to TRAIL therapy.
Using comprehensive loss-of-function and gain-of-function approaches, a preclinical animal model and human normal and pancreatic cancer tissues, the present studies uncovered different effects of cytoplasmic and nuclear PARP-1 expression on pancreatic cancer tumorigenesis and their sensitivity to death receptor 5-activated apoptosis. Mechanistic studies revealed that increased cytoplasmic PARP-1, distinct from wild-type PARP-1, was recruited into the death-inducing signaling complex (DISC) and associated with increased and sustained activation of Src survival signals. Our finding of a novel function of cytoplasmic PARP-1 in regulating TRAIL-induced apoptosis supports strategies to target cytoplasmic PARP-1, by innovative application of the readily available PARP inhibitors or developing new cytoplasmic PARP-1 antagonists, to enhance the efficacy of TRAIL therapy for TRAIL-resistant pancreatic cancers.
Materials and Methods
Cell culture and reagents
The human pancreatic cancer cell lines BxPc-3, MiaPaCa-2, and PANC-1 were from the American Type Culture Collection. BxPc-3 cells were grown in RPMI-1640 and MiaPaCa-2 and PANC-1 cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM, Invitrogen) supplemented with penicillin (5 units/mL), streptomycin (5 mg/mL), and 10% heat-inactivated fetal bovine serum.
Plasmid mutation and lentiviral infection
Construct for wild type PARP-1 was produced by polymerase chain reaction amplification of the full-length coding sequence of PARP-1. The construct was packaged into lentivirus-like particles pseudotyped with the vesicular stomatitis virus glyco-protein as we previously described.22 Point mutations in the sequence of the lentiviral PARP-1 were made at R208 and K222, which were in the PARP-1 nuclear localization domain,23 using the QuikChange II Site-Directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA). The PARP-1 mutants (PARP-1 R208Q and PARP-1 K222I) were confirmed by sequencing analysis. Stably transduced cells were selected with puromycin (2 μg/mL).
Immunofluorescent staining
To detect the PARP-1 cellular location, cells grown on 18 mm × 18 mm glass coverslips in a 6-well plates were washed with ice-cold PBS, fixed with 3% (w/v) paraformaldehyde, and blocked with 1% BSA. Immunofluorescent staining was performed as we described previously,24 using PARP-1 (Santa Cruz Biotech, SC-7150) or DR5 (Prosci,#2019) with Alexa Flour® 488 anti-mouse IgG or Alexa Flour® 594 anti-rabbit IgG (10 μg/mL) anti-obodies. DAPI (4′, 6-diamidino-2-phenylindole) was used to identify nuclei.
Assessment of apoptosis
DR5 agonist antibody TRA-8 was generated as previously reported.25 Apoptosis was determined by Annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) staining (BD Biosciences) and analyzed by flow cytometry (BD Biosciences) as we previously reported.13
Western blot analysis
Proteins were extracted, quantified with a BCA protein assay kit (Thermo Scientific), separated by SDS-PAGE, and transferred to Immobilon P membranes (Millipore) as described previously.13,26 Membranes were blocked in 5% non-fat milk and incubated with primary antibodies overnight at 4 °C and then with horseradish peroxidase–conjugated secondary antibodies for 1 h at room temperature. Signals were detected using the Immobilon Western Chemiluminescent Horseradish Peroxidase Substrate Detection Kit (Millipore). The antibodies used were commercially available, including Src (Cells Signaling, 2108S, 1:1000) and p-Src (Cells Signaling, 6943S, 1:2000), caspase-3 (Enzo Life, ADI-AAP-113-F, 1:2000), caspase-8 (BD Bioscience, 551242, 1:2000), PARP-1 (Santa Cruz Biotech, SC-7150, 1:2000), and GAPDH (Fitzgerald, 10R-G109a,1:20000), and DR5 (Prosci,#2019,1:1000).
Immunoprecipitation
DISC associated complex was immunoprecipitated as we reported.13 In brief, 500 micrograms of extracted proteins was incubated with 1 μg of anti-DR5 antibody for 1 h and subsequently incubated with 50 μL of 1:1 slurry of protein G-agarose beads (Invitrogen) overnight at 4 °C. Beads were then washed, and 20 μL 2 × Laemmli sample buffer was added to the beads followed by heating at 95 °C for 5 min and chilling on ice. After brief centrifugation, proteins in the supernatant were analyzed by Western blotting with specific antibodies.
Mouse xenograft model
The animal protocol was approved by the Institutional Animal Care and Use Committee at the University of Alabama at Birmingham (Birmingham, AL). Male athymic nu/nu mice (6 weeks old, NCI-Frederick) were used for tumor inoculation as we described.13,26 Briefly, BxPc-3-vector, BxPc-3-PARP-1-WT, or BxPc-3-PARP-1-K222I cells were inoculated subcutaneously into the flanks of mice (2 × 106 cells in Matrigel/site). Three weeks after tumor inoculation, mice were divided into 2 groups (n = 8–9/group), mice in the treatment group were intraperitoneally injected with TRA-8 once a week for 6 weeks (200 μg/mouse), and mice in the control group were injected with 0.9% sodium chloride. Tumor size was measured every week, and volumes were determined using the formula volume = length×width2/2. At the end of the experiment, tumors were removed from mice, half of each tumor was homogenized for Western blot analysis and the other half was fixed in 4% paraformaldehyde and embedded in paraffin.
Human pancreatic tissues
The expression of PARP-1 in human pancreatic tissues was determined using the Pancreatic Cancer and Normal Tissue Arrays with characterization of clinical stages and tumor grades (PA804, PA805a and PA961c, US Biomax, https://www.biomax.us/Pancreas). A total of 25 normal (normal) pancreatic tissue samples and 193 pancreatic cancer at grade 1 (G1), grade 2 (G2) and grade 3 (G3) were analyzed by immunohistochemical staining using antibody for PARP-1 as we previous described.13
Immunohistochemistry staining
Immunohistochemical staining of PARP-1 was performed on human pancreatic cancer and normal tissues, as well as mouse xenografted tumors. Paraffin embedded human tissue microarray slides or xenograft tumor sections were deparaffinized, rehydrated, and heated in 10 μmol/L citrate buffer (pH 6.0), and subsequentially incubated with anti–PARP-1 antibody (Santa Cruz Biotech, SC-7150) followed by a horseradish peroxidase-conjugated secondary antibody and diaminobenzidine. The sections were counterstained with hematoxylin. A second anti-PARP-1 antibody (Abcam, ab110915) was also used to confirm the results.
To quantify the PARP-1 nuclear and cytoplasmic localization in human pancreatic tissues, stained slides were digitalized and evaluated by three independent investigators. The percentage of positive cells that expressed cytoplasmic PARP-1 in the normal and grade 1, 2 and 3 pancreatic cancer samples were calculated.27
TUNEL staining
TUNEL staining was conducted on tumor sections (DeadEnd Fluorometric TUNEL System; Promega) to determine cell death and DAPI staining was used to identify nuclei.13 Stained specimens were examined microscopically (Leica M165 FC). For quantitative analysis, cell numbers were counted under a microscope (×200). Four fields in each slide were counted and the percentage of apoptotic cells was determined.
Statistical analysis
Results are expressed as means ± SD. Differences between two groups were identified with Student’s t-test. For multiple groups, one-way analysis of variance and Student–Newman– Keuls tests was performed to identify differences. Significance was defined as p < 0.05.
Results
Increased cytoplasmic PARP-1 expression in human pancreatic cancer tissues
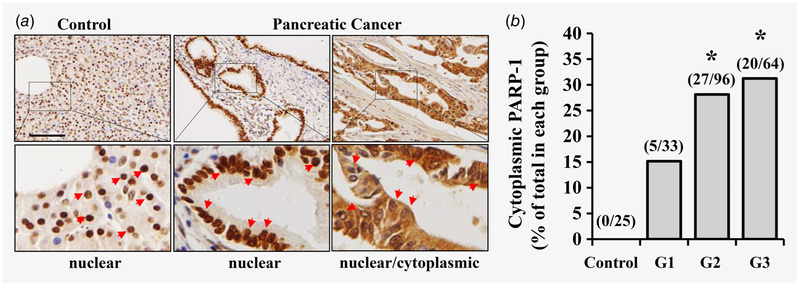
Our recent studies have demonstrated that inhibition of PARP-1 sensitizes TRAIL-resistant pancreatic cancer cells to TRA-8-induced apoptosis,13 suggesting a previously unknown function of PARP-1 in regulating the extrinsic apoptotic pathways in the cytoplasm, a cellular location that is different from the known PARP-1 function in repairing damaged DNA in the nucleus. To evaluate the cytoplasmic function of PARP-1 in pancreatic cancer pathogenesis, we first determined PARP-1 expression and localization in human pancreatic tissues and pancreatic tumor samples. A total of 193 tumor samples from grade 1, 2 and 3 pancreatic cancers (G1, G2 and G3) and 25 normal pancreatic tissue samples were analyzed by immunohistochemical staining for PARP-1. As shown in Figure 1, PARP-1 expression was exclusively identified in the nucleus, but not cytoplasm, in the normal pancreatic tissues (Fig. 1a, Control). In contrast, PARP-1 expression was identified in both nuclear and cytoplasmic locations in pancreatic cancer tissues (Fig. 1a, Pancreatic cancer). Comparison of the frequency of cytoplasmic localization of PARP-1 in grade 1, 2 and 3 pancreatic cancers revealed greater prevalence of cytoplasmic PARP-1 in grade 2 and grade 3 pancreatic tumors, compared to those in normal pancreatic tissues and grade 1 pancreatic tumors (Fig. 1b).
Figure 1.
PARP-1 expression in human normal and pancreatic cancer tissues. Expression of PARP-1 in normal and pancreatic cancer tissues was determined by immunohistochemical staining using anti-PARP-1 antibody. (a) Representative images of stained sections from normal (Control) and pancreatic cancer tissues (Pancreatic Cancer) are shown. Higher magnification of the boxed areas are shown below each image. Red arrows indicate nuclear or nuclear/cytoplasmic PARP-1 expression. Scale bars = 100 μm. (b) Quantitative analysis of percentage of tissues that express cytoplasmic PARP-1 in the normal (Control) and grade 1 (G1), grade 2 (G2) and grade 3 (G3) pancreatic cancers. The numbers in the brackets are the numbers of the samples that are positive for cytoplasmic PARP-1 and total samples examined in each group (*p < 0.05, compared to the Control).
Overexpression of cytoplasmic PARP-1 inhibits TRA-8-induced apoptosis in TRA-8-sensitive pancreatic cancer cells
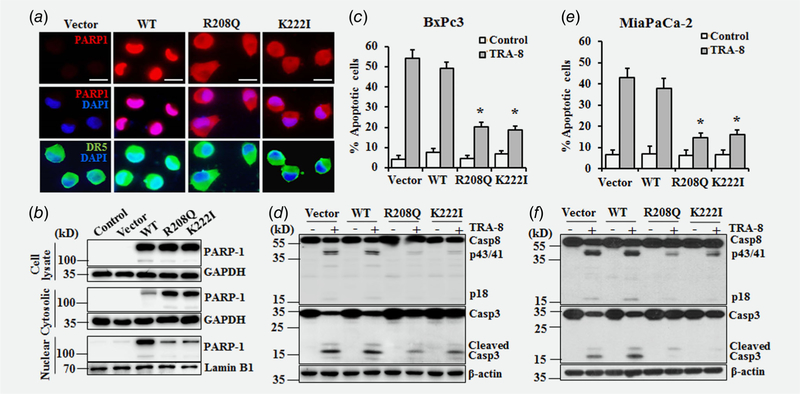
We have recently demonstrated the TRAIL-sensitive BxPc-3 and MiaPaCa-2 pancreatic cancer cells express markedly lower PARP-1 protein, compared to those in TRAIL-resistant PANC-1 and Suit-2 cells, suggesting a role of PARP-1 in regulating TRAIL resistance of pancreatic cancer cells.13 To deter mine the functions of the nuclear and cytoplasmic PARP-1 in regulating pancreatic cancer sensitivity to TRA-8-induced apoptosis, we generated lentiviruses carrying wild-type (WT) PARP-1 or PARP-1 mutants (PARP-1 R208Q and PARP-1 K222I), which contain point mutation in the PARP-1 nuclear localization domain that lead to increased cytoplasmic expression of PARP-1.23 Stable transfectants of the BxPc-3 cells with the WT PARP-1 demonstrated increased PARP-1 expression in the nucleus, compared to those in BxPc-3 cells infected with lentiviruses carrying the control vectors (Fig. 2a, WT & Vector). In contrast, stably transfectants of BxPc-3 cells with the cytoplasmic PARP-1 mutants exhibited increased PARP-1 expression in the cytoplasm (Fig. 2a, R208Q, K222I). Of note, overexpression of WT or cytoplasmic mutants of PARP-1 did not affect the expression or localization of DR5 (Fig. 2a, DR5). Western blot analysis of the total protein extracts demonstrated increased total PARP-1 expression in WT and cytoplasmic mutants of PARP-1-overexpressed cells, compared to those in non-infected (Control) or vector-infected (Vector) BxPc-3 cells (Fig. 2b, Cell lysate). Further analysis of PARP-1 expression in the cytoplasmic and nuclear fractions confirmed a marked increase of cytoplasmic PARP-1 in the PARP-1 R208Q and K222I-overexpressed BxPc-3 cells (Fig. 2b, Cytoplasmic), while the majority of PARP-1 expression was identified in the nuclear fraction of cells overexpres-sing WT PARP-1 (Fig. 2b, Nuclear).
Figure 2.
Effects of nuclear and cytoplasmic PARP-1 on TRA-8-induced apoptosis in sensitive pancreatic cancer cells. (a,b) BxPc-3 cells were infected with lentiviruses carrying wild-type (WT), cytoplasmic mutant PARP-1 (R208Q and K222I), or control vector (Vector). Stable transfectants were selected by puromycin. (a) The expression and localization of PARP-1 was determined by immunofluorescence staining (PARP1, red). Immunofluorescence staining of DR5 expression (DR5, green) and DAPI staining for nuclei (DAPI, blue) are also shown. Scale bars = 100 μm. (b) Western blotting analysis of PARP-1 in total, cytoplasmic and nuclear protein extracts. The expression of GAPDH or Lamin B was used as a loading control. (c–f) TRA-8-induced apoptosis in stably infected BxPc-3 (c, d) and MiaPaCa-2 (e, f) cells. (c and e) Apoptosis of cells treated with vehicle (Control) or TRA-8 (1 μg/mL) for 24 h, as determined by Annexin V and PI staining analyzed by flow cytometry. Results shown are means ± SD of three experiments performed in duplicate (*p < 0.01, compared to the TRA-8-treated cells in the Vector group). (d and f) Western blot analysis of the expression of caspase-8 (Casp8) and caspsae-3 (Casp3). The expression of β-actin was used as a loading control. Representative blots of three independent experiments are shown.
The effect of nuclear and cytoplasmic PARP-1 on TRA-8-induced apoptosis was determined in two TRA-8 sensitive pancreatic cancer cells, BxPc-3 and MiaPaCa-2 that express low endogenous PARP-1. Overexpression of WT PARP-1 in BxPc-3 cells, mostly localized in the nucleus, did not significantly inhibit TRA-8-induced apoptosis, which was similar to TRA-8-induced apoptosis in BxPc-3 cells carrying the control vector (Fig. 2c, Vector, WT). Consistent with this, WT PARP-1 did not affected TRA-8-induced cleavage and activation of casepase-8 and caspase-3 (Fig. 2d, Vector, WT). Significant inhibition of TRA-8-induced apoptosis was demonstrated in BxPc-3 cells overexpressing the cytoplasmic PARP-1 mutants (Fig. 2c, R208Q, K222I), which was associated with inhibition of TRA-8-induced activation of caspase-8 and caspase-3 (Fig. 2d, R208Q, K222I). Overexpression of cytoplasmic PARP-1, but not WT PARP-1, inhibited TRA-8-induced apoptosis (Fig. 2e) and activation of caspase-8 and caspase-3 (Fig. 2f) in the MiaPaCa-2 cells. Results from these studies support a direct effect of cytoplasmic PARP-1, but not nuclear PARP-1, on TRA-8-induced apoptosis.
Cytoplasmic PARP-1 increases the resistance of pancreatic cancer to TRA-8 therapy in vivo
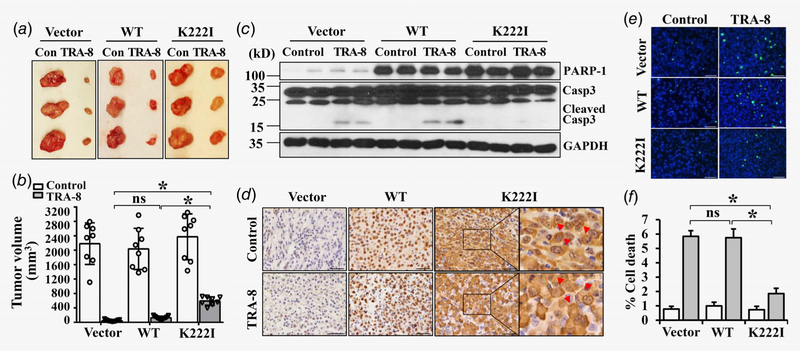
The effects of nuclear and cytoplasmic PARP-1 on pancreatic cancer tumorigenesis and their sensitivity to TRAIL therapy was further determined in a mouse xenograft model, using BxPc-3 cells stably overexpressing Vector, WT PARP-1 and PARP-1 cytoplasmic mutant (K222I). We did not find a significant difference in tumorigenesis of the BxPc-3 cells overex-pressing Vector, WT or K222I mutant PARP-1 (Fig. 3a, Con; Fig. 3b, Control). Consistently with its apoptotic effects in vitro, TRA-8 therapy markedly reduced tumor sizes in the sensitive BxPc-3 cells expressing Vector (Figs. 3a and 3b, Vector). Overexpression of WT PARP-1 did not affect the sensitivity to TRA-8 treatment (Figs. 3a and 3b, WT); whereas overexpression of cytoplasmic PARP-1 rendered BxPc-3 cells more resistant to TRA-8 therapy (Figs. 3a and 3b, K222I). Western blot analysis confirmed increased expression of PARP-1 in isolated tumors derived from BxPc-3 cells with WT or cytoplasmic mutant PARP-1; which was not affected by TRA-8 treatment (Fig. 3c, PARP-1). Immunohistochemical analysis of PARP-1 localization in tumors from each group further verified the nuclear (Fig. 3d, Vector, WT) and cytoplasmic localization (Fig. 3d, K222I, red arrows) of PARP-1, which was not affected by TRA-8 treatment. Consistent with larger tumor sizes, inhibition of TRA-8-induced caspase-3 activation was demonstrated in tumors derived from BxPc-3 cells with PARP-1 cytoplasmic mutant (Fig. 3c, TRA-8 groups, cleaved Casp3). Furthermore, TUNEL staining of tumor sections revealed that cytoplasmic PARP-1 overexpression significantly reduced TRA-8-induced cell death, compared to the Vector and WT PARP-1 (Figs. 3e and 3f). Altogether, these studies provide in vivo evidence that support a causative role of cytoplasmic PARP-1 in regulating the resistance of pancreatic cancer cells to TRA-8-induced apoptosis that contribute to the sensitivity of pancreatic cancer to TRAIL therapy.
Figure 3.
Cytoplasmic PARP-1 increases resistance of sensitive pancreatic cancer to TRA-8 therapy in mice. BxPc-3 cells stably infected with Vector, wild-type PARP-1 (WT) or the PARP-1 cytoplasmic mutant (K222I) were injected into nude mice, which were then subjected to control vehicle (Control, Con) or TRA-8 treatment for 6 weeks. (a) Representative tumors; and (b) Tumor volumes in each group are shown (n = 8–9/ group, ns = not significant, *p < 0.01). (c) Western blot analysis of the expression of PARP-1 and activation of caspsase-3 (Casp3) in representative tumors in each group. The expression of GAPDH was used as a loading control. (d) Immunohistochemical staining of PARP-1 in representative tumor sections from each group (Scale bar = 50 μm). Higher magnification of the boxed area are shown to the right. Red arrows indicate cytoplasmic localization of PARP-1 in tumors with the K222I PARP-1 mutants. (e and f) Cell death in tumors from each group was analyzed by TUNEL staining. (e) Representative TUNEL staining images from each group (Scale bar = 50 μm). (f) Quantitative analysis of TUNEL positive cells as percentage of total cells in the tumor sections (n = 5, *p < 0.01).
Inhibition of PARP activity attenuates the inhibitory effects of cytoplasmic PARP-1 on TRA-8-induced apoptosis
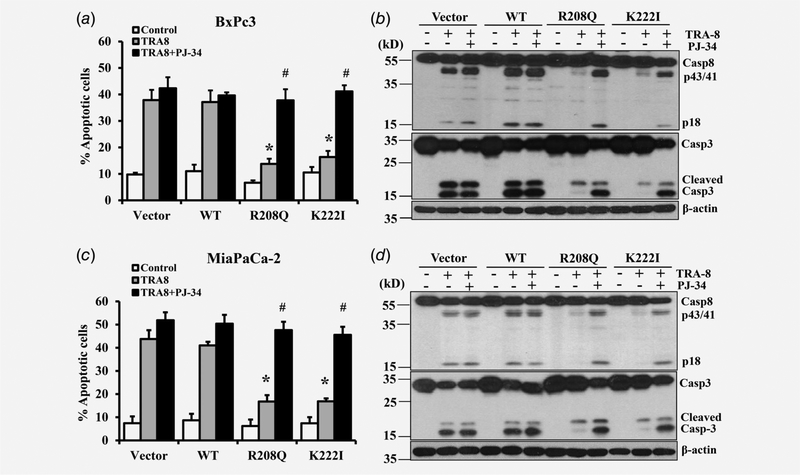
To understand the molecular mechanisms underlying cytoplasmic PARP-1 regulation of TRA-8-induced apoptosis, we first evaluated whether the poly(ADP-ribosyl)ation activity of PARP-1 was required. With the use of PJ-34, a pharmacologic inhibitor of PARP activity, we demonstrated that inhibition of PARP activity restored TRA-8-induced apoptosis in the cytoplasmic PARP-1 overexpressing BxPc-3 cells as well as MiaPaCa-2 cells (Figs. 4a and 4c, R208Q, K222I black bars). Consistently, cytoplasmic PARP-1 inhibited TRA-8-induced activation of caspase-8 and caspase-3 in BxPc-3 and MiaPaCa-2 cells (Figs. 4b and 4d, R208Q, K222I with TRA-8), which was abolished by PJ-34 (Figs. 4b and 4d, R208Q, K222I with TRA-8 + PJ-34). Of note, PJ-34 did not affect TRA-8-induced apoptosis in BxPc-3 or MiaPaCa-2 cells stably expressing Vector or WT PARP-1, but only blocked the inhibitive effects of cytoplasmic PARP-1 on TRA-8-induced apoptosis. These results support that poly(ADP-ribosyl)ation activity is important for the function of cytoplasmic PARP-1 in inhibiting TRA-8-induced apoptosis.
Figure 4.
Inhibition of PARP activity on the effects of cytoplasmic PARP-1 on TRA-8-induced apoptosis. BxPc-3 (a, b) and MiaPaCa-2 (c, d) cells stably infected with Vector, wild-type PARP-1 (WT) or PARP-1 cytoplasmic mutants (R208Q, K222I) were treated with vehicle (Control), TRA-8 (1 μg/mL), or TRA-8 with PARP inhibitor (PJ-34, 20 μM) for 24 h. (a and c) Apoptosis was analyzed by flow cytometry. Results shown are means ± SD of three experiments performed in duplicates (*p < 0.01, compared to the TRA-8-treated cells in the Vector group or WT group; #, no difference compared to TRA-8 + PJ-34-treated cells in the Vector group or WT group). (b and d) Western blot analysis of the expression of caspase-8 (Casp8) and caspsae-3 (Casp3) in each group. The expression of β-actin was used as a loading control. Representative blots of three independent experiments are shown.
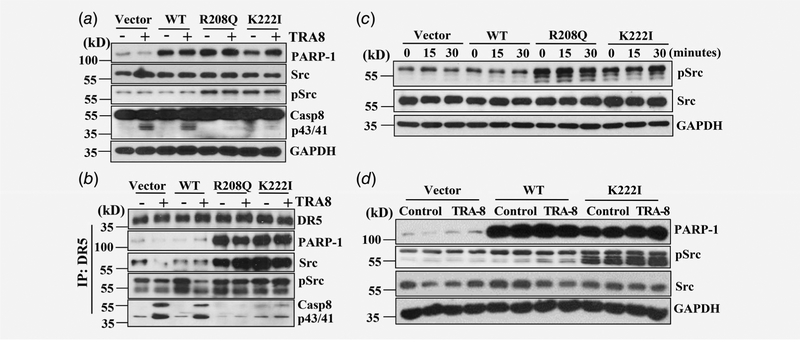
Cytoplasmic PARP-1 promotes activation of Src survival signaling
We have previously shown that activation of survival signals, such as Src kinase signaling, contributed to death receptor-mediated cell survival. Accordingly, we determine whether cytoplasmic PARP-1 may affect the Src survival signals. In addition to inhibiting TRA-8-activated apoptotic signals, such as caspase-8, cytoplasmic PARP-1 markedly induced activation of Src, as evident by increased phosphorylation of Src in BxPc-3 cells overexpres-sing the PARP-1 R208Q and K222I cytoplasmic mutants (Fig. 5a, pSrc). Analysis of the DR5-associated DISC revealed that overex-pressing cytoplasmic PARP-1, but not the WT PARP-1, led to increased recruitment of PARP-1 in the DR5-associated DISC (Fig. 5b, PARP-1). Furthermore, increased recruitment of Src and activation of Src in the DISC was demonstrated in the BxPc-3 cells overexpressing cytoplasmic PARP-1 mutants (Fig. 5b, Src, pSrc). Importantly, TRA-8 treatment decreased Src activation in BxPc-3 cells overexpressing the Vector or WT PARP-1; whereas cytoplasmic PARP-1 overexpression sustained Src activation regardless of TRA-8 treatment (Fig. 5b, pSrc), suggesting that cytoplasmic PARP-1-induced sustained Src activation in the DISC may contributed to its inhibitory effects on TRA-8-induced apoptosis. Consistent with this, total Src expression was not affected by PARP-1 overexpression (Fig. 5c, Src), however, increased Src activation was observed in the BxPc-3 cells overex-pressing the cytoplasmic PARP-1 at basal level (Fig. 5c, time 0); and this activation was sustained in response to TRA-8 treatment (Fig. 5c, 15 and 30 min). Analysis of proteins extracted from in vivo xenografted tumors further demonstrated increased Src activation in tumors derived from BxPc-3 cells overexpressing cytoplasmic PARP-1, compared to tumors derived from BxPc-3 cells expressing Vector or WT PARP-1; while total Src expression was unaffected (Fig. 5d). Similar to the in vitro observations, increased Src activation was sustained in the cytoplasmic PARP-1 overexpressing tumors after TRA-8 treatment (Fig. 5d).
Figure 5.
Effects of nuclear and cytoplasmic PARP-1 expression on TRA-8-induced survival signaling in the DISC. BxPc-3 cells were stably infected with Vector, wild-type PARP-1 (WT) or PARP-1 cytoplasmic mutants (R208Q, K222I). (a and b) Cells were exposed to vehicle or TRA-8 (1 μg/mL) for 30 min. The expression and activation of survival signal, Src, and apoptotic signal, caspase-8 (Casp8), was determined by Western blot analysis (a) in cell lysates; or (b) in DR5-associated complex immunoprecipitated with DR5 antibody. Representative blots from three independent experiments are shown. (c) Western blot analysis of Src activation in response to TRA-8 treatment at different time points. Representative results from three independent experiments are shown. (d) Western blot analysis of Src activation in representative xenografted tumors derived from BxPc-3 cells overexpressing Vector, wild-type (WT) or cytoplasmic mutant PARP-1 (K222I). The expression of GAPDH was used as a loading control.
PARP-1 knockdown inhibits activation of survival signal and induces apoptotic signaling
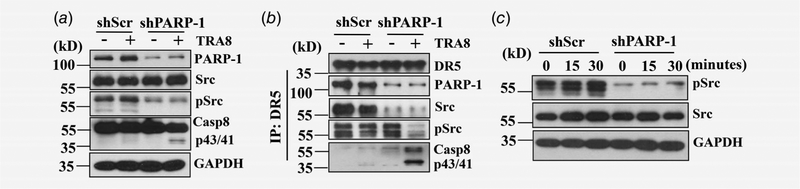
To further determine whether PARP-1 is required for TRA-8-induced activation of Src or caspase-8, we used shRNA for PARP-1 (shPARP-1) to generate PARP-1 knockdown in PANC-1 pancreatic cancer cells, which we have shown previously express high levels of PARP-1 and are resistant to TRA-8-induced apoptosis.13 Compared to PANC-1 cells infected with control scrambled shRNA (shScr), decreased PARP-1 expression was associated with TRA-8-induced inhibition of Src activation and increased caspase-8 activation (Fig. 6a). In contrast to the effects of cytoplasmic PARP-1 overexpression, knockdown of PARP-1 blocked DR5-associated DISC recruitment of Src; and resulted in decreased Src activation and increased caspase-8 activation upon TRA-8 treatment (Fig. 6b). Consistent with this, inhibition of Src activation was demonstrated in cell lysates from the PARP-1 knockdown PANC-1 cells (Fig. 6c), supporting an essential role of PARP-1 expression in regulating Src activation.
Figure 6.
Effects of PARP-1 knockdown on activation of survival and apoptotic signals. TRA-8 resistant PANC-1 cells were stably infected with shRNA for PARP-1 (shPARP-1) or control scrambled shRNA (shScr). (a) Cells were exposed to control or TRA-8 (1 μg/mL) for 30 min. The expression and activation of survival signal, Src, and apoptotic signal, caspase-8 (Casp8), was determined by Western blot analysis in (a) cell lysates; or (b) in DR5-associated complex immunoprecipitated by DR5 antibody. Representative blots from three independent experiments are shown. (c) Western blot analysis of Src activation in response to TRA-8 treatment at different time point. The expression of GAPDH was used as a loading control. Representative results from three independent experiments are shown.
Discussion
The present studies uncovered a new role of cytoplasmic expression of PARP-1 in regulating DR5-activated apoptosis in pancreatic cancer cells in vitro as well as regulating the sensitivity of pancreatic tumors to TRA-8 therapy in vivo. These results have provided the first evidence for the novel functions of cytoplasmic PARP-1 in regulating extrinsic apoptosis pathways, which is distinct from the known nuclear DNA repairing function of PARP-1 and its effect on the intrinsic apoptosis pathways.
The link between PARP-1 and the intrinsic apoptosis has been well documented. PARP-1 is a highly abundant nuclear protein that is activated when DNA is damaged28; and the majority of studies on PARP-1 have focused on its function in the nucleus, including DNA repair, chromatin remodeling, transcriptional regulation.28,29 The function of PARP-1 is essential for the repair of single-strand DNA breaks through the base excision repair mechanism,28 and for the maintenance of genomic integrity and survival in response to genotoxic insults.30–33 Accordingly, PARP-1 inhibition has been shown to be an effective therapeutic strategy for cancers associated with mutations in double-strand DNA repair genes or combining PARP inhibitors with DNA damaging agents, such as chemotherapy reagents or radiation. Several phase I and II clinical trials are ongoing with PARP inhibitors in combination with DNA-damaging agents such as platins, cyclophosphamide, ionizing radiation, gemcitabine and topoisomerase I poisons.28 All these investigations are targeting the nuclear DNA repair function of PARP-1, which induces the intrinsic apoptosis pathways. By inhibiting PARP-1 using a pharmacological inhibitor or siRNA, we recently demonstrated that PARP-1 inhibition did not affect pancreatic cancer cell survival at basal conditions but promoted death receptor-induced apoptosis.13 These previous studies suggested a role of PARP-1 in regulating extrinsic apoptotic pathways that might contribute to pancreatic cancer TRAIL resistance. However, it was entirely unknown whether the function of PARP-1 in regulating death receptor-activated apoptosis was mediated by cytoplasmic PARP-1 or nuclear PARP-1.
The findings in the present studies highlight the cytoplasmic localization and function of PARP-1 in regulating DR5-activated extrinsic apoptosis pathways. Using human normal and pancreatic cancer tissues, we determined increased cytoplasmic localization of PARP-1 in pancreatic cancers, while only nuclear PARP-1 expression was identified in the normal human pancreatic tissues, suggesting a role of the cytoplasmic PARP-1 in pancreatic tumor pathogenesis and progression. Previous analysis of PARP-1 protein expression in normal, preneoplastic and pancreatic tumor indicated that PARP-1 was highly expressed in acinar cells in normal and cancer tissues, but very low or undetectable levels in the ductal cells.34 Immunohistochemical assessment of PARP expression in a population-based cohort of 178 adenocarcinomas of the pancreas found that low-level nuclear expression of PARP was associated with a poor prognosis.27 Furthermore, a recent report showed that cytoplasmic PARP-1 was correlated with tumor aggressiveness, higher risk for relapse and patient death of human breast cancer.35 Accordingly, our observations are consistent with these reports, suggesting cytoplasmic PARP-1 expression may be used as an indicator to predict pancreatic cancer progression.
Although cytoplasmic localization of PARP-1 has been identified in neurons,36 smooth muscle cells,37 and cancer cells,38 the cytoplasmic functions of PARP-1 are largely unknown and its role in pancreatic cancer pathogenesis and resistance has never been explored. With the use of gain-of-function expression of wild-type or cytoplasmic mutants of PARP-1, we discovered a unique role of cytoplasmic PARP-1 in regulating DR5-induced apoptosis. Importantly, wild-type PARP-1 did not significantly inhibit DR5-induced apoptosis, which was consistent with the observation that only a small amount of the WT PARP-1 localized to the cytoplasmic fraction while the majority of WT PARP-1 was determined in the nucleus. The in vivo studies using mouse xenograft model of pancreatic cancers further demonstrated that cytoplasmic PARP-1, but not wild-type PARP-1 (nuclear) rendered TRA-8-sensitive pancreatic tumors more resistant to TRA-8 therapy by inhibiting TRA-8-induced apoptosis. Therefore, results from both in vitro and in vivo studies support the notion that the cellular localizations of PARP-1 determine its functions in regulating distinct cellular functions, including the intrinsic or extrinsic apoptotic pathways.
It remains not fully clear how PARP-1 expression and cellular localization are regulated. Point mutations in the PARP-1 nuclear localization domain, such as R208 or K222, have been shown to lead to increased cytoplasmic expression of PARP-1.23 In breast cancer cells, gain-of-function mutant p53 was associated with increased nuclear localization of PARP-1; and mutant p53 depletion increased cytoplasmic and decreased chromatin-associated nuclear PARP-1 protein.39 In addition, HIV-1 viral protein R-glucocorticoid receptor complex has been shown to interact with PARP-1, which prevents PARP-1 nuclear localization.40 We found that increased cytoplasmic PARP-1 was recruited into the DR5-associated DISC, an important cellular complex that controls downstream apoptotic and survival signals. As inhibition of PARP-1 enzymatic activity blocked the inhibitory effects of cytoplasmic PARP-1 on TRA-8-induced apoptosis, it is postulated that cytoplasmic PARP-1 may function in the DR5-associated DISC via pADPr modification of DISC proteins. We previously found that PARP-1 mediates pADPr modification of caspase-8, thereby inhibiting caspase-8 activation.13 Consistently, the present studies demonstrated a direct regulation of cytoplasmic PARP-1 on TRA-8-induced activation of caspase-8 and the downstream effector caspase-3. In contrast, wild-type PARP-1, which was mainly localized in the nucleus, was not recruited into the DR5-associated DISC, and had no effects on TRA-8-indiced activation of caspase-8 and caspase-3. Accordingly, inhibiting the activation of the apoptotic signal, caspase-8, in the DISC may contribute to the inhibitory effects of cytoplasmic PARP-1 on TRA-8-induced apoptosis.
In addition to the inhibition on the apoptotic signals, resistance to TRAIL-mediated apoptosis is also associated with aberrant activation of survival pathways.41 Recruitments of the survival signals into the DISC, such as the enzymatically inactive homolog of caspase-8, FLICE-like inhibitor protein, or Src kinase, may convert death receptor-activated apoptotic signals into survival signals.42 The Src family of kinases has a critical role in cell proliferation, survival and angiogenesis during tumor development.43 We have previously reported that activation of Src signaling contributes to the Fas death receptor-induced survival of pancreatic cancer cells.22 Increased Src activation has been associated with TRAIL resistance in prostate cancer and hepatic carcinoma.42 In the wild-type PARP-1 overexpressing BxPc-3 pancreatic cancer cells, we found TRA-8 treatment inhibited activation of Src survival signal and induced activation of apoptotic signal caspase-8 in the DISC, thus leading to apoptosis of the cells. Recruitment of cytoplasmic PARP-1 into the DR5-associated DISC, however, resulted in increased and sustained activation of Src survival signal and decreased caspase-8, thus conferring resistance of the sensitive BxPc-3 cells to TRA-8-induced apoptosis. Conversely, knockdown of PARP-1 attenuated Src activation in the DR5-associated DISC, suggesting that PARP-1 recruitment in the DISC is essential for the activation of Src in the DISC. Therefore, activation of the Src survival signaling in the DR5-assocaited DISC contributes to the inhibitory effects of cytoplasmic PARP-1 on TRA-8-induced apoptosis of pancreatic cancer cells in vitro, and the sensitivity of pancreatic tumors to TRA-8 therapy in vivo.
In summary, our studies have demonstrated increased cytoplasmic expression and localization of PARP-1 in human pancreatic tumors; and uncovered a new critical function of cytoplasmic PARP-1 in promoting resistance of pancreatic cancer cells to TRA-8-induced extrinsic apoptosis in vitro and TRA-8 therapy of pancreatic tumors in vivo. Furthermore, we have elucidated a novel function of cytoplasmic PARP-1, distinct from the known function of nuclear PARP-1, in promoting DISC-recruitment and activation of Src survival signaling and inhibition of the apoptotic mediator, caspase-8, thus conferring resistance of the pancreatic cancer cells to TRA-8-induced apoptosis. These results support the potential use of cytoplasmic PARP-1 as an indicator to predict pancreatic tumor progression and TRAIL sensitivity; and support an innovative application of the currently available PARP inhibitors or designing new inhibitors targeting cytoplasmic PARP-1 to enhance the efficacy of TRAIL therapy for TRAIL-resistant pancreatic cancers.
What’s new?
Poly(ADP-ribose) polymerases (PARP) play important roles in repairing damaged DNA during intrinsic cell death. Recently, PARP-1 was also linked to death receptor (DR)-activated extrinsic apoptosis. This study uncovers a new role of cytoplasmic expression of PARP-1 in regulating DR5-activated apoptosis in pancreatic cancer cells in vitro as well as regulating the sensitivity of pancreatic tumors to TRA-8 therapy in vivo. The findings highlight cytoplasmic PARP-1 as a potential new marker for pancreatic cancer progression and support an innovative application of currently available PARP inhibitors or the design of new inhibitors targeting cytoplasmic PARP-1 to enhance the efficacy of DR targeted therapy.
Grant sponsor:
US Department of Veterans Affairs, Office of Research and Development;
Grant numbers: BX002296, BX003617
Footnotes
Conflict of interest: The authors have no potential conflicts of interest to disclose.
References
- 1.Raimondi S, Maisonneuve P, Lowenfels AB. Epidemiology of pancreatic cancer: an overview. Nat Rev Gastroenterol Hepatol 2009;6:699–708. [DOI] [PubMed] [Google Scholar]
- 2.Hartman DJ, Krasinskas AM. Assessing treatment effect in pancreatic cancer. Arch Pathol Lab Med 2012;136:100–9. [DOI] [PubMed] [Google Scholar]
- 3.Saif MW. Controversies in the adjuvant treatment of pancreatic adenocarcinoma. JOP 2007;8: 545–52. [PubMed] [Google Scholar]
- 4.Moore MJ, Goldstein D, Hamm J, et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada clinical trials group. J Clin Oncol 2007; 25:1960–6. [DOI] [PubMed] [Google Scholar]
- 5.Conroy T, Desseigne F, Ychou M, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med 2011;364:1817–25. [DOI] [PubMed] [Google Scholar]
- 6.Schutze S, Tchikov V, Schneider-Brachert W. Regulation of TNFR1 and CD95 signalling by receptor compartmentalization. Nat Rev Mol Cell Biol 2008;9:655–62. [DOI] [PubMed] [Google Scholar]
- 7.Fulda S, Debatin KM. Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy. Oncogene 2006;25:4798–811. [DOI] [PubMed] [Google Scholar]
- 8.Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 2009;361: 123–34. [DOI] [PubMed] [Google Scholar]
- 9.Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol 2012; 13:411–24. [DOI] [PubMed] [Google Scholar]
- 10.Goncalves A, Finetti P, Sabatier R, et al. Poly (ADP-ribose) polymerase-1 mRNA expression in human breast cancer: a meta-analysis. Breast Cancer Res Treat 2011;127:273–81. [DOI] [PubMed] [Google Scholar]
- 11.Fogelman DR, Wolff RA, Kopetz S, et al. Evidence for the efficacy of Iniparib, a PARP-1 inhibitor, in BRCA2-associated pancreatic cancer. Anticancer Res 2011;31:1417–20. [PubMed] [Google Scholar]
- 12.Melisi D, Ossovskaya V, Zhu C, et al. Oral poly(ADP-ribose) polymerase-1 inhibitor BSI-401 has antitumor activity and synergizes with oxaliplatin against pancreatic cancer, preventing acute neurotoxicity. Clin Cancer Res 2009;15:6367–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan K, Sun Y, Zhou T, et al. PARP-1 regulates resistance of pancreatic cancer to TRAIL therapy. Clin Cancer Res 2013;19:4750–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahmood Z, Shukla Y. Death receptors: targets for cancer therapy. Exp Cell Res 2010;316:887–99. [DOI] [PubMed] [Google Scholar]
- 15.Dimberg LY, Anderson CK, Camidge R, et al. On the TRAIL to successful cancer therapy? Predicting and counteracting resistance against TRAIL-based therapeutics. Oncogene 2013;32:1341–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holoch PA, Griffith TS. TNF-related apoptosis-inducing ligand (TRAIL): a new path to anti-cancer therapies. Eur J Pharmacol 2009;625:63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kindler HL, Richards DA, Garbo LE, et al. A randomized, placebo-controlled phase 2 study of ganitumab (AMG 479) or conatumumab (AMG 655) in combination with gemcitabine in patients with metastatic pancreatic cancer. Ann Oncol 2012;23:2834–42. [DOI] [PubMed] [Google Scholar]
- 18.Rajeshkumar NV, Rasheed ZA, Garcia-Garcia E, et al. A combination of DR5 agonistic monoclonal antibody with gemcitabine targets pancreatic cancer stem cells and results in long-term disease control in human pancreatic cancer model. Mol Cancer Ther 2010;9:2582–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiezorek J, Holland P, Graves J. Death receptor agonists as a targeted therapy for cancer. Clin Cancer Res 2010;16:1701–8. [DOI] [PubMed] [Google Scholar]
- 20.Hinz S, Trauzold A, Boenicke L, et al. Bcl-XL protects pancreatic adenocarcinoma cells against. Oncogene 2000;19:5477–86. [DOI] [PubMed] [Google Scholar]
- 21.Satoh K, Kaneko K, Hirota M, et al. Tumor necrosis factor-related apoptosis-inducing ligand and its receptor expression and the pathway of apoptosis in human pancreatic cancer. Pancreas 2001;23: 251–8. [DOI] [PubMed] [Google Scholar]
- 22.Yuan K, Jing G, Chen J, et al. Calmodulin mediates Fas-induced FADD-independent survival signaling in pancreatic cancer cells via activation of Src-extracellular signal-regulated kinase (ERK). J Biol Chem 2011;286:24776–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schreiber V, Molinete M, Boeuf H, et al. The human poly(ADP-ribose) polymerase nuclear localization signal is a bipartite element functionally separate from DNA binding and catalytic activity. EMBO J 1992;11:3263–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Byon CH, Sun Y, Chen J, et al. Runx2-upregulated receptor activator of nuclear factor kappaB ligand in calcifying smooth muscle cells promotes migration and osteoclastic differentiation of macrophages. Arterioscler Thromb Vasc Biol 2011;31:1387–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ichikawa K, Liu W, Zhao L, et al. Tumoricidal activity of a novel anti-human DR5 monoclonal antibody without hepatocyte cytotoxicity. Nat Med 2001;7:954–60. [DOI] [PubMed] [Google Scholar]
- 26.Pawar P, Ma L, Byon CH, et al. Molecular mechanisms of tamoxifen therapy for cholangiocarcinoma: role of calmodulin. Clin Cancer Res 2009; 15:1288–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klauschen F, von WM, Stenzinger A, et al. High nuclear poly-(ADP-ribose)-polymerase expression is prognostic of improved survival in pancreatic cancer. Histopathology 2012;61:409–16. [DOI] [PubMed] [Google Scholar]
- 28.Rouleau M, Patel A, Hendzel MJ, et al. PARP inhibition: PARP1 and beyond. Nat Rev Cancer 2010;10:293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krietsch J, Rouleau M, Pic E, et al. Reprogramming cellular events by poly(ADP-ribose)-binding proteins. Mol Aspects Med 2013;34:1066–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Murcia JM, Niedergang C, Trucco C, et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci USA 1997;94:7303–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samper E, Goytisolo FA, Menissier-de MJ, et al. Normal telomere length and chromosomal end capping in poly(ADP-ribose) polymerasedeficient mice and primary cells despite increased chromosomal instability. J Cell Biol 2001;154:49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trucco C, Oliver FJ, de MG, et al. DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res 1998;26:2644–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vodenicharov MD, Sallmann FR, Satoh MS, et al. Base excision repair is efficient in cells lacking poly(ADP-ribose) polymerase 1. Nucleic Acids Res 2000;28:3887–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez-Bosch N, Iglesias M, Munne-Collado J, et al. Parp-1 genetic ablation in Ela-myc mice unveils novel roles for Parp-1 in pancreatic cancer. J Pathol 2014;234:214–27. [DOI] [PubMed] [Google Scholar]
- 35.von MG, Muller BM, Loibl S, et al. Cytoplasmic poly(adenosine diphosphate-ribose) polymerase expression is predictive and prognostic in patients with breast cancer treated with neoadjuvant chemotherapy. J Clin Oncol 2011;29:2150–7. [DOI] [PubMed] [Google Scholar]
- 36.Cookson MR, Ince PG, Usher PA, et al. Poly (ADP-ribose) polymerase is found in both the nucleus and cytoplasm of human CNS neurons. Brain Res 1999;834:182–5. [DOI] [PubMed] [Google Scholar]
- 37.Ertsey R, Chapin CJ, Kitterman JA, et al. Ontogeny of poly(ADP-ribose) polymerase-1 in lung and developmental implications. Am J Respir Cell Mol Biol 2004;30:853–61. [DOI] [PubMed] [Google Scholar]
- 38.Rossi MN, Carbone M, Mostocotto C, et al. Mitochondrial localization of PARP-1 requires interaction with mitofilin and is involved in the maintenance of mitochondrial DNA integrity. J Biol Chem 2009;284:31616–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polotskaia A, Xiao G, Reynoso K, et al. Proteome-wide analysis of mutant p53 targets in breast cancer identifies new levels of gain-of-function that influence PARP, PCNA, and MCM4. Proc Natl Acad Sci USA 2015;112:E1220–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muthumani K, Choo AY, Zong WX, et al. The HIV-1 Vpr and glucocorticoid receptor complex is a gain-of-function interaction that prevents the nuclear localization of PARP-1. Nat Cell Biol 2006;8:170–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ricci MS, Jin Z, Dews M, et al. Direct repression of FLIP expression by c-myc is a major determinant of TRAIL sensitivity. Mol Cell Biol 2004;24: 8541–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Azijli K, Weyhenmeyer B, Peters GJ, et al. Non-canonical kinase signaling by the death ligand TRAIL in cancer cells: discord in the death receptor family. Cell Death Differ 2013;20:858–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim LC, Song L, Haura EB. Src kinases as therapeutic targets for cancer. Nat Rev Clin Oncol 2009;6:587–95. [DOI] [PubMed] [Google Scholar]