Abstract
Background: Radiotherapy is one of the most common treatments for esophageal squamous cell carcinoma (ESCC). Radioresistance is a major obstacle that limits the efficacy of radiotherapy. H19 has been considered as a factor affecting radioresistance, whereas the specific mechanism of H19 in ESCC radioresistance remains to be further elucidated.
Purpose: The objective of this study was to identify the relationship between H19 and radioresistance. The findings are expected to provide new insights into the treatment of radioresistant ESCC.
Methods: The expression levels of H19 in ESCC was analyzed using the online database starBase. The Oncomine database was used to further verify the association between H19 expression and patient age, gender, and tumor stage. The overall survival rates of ESCC patients were analyzed using the KM plotter database. Clonogenic survival was conducted to identify the value of survival fraction. The optical density values were obtained via MTS assays. Cells migration and stemness were observed through Transwell and sphere formation assays. The expression levels of H19, miR-22-3p and WNT1 were analyzed using qPCR.
Results: In our study, we firstly screened the H19 according to the online database starBase, and then the Oncomine database and KM plotter database showed that H19 expression was significantly upregulated in the ESCC tissues and associated with poor prognosis. Secondly, an ESCC radioresistant cell line, KYSE150R was established. Clonogenic survival showed that radiation decreased the value of survival fraction. MTS assays suggested that optical density values in KYSE150R cells were significantly higher than that in KYSE150 cells. Transwell and sphere formation assays showed radiation enhanced cell migration and stemness in ESCC cells. In addition, qPCR showed that H19 was upregulated in KYSE150R cells, and survival fraction assays showed that knockdown of H19 decreased the survival fraction values. MTS assays, migration and invasion assays suggested that H19 inhibited cells proliferation, migration and stemness in radioresistant KYSE150 cells. Moreover, qPCR assay showed that miR-22-3p expression levels was downregulated, but WNT1 was upregulated in KYSE150R cells as well as protein levels. Luciferase activity assay further showed that miR-22-3p inhibits the WNT1 expression.
Conclusion: Our results demonstrate that H19 knockdown downregulates the WNT1 via upregulating miR-22-3p expression, which leads to the inhibition of cells proliferation, migration and stemness in the radioresistant ESCC cells.
Keywords: ESCC, radioresistance, KYSE150R, H19, miR-22-3p, WNT1
Introduction
Esophageal cancer (EC) is a malignant cancer that occurs in the esophageal epithelial tissue, and is common tumor and one of leading causes of mortality worldwide.1 Based on the differences in histology, EC is divided into two main subtypes: Esophageal adenocarcinoma (EA) and Esophageal squamous cell carcinoma (ESCC).2 Generally, ESCC accounts for 90% of EC and is the predominant histological type versus EC in China.3 For ESCC treatments, radiation therapy is one of the commonly used methods, which can be used alone or in combination with chemotherapy and surgery.4 Despite great advances in radiotherapy technology, tumor recurrence and metastasis caused by radioresistant cancer cells lead to the increased mortality and the poor overall 5-year survival rate in ESCC patients.5 Currently, few clinical approaches are able to predict the effects of radiation therapy in patients with cancer or its effect on the radiosensitivity of cancer cells. Thus, it is significant to investigate mechanisms that promote radiation resistance in ESCC. Meanwhile, identifying more biomarkers that involved in ESCC are expected to improve the efficacy of radiotherapy.
Cancer stem cells (CSCs), accounting for a small part of the tumor mass, are a small proportion of tumor cells with the capabilities of self-renewal and differentiation.6 CSCs have been identified as the main contributors for the recurrence and metastasis in tumors.7–10 In addition, the correlation between lncRNAs and CSCs-related transcriptional factors expression (including OCT4, SOX2 and NANOG) has been unveiled.11,12 More importantly, lncRNAs are associated with stemness, such as, B4GALT1-AS1 promotes colon cancer cell stemness.13 NEAT1 enhances the radioresistance of cervical cancer.14 FAM83H-AS1 contributes to the radioresistance, proliferation, and metastasis in ovarian cancer.15
Using bioinformatic analysis, it showed the aberrant expression of H19 in ESCC tissues and the correlation between H19 and poor prognosis. H19, an imprinted lncRNA, is one of the first identified lncRNAs.16 Accumulating evidence reveals that the dysregulated H19 acts as the tumorigenic and anti-tumorigenic roles in many tumors, such as bladder cancer,17 breast cancer.18 Huang et al have reported that H19 are associated with cell invasion and metastasis in EC.19 It has been found that H19 plays some certain role in the alteration of radio/chemoresistance in hepatocellular carcinoma cells.6 H19 also is associated with poor prognosis in breast cancer patients and promotes cancer stemness.20 In addition, H19 interacted with miR-130a-3p and miR-17-5p to modify radioresistance.21
Many reports have revealed that the implication of Wnt signaling in various cancers.22,23 Meanwhile, researchers also revealed its roles in radioresistance, such as LIG4 mediates Wnt signalling-induced radioresistance;24 microRNA-324-3p regulates nasopharyngeal carcinoma radioresistance by directly targeting WNT2B;25 The inhibitory role of miR-301a in cell migration and the facilitating role of miR-301a in radiosensitivity have been found in radioresistant‑ESCC cells.26 Wnt proteins are involved in the differentiation of stem cells via the Wnt/β-catenin signaling pathway27 or affect stem cell fate.28 The activation of Wnt/β-catenin signaling pathways is required for human embryonic stem cells.29 Moreover, Wnt pathway participated in inhibition of mouse embryonic stem cell differentiation.30 However, little is known about the relationship between H19 and Wnt signaling in in radioresistant ESCC cells.
In this current study, we aimed to elucidate the role of H19 in radioresistant ESCC cells. We firstly found that H19 is upregulated in the ESCC cells and associated with poor prognosis. Then, we established radio-resistant KYSE150 cells line, KYSE150R. The results showed that radiation enhanced cells migration and stemness in ESCC cells. Furthermore, we observed that knockdown of H19 inhibits cells proliferation, migration, and stemness in radioresistant KYSE150 cells. In addition, knockdown of H19 downregulates WNT1 expression. Overexpression of miR-22-3p inhibits WNT1 expression. Our findings suggested that H19 promotes cells proliferation, migration, and stemness in the ESCC radioresistant cells.
Materials and methods
Bioinformatic analysis
The expression levels of H19 in ESCC was analyzed using the online database starBase (http://starbase.sysu.edu.cn/). The publicly available Oncomine database (http://www.oncomine.com) was utilized to further verify the association between H19 expression and patient age, gender, and tumor stage. The overall survival rates of ESCC patients were analyzed using the KM plotter database (http://kmplot.com/). Additionally, the starBase database was also used to assess the combination between H19, miR-22-3p and WNT1 in ESCC cells. The binding sites of miR-22-3p in WNT1 were predicted using the bioinformatics analysis tool TargetScan.
Establishment of a radioresistant ESCC cell line and cell culture
ESCC cell line KYSE150 was purchased from the Cell Bank of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China). KYSE150 cells were cultured in RPMI-1640 medium (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS, Hyclone, Logan, USA). After reaching 50 to 60% confluence, KYSE150 cells were exposed to 2 Gy X-ray irradiation (2.5 Gy/min) using a Varian-6/100 Linear Accelerator (Varian Medical Systems, Inc., Palo Alto, CA, USA). Next, the medium was changed into the RPMI-1640 culture medium (Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS, Hyclone, Logan, USA). The culture medium was changed every two days. 15% FBS was employed when numerous dead cells were observed. Cells were further maintained in a humidified incubator with 5% CO2 (v/v). After reaching 70% to 80% confluence, cells were passaged. Repeat the above process until the total radiation reached 60 Gy. The obtained cells were designated KYSE150R cells, and further cultured for ≥2 weeks before the subsequent experiments.
H19 knockdown assays
Specific small interfering RNA (siRNAs) for H19 and negative control siRNA (NC-si) were obtained from RiboBio (Guangzhou, China). KYSE150R cells were planted into 12-well plates (1x106/well) and incubated for 12 h. H19-si or NC-si was transfected into cells using X-treme GENE transfection reagent (Roche, Basel, Switzerland) according to the manufacturer’s instructions. After 48 h transfection, total RNA was isolated and used for analysis of the mRNA and protein expression. The sequences of H19-si in this study were as follows, siRNA GACACCAUCGGAACAGCAG and NC-si 5ʹ-UUCUCCGAACGUGUCACGU-3ʹ (sense).
Clonogenic survival assays
KYSE150 and KYSE150R cells transfected with H19-si or untransfected with H19-si were plated into 6-well plates ((2x106/well). Cells were incubated at 37 °C in a humidified incubator with 5% CO2. After incubation for 24 h, cells were irradiated with 0, 2, 4, 6, or 8 Gy using X-ray irradiation. After incubation for 48 h, cells were washed twice with PBS, fixed in 70% methanol, and stained with 0.1% crystal violet. Colonies containing more than 50 cells were counted. The survival fraction of each dose was determined using the equation survival fraction = colonies counted/(cells seeded × PE) ×100%.
Cell proliferation assays
To determine the proliferative ability of cells, the MTS assays were performed. Cells were seeded into 96-well plates (5x103/well) containing 100 µL of 10% FBS/medium and incubated at 37 °C with 5% CO2. After incubation for 1 to 7 days, the MTS assays were conducted according to the manufacturer’s instructions. The cell viability was determined by measuring the optical density (OD) at 490 nm wavelength using a spectrophotometer FP-6500 (JASCO Corp.).
Transwell assays
Transwell assays were performed in 6.5 mm Transwell chambers with 8 μm pores (Corning Costar, Corning, NY, USA) to evaluate the migration ability of cells according to the manufacturer’s instructions. The upper chamber was loaded with cells in serum-free medium. The lower chamber was filled with medium containing 10% FBS. After incubation for 24 h, cells that had invaded through the membrane were fixed with methanol and stained with 0.1% crystal violet (Sigma-Aldrich, St. Louis, MO, USA). The number of cells in the lower chamber was counted using an inverted microscope (×100 magnification).
qPCR analysis
Total RNA was extracted from cells with Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. cDNA synthesis was performed using the high capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). GAPDH and U6 were used as the internal control. All qPCR assays were performed with a CFX Connect™ Real-Time PCR Detection System (BIO-RAD Laboratories, Inc., California, USA), and the fold changes were calculated by the relative quantification (2−ΔΔCt) method. The primers were shown in Table 1.
Table 1.
The primers used for qPCR were listed
| Gene | Primer sequence (5ʹ-3ʹ) | |
|---|---|---|
| H19 | Forward | TTACTGCTGCGTTTTATGTTGGG |
| Reverse | GCTGGCCGATGTGATGACTA | |
| miR-22-3p | Forward | GGGAAGCTGCCAGTTGAAG |
| Reverse | GTGCGTGTCGTGGAGTCG | |
| WNT1 | Forward | TTCAGACACGAGAGATGGAACT |
| Reverse | CCAGCCTTCACTTGCTGAG | |
| OCT4 | Forward | GTGTTCAGCCAAAAGACCATCT |
| Reverse | GGCCTGCATGAGGGTTTCT | |
| SOX2 | Forward | CTCGTGCAGTTCTACTCGTCG |
| Reverse | AGCTCTCGGTCAGGTCCTTT | |
| NANOG | Forward | TCCCGAGAAAAGATTAGTCAGCA |
| Reverse | AGTGGGGCACCTGTTTAACTT | |
| GAPDH | Forward | GGGAGCCAAAAGGGTCAT |
| Reverse | GAGTCCTTCCACGATACCAA | |
Sphere-forming assays
Cells were digested using 0.25% trypsin (Sigma); washed twice using calcium/magnesium-free phosphate-buffered saline (PBS); suspended in serum-free DMEM-F12 medium supplemented with 1% penicillin-streptomycin solution, 20 ng/mL epidermal growth factor, 10 ng/mL basic fibroblast growth factor, and 2% B27; and then seeded in a low-attachment T25 flask (Corning). Cells were collected after 5 days, and then digested using Accutase enzyme and seeded into 96-well plates. Cells were cultivated for 7–14 days, and the spheres were then assessed using a microscope.
Western bolt analysis
Cells were grown in 6-well plates (1x106) and transfected with the indicated plasmids and siRNA using Lipofectamine 2000 (Thermo Fisher Scientific). Cells were washed with precooling phosphate-buffered saline (PBS), and cells were collected and then lysed for 20 mins in cold lysis buffer (Beyotime Institute of Biotechnology). Extracts were clarified by centrifugation (12,000 rpm/min) for 30 mins at 4 °C. Protein samples were resolved by electrophoresing on 10% polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to polyvinylidene fluoride (PVDF). Membranes were blocked in Tris-buffered saline (TBS) containing 5% nonfat dry milk and 0.05% Tween-20 for 20 mins with shaking and then incubated with primary antibody following the manufacturer’s instructions for 2 hrs at room temperature. Samples were then washed with TBST (TBS-0.1% Tween-20) and incubated for 1 hr with horseradish peroxidase-conjugated secondary antibody (1:10,000 dilution). Protein bands were visualized using enhanced chemiluminescent substrate according to the manufacturer’s protocol (Millipore, Billerica, MA, USA). The primary antibodies were WNT1 and β-actin from ABclonal Biotechnology (Wuhan, P.R. China).
Luciferase activity assays
The DNA sequences of 3ʹ-UTR-wide type of WNT1 (WNT1-WT) and the corresponding mutant vector 3ʹ-UTR-mutant type of WNT1 (WNT1-MUT) were separately synthesized and cloned into the pGL-3 basic vector (Promega, Madison, WI, USA). Cells were planted into 24-well plates, and then cells were co-transfected with WNT1-WT or WNT1-MUT plasmids, miR-22-3p mimic or miR-NC (RiboBio, Guangzhou, China), and pRL-TK plasmids (Promega, Madison, WI, USA). Cells were collected and then luciferase activity were measured using the Dual-Luciferase Reportor Assay System (Promega, Madison, WI, USA).
Statistical analysis
Each experiment was repeated in triplicate. Unless other noted, all data were presented as the mean ± SD. The SPSS version 17.0 (SPSS, Chicago, IL, USA) was used to perform all statistical analyses. Unless otherwise mentioned, one-way ANOVA was used for analyzing the difference in multiply group (>2). Two-way ANOVA was used for analyzing the differences in luciferase activity assay. P<0.05 was considered statistically significant.
Results
H19 is upregulated in the ESCC and associated with poor prognosis
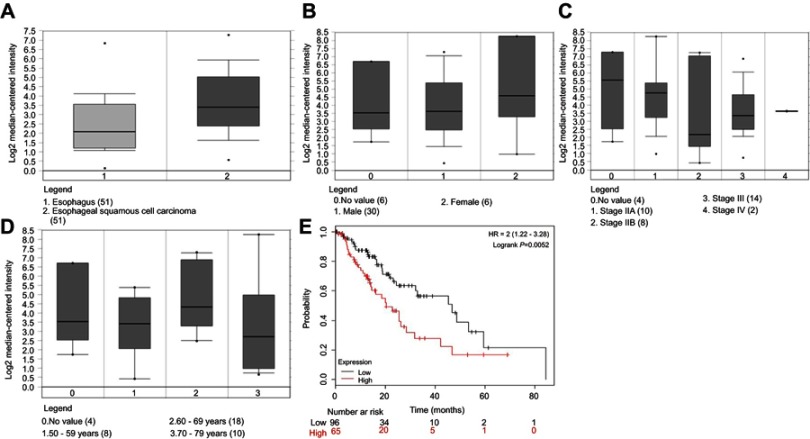
To seek more the ESCC related lncRNAs, the online database starBase was firstly used. We found that H19 was upregulated (Figure 1A). Based on the Oncomine database, we found that H19 expression had no effects on gender, clinical stage IV, and age (Figure 1B–D). However, data retrieved from the KM plotter database showed that H19 expression was associated with a poor prognosis in ESCC patients (Figure 1E, P=0.0052). Collectively, these results suggest that H19 is upregulated in the ESCC and associated with poor prognosis.
Figure 1.
H19 is upregulated in the ESCC and associated with poor prognosis. (A) Online database starBase suggested that H19 was upregulated. (B–D) Oncomine database showed that H19 expression had no effects on gender, clinical stage IV, and age. (E) The KM plotter database showed that H19 expression was associated with a poor prognosis in ESCC patients.
Radiation enhanced cells migration and stemness
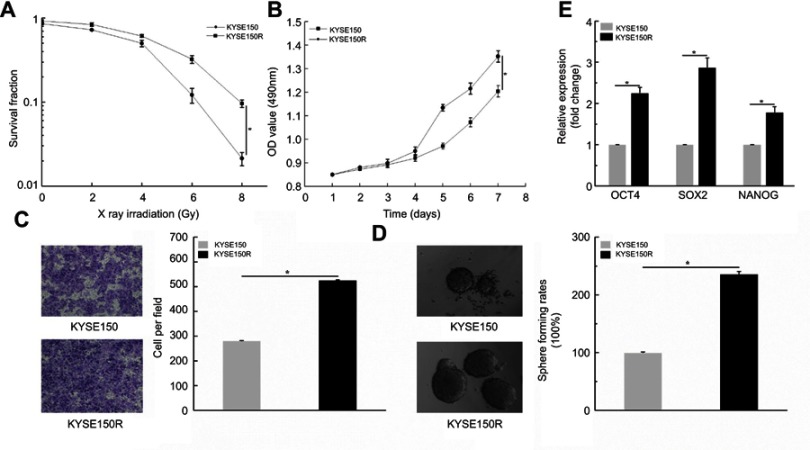
To explore the effect of radiation on ESCC cells, we established the resistant cells in KYSE150 and KYSE510 (failed). Therefore, we conducted clonogenic survival and cell viability assays to verify the resistant KYSE150 cells, KYSE150R. The results showed that radiation decreased the value of survival fraction with increasing of radiation doses (Figure 2A, P< 0.05). Subsequently, MTS assays were conducted to analyze cells viability. We observed that the OD values in KYSE150R cells were significantly higher than that in KYSE150 cells. (Figure 2B, P< 0.05). Taken together, we successfully constructed the KYSE150R cells. Therefore, we further performed related functional experiments. Transwell assays were performed. The results showed that cells migration ability was higher in KYSE150R (Figure 2C, P<0.05). Then, sphere formation assays were conducted. The results suggested that the sphere formation ability was enhanced in KYSE150R cells (Figure 2D). Furthermore, qPCR assays were performed in KYSE150R and KYSE150 cells. We found that the stemness-associated genes (OCT4, SOX2 and NANOG) were significantly higher in KYSE150R cells. (Figure 2E, P<0.05). Conjointly, these results indicate that radiation enhanced cells migration and stemness in ESCC cells.
Figure 2.
Radiation enhanced cells migration and stemness. (A) Radiation decreased the value of survival fraction with increasing of radiation doses. (B) MTS assays showed that the OD values in KYSE150R cells were significantly higher than that in KYSE150 cells. (C) Transwell assays indicated that cells migration ability was higher in KYSE150R. (D) Sphere formation assays showed that sphere formation ability was enhanced in KYSE150R cells. (E) qPCR assays implied that stemness-associated genes (OCT4, SOX2 and NANOG) were significantly higher in KYSE150R cells. *P<0.05.
Knockdown of H19 inhibits cells proliferation, migration, and stemness in radioresistant KYSE150 cells
To explore the role of H19 in ESCC radioresistant cells, qPCR assays were performed. We found that H19 expression was significantly upregulated in KYSE150R cells. (Figure 3A, P<0.05). Then, survival fraction assays were conducted. H19-si was transfected into KYSE150R. We found that knockdown of H19 decreased the survival fraction values with the increasing of radiation doses (Figure 3B, P<0.05). MTS and transwell assays were conducted. The results suggested that knockdown of H19 inhibited cells proliferation and migration (Figure 3C, D, P<0.05). In addition, the sphere formation assays were conducted. The results showed that knockdown of H19 markedly decreased the sphere formation ability of KYSE150R cells (Figure 3E, P<0.05). Moreover, the, the levels of the stemness-associated genes were identified using qPCR. We found that knockdown of H19 restrained OCT4, SOX2, and NANOG expression levels (Figure 3F, P<0.05). Taken together, these results revealed that knockdown of H19 inhibits cells proliferation, migration, and stemness in radioresistant KYSE150 cells.
Figure 3.
Knockdown of H19 inhibits cells proliferation, migration, and stemness in radioresistant KYSE150 cells. (A) qPCR assays showed that H19 expression was significantly upregulated in KYSE150R cells. (B) knockdown experiment suggested that downregulation of H19 decreased the survival fraction values with the increasing of radiation doses. (C and D) MTS and transwell assays indicated that knockdown of H19 inhibited cells proliferation and migration ability. (E) Sphere formation assays showed that knockdown of H19 markedly decreased the sphere formation ability of KYSE150R cells. (F) qPCR assays implied that knockdown of H19 restrained OCT4, SOX2, and NANOG expression levels. *P<0.05.
Knockdown of H19 downregulates WNT1 expression
Based on the online starBase database, data showed that H19 and WNT 3ʹUTR contain the binding sites of miR-22-3p (Figure 4A). Knockdown experiment was conducted. H19-si was transfected into KYSE150R cells and the results showed that knockdown of H19 increased the levels of miR-22-3p expression (Figure 4B, P< 0.05). In addition, we further analyzed the association between H19 and WNT1. The results showed that knockdown of H19 significantly decreased WNT1 expression at both the mRNA and protein levels in KYSE150R cells (Figure 4C, D). Taken together, knockdown of H19 upregulates miR-22-3p, while downregulates WNT1 expression.
Figure 4.
Knockdown of H19 downregulates WNT1 expression. (A) Online starBase database showed that H19 and WNT 3ʹUTR contain the binding sites of miR-22-3p. (B) Knockdown experiment suggested that downregulation of H19 increased the levels of miR-22-3p expression (C, D) Knockdown of H19 significantly decreased WNT1 expression at both the mRNA and protein levels in KYSE150R cells. *P<0.05.
Overexpression of miR-22-3p inhibits WNT1 expression
To further explore the relationship of miR-22-3p and WNT1, the bioinformatics analysis tool TargetScan was used. The results showed that WNT1 3ʹUTR contained the binding sites of miR-22-3p (Figure 5A). Then, qPCR assays were conducted and the results showed that miR-22-3p expression levels were downregulated, but WNT1 upregulated in KYSE150R cells. (Figure 5B, C, P< 0.05). Moreover, Western blot assays showed WNT1 protein levels were upregulated in KYSE150R. (Figure 5D). Luciferase activity assay showed a significant reduction after co-transfection of miR-22-3p mimic in WNT1-WT group, but not in the WNT1-MUT (Figure 5E, P<0.05). Furthermore, miR-22-3p mimic and NC were transfected into KYSE150 and KYSE150R cells, separately. qPCR and Western blot assays were performed to exam the WNT1 expression. The findings suggested that overexpression of miR-22-3p inhibits WNT1 expression (Figure 5F, G). Collectively, our results indicated that miR-22-3p targets WNT1 expression.
Figure 5.
Overexpression of miR-22-3p inhibits WNT1 expression. (A) TargetScan showed that WNT1 3ʹUTR contained the binding sites of miR-22-3p (B, C) qPCR assays suggested that miR-22-3p expression levels were downregulated, but WNT1 upregulated in KYSE150R cells. (D) Western blot assays showed WNT1 protein levels were upregulated in KYSE150R. (E) Luciferase activity assay showed a significant reduction after co-transfection of miR-22-3p mimic in WNT1-WT group, but no significant changes in the WNT1-MUT. (F, G) qPCR and Western blot assays showed overexpression miR-22-3p inhibits WNT1 expression. *P<0.05.
Discussion
Despite radiotherapy is a common treatment for ESCC, radioresistance always occurs and limits the use of radiotherapy. Resistance to radiation therapy leads to high tumor recurrence rates, cancer metastasis, and poor prognosis, and therefore clarifying the mechanism of radioresistance is essential to control tumor growth during radiation therapy.31 In this study, the upregulation of H19 in the ESCC tissues and the association between H19 and poor prognosis were obtained using the Oncomine database. Although the mechanisms of tumor radioresistance are complicated, studies with respect to the improvement of radiosensitization have achieved continuous progression.32
LncRNAs regulate the transcription of genes associated with DNA damage response, which is closely correlated with sensitivity to radiation therapy. The potential associations between lncRNAs and stemness have also been identified. For example, lncRNA HOTAIR regulates cell viability and radiosensitivity through inhibiting miR-218 in colorectal cancer.33 H19 is aberrantly expressed in various cancers and plays an pivotal role in enhancing stemness of cancer cells.34 CASC11 increases cancer cell stemness and predicts postoperative survival in small cell lung cancer. Moreover, previous evidence has revealed that CSCs play important roles in cancer therapy.10,35–40
In addition, the interactions between lncRNAs and miRNAs in tumor cells are implicated in tumor progression or regression.41 H19 has been reported to target miR-194-5p, which was essential for development of colorectal adenocarcinoma.42 Moreover, miRNAs have been widely identified involved in radiosensitivity. NRF2 as a new potential molecular target whose inhibition might represent a novel radiosensitizing in rhabdomyosarcoma.43 microRNA-153-3p enhances cell radiosensitivity by targeting BCL2 in human glioma.44 microRNA-16-5p enhances radiosensitivity in prostate tumor cells.45 miR-122 has been found as a tumor suppressor to influence the radioresistance in breast cancer.46 In addition,
One study showed that microRNA 301a targets WNT1 to suppress cell proliferation and migration and enhance radiosensitivity in esophageal cancer cells.
In this study, we firstly screened the H19 according to the online database starBase. Then, Oncomine database and KM plotter database were used and the data showed that H19 is upregulated in the ESCC and associated with poor prognosis. To identify the role of radiation in the resistant KYSE150 cells. The resistant KYSE150 cells, KYSE150R was firstly established. Clonogenic survival showed that radiation decreased the value of survival fraction. MTS assays suggested that OD values in KYSE150R cells were significantly higher than that in KYSE150 cells. Subsequently, we further performed related functional experiments. Transwell and sphere formation assays showed that cells migration and sphere formation ability were higher in KYSE150R, suggesting that radiation enhanced cells migration and stemness in ESCC cells. To figure out the role of H19 in ESCC radioresistant cells, qPCR firstly showed that H19 expression was significantly upregulated in KYSE150R cells. Furthermore, survival fraction assays showed that knockdown of H19 decreased the survival fraction values. A series of functional experiments indicated that H19 inhibited cells proliferation, migration and stemness in radioresistant KYSE150 cells. Moreover, starBase database suggested that H19 and WNT1 3ʹUTR contained the miR-22-3p binding sites. In KYSE150R cells, qPCR assays showed that miR-22-3p expression levels was downregulated, but WNT1 upregulated as well as protein levels after transfected with H19-si. Moreover, luciferase activity assay further showed that miR-22-3p inhibits the WNT1 expression. Our findings may implicate that knockdown of H19 downregulates the WNT1 via upregulating miR-22-3p expression, which could contribute to the inhibition of cells proliferation, migration and stemness in the radioresistant ESCC cells.
In conclusion
Our preliminary findings demonstrate that H19 promotes cells proliferation, migration, and stemness in the ESCC radioresistant cells. The more specified mechanisms remain to be elucidated and therefore additional researches with a large number of clinical samples are required for the application of targeted therapy for ESCC radioresistance. We will further investigate the potential role of H19 in ESCC radioresistance.
Acknowledgment
This work was supported by grants from The Natural Science Foundation of Anhui Province (NO. 1808085MH266).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Domper Arnal MJ, Ferrandez Arenas A, Lanas Arbeloa A. Esophageal cancer: risk factors, screening and endoscopic treatment in Western and Eastern countries. World J Gastroenterol. 2015;21(26):7933–7943. doi: 10.3748/wjg.v21.i26.7933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang W, Ma J, Zhou W, et al. Molecular mechanisms and clinical implications of miRNAs in drug resistance of esophageal cancer. Expert Rev Gastroenterol Hepatol. 2017;11(12):1. doi: 10.1080/17474124.2017.1372189 [DOI] [PubMed] [Google Scholar]
- 3.Hinrichs CS, van Way CW. Esophageal cancer. Curr Surg. 2002;59(1):12–17. [DOI] [PubMed] [Google Scholar]
- 4.Baskar R, Itahana K. Radiation therapy and cancer control in developing countries: can we save more lives? Int J Med Sci. 2017;14(1):13–17. doi: 10.7150/ijms.17288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Bras GF, Farooq MH, Falk GW, Andl CD. Esophageal cancer: the latest on chemoprevention and state of the art therapies. Pharmacol Res. 2016;113(Pt A):236–244. doi: 10.1016/j.phrs.2016.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma H, Yuan L, Li W, Xu K, Yang L. The LncRNA H19/miR-193a-3p axis modifies the radio-resistance and chemotherapeutic tolerance of hepatocellular carcinoma cells by targeting PSEN1. J Cell Biochem. 2018. doi: 10.1002/jcb.26883 [DOI] [PubMed] [Google Scholar]
- 7.Qiu H, Fang X, Luo Q, Ouyang G. Cancer stem cells: a potential target for cancer therapy. Cell Mol Life Sci. 2015;72(18):3411. doi: 10.1007/s00018-015-1920-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharifzad F, Ghavami S, Mardpour S, et al. Glioblastoma cancer stem cell biology: potential theranostic targets. Drug Resist Updat. 2019;42:35–45. doi: 10.1016/j.drup.2018.03.003 [DOI] [PubMed] [Google Scholar]
- 9.Toden S, Kunitoshi S, Cardenas J, et al. Cancer stem cell-associated miRNAs serve as prognostic biomarkers in colorectal cancer. JCI Insight. 2019;4:6. doi: 10.1172/jci.insight.122686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dzobo K, Senthebane DA, Rowe A, et al. Cancer stem cell hypothesis for therapeutic innovation in clinical oncology? taking the root out, not chopping the leaf. Omics. 2016;20(12):681–691. doi: 10.1089/omi.2016.0152 [DOI] [PubMed] [Google Scholar]
- 11.Fu Z, Chen C, Zhou Q, et al. LncRNA HOTTIP modulates cancer stem cell properties in human pancreatic cancer by regulating HOXA9. Cancer Lett. 2017;410:68. doi: 10.1016/j.canlet.2017.09.019 [DOI] [PubMed] [Google Scholar]
- 12.Pan Y, Li C, Chen J, et al. The emerging roles of long noncoding RNA ROR (lincRNA-ROR) and its possible mechanisms in human cancers. Cell Physiol Biochem. 2016;40(1–2):219–229. doi: 10.1159/000452539 [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Fang Z, Guo X, et al. lncRNA B4GALT1-AS1 promotes colon cancer cell stemness and migration by recruiting YAP to the nucleus and enhancing YAP transcriptional activity. J Cell Physiol. 2019. [DOI] [PubMed] [Google Scholar]
- 14.Han D, Wang J, Cheng G. LncRNA NEAT1 enhances the radio-resistance of cervical cancer via miR-193b-3p/CCND1 axis. Oncotarget. 2018;9(2):2395–2409. doi: 10.18632/oncotarget.23416 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Dou Q, Xu Y, Zhu Y, Hu Y, Yan Y, Yan H. LncRNA FAM83H-AS1 contributes to the radioresistance, proliferation, and metastasis in ovarian cancer through stabilizing HuR protein. Eur J Pharmacol. 2019;852:134–141. doi: 10.1016/j.ejphar.2019.03.002 [DOI] [PubMed] [Google Scholar]
- 16.Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13(3):313–316. doi: 10.1261/rna.351707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lv M, Zhong Z, Huang M, Qiang T, Rong J, Chen J. lncRNA H19 regulates epithelial–mesenchymal transition and metastasis of bladder cancer by miR-29b-3p as competing endogenous RNA. Biochim Biophys Acta. 2017;1864:10. [DOI] [PubMed] [Google Scholar]
- 18.Zhou ZR, Yang ZZ, Yu XL, Guo XM. Highlights on molecular targets for radiosensitization of breast cancer cells: current research status and prospects. Cancer Med. 2018;7(7):3110–3117. doi: 10.1002/cam4.1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang C, Cao L, Qiu L, et al. Upregulation of H19 promotes invasion and induces epithelial-to-mesenchymal transition in esophageal cancer. Oncol Lett. 2015;10(1):291. doi: 10.3892/ol.2015.3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shima H, Kida K, Adachi S, Yamada A. Lnc RNA H19 is associated with poor prognosis in breast cancer patients and promotes cancer stemness. Breast Cancer Res Treat. 2018;170(3):507–516. doi: 10.1007/s10549-018-4793-z [DOI] [PubMed] [Google Scholar]
- 21.Aalijahan H, Ghorbian S. Long non-coding RNAs and cervical cancer. Exp Mol Pathol. 2019;106:7–16. doi: 10.1016/j.yexmp.2018.11.010 [DOI] [PubMed] [Google Scholar]
- 22.Bengoa-Vergniory N, Gorrono-Etxebarria I, Lopez-Sanchez I, Marra M, Di Chiaro P, Kypta R. Identification of noncanonical Wnt receptors required for wnt-3a-induced early differentiation of human neural stem cells. Mol Neurobiol. 2017;54(8):6213–6224. doi: 10.1007/s12035-016-0151-5 [DOI] [PubMed] [Google Scholar]
- 23.Bengoa-Vergniory N, Kypta RM. Canonical and noncanonical Wnt signaling in neural stem/progenitor cells. Cell Mol Life Sci. 2015;72(21):4157–4172. doi: 10.1007/s00018-015-2028-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jun S, Jung YS, Han NS, et al. LIG4 mediates Wnt signalling-induced radioresistance. Nat Commun. 2016;7:10994. doi: 10.1038/ncomms10994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li G, Liu Y, Su Z, et al. MicroRNA-324-3p regulates nasopharyngeal carcinoma radioresistance by directly targeting WNT2B. Eur J Cancer. 2013;49(11):2596–2607. doi: 10.1016/j.ejca.2013.03.001 [DOI] [PubMed] [Google Scholar]
- 26.Su H, Wu Y, Fang Y, et al. MicroRNA‑301a targets WNT1 to suppress cell proliferation and migration and enhance radiosensitivity in esophageal cancer cells. Oncol Rep. 2019;41(1):599–607. doi: 10.3892/or.2018.6799 [DOI] [PubMed] [Google Scholar]
- 27.Foulquier S, Daskalopoulos EP, Lluri G, KCM Hermans, Deb A, Blankesteijn WM. WNT signaling in cardiac and vascular disease. Pharmacol Rev. 2018;70(1):68–141. doi: 10.1124/pr.117.013896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Camp JK, Beckers S, Zegers D, Van Hul W. Wnt signaling and the control of human stem cell fate. Stem Cell Rev. 2014;10(2):207–229. doi: 10.1007/s12015-013-9486-8 [DOI] [PubMed] [Google Scholar]
- 29.Dzobo K, Vogelsang M, Parker MI. Wnt/beta-catenin and MEK-ERK signaling are required for fibroblast-derived extracellular matrix-mediated endoderm differentiation of embryonic stem cells. Stem Cell Rev. 2015;11(5):761–773. doi: 10.1007/s12015-015-9598-4 [DOI] [PubMed] [Google Scholar]
- 30.Wang Q, Song JW, Liu Y, Zhao XX. Involvement of Wnt pathway in ethanol-induced inhibition of mouse embryonic stem cell differentiation. Alcohol. 2017;58:13–18. doi: 10.1016/j.alcohol.2016.11.006 [DOI] [PubMed] [Google Scholar]
- 31.Chi HC, Tsai CY, Tsai MM, Yeh CT, Lin KH. Roles of long noncoding rnas in recurrence and metastasis of radiotherapy-resistant cancer stem cells. Int J Mol Sci. 2017;18(9):1903. doi: 10.3390/ijms18091903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dongping W, Qiang Z, Schreiber JS, et al. Targeting mcl-1 for radiosensitization of pancreatic cancers. Transl Oncol. 2015;8(1):47–54. doi: 10.1016/j.tranon.2014.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li P, Zhang X, Wang L, et al. lncRNA HOTAIR contributes to 5FU resistance through suppressing miR-218 and activating NF-κB/TS signaling in colorectal cancer. Mol Ther Nucleic Acids. 2017;8:356–369. doi: 10.1016/j.omtn.2017.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gallula J, Schneider T, Barkali M. Oncofetal H19 RNA promotes tumor metastasis. Biochim Biophys Acta Mol Cell Res. 2014;1843(7):1414–1426. doi: 10.1016/j.bbamcr.2014.03.023 [DOI] [PubMed] [Google Scholar]
- 35.Eun K, Ham SW, Kim H. Cancer stem cell heterogeneity: origin and new perspectives on CSC targeting. BMB Rep. 2017;50(3):117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peiris-Pagès M, Martinez-Outschoorn UE, Pestell RG, Sotgia F, Lisanti MP. Cancer stem cell metabolism. Breast Cancer Res. 2016;18(1):55. doi: 10.1186/s13058-016-0712-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaheenah D, Laura A, Massimo C. Cancer stem cells: implications for cancer therapy. Oncology. 2014;28(12):1101. [PubMed] [Google Scholar]
- 38.Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23(10):1124–1134. doi: 10.1038/nm.4409 [DOI] [PubMed] [Google Scholar]
- 39.Vlashi E, Pajonk F. Cancer stem cells, cancer cell plasticity and radiation therapy. Semin Cancer Biol. 2015;31:28–35. doi: 10.1016/j.semcancer.2014.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang T, Shigdar S, Gantier MP, et al. Cancer stem cell targeted therapy: progress amid controversies. Oncotarget. 2015;6(42):44191–44206. doi: 10.18632/oncotarget.6176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng W, Shangwei N, Yunpeng Z, et al. Identification of lncRNA-associated competing triplets reveals global patterns and prognostic markers for cancer. Nucleic Acids Res. 2015;43(7):3478. doi: 10.1093/nar/gkv233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li CF, Li YC, Wang Y, Sun LB. The effect of LncRNA H19/miR-194-5p axis on the epithelial-mesenchymal transition of colorectal adenocarcinoma. Cell Physiol Biochem. 2018;50(1):196–213. doi: 10.1159/000493968 [DOI] [PubMed] [Google Scholar]
- 43.Marampon F, Codenotti S, Megiorni F, et al. NRF2 orchestrates the redox regulation induced by radiation therapy, sustaining embryonal and alveolar rhabdomyosarcoma cells radioresistance. J Cancer Res Clin Oncol. 2019;145(4):881–893. doi: 10.1007/s00432-019-02851-0 [DOI] [PubMed] [Google Scholar]
- 44.Sun D, Mu Y, Piao H. MicroRNA-153-3p enhances cell radiosensitivity by targeting BCL2 in human glioma. Biol Res. 2018;51(1):56. doi: 10.1186/s40659-018-0176-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang F, Mao A, Tang J, et al. microRNA-16-5p enhances radiosensitivity through modulating Cyclin D1/E1-pRb-E2F1 pathway in prostate cancer cells. J Cell Physiol. 2018. [DOI] [PubMed] [Google Scholar]
- 46.Perez-Anorve IX, Gonzalez-De la Rosa CH, Soto-Reyes E, et al. New insights into radioresistance in breast cancer identify a dual function of miR-122 as a tumor suppressor and oncomiR. Mol Oncol. 2019;234(8):13182–13190. [DOI] [PMC free article] [PubMed] [Google Scholar]