Abstract
Background
Non-small cell lung cancer (NSCLC) is one of the most important causes of death worldwide. Most patients are diagnosed in the advanced stage and have a poor prognosis. This study was to investigate the expression and significance of CENPE in NSCLC.
Method
Collecting information about CENPE in the Oncoming database, and perform a further analysis of the data in the current database to conduct a meta-analysis for its functional role in NSCLC. Patient life cycle analysis using Kaplan-Meier Plotter and GEPIA databases are used to perform patient survival analysis.
Result
A total of 12 studies involved the expression of CENPE in NSCLC cancer tissues and normal tissues, including 1195 samples. CENPE was highly expressed in NSCLC cell carcinoma compared with the control group (P < 0.05). Moreover, the expression of CENPE was correlated with the overall survival rate of CENPE. The overall survival rate of patients with high expression of CENPE was poor, and the prognosis of patients with low expression of CENPE was better (P<0.05).
Conclusion
We propose high expression of CENPE in NSLCL tissue is related to the prognosis of NSCLC, which may provide important basis for the development of tumor drugs.
Keywords: Non-small cell lung cancer, CENPE, Oncomine
1. Introduction
Non-small cell lung cancer poses a serious threat to health and is one of the most fatal malignant tumors, causing enormous economic burden on society [1]. Although many new treatments have been discovered in recent years, most of them are found in the middle and late stages, and the prognosis has not been significantly improved [2,3]. Studying the molecular mechanism of NSCLC development is conducive to the discovery of new molecular targets, and the development of new treatments is extremely important for reducing patient suffering and prolonging patient survival [4].
The Oncomine database is currently the world’s largest oncogene array database and integrated data mining platform. To date, the database has collected 715 gene expression data sets, sample data from 86,733 cancer tissues and normal tissues. The Oncomine database can be used to compare differential expression of common cancer types and their normal adjacent tissues. It can also explore various cancer subtypes and clinical and pathological analysis based on differential expression sorting and co-expression analysis. Differentially expressed genes, identifying the target genes, and then determining the research direction, not only save scientific research costs, but also gain more comprehensive information.
The kinesin superfamily is a kind of microtubule-based molecular motor protein, which mediates a variety of functions, and its abnormal expression plays an important role in the occurrence and development of tumors [5,6]. The CENPE (KIF10) gene belongs to the KIF family and has been found to play an important role in the process of mitotic cytoplasmic separation [7]. The loss of CENPE expression can lead to increased frequency of chromosome misalignment, subsequent delayed mitotic progression in normal cells [8,9]. Although previous studies have shown that CENPE is expressed in a variety of tumor tissues and cells, there is little systematic study in non-small cell lung cancer tissues.
This study utilized the Oncomine database and Kaplan-Meier Plotters database to analyze the expression and prognosis of CENPE in NSCLC. Through a secondary analysis of the possible relationship between CENPE and NSCLC, it provides clues and evidence for further study on the mechanism of CENPE during development of NSCLC.
2. Materials and methods
2.1. The extraction data
Oncomine database is a gene array database and integrated data mining platform. In this database, you can set the conditions for filtering and mining data according to your needs. In this study, we set the screening criteria: “Cancer Type: Lung cancer”; “Gene: CENPE”; “Data Type: mRNA and DNA copy number”; Analysis Type: Cancer vs Normal Analysis”; The threshold setting condition (P value < 0.01, fold change > 2, gene rank = top 10%). Select the histogram to display the result.
2.2. Patient survival analysis
Online survival analysis was performed using the NSCLC dataset from the Kaplan Meier Plotter database. The screening conditions are as follows: “Cancer: Lung Cancer”; “Gene: CENPE “; “Survival: OS”.
2.3. Statistical methods
Differences in CENPE expression between normal tissue and NSCLC groups were analyzed by t-test. All data were statistically analyzed using SPSS 16.0. The difference between the two sides was P < 0.05.
3. Results
3.1. Expression of CENPE in common tumor types
A total of 451 different types of studies were collected in the Oncomine database (Figure 1). There were 49 studies with statistically significant differences in CENPE expression, 45 with increased CENPE expression, and 4 with reduced expression.
Figure 1.
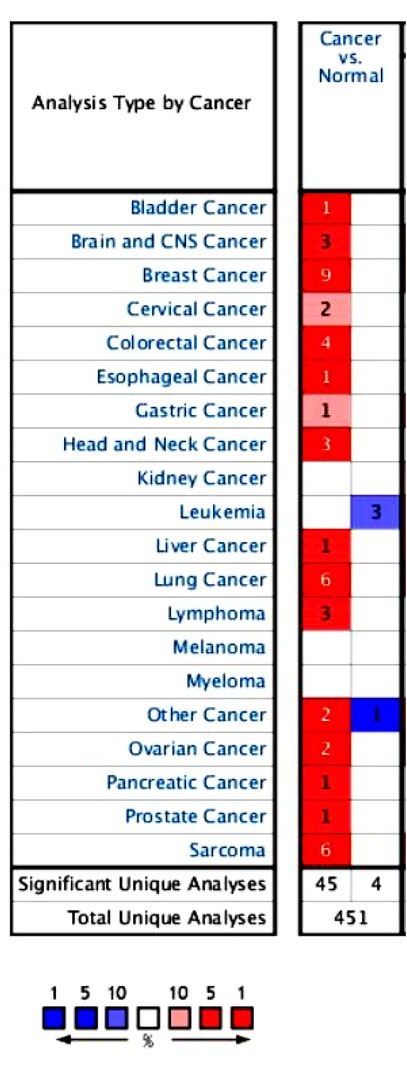
Expression of CENPE in common tumor types in the Oncomine database
3.2. Expression of CENPE in NSCLC
In the Oncomine database, we found that a total of 12 studies have involved CENPE expression in NSCLC tissues and normal tissues (Figure 2), a total of 1195 samples, including lung adenocarcinoma, squamous cell carcinoma and large cell lung cancer and normal lung tissue comparison. Articles were published separately in Nat Med [4], Proc Natl Acad Sci [10,11], PLos One [12,13], Cancer Res [14,15], Genome Res [16], Am J Pathol [17], BMC Genomics [18], Bioinformatics [19], Clin Cancer Res [20]. In a meta-analysis of 12 studies in the Oncomine database, the CENPE gene was ranked as 502.0 in all differentially expressed genes, P = 8.97E-5, suggesting that CENPE is highly expressed in NSCLC.
Figure 2.
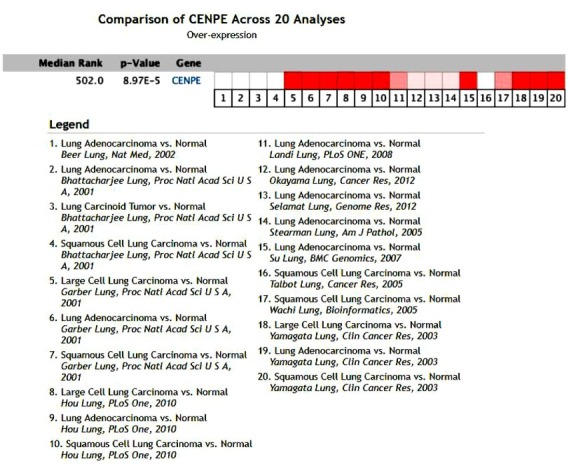
Expression of CENPE in NSCLC in the Oncomine database
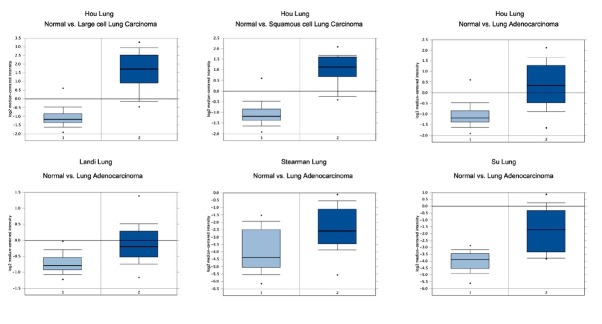
3.3. Differences in expression of CENPE in different NSCLC research arraies
Figure 3 shows the expression of CENPE in different NSCLC research arraies in the Oncomine database. In these four studies, the expression of CENPE in NSCLC was higher than that in the normal group (P < 0.001).
Figure 3.
Differences in expression of CENPE in different NSCLC research arraies
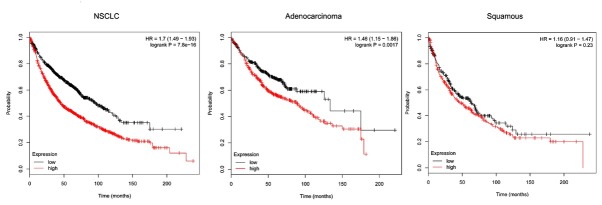
3.4. Correlation of CENPE and prognosis of patients with NSCLC
Kaplan-Meier Plotter data showed that CENPE expression levels had a significant impact on overall patient survival. The overall survival time of patients with CENPE high expression group was significantly lower than that of the low expression group. Further subgroup analysis found that CENPE expression levels had a significant effect on the prognosis of patients with lung adenocarcinoma, whereas in squamous cell carcinoma patients, the expression level had no significant effect on prognosis (Figure 4).
Figure 4.
Correlation of CENPE and prognosis of patients with NSCLC
4. Discussion
Lung cancer is one of the most malignant tumors with the highest global morbidity and mortality. Epidemiological statistics have found that the incidence of lung adenocarcinoma has replaced lung squamous cell carcinoma as the highest incidence of NSCLC [21]. For a long time, the prognosis of NSCLC was poor. Until the past ten years, with the advancement of molecular biology technology, a number of lung adenocarcinoma drive genes such as EGFR, KARS, and ALK have been discovered [22], and some target therapeutic drugs have been developed and improved the prognosis of patients with lung adenocarcinoma [23]. However, mutations of these drive genes are only present in some patients with lung adenocarcinoma. Therefore, finding key molecules or targets during occurrence and development of NSCLC has important theoretical and clinical significance for the development of new targeted drugs for the treatment of lung cancer.
Kinesin is a kind of motor protein that can drive the cargo molecules carried by itself along the microtubules by the energy of ATP hydrolysis. It is involved in the transport of intracellular substances and participates in vesicles, organelles, chromosomes and RNA binding proteins. Kinesin plays an important role in intracellular transport and mitosis. So far, 45 kinesins have been found in humans and mice. They belong to 14 protein families. According to the position of the motor domain on the heavy chain, the kinesin is divided into three types: the motor domain of C-kinesin is at the c-terminus, M-kinesin is at the middle, and the motor domain of N-kinesin is at the N-terminus [24]. Most of the kinesin is N-kinesin. CENPE (KIF10) is an essential plus end-directed microtubule motor and acts to align chromosomes on the metaphase plate [24,25]. The study found that CENPE is highly expressed in a variety of tumors such as glioma [26], breast cancer [27] and gastric cancer [28]. CENPE overexpression in esophageal adenocarcinoma is significantly associated with tumor grade, invasion, and prognosis in esophageal adenocarcinoma [7]. In vivo and in vitro experiments have demonstrated that genetic deletion or pharmacological inhibition of CENPE significantly inhibits the proliferation of prostate cancer cells [29].
For the first time, we found the prognostic value of CENPE in NSCLC through the KM plotter database. The results showed that the expression of CENPE was clearly correlated with the overall survival rate of NSCLC. The overall survival time of the high expressed CENPE of patient was significantly reduced. Further subgroup analysis found that CENPE expression level had a significant effect on the prognosis of patients with lung adenocarcinoma, while in squamous cell carcinoma patients, its expression level had no significant effect on prognosis. The high expression of CENPE may affect the occurrence of tumors, perhaps due to the excessive expression of the kinesin family can affect the normal progression of mitosis, and lead to the occurrence of division or abnormal division, resulting in aneuploid cells, and ultimately cause tumors. All of our data comes from gene chips, the research method is consistent, and contains the largest sample size to date, removing the error caused by the sample size problem and increasing the credibility of the conclusion.
In summary, through the deep exploration of CENPE related information in NSCLC, we propose that CENPE is highly expressed in NSCLC tissues and is associated with the prognosis of NSCLC. Using the database for large sample analysis can avoid the error caused by the small sample size of a single study, and provide an important theoretical basis for clinical treatment. The specific mechanism of action of CENPE in the development of NSCLC disease will require further experiments to prove it in the future.
Footnotes
Conflict of interest
Conflicts of interest: The authors have no conflicts of interest to declare.
References
- [1].Zhou Q., Fan Y., Wu N., Huang Y., Wang Y., Li L.. Demonstration program of population-based lung cancer screening in China: Rationale and study design. Thorac. Cancer. 2014;5:197–203. doi: 10.1111/1759-7714.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Chen W., Zheng R., Baade P., Zhang S., Zeng H., Bray F.. Cancer statistics in China, 2015. Ca. Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- [3].Eberhardt W., Ruysscher D., Weder W., Péchoux C., Leyn P., Hoffmann H.. 2nd ESMO Consensus Conference in Lung Cancer: locally advanced stage III non-small-cell lung cancer. Ann. Oncol. 2015;26:1573–1588. doi: 10.1093/annonc/mdv187. [DOI] [PubMed] [Google Scholar]
- [4].Bergot E., Levallet G., Campbell K., Dubois F., Lechapt E., Zalcman G.. Predictive biomarkers in patients with resected non-small cell lung cancer treated with perioperative chemotherapy. Eur. Respir. Rev. 2013;22:565–576. doi: 10.1183/09059180.00007113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hirokawa N., Tanaka Y.. Kinesin superfamily proteins (KIFs): Various functions and their relevance for important phenomena in life and diseases. Exp. Cell Res. 2015;334:16–25. doi: 10.1016/j.yexcr.2015.02.016. [DOI] [PubMed] [Google Scholar]
- [6].Zou J., Duan Z., Wang J., Sokolov A., Xu J., Chen C.. Kinesin family deregulation coordinated by bromodomain protein ANCCA and histone methyltransferase MLL for breast cancer cell growth, survival and tamoxifen resistance. Mol. Cancer Res. 2014;12:539–549. doi: 10.1158/1541-7786.MCR-13-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhu X., Luo X., Feng G., Huang H., He Y., Ma W.. CENPE expression is associated with its DNA methylation status in esophageal adenocarcinoma and independently predicts unfavorable overall survival. PloS One. 2019;14:e0207341. doi: 10.1371/journal.pone.0207341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wood K.W., Chua P., Sutton D., Jackson J.R.. Centromere-associated protein E: a motor that puts the brakes on the mitotic checkpoint. Clin. Cancer Res. 2008;14:7588–7592. doi: 10.1158/1078-0432.CCR-07-4443. [DOI] [PubMed] [Google Scholar]
- [9].Tame M.A., Raaijmakers J.A., Afanasyev P., Medema R.H.. Chromosome misalignments induce spindle‐positioning defects. Embo. Reports. 2016;17:317–325. doi: 10.15252/embr.201541143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bhattacharjee A., Richards W.G., Staunton J., Li C., Monti S., Vasa P.. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc. Natl. Acad. Sci. U. S. A. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Garber M.E., Troyanskaya O.G., Schluens K., Petersen S., Thaesler Z., Pacyna-Gengelbach M.. Diversity of gene expression in adenocarcinoma of the lung. Proc. Natl. Acad. Sci. U. S. A. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hou J., Aerts J., den Hamer B., van Ijcken W., den Bakker M., Riegman P.. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PloS One. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Landi M.T., Dracheva T., Rotunno M., Figueroa J.D., Liu H., Dasgupta A.. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PloS One. 2008;3:e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Okayama H., Kohno T., Ishii Y., Shimada Y., Shiraishi K., Iwakawa R.. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72:100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- [15].Talbot S.G., Estilo C., Maghami E., Sarkaria I.S., Pham D.K., Oc P. Gene expression profiling allows distinction between primary and metastatic squamous cell carcinomas in the lung. Cancer Res. 2005;65:3063–3071. doi: 10.1158/0008-5472.CAN-04-1985. [DOI] [PubMed] [Google Scholar]
- [16].Selamat S.A., Chung B.S., Girard L., Zhang W., Zhang Y., Campan M.. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. 2012;22:1197–1211. doi: 10.1101/gr.132662.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Stearman R.S., Dwyer-Nield L., Zerbe L., Blaine S.A., Chan Z., Bunn P.A.. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am. J. Pathol. 2005;167:1763–1775. doi: 10.1016/S0002-9440(10)61257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Su L.J., Chang C.W., Wu Y.C., Chen K.C., Lin C.J., Liang S.C.. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics. 2007;8:140. doi: 10.1186/1471-2164-8-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wachi S., Yoneda K., Wu R.. Interactome-transcriptome analysis reveals the high centrality of genes differentially expressed in lung cancer tissues. Bioinformatics. 2005;21:4205–4208. doi: 10.1093/bioinformatics/bti688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yamagata N., Shyr Y., Yanagisawa K., Edgerton M., Dang T.P., Gonzalez A.. A training-testing approach to the molecular classification of resected non-small cell lung cancer. Clin. Cancer Res. 2003;9:4695–4704. [PubMed] [Google Scholar]
- [21].Detterbeck F.C., Boffa D.J., Kim A.W., Tanoue L.T.. The Eighth Edition Lung Cancer Stage Classification. Chest. 2017;151:193–203. doi: 10.1016/j.chest.2016.10.010. [DOI] [PubMed] [Google Scholar]
- [22].Sakashita S., Sakashita M., Sound T.M.. Genes and pathology of non-small cell lung carcinoma. Semin. Oncol. 2014;41:28–39. doi: 10.1053/j.seminoncol.2013.12.008. [DOI] [PubMed] [Google Scholar]
- [23].Zhang J., Yu J., Sun X., Meng X.. Epidermal growth factor receptor tyrosine kinase inhibitors in the treatment of central nerve system metastases from non-small cell lung cancer. Cancer Lett. 2014;351:6–12. doi: 10.1016/j.canlet.2014.04.019. [DOI] [PubMed] [Google Scholar]
- [24].Hirokawa N., Noda Y., Tanaka Y., Niwa S.. Kinesin superfamily motor proteins and intracellular transport. Nature Reviews: Molecular Cell Biology. 2009;10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- [25].Kapoor T.M., Lampson M.A., Hergert P., Cameron L., Cimini D., Salmon E.D.. Chromosomes Can Congress to the Metaphase Plate Before Biorientation. Science. 2006;311:388–391. doi: 10.1126/science.1122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rahane C.S., Kutzner A., Heese K.. A cancer tissue-specific FAM72 expression profile defines a novel glioblastoma multiform (GBM) gene-mutation signature. J. Neurooncol. 2019;141:57–70. doi: 10.1007/s11060-018-03029-3. [DOI] [PubMed] [Google Scholar]
- [27].Yang K., Gao J., Luo M.. Identification of key pathways and hub genes in basal-like breast cancer using bioinformatics analysis. Onco Targets Ther. 2019;12:1319–1331. doi: 10.2147/OTT.S158619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Dun B., Sharma A., Xu H., Liu H., Bai S., Zeng L.. Transcriptomic changes induced by mycophenolic acid in gastric cancer cells. Am. J. Transl. Res. 2013;6:28–42. [PMC free article] [PubMed] [Google Scholar]
- [29].Liang Y., Ahmed M., Guo H., Soares F., Hua J.T., Gao S.. LSD1-Mediated Epigenetic Reprogramming Drives CENPE Expression and Prostate Cancer Progression. Cancer Res. 2017;77:5479–5490. doi: 10.1158/0008-5472.CAN-17-0496. [DOI] [PubMed] [Google Scholar]