Abstract
Ubiquitin specific peptidase 5 (USP5) is a ubiquitous expressed deubiquitinating enzyme (DUB). It has been shown involved in DNA repair, apoptosis, inflammation, and tumor cell growth. However, the function and molecular mechanism of USP5 in colorectal cancer (CRC) are still unclear. In the present study, we asked how it affected the growth of colorectal cancer cells.
Methods: A shRNA-based high-content screening was performed to identify DUBs affecting the growth of CRC cells. CCK-8 assay and xenografts were used to assess CRC cell growth, survival and tumorigenesis. RT-qPCR, immunoblotting and immunohistochemistry were carried out to quantitate USP5 expression in CRC tissues and cell lines. Immunoprecipitation and mass spectrometry analysis were performed to identify USP5-interacting proteins. Cycloheximide chase was performed to assess Tu translation elongation factor (TUFM) stability. Dual luciferase reporter assay was utilized for USP5 promoter analysis.
Results: We found that USP5 was highly expressed in a group of primary CRC tissues, and the increased USP5 was correlated with clinical stages and shorter overall survival. While USP5 knockdown effectively inhibited CRC cell growth, overexpressed USP5 promoted the growth of CRC cells and made them more resistant to doxorubicin (DOX). TUFM was discovered as a substrate of USP5. USP5 deubiquitinated TUFM and increased its level in CRC cells. Enforced expression of TUFM was able to alleviate the growth inhibition induced by USP5 knockdown. Further analyses showed that EBF transcription factor 1 (EBF1) was a major regulator for USP5 transcription, and DOX inhibited EBF1-USP5-TUFM axis in CRC cells.
Conclusions: USP5 was required for CRC cells and promoted their growth and resistance to chemotherapeutics. TUFM was a USP5 deubiquitinating substrate that mediated the cellular effects of USP5. The transcription of USP5 was regulated by EBF1. Thus, targeting EBF1-USP5-TUFM axis is a potential novel strategy for CRC treatment.
Keywords: Deubiquitinase, USP5, Colorectal cancer, TUFM, EBF1
Introduction
Ubiquitination is a major mechanism to modify proteins post-translationally. In addition to tag proteins for proteasomal degradation, it also regulates the activities and intracellular locations of many proteins 1. Ubiquitination-mediated degradation of tumor suppressors and the resistance of oncoproteins to modification and degradation are critical in the initiation and development of many tumors 2. Noteworthily, the ubiquitination process, catalyzed by the sequential actions of E1, E2, and E3 to add mono-, multi-, or poly-ubiquitin to target proteins, can be reversed by the action of deubiquitinating enzymes (DUBs), which remove ubiquitin from target proteins and polyubiquitin chains 3. Importantly, they display specificity for both the types of ubiquitin chains and the substrates. Therefore, DUBs play critical roles in cell growth, death and transformation 4. It has been proposed that the characterized catalytic domains and active sites made them the more desirable targets for druggable inhibitors 5.
There are ~100 DUBs in mammalian cells that can be classified into 6 subfamilies: ubiquitin-specific proteases, ubiquitin C-terminal hydrolases, ovarian tumor proteases, Josephins, JAMMs, and MINDYs 6. Except the JAMMs subfamily, DUBs are all thiol proteases 7. Ubiquitin specific peptidase 5 (USP5) belongs to the largest ubiquitin-specific protease (USP) subfamily (~58 members) 8, and has a zinc finger at its N-terminal portion and two ubiquitin-associated domains in the large catalytic region that mediate polyubiquitin binding 9. Structural and biochemical analysis indicated that USP5 cleaved preferentially branched ubiquitins, including unanchored polyubiquitin chains 10. USP5 may affect cancer development and progression through its action on multiple substrates. It has been shown that USP5 deubiquitinated and stabilized FoxM1, which promoted the recruitment of β-catenin to promoters and activated the Wnt signaling pathway 11, 12. Increased expression of FoxM1 and activation of Wnt pathway have been found contribute to tumorigenesis in many tissue types, including liver, prostate, brain, breast, lung, colon, glioma, and pancreatic tumors 13. In pancreatic cancer cells, knockdown of USP5 led to G1/S block, likely through altering cell cycle regulators 14, whereas USP5 notably prevented the polyubiquitination of c-Maf by E3 ligase Herc4 in multiple myeloma 13. Interestingly, the Helicobacter pylori protein Hpn induced apoptosis in hepatocellular carcinoma through suppressing USP5 expression and activating the p53 pathway 15. It is likely that the reduced USP5 led to accumulation of unanchored polyubiquitin that competed with ubiquitinated p53 for proteasomal recognition, leading to activation of p53 10.
In the present study, we showed that USP5 was important for the growth of colorectal cancer cells in culture and in mice. It conferred CRC cells more resistant to chemotherapeutics, and was highly expressed in many primary CRC tissues, which correlated with disease stage and overall survival of CRC patients. TUFM was identified as a substrate of USP5, and thereby regulated at protein level by USP5. Furthermore, enforced expression of TUFM was able to alleviate growth inhibition induced by USP5 knockdown, indicating that it is an important mediator for the action of USP5 in CRC cells. In addition, we found that EBF transcription factor 1 (EBF1) was a major regulator of USP5 transcription These results indicated that the EBF1-USP5-TUFM axis might be a novel target for the treatment of CRC.
Materials and Methods
Cells, tissues and chemicals
CRC cell lines HT-29, HCT116, Lovo, RKO, SW480, SW620 and SW948 were purchased from American Type Culture Collection (Manassas, VA). HEK293T cell line was kindly provided by Dr. Huashun Li from Tongji University, Shanghai, China. The cells were maintained in DMEM supplemented with 10% fetal calf serum, 100 μg/ml of penicillin, and 100 units/ml of streptomycin. The primary CRC tissues and para-cancerous normal tissues were collected from the Department of Colorectal Surgery, Xinhua Hospital, Shanghai Jiaotong University School of Medicine. The collection and use of human tissues for this study were approved by the Ethics Committee of Xinhua Hospital and informed consent was obtained for all the collections. DOX was purchased from Sigma-Aldrich (St. Louis, MO).
shRNA-based screening
The shRNA library (Table S1) targeting nuclear exporting signal-containing DUBs was purchased from Shanghai GeneChem (Shanghai, China). High-content screening (HCS) was carried out to identify DUBs whose knockdown affected the growth of CRC cells HCT116 according to the manufacturer's instruction.
Preparation of shRNA lentivirus
The additional lentivirus-delivered shRNAs against USP5 (shUSP5) and the negative control (shNC) were purchased from Shanghai GeneChem (Shanghai, China). The targeting sequences of shUSP5#1, shUSP5#2 and shUSP5#3 were 5′-CTTTGCCTTCATTAGTCACAT-3′, 5′-GACCACACGATTTGCCTCATT-3′ and 5′-GATAGACATGAACCAGCGGAT-3′, respectively. The viral particles were prepared with a standard protocol as described previously 16.
Plasmids construction and gene transfection
The human USP5, USP13, TUFM and EBF1 cDNAs were generated and cloned into pcDNA3.1 vector with a Myc or Flag tag as previously described 17. The catalytically inactive mutant of USP5 (USP5-C335A) was constructed according to previous study 18. The siRNAs against TUFM (siTUFM) and the negative control (siNC) were purchased from Guangzhou Ribobio (Guangzhou, China). The targeting sequences of siTUFM#1, siTUFM#2 and siTUFM#3 were 5′-CGACAAGCCACATGTGAAT-3′, 5′-GAGCTCCTAGGACATAGCA-3′, and 5′-GATGGCAACCGGACTATTG-3′, respectively. The siRNAs against the transcription factors E2F1, E2F4, E2F6, EBF1, FOXC1, KLF5, SP1 and TFAP2C were synthesized by GenePharma (Suzhou, China) as described previously 19-24. The sequences of siE2F1, siE2F4, siE2F6, siEBF1, siFOXC1, siKLF5, siSP1 and siTFAP2C were 5′-GUCACGCUAUGAGACCUCATT-3′, 5′-GCGGCGGAUUUACGACAUUTT-3′, 5′-GGAACUUUCUGACUUAUCATT-3′, 5′-CCUCAAAUGUAACCAAAAUTT-3′, 5′-GCAGUAAUUGCUGUUGCUUGUUGTC-3′, 5′-CGAUUACCCUGGUUGCACATT-3′, 5′-GGAUGGUUCUGGUCAAAUATT-3′ and 5′-CCACACUGGAGUCGCCGAAUATT-3′, respecttively. And three additional independent siRNAs against EBF1 (siEBF1#1, siEBF1#2 and siEBF1#3) were purchased from GenePharma. (Suzhou, China). The sequences of siEBF1#1, siEBF1#2 and siEBF1#3 were 5'-CCCACCAUCGAUUAUGGUUTT-3', 5'-GGGAUGAUGGGCGUGAAUUTT-3' and 5'-GCAUGAUUGUUCCUCCUAUTT-3', respectively. Plasmids or siRNAs were transiently transfected into HEK293T or CRC cells by Lipofectamine® 2000 (Invitrogen) according to the manufacturer's instruction.
Immunoblotting analysis
The primary tissue lysates and whole cell lysates were prepared for immunoblotting as described previously 25. Equal amounts of total proteins (30 µg) were subjected to SDS-PAGE and immunoblotting analysis with specific antibodies. The primary antibody against Cyclin D1 was purchased from Cell Signaling Technology (Danvers, MA). Anti-USP5 antibody was obtained from Proteintech Group (Wuhan, China). Anti-TUFM and anti-EBF1 antibodies were purchased from ABclonal Biotechnology (Hubei, China). Anti-Flag, Myc and HA antibodies were purchased from Medical & Biological Laboratories (Tokyo, Japan). Anti-GAPDH antibody was purchased from Abgent Biotechnology (Suzhou, China). Horseradish peroxidase-conjugated anti-mouse and anti-rabbit IgG antibodies were purchased from Beyotime Biotechnology (Nantong, China).
Cell growth and viability
CRC cells HCT116 seeded in 24-well plates (8000 cells per well) were infected with indicated lentivirus, or transfected with indicated plasmids or siRNAs. After cultured for additional 1 to 5 days, cell viability was evaluated by CCK-8 staining as described previously 26.
Xenograft studies
HCT116 infected with lentivirus expressing shUSP5 or scramble were injected subcutaneously into the right flanks of nude mice (Shanghai SLAC Laboratory Animal, Shanghai, China) (5 million cells/site/mouse). Tumor volumes were monitored every other day after they became palpable. At the end of the experiment, tumors were excised for further evaluation. This animal study was approved by the Review Board of Animal Care and Use of Suzhou Institute of Systems Medicine.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted using RNAiso Plus (Takara Bio Group, Japan) as described previously 26. cDNA was synthesized from equal quantities of total RNA using the PrimeScriptTM RT reagent Kit with gDNA Eraser (Takara Bio Group, Japan) according to the manufacturer's instructions. To determine the mRNA levels of USP5 and TUFM, qRT-PCRs were performed with SYBR Green qPCR Master Mix (Takara Bio Group, Japan) using the Roche LightCycler® 480II real-time PCR system (Roche, Basel, Switzerland). The primers used were as follows: USP5, forward 5'-CCACGAACAATAGTTTAGAACG-3' and reverse 5'-AGGTCCCACTGGCACAGA-3'; GAPDH, forward 5'-GCACCGTCAAGGCTGAGAAC-3' and reverse 5'-TGGTGAAGACGCCAGTGGA-3'.
Immunohistochemistry analysis
Paraffin embedded slides of human colorectal cancers were first deparaffinized and rehydrated. Following antigen retrieve and blocking with 10% normal horse serum for 10 minutes, they were incubated with the anti-USP5 antibody (Santa Cruz Biotechnology) overnight at 4°C. After extensive washing, the biotin-conjugated secondary antibody diluted with TBS containing 10% serum and 1% BSA was applied to the slides, which were then incubated at room temperature for 10 minutes and rinsed with cold TBS before streptavidin-peroxidase and 3,3'-Diaminobenzidine (Invitrogen) were added. The slides were also stained with Hematoxylin and eosin and mounted for microscopic analysis. USP5 staining in the tumor and normal tissues was scored on a semi-quantitative score (0, negative; 1, weak; 2, moderate; and 3, strong). Two histopathologists were blindly assigned to review the slides and score the staining.
Co-immunoprecipitation (Co-IP) analysis
Whole cell lysates were prepared for co-immunoprecipitation as described previously 16. They were incubated with a specific primary antibody overnight at 4°C and then mixed with protein A/G-Sepharose beads (Santa Cruz Biotechnology) for 4 hours. After extensive washing, the beads were boiled in 2×SDS-PAGE loading buffer for 5 minutes and analyzed by immunoblotting with specific antibodies.
Immunoprecipitation-coupled mass spectrometry (IP-MS)
HEK293T cells were transfected with vector or Myc-USP5-expressing plasmids by Lipofectamine® 2000 (Invitrogen). Twenty-four hours later, cell lysates were prepared for immunoprecipitation with anti-Myc or control antibodies as described above. The eluated proteins were separated by SDS-PAGE, and visualized by silver staining as described previously 27. A number of anti-Myc immunoprecipitation-specific protein bands were excised from the gel, and processed for LC-MS/MS analysis as described previously 28. To identify the peptides and proteins, the LC-MS/MS spectra were collected and subjected to comparison with the UniProt human proteome database using Proteome Discoverer 1.4 (Thermo Fisher). Proteins were included for analysis when represented by at least two unique peptides.
Cycloheximide chase assay
To evaluate whether USP5 stabilized TUFM protein, HCT116 cells were transfected with plasmids expressing Myc-USP5 or vector by Lipofectamine® 2000 (Invitrogen). Twenty-four hours later, cells were treated with 50 μg/ml cycloheximide (CHX, Sigma-Aldrich) for indicated times and lysed for immunoblotting analysis as described previously 17.
Construction of the truncated USP5 regulatory regions
Genomic DNA was extracted from HCT116 cells according to the manufacturer's protocol (Qiagen). The regulatory sequences of USP5 were predicted by the UCSC Genome Browser website, and different truncated USP5 regulatory regions were amplified by PCR. The primers used for PCR amplification of truncated USP5 regulatory regions were as shown in supplementary Table S2, and the bold sequence indicated the protection bases and restriction enzyme cutting sites. These fragments were then inserted into the pGL4 vector (Promega, Madison, WI) as described previously 16.
Dual-luciferase reporter assays
HCT116 cells were transfected with the fragments of USP5 regulatory sequences along with the internal control vector Renilla by Lipofectamine® 2000 (Invitrogen) according to the manufacturer's instruction. Thirty-six hours later, cells were prepared for luciferase assays using Dual-Luciferase® Reporter Assay System (Promega, Madison, WI, USA) as described previously 29. Firefly luciferase activity was normalized to the Renilla expression for each sample.
Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was performed according to the manufacturer's instructions (Millipore, American Massachusetts) as described previously 16. Briefly, HCT116 cells were firstly fixed with 1% formaldehyde, and then cells were lysed and sonication to shear genomic DNA. After centrifugation, the supernatants were incubated with anti-EBF1 antibody or anti-rabbit IgG for 24 hours at 4 °C, followed by precipitation with protein A beads. The fragment -230/-160 of USP5 promoter regulatory region was determined by qRT-PCR. The primers used were as follows: forward 5'-GCTGCTCTACGTGCGCTC-3' and reverse 5'-GCTCCTAAGGCAATTGAT-3'.
Statistical analysis
The student's t test was used for comparing two groups in the studies. The patient survival time was examined by Kaplan-Meier curve analysis and compared by log-rank test as described previously 30. The association between USP5 expression and patient clinicopathological parameters was evaluated by the Chi square (χ2) analysis. All statistical tests were two-sided, and a p value < 0.05 was considered statistically significant.
Results
USP5 regulated colorectal cancer cell growth
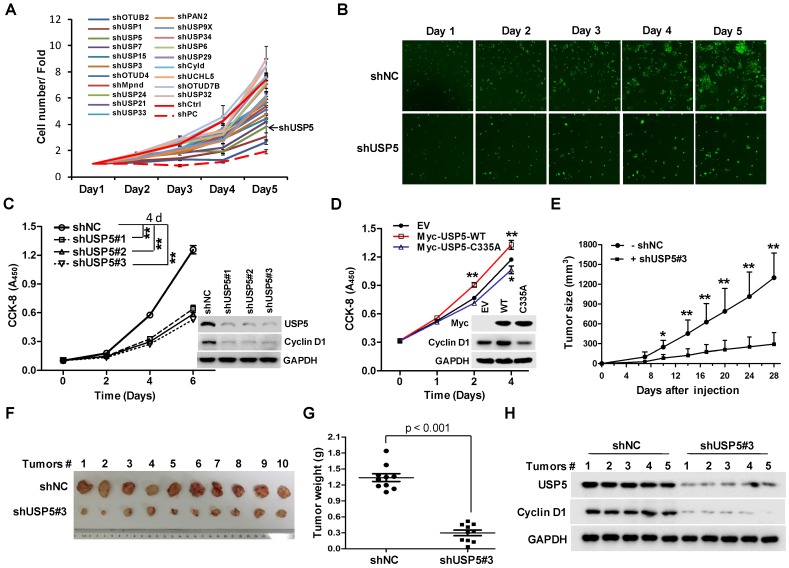
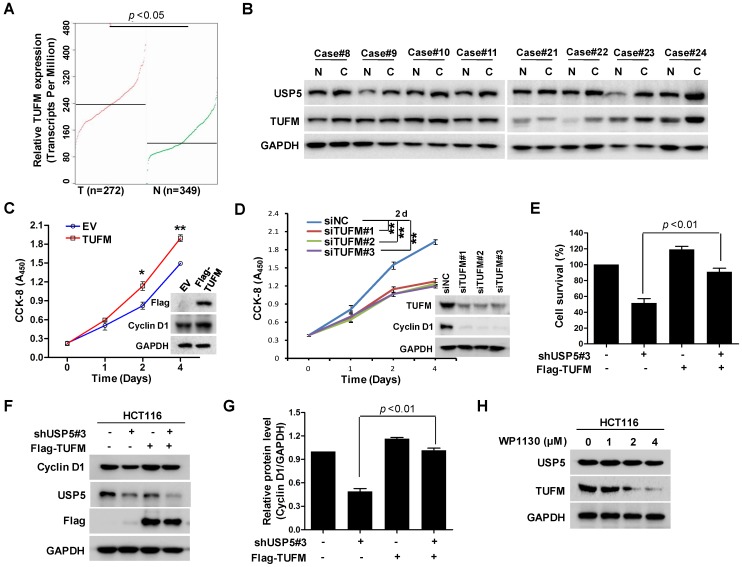
To identify DUBs involved in the growth of CRC cells, a lentivirus library expressing various shRNAs targeting DUBs was utilized in a high-content screening (HCS). The detail information of the library was listed in supplementary Table S1. A number of shRNAs apparently suppressed the growth of HCT116 cells (Figure 1A). As shown in Figure 1B, shRNA targeting USP5 markedly decreased GFP fluorescence, an indicative of cell growth in this screening system. To verify the result, HCT116 cells were infected with 3 different lentiviruses expressing USP5-targeting shRNA#1, shRNA#2, and shRNA#3 respectively. They all reduced cell growth significantly and decreased USP5 level, accompanied by the downregulation of Cyclin D1 (Figure 1C). USP5 knockdown had similar effects on the growth of CRC cell lines RKO and HT-29 (Figure S1). Consistent with the results, enforced expression of wild-type USP5 promoted cell growth as assessed by CCK-8 (Figure 1D), and decreased the drug sensitivity of doxorubicin (DOX) on CRC cells (Figure S2), whereas, expression of a catalytically inactive mutant of USP5 (USP5-C335A) had no effects (Figure 1D). We then examined whether USP5 knockdown affected tumor growth in vivo. HCT116 cells expressing shUSP5#3 or shNC were inoculated subcutaneously into nude mice. As shown in Figure 1E, USP5 knockdown significantly slowed xenografted tumor growth. At the end of the experiment (day 28), the average weight of tumors derived from USP5 knockdown cells was reduced to 23% of that from the control group (Figure 1F & 1G). In the shUSP5#3-expressing xenografts, the levels of both USP5 and Cyclin D1 were significantly lower than that of the controls (Figure 1H). These results indicated that USP5 was an important regulator for CRC cell growth.
Figure 1.
USP5 regulates colorectal cancer cell growth. A & B. The lentivirus-delivered shRNAs against twenty DUBs and controls were constructed. HCT116 cells were infected with different lentivirus-delivered shRNAs, and cell number was counted from 0 to 5 days. The cell growth curve was made (A), and the photos were taken (B). C. HCT116 cells were stably infected with lentiviral shUSP5#1, shUSP5#2, shUSP5#3 or control, followed by immunoblotting and CCK-8 staining at day 0, 2, 4 and 6. Immunoblotting assay was also performed against USP5, Cyclin D1 and GAPDH at day 6. *p<0.01. D. HCT116 cells were transfected with plasmids expressing wild-type Myc-USP5 (Myc-USP5-WT), the catalytically inactive mutant of USP5 (Myc-USP5-C335A) or empty vector (EV), followed by CCK-8 staining at day 0, 1, 2 and 4. Immunoblotting was also performed to detect the expression levels of Myc-USP5, Cyclin D1 and GAPDH at day 4. *p<0.05, **p<0.01. E. HCT116 cells stably infected with lentiviral shUSP5#3 were subcutaneously injected into the right flank of each mouse. When tumors were palpable after seven days, tumor sizes were monitored twice a week for continuously three weeks. *p<0.05, **p<0.01. F. Tumors were excised from nude mice at the end of the experiment. G. Tumor weight was measured at the end of the experiment. H. The excised tumors were prepared for immunoblotting against USP5 and Cyclin D1. GAPDH was used as a loading control.
Increased USP5 expression in primary colorectal cancers
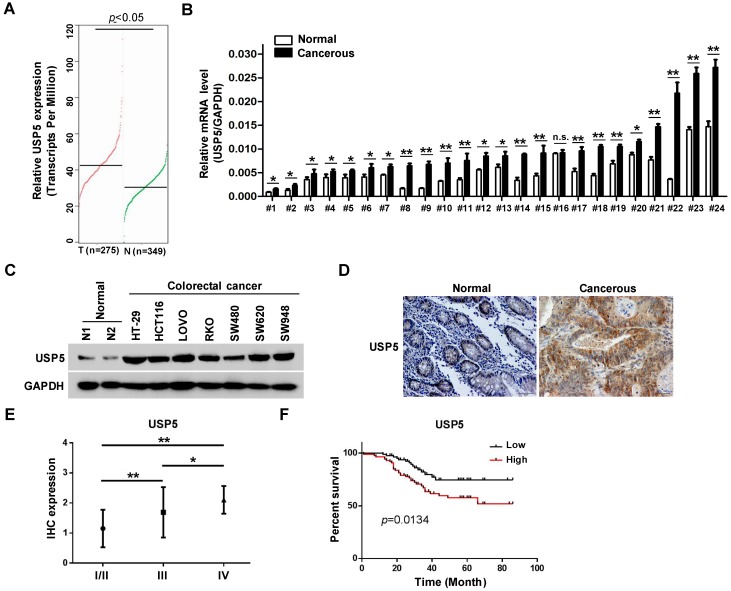
The effects of USP5 on CRC cells propelled us to explore its expression in primary CRC tissues. In the public GEPIA RNAseq database, USP5 expression was significantly higher in colorectal adenocarcinoma than the normal controls (Figure 2A). We assessed USP5 mRNA levels in twenty-four pairs of CRC tissues and non-cancerous tissues by qRT-PCR. As shown in Figure 2B, USP5 mRNA was significantly high in all but one pair (#16) of the tissues. We were also able to examine USP5 expression by immunoblotting in a number of CRC cell lines. As shown in Figure 2C, USP5 protein was highly expressed in the CRC cell lines examined compared with the normal tissues.
Figure 2.
USP5 is highly expressed and a negative index for patient survival in colorectal cancer. A. USP5 expression levels in normal or colorectal cancer tissues retrieved from GEPIA database (http://gepia.cancer-pku.cn). B. Primary colorectal cancer tissues and paired para-cancerous tissues were analyzed for USP5 expression by qRT-PCR. *p<0.05, **p<0.01. C. The expression of USP5 examined by immunoblotting in seven colorectal cancer cell lines and two normal colon tissues. GAPDH was used as a loading control. D. Representative fields of human colorectal cancer tissue arrays stained by using an anti-USP5 immunohistochemistry E. The expression of USP5 in tumors of different clinical stages according to the immunohistochemical staining. *p<0.05, **p<0.01. F. The overall survival time of colorectal cancer patients was calculated by the Kaplan-Meier estimates. All patients were classified into two groups based on the USP5 expression levels.
Use was made of tissue arrays that contained 169 CRC tumor tissues and paired non-cancerous tissues as controls. The clinicopathological conditions of these patients were summarized in Table 1. Immunohistochemical staining with a specific antibody showed that USP5 was highly expressed in human CRC tissues compared with the controls, as illustrated in Figure 2D and Table 2. USP5 expression level was correlated with clinical stages of CRC, and stage IV tumors had the highest level of USP5 (Figure 2E; Table 2). Furthermore, the overall survival of patients with CRC expressing high-level USP5 was significantly shorter than these with low USP5 expression (Figure 2F), indicating that the level of USP5 was an informative prognostic factor for patients with CRC. A multivariate analysis has been also carried out for cohort shown in Figure 2E and 2F to compare USP5 expression with age, gender, and tumor stage. Between the USP5 high and low groups, there were significant differences in age and tumor stages, but no difference in gender (Table S3).
Table 1.
Case information
| Clinical parameters | Case (%) | |
|---|---|---|
| Gender | Male | 91 (53.8) |
| Female | 78 (46.2) | |
| Age | ≤60 | 79 (46.7) |
| >60 | 90 (53.3) | |
| Stage | I | 13 (7.7) |
| II | 66 (39.1) | |
| III | 71 (42.0) | |
| IV | 19 (11.2) | |
| T | 1 | 3 (1.8) |
| 2 | 15 (8.9) | |
| 3 | 57 (33.7) | |
| 4 | 94 (55.6) | |
| N | 0 | 85 (50.3) |
| 1 | 55 (32.5) | |
| 2 | 29 (17.2) | |
| M | 0 | 148 (87.6) |
| 1 | 21 (12.4) | |
| Pathology | I | 31 (18.3) |
| II | 125 (74.0) | |
| III | 13 (7.7) | |
Table 2.
IHC expression of USP5
| Tissues | Cases(N) | Score of USP5 expression | p | |||
|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | |||
| CRC | 169 | 17 | 66 | 73 | 13 | < 0.0001 |
| NT | 169 | 99 | 65 | 5 | 0 | |
| Stage | ||||||
| I | 13 | 2 | 9 | 2 | 0 | |
| II | 66 | 5 | 47 | 11 | 3 | |
| III | 71 | 10 | 9 | 45 | 7 | |
| IV | 19 | 0 | 1 | 15 | 3 | |
| Grade# | ||||||
| Low | 150 | 17 | 65 | 58 | 10 | 0.0103 |
| High | 19 | 0 | 1 | 15 | 3 | |
#Low grade stands for Stage I, II and III; high grade stands for stage IV.
USP5 interacts with TUFM
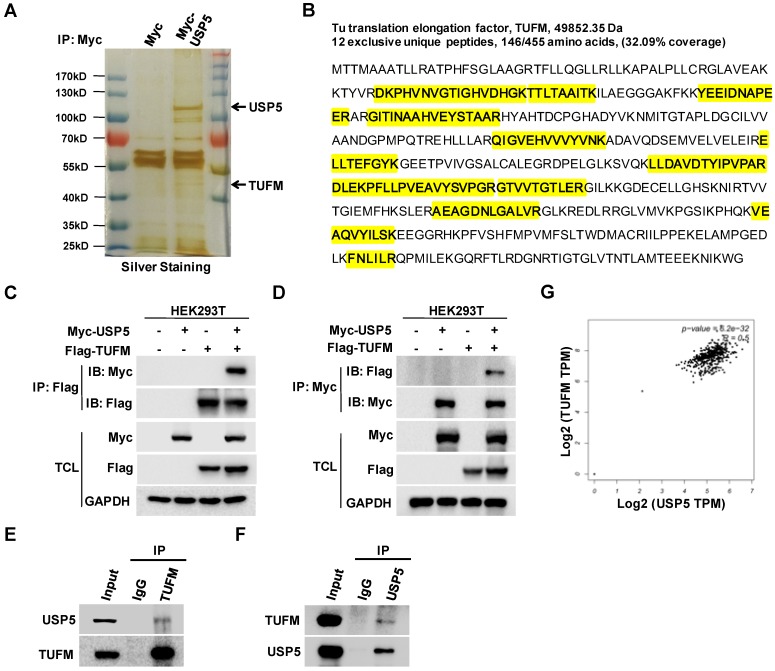
To explore how USP5 affected the growth of CRC cells, we expressed Myc-USP5 in HEK293T cells and pulled down USP5 with an anti-Myc antibody. The immunoprecipitated proteins were subjected to SDS-PAGE, silver staining, and LC-MS/MS analysis. A number of peptides were identified through searching the database, including 12 unique peptides of TUFM (Figure 3A & 3B). To confirm their interactions, cells were transfected with plasmids expressing Myc-USP5 and Flag-TUFM for reciprocal co-immunoprecipitation assay. As shown in Figure 3C & 3D, anti-Myc or anti-Flag antibody brought down TUFM or USP5 respectively. Moreover, anti-USP5 and anti-TUFM antibodies were able to immunoprecipitate endogenous TUFM and USP5 respectively in HCT116 cells (Figure 3E & 3F), indicating that these two proteins interacted in CRC cells. Additionally, it was found from the GEPIA database that the expression of USP5 and TUFM was highly correlated (R=0.5) in CRCs (Figure 3G), suggesting that their interaction may play important roles in primary CRCs.
Figure 3.
USP5 interacts with TUFM. A. HEK293T cells transfected with vector or Myc-USP5 plasmid were lysed and immunoprecipitated with anti-Myc antibody. The immunoprecipites were separated on SDS-PAGE and visualized with silver staining. Differential bands indicated were cut for LC-MS/MS analysis. B. Twelve peptides identified by LC-MS/MS (highlighted in yellow) were fragments of TUFM. C & D. Myc-USP5 and Flag-TUFM-expressing plasmids were transfected into HEK293T for 24 hours. Reciprocal co-immunoprecipitation and immunoblotting were performed by using anti-Flag (C) and anti-Myc antibodies (D). E & F. Whole cell lysates of HCT116 cells were subjected to reciprocal co-immunoprecipitation assays were performed by using anti-TUFM (E) or anti-USP5 antibody (F). G. The correlation between USP5 and TUFM based on data retrieved from GEPIA (http://gepia.cancer-pku.cn).
USP5 stabilizes TUFM through ubiquitin-proteasome pathway
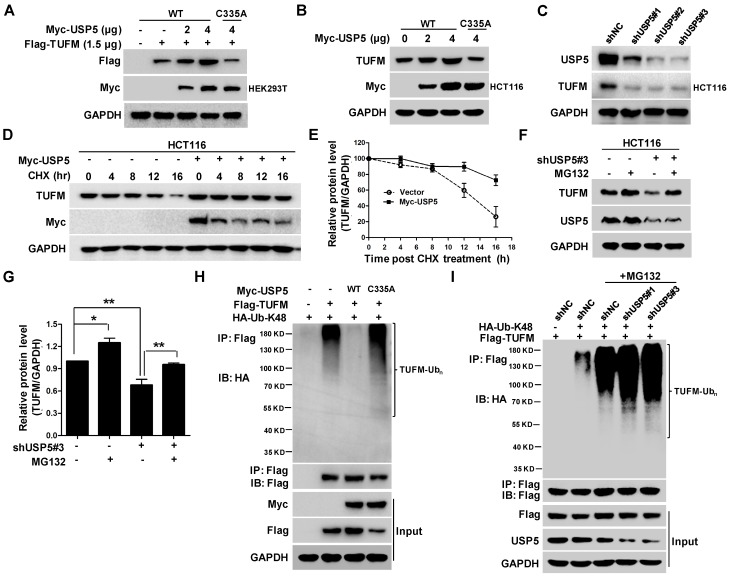
To examine whether USP5 could act as a DUB to affect the ubiquitination status of TUFM, we co-transfected indicated amounts of plasmids expressing wild-type USP5 (USP5-WT) or, mutated USP5 (USP5-C335A) and TUFM into HEK293T cells and examined their protein levels by immunoblotting. As shown in Figure 4A, increased expression of USP5-WT led to dose-dependent elevation of TUFM levels in the cells. Enforced expression of USP5-WT also increased the level of endogenous TUFM in HCT116 cells (Figure 4B), whereas USP5 knockdown by shRNAs led to reduction of TUFM (Figure 4C). However, the catalytically inactive mutant of USP5 could not increase the expression levels of exogenous and endogenous TUFM (Figure 4A & 4B). Furthermore, the half-life of TUFM was markedly prolonged with the enforced expression of USP5 in the CHX chase assay (Figure 4D & 4E). The levels of TUFM in 3 different CRC cells were moderately increased after exposed to proteasome inhibitor MG132 (Figure S3A & S3B), and the decrease of TUFM in HCT116 cells following the USP5 knockdown was largely prevented by MG132 (Figure 4F & 4G). These results indicated that the proteasomal degradtion of TUFM was actively regulated by deubiquitinating action of USP5 in the cells.
Figure 4.
USP5 stabilizes TUFM through deubiquitination. A. Myc-USP5-WT, Myc-USP5-C335A and Flag-TUFM were co-transfected into HEK293T cells. Twenty-four hours later, cells were prepared for immunoblotting analysis using anti- Flag, Myc and GAPDH antibodies. B. HCT116 cells were transfected with increased amounts of plasmids Myc-USP5-WT and Myc-USP5-C335A, and then analyzed by immunoblotting with antibodies against TUFM, Myc and GAPDH. C. After infected with lentiviruses expressing shUSP5#1, shUSP5#2, shUSP5#3 or control for 3 days, HCT116 cells were lysed and analyzed by immunoblotting against USP5 and TUFM. GAPDH was used as a loading control. D. After transfected with Myc-USP5 or control vector for 24 hours, HCT116 cells were exposed to CHX for indicated time and analyzed for TUFM and Myc-USP5 levels by immunoblotting. GAPDH was used as a loading control. E. Quantitative and statistical analyses of data from Figure D (mean +/- SD). F. Following infected with lentiviruses expressing shUSP5#3 or shNC for 36 hours, HCT116 cells were treated with 10 μM of MG132 for 12 hours and then analyzed for the levels of TUFM and USP5 by immunoblotting. GAPDH was used as a loading control. G. Quantitative and statistical analyses of data from Figure F (mean +/- SD). H. Plasmids expressing Myc-USP5-WT, Myc-USP5-C335A, Flag-TUFM or HA-Ub-K48 were transfected into HEK293T cells for 24 hours. The cells were then lysed, immunoprecipitated with anti-Flag antibody, and immnublotted with anti-HA antibody as indicated. The cell lysates were also directly immunoblotted with anti- Flag and Myc antibodies. GAPDH was used as a loading control. I. HCT116 cells were transfected with plasmids Flag-TUFM, HA-Ub-K48, or infected with lentiviruses expressing shUSP5#1, shUSP5#3 or shNC for 48 hours. After treated with 20 μM of MG132 for 6 hours, the cells were lysed, immunoprecipitated with anti-Flag antibody, and immunoblotted with anti-HA antibody to detect the ubiquitination of TUFM. The cell lysates were also directly immunoblotted with antibodies against Flag, USP5 and GAPDH.
To further assess whether deubiquitination of TUFM by USP5 was responsible for the increased stability of TUFM, we examined the ubiquitination status of TUFM. In HEK293T cells transfected with plasmids expressing Flag-TUFM and Myc-Ub, the immunoprecipitated TUFM was heavily ubiquitinated, especially in the presence of the proteasome inhibitor MG132 (Figure S3C). When wild-type USP5 was also co-transfected in the system, TUFM ubiquitination was markedly decreased (Figure 4H). However, co-transfection of USP5-C335A did not decrease TUFM ubiquitination (Figure 4H). Morevoer, knockdown of USP5 enhanced the ubiquitination of TUFM (Figure 4I). Taken together, these results indicated that USP5 likely directly deubiquitinated TUFM and increased its stability. Noteworthily, while similar ubiquitinated species were detected when mutanted ubiquitin that contained only one lysine (K-48) was expressed, they were largely disappeared in cells transfected with K48R ubiquitin-expressing plasmids (Figure S3D), indicating that TUFM was polyubiquitinated by K48-linked chain and subsequently degraded.
TUFM regulates colorectal cancer cell growth and is regulated by USP5
TUFM is a mitochondria protein widely expressed in different tissues including the colon. It has been reported that TUFM expression assessed by immunohistochemistry was markedly increased in many CRC tissues and was a stage-independent unfavourable prognostic indicator 31. It has been also found that upregulated TUFM played significant roles in the transformation from colorectal normal mucosa to carcinoma through adenoma 32. As shown in Figure 5A, the public cancer database TCGA showed that TUFM was highly expressed in CRC tumors, which was consistent with previous study 31. Eight representative pairs of tumor tissues (in which USP5 mRNA was significantly high in tumor tissues) from Figure 2B were subjected to immunoblotting analysis against TUFM and USP5. As shown in Figure 5B, TUFM along with USP5 were both highly expressed in CRC tissues. Furthermore, overexpression or knockdown of TUFM promoted or inhibited CRC cell growth and regulated the expression of Cyclin D1 (Figure 5C & 5D), whereas overexpressed TUFM reversed shUSP5-induced cell growth inhibition and Cyclin D1 downregulation in CRC cells (Figure 5E-5G). Use was also made of small molecular USP5 inhibitor WP1130. As shown in Figure 5H, it downregulated the expression of TUFM dose-dependently. Interestingly, anti-cancer drug DOX markedly suppressed USP5/TUFM expression in CRC cells (Figure S4), suggesting that targeting USP5-TUFM was a novel strategy for CRC treatment.
Figure 5.
TUFM regulates colorectal cancer cell growth and is regulated by USP5. A. TUFM expression in normal and colorectal cancer tissues (http://gepia.cancer-pku.cn). B. Immunoblotting analysis of USP5 and TUFM in 8 pairs of primary colorectal cancer and non-cancerous tissues. GAPDH was used as a loading control. C. The viability of HCT116 cells transfected with Flag-TUFM-expressing plasmid or empty vector (EV) were assessed by CCK-8 staining at day 0, 1, 2 and 4. Immunoblotting was performed to determine the expression levels of Flag-TUFM, Cyclin D1 and GAPDH at day 4. *p<0.05, **p<0.01. D. The viability of HCT116 cells transfected with siTUFM#1, siTUFM#2, siTUFM#3 or control were measured by CCK-8 staining at day 0, 2, 4 and 6. Immunoblotting assay was performed to examine the expression of TUFM, Cyclin D1 and GAPDH at day 4. *p<0.01. E & F. HCT116 cells infected with lentiviruses expressing shUSP5#3 or transfected with Flag-TUFM-expressing vectors were assessed by CCK-8 staining (E) and Immunoblotting analyses (F). G. Quantitative and statistical analysis of Cyclin D1 expression from F. H. HCT116 cells treated with indicated concentrations of WP1130 for 12 hours were examined by immunoblotting against USP5, TUFM and GAPDH.
EBF1 regulates USP5 expression
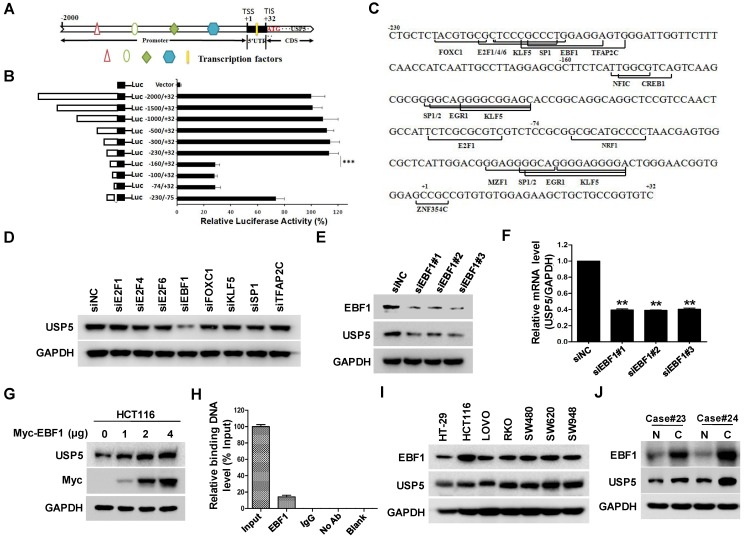
The effect of USP5 on CRC cell growth propelled us to examine the regulation of USP5 expression by using luciferase reporter driven by various fragments of USP5 promoter region. As shown in Figure 6A & B, the construct containing -230/+32 fragment expressed high level of luciferase activity, whereas the one harboring -160/+32 sequence only expressed basal level luciferase, indicating that the -230/-160 fragment was a major region responsible for activating USP5 transcription. Further analyses revealed that the region contained putative binding sites for transcription factors, including E2F1, E2F4, E2F6, EBF1, FOXC1, KLF5, SP1 and TFAP2C (Figure 6C). After silencing them individually with siRNAs in HCT116, we found that only EBF1 knockdown significantly down-regulated USP5 (Figure 6D). The result was further validated with additional siRNAs targeting EBF1 and by the finding that overexpressed EBF1 promoted USP5 expression (Figure 6E-6G). CHIP assay with anti-EBF1 antibody also indicated that EBF1 bound directly to USP5 promoter (Figure 6H). Noteworthily, EBF1 and USP5 were co-expressed in the CRC cell lines and some primary CRC tissues (Figure 6I & 6J). Taken together, these data showed that EBF1 was an important regulator of USP5 expression.
Figure 6.
EBF1 regulates USP5 expression. A. The schematic diagram of the USP5 promoter region (http://genome.ucsc.edu/). TSS: transcription start site; TIS: translation initial site. B. Different truncated fragments of USP5 promoter were cloned into pGL4 reporter. The luciferase activities in transfected cells were measured by using the dual luciferase reporter assays. C. The predicted binding sites for transcription factors in -230/+32 region of USP5 promoter (http://jaspar.binf.ku.dk/). D. After transfected with indicated siRNAs targeting the transcription factors for 72 hours, USP5 in HCT116 cells was analyzed by immunoblotting. GAPDH was used as a loading control. E & F. After transfected with siEBF1#1, siEBF1#2, siEBF1#3 or siNC for 72 hours, EBF1 and USP5 levels in HCT116 cells were assessed by immunoblotting (E) and qRT-PCR (F). **p<0.01. G. HCT116 cells were transfected with indicated amounts of Myc-EBF1-expressing plasmids and then analyzed by immunoblotting against USP5, Myc and GAPDH. H. ChIP assay with anti-EBF1 antibody was performed. The -230/-160 fragment in USP5 promoter region was preferentially pulled down in HCT116 cells. I. The expression of EBF1 and USP5 in seven colorectal cancer cell lines was examined by immunoblotting. J. The expression of EBF1 and USP5 in 2 representative fresh primary colorectal cancer tissues and individual normal tissues were assessed by immunoblotting. GAPDH was used as a loading control.
Doxorubicin inhibits EBF1-USP5-TUFM axis in colorectal cancer cells
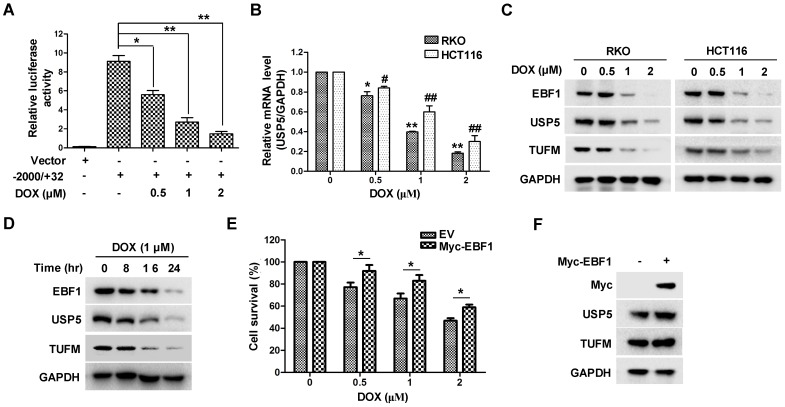
The important role of USP5 in CRC cell growth led us to examine whether its expression was involved in the anti-tumor action of chemotherapeutics. As shown in Figure 7A & 7B, the anti-cancer drug doxorubicin (DOX) significantly suppressed USP5 promoter-driven luciferase activity and decreased USP5 mRNA levels in CRC cells. We then examined the effects of DOX on the expressions of EBF1 and TUFM by immunoblotting. In addition to the decreasing USP5 expression, DOX also induced reduction of EBF1 and TUFM in HCT116 and RKO cells both dose- and time-dependently (Figure 7C & 7D). Noteworthily, enforced expression of EBF1 increased the levels of USP5 and TUFM, and decreased the sensitivity of CRC cells to the cytotoxic action of DOX (Figure 7E & 7F). These results underlined the importance of the EBF1-USP5-TUFM axis in CRC cells and indicated that targeting this axis could be an effective strategy for CRC therapy.
Figure 7.
Doxorubicin inhibits EBF1-USP5-TUFM axis. A. The luciferase reporter driven by the USP5 promoter (-2000/+32) was transfected into HCT116 cells for 24 hours. After treated with indicated concentrations of doxorubicin (DOX) for 12 hours, the luciferase activity in the cells was assessed by using the Dual-Luciferase reporter assay system. B. RKO and HCT116 cells were treated with DOX at indicated concentrations for 24 hours. The levels of USP5 mRNA were quantitated by qRT-PCR. C. Immunoblotting of EBF1, USP5, TUFM and GAPDH in RKO and HCT116 cells treated with indicated concentrations of DOX for 24 hours. D. Immunoblotting of EBF1, USP5, TUFM and GAPDH in RKO cells exposed to 1 μM of DOX for indicated times. E & F. Empty vector (EV) or Myc-EBF1-expressing plasmids were transfected into HCT116 cells. Twenty-four hours later, the cells were treated with indicated concentrations of DOX overnight and then evaluated by CCK-8 assay (E) or prepared for immunoblotting analysis (F).
Discussion
As ubiquitination is involved in most if not all cellular processes, growing evidence indicates that DUBs also play important roles in these processes, particularly in cell cycle, apoptosis, and transformation 33. While germline and somatic mutations of DUBs that drive tumor development are not common, the expression of many DUBs is altered in various cancers 34. Moreover, their roles in different tumors appeared varied significantly 35. Therefore, identifying the functional DUBs in particular cancer and finding their substrates are essential for exploring their mechanisms of action and developing novel therapeutic strategies. Our present studies found that upregulated USP5 was required for CRC cell growth, conferred drug resistance, and correlated with CRC stages and the overall survival of CRC patients. Giving the readily feasibility to inhibit thiol protease by small molecules, these results indicated that targeting USP5 could be an effective anti-CRC strategy.
USP5 is characterized by containing two UBA domain, each contains ~ 45 amino acid residues that form a compact three-helix bundle 36. It is believed that the hydrophobic surface of UBA domain interacts with the hydrophobic surface on the five-stranded β-sheet of ubiquitin, which is responsible for recognizing the ubiquitin chain 37. Interestingly, USP5 and USP13 share approximately 80% similarity and the same domain architecture 38. In our co-IP/MS studies, USP13 was also pulled down by anti-USP5 antibody (data not shown), likely through the interactions of their UBAs. Their interactions are likely functional important as it has been shown recently that both USP5 and USP13 were recruited to heat-induced stress granules in the cells and regulated granules formation through their deubiquitylating activities 39. The stress granules are transient cytoplasmic foci that contain translation-stalled mRNAs and RNA-binding proteins, presumably modulating mRNA translation to accomodate stress responses 40. Interestingly, we have found in the present study that USP5 binds to TUFM and regulates its level through deubiquitination. As translation elongation factors from bacteria, mitochondria and chloroplasts all have a N-terminal domain similar to UBA 41, it is likely the interaction between the domain and UBAs are responsible for USP5 and TUFM binding. It will also be interesting to further examine whether the elongation factors are recruited to the stress granules. However, our preliminary data indicated that USP13 alone did not regulate TUFM stabilization (Figure S5).
Although multiple molecules have been found associated with the effects of USP5 on various cancer cells, including hepatocellular carcinoma 42, glioma 43, myeloma 44 and pancreatic cancer 12, there have been little clues on how it acts on CRC cells. We found in this study that TUFM was a major mediator of USP5 knockdown-induced growth inhibition. Consistent with the finding, both USP5 and TUFM expression were increased in CRC and correlated with the prognosis of patients. Consistent with these findings, it has been shown that the resveratrol analogue HS-1793 exhibited anti-tumor activity in breast cancer and inhibited the expression of major mitochondrial biogenesis-regulating proteins, including TUFM, leading to a block in normal mitochondrial function and sensitized tumor cells to cell death 45. In an subset of diffuse large B-cell lymphomas that possessed the oxidative phosphorylation RNA profile, TUFM knockdown selectively induced cytotoxicity 46. Interestingly, we also found that enforced expression of TUFM alone downregulated USP5 level in CRC cells (Figure 5F), suggesting that overexpressed TUFM may exert a negative feedback role to reduce deubiquitination. Taken together, these studies provided new clues for understanding CRC and indicated that TUFM could be an effective target for therapeutic intervention of CRC.
Supplementary Material
Supplementary figures and tables.
Acknowledgments
This work was supported by National Natural Science Foundation of China (81572378), Natural Science Foundation of Jiangsu Province (BK20171231), CAMS Initiative for Innovative Medicine (CAMS-I2M, 2016-I2M-1-005), and the Special Research Fund for Central Universities, Peking Union Medical College (2016ZX310194).
Avalibility of data and materials
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
Authors' contributions
X.X, A.H, X.C, K.H X.H and Q.W performed the experiments. X.X, K.H and Y.Y wrote and edited the manuscript. X.X., L.C., and Y.Y designed the research project.
Ethics approval and consent to participate
This study was approved by the Review Board and Ethical Committee of Suzhou Institute of Systems Medicine. This study conforms to the Declaration of Helsinki.
Consent for publication
Written informed consent for publication of clinical details and/or clinical images was obtained from the patients.
Abbreviations
- CRC
Colorectal cancer
- USP5
Ubiquitin specific peptidase 5
- TUFM
Tu translation elongation factor
- EBF1
EBF transcription factor 1
- CCK-8
Cell Counting Kit-8
- LC/MS/MS
Liquid chromatography tandem mass spectrometry
- DOX
Doxorubicin
- DUB
Deubiquitinase
- HCS
High-content screening
- Co-IP
Co-immunoprecipitation
- GEPIA
Gene Expression Profiling Interactive Analysis
References
- 1.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–29. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 2.Kitagawa K, Kotake Y, Kitagawa M. Ubiquitin-mediated control of oncogene and tumor suppressor gene products. Cancer Sci. 2009;100:1374–81. doi: 10.1111/j.1349-7006.2009.01196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–63. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 4.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK. et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–86. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Fraile JM, Quesada V, Rodriguez D, Freije JM, Lopez-Otin C. Deubiquitinases in cancer: new functions and therapeutic options. Oncogene. 2012;31:2373–88. doi: 10.1038/onc.2011.443. [DOI] [PubMed] [Google Scholar]
- 6.Lee HJ, Kim MS, Shin JM, Park TJ, Chung HM, Baek KH. The expression patterns of deubiquitinating enzymes, USP22 and Usp22. Gene Expr Patterns. 2006;6:277–84. doi: 10.1016/j.modgep.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 7.Harhaj EW, Dixit VM. Deubiquitinases in the regulation of NF-kappaB signaling. Cell Res. 2011;21:22–39. doi: 10.1038/cr.2010.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumari N, Jaynes PW, Saei A, Iyengar PV, Richard JLC, Eichhorn PJA. The roles of ubiquitin modifying enzymes in neoplastic disease. Biochim Biophys Acta Rev Cancer. 2017;1868:456–483. doi: 10.1016/j.bbcan.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 9.Reyes-Turcu FE, Shanks JR, Komander D, Wilkinson KD. Recognition of polyubiquitin isoforms by the multiple ubiquitin binding modules of isopeptidase T. J Biol Chem. 2008;283:19581–92. doi: 10.1074/jbc.M800947200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dayal S, Sparks A, Jacob J, Allende-Vega N, Lane DP, Saville MK. Suppression of the deubiquitinating enzyme USP5 causes the accumulation of unanchored polyubiquitin and the activation of p53. J Biol Chem. 2009;284:5030–41. doi: 10.1074/jbc.M805871200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, Li Y, Xue J, Gong A, Yu G, Zhou A. et al. Wnt-induced deubiquitination FoxM1 ensures nucleus beta-catenin transactivation. EMBO J. 2016;35:668–84. doi: 10.15252/embj.201592810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li XY, Wu HY, Mao XF, Jiang LX, Wang YX. USP5 promotes tumorigenesis and progression of pancreatic cancer by stabilizing FoxM1 protein. Biochem Biophys Res Commun. 2017;492:48–54. doi: 10.1016/j.bbrc.2017.08.040. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Z, Tong J, Tang X, Juan J, Cao B, Hurren R. et al. The ubiquitin ligase HERC4 mediates c-Maf ubiquitination and delays the growth of multiple myeloma xenografts in nude mice. Blood. 2016;127:1676–86. doi: 10.1182/blood-2015-07-658203. [DOI] [PubMed] [Google Scholar]
- 14.Kaistha BP, Krattenmacher A, Fredebohm J, Schmidt H, Behrens D, Widder M. et al. The deubiquitinating enzyme USP5 promotes pancreatic cancer via modulating cell cycle regulators. Oncotarget. 2017;8:66215–66225. doi: 10.18632/oncotarget.19882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Wang WM, Zou LY, Li L, Feng L, Pan MZ, Ubiquitin specific peptidase 5 mediates Histidine-rich protein Hpn induced cell apoptosis in hepatocellular carcinoma through P14-P53 signaling. Proteomics; 2017. p. 17. [DOI] [PubMed] [Google Scholar]
- 16.Xu X, Han K, Tang X, Zeng Y, Lin X, Zhao Y. et al. The Ring Finger Protein RNF6 Induces Leukemia Cell Proliferation as a Direct Target of Pre-B-cell Leukemia Homeobox 1. J Biol Chem. 2016;291:9617–28. doi: 10.1074/jbc.M115.701979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen G, Xu X, Tong J, Han K, Zhang Z, Tang J. et al. Ubiquitination of the transcription factor c-MAF is mediated by multiple lysine residues. Int J Biochem Cell Biol. 2014;57:157–66. doi: 10.1016/j.biocel.2014.10.024. [DOI] [PubMed] [Google Scholar]
- 18.Scott D, Layfield R, Oldham NJ. Ion mobility-mass spectrometry reveals conformational flexibility in the deubiquitinating enzyme USP5. Proteomics. 2015;15:2835–41. doi: 10.1002/pmic.201400457. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Jove Navarro M, Basset C, Arcondeguy T, Touriol C, Perez G, Prats H. et al. Api5 contributes to E2F1 control of the G1/S cell cycle phase transition. PLoS One. 2013;8:e71443. doi: 10.1371/journal.pone.0071443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goto Y, Hayashi R, Kang D, Yoshida K. Acute loss of transcription factor E2F1 induces mitochondrial biogenesis in HeLa cells. J Cell Physiol. 2006;209:923–34. doi: 10.1002/jcp.20802. [DOI] [PubMed] [Google Scholar]
- 21.Jimenez MA, Akerblad P, Sigvardsson M, Rosen ED. Critical role for Ebf1 and Ebf2 in the adipogenic transcriptional cascade. Mol Cell Biol. 2007;27:743–57. doi: 10.1128/MCB.01557-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong XP, Yao J, Luo W, Feng FK, Ma JT, Ren YP. et al. The expression and functional role of a FOXC1 related mRNA-lncRNA pair in oral squamous cell carcinoma. Mol Cell Biochem. 2014;394:177–86. doi: 10.1007/s11010-014-2093-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia L, Zhou Z, Liang H, Wu J, Shi P, Li F. et al. KLF5 promotes breast cancer proliferation, migration and invasion in part by upregulating the transcription of TNFAIP2. Oncogene. 2016;35:2040–51. doi: 10.1038/onc.2015.263. [DOI] [PubMed] [Google Scholar]
- 24.Hirata Y, Masuda Y, Kakutani H, Higuchi T, Takada K, Ito A. et al. Sp1 is an essential transcription factor for LPS-induced tissue factor expression in THP-1 monocytic cells, and nobiletin represses the expression through inhibition of NF-kappaB, AP-1, and Sp1 activation. Biochem Pharmacol. 2008;75:1504–14. doi: 10.1016/j.bcp.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 25.Han K, Xu X, Chen G, Zeng Y, Zhu J, Du X. et al. Identification of a promising PI3K inhibitor for the treatment of multiple myeloma through the structural optimization. J Hematol Oncol. 2014;7:9. doi: 10.1186/1756-8722-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu X, Wang J, Han K, Li S, Xu F, Yang Y. Antimalarial drug mefloquine inhibits nuclear factor kappa B signaling and induces apoptosis in colorectal cancer cells. Cancer Sci. 2018;109:1220–1229. doi: 10.1111/cas.13540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan JX, Wait R, Berkelman T, Harry RA, Westbrook JA, Wheeler CH. et al. A modified silver staining protocol for visualization of proteins compatible with matrix-assisted laser desorption/ionization and electrospray ionization-mass spectrometry. Electrophoresis. 2000;21:3666–72. doi: 10.1002/1522-2683(200011)21:17<3666::AID-ELPS3666>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 28.Zhou Y, Xiong L, Zhang Y, Yu R, Jiang X, Xu G. Quantitative proteomics identifies myoferlin as a novel regulator of A Disintegrin and Metalloproteinase 12 in HeLa cells. J Proteomics. 2016;148:94–104. doi: 10.1016/j.jprot.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 29.Xu X, Han K, Zhu J, Mao H, Lin X, Zhang Z. et al. An inhibitor of cholesterol absorption displays anti-myeloma activity by targeting the JAK2-STAT3 signaling pathway. Oncotarget. 2016;7:75539–75550. doi: 10.18632/oncotarget.12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng Y, Xu X, Wang S, Zhang Z, Liu Y, Han K. et al. Ring finger protein 6 promotes breast cancer cell proliferation by stabilizing estrogen receptor alpha. Oncotarget. 2017;8:20103–20112. doi: 10.18632/oncotarget.15384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi H, Hayes M, Kirana C, Miller R, Keating J, Macartney-Coxson D. et al. TUFM is a potential new prognostic indicator for colorectal carcinoma. Pathology. 2012;44:506–12. doi: 10.1097/PAT.0b013e3283559cbe. [DOI] [PubMed] [Google Scholar]
- 32.Xi HQ, Zhang KC, Li JY, Cui JX, Zhao P, Chen L. Expression and clinicopathologic significance of TUFM and p53 for the normal-adenoma-carcinoma sequence in colorectal epithelia. World J Surg Oncol. 2017;15:90. doi: 10.1186/s12957-017-1111-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song L, Rape M. Reverse the curse-the role of deubiquitination in cell cycle control. Curr Opin Cell Biol. 2008;20:156–63. doi: 10.1016/j.ceb.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D'Arcy P, Wang X, Linder S. Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacol Ther. 2015;147:32–54. doi: 10.1016/j.pharmthera.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 35.McClurg UL, Robson CN. Deubiquitinating enzymes as oncotargets. Oncotarget. 2015;6:9657–68. doi: 10.18632/oncotarget.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mueller TD, Feigon J. Solution structures of UBA domains reveal a conserved hydrophobic surface for protein-protein interactions. J Mol Biol. 2002;319:1243–55. doi: 10.1016/S0022-2836(02)00302-9. [DOI] [PubMed] [Google Scholar]
- 37.Raasi S, Orlov I, Fleming KG, Pickart CM. Binding of polyubiquitin chains to ubiquitin-associated (UBA) domains of HHR23A. J Mol Biol. 2004;341:1367–79. doi: 10.1016/j.jmb.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 38.Laddaga M, Calderazzi A, Sargenti A. [Future possibilites and usefulness of telecobalt radiation alone in the preoperative treatment of uterine neoplasms. Anatomo-clinical contribution] Radiol Med. 1968;54:666–89. [PubMed] [Google Scholar]
- 39.Xie X, Matsumoto S, Endo A, Fukushima T, Kawahara H, Saeki Y, Deubiquitylases USP5 and USP13 are recruited to and regulate heat-induced stress granules through their deubiquitylating activities. J Cell Sci; 2018. p. 131. [DOI] [PubMed] [Google Scholar]
- 40.Oshiumi H, Mifsud EJ, Daito T. Links between recognition and degradation of cytoplasmic viral RNA in innate immune response. Rev Med Virol. 2016;26:90–101. doi: 10.1002/rmv.1865. [DOI] [PubMed] [Google Scholar]
- 41.Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell. 2004;16:3496–507. doi: 10.1105/tpc.104.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Wang WM, Lu YF, Feng L, Li L, Pan MZ. et al. Usp5 functions as an oncogene for stimulating tumorigenesis in hepatocellular carcinoma. Oncotarget. 2017;8:50655–50664. doi: 10.18632/oncotarget.16901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Izaguirre DI, Zhu W, Hai T, Cheung HC, Krahe R, Cote GJ. PTBP1-dependent regulation of USP5 alternative RNA splicing plays a role in glioblastoma tumorigenesis. Mol Carcinog. 2012;51:895–906. doi: 10.1002/mc.20859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang S, Juan J, Zhang Z, Du Y, Xu Y, Tong J. et al. Inhibition of the deubiquitinase USP5 leads to c-Maf protein degradation and myeloma cell apoptosis. Cell Death Dis. 2017;8:e3058. doi: 10.1038/cddis.2017.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeong SH, Song IS, Kim HK, Lee SR, Song S, Suh H. et al. An analogue of resveratrol HS-1793 exhibits anticancer activity against MCF-7 cells via inhibition of mitochondrial biogenesis gene expression. Mol Cells. 2012;34:357–65. doi: 10.1007/s10059-012-0081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Norberg E, Lako A, Chen PH, Stanley IA, Zhou F, Ficarro SB. et al. Differential contribution of the mitochondrial translation pathway to the survival of diffuse large B-cell lymphoma subsets. Cell Death Differ. 2017;24:251–262. doi: 10.1038/cdd.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.